Abstract
Post-translational modificationsof the histone octamer play important roles in regulating responses to DNA damage. Here, we reveal that Saccharomyces cerevisiae Rtt109p promotes genome stability and resistance to DNA damaging agents, and that it does this by functionally cooperating with the histone chaperone Asf1p to maintain normal chromatin structure. Furthermore, we show that, as for Asf1p, Rtt109p is required for histone H3 acetylation on lysine 56 (K56) in vivo. Moreover, we reveal that Rtt109p directly catalyzes this modification in vitro in a manner that is stimulated by Asf1p. These data establish Rtt109p as a member of a new class of histone acetyl-transferases, and show that its actions are critical for cell survival in the presence of DNA damage during S-phase.
Results
Regulation of retrotransposition (RTT) by the Saccharomyces cerevisiae Ty1 transposon is intimately linked to the DNA damage response (DDR), as many proteins are known to play roles in both pathways (1). We therefore reasoned that uncharacterized RTT genes might represent novel DDR factors. While not having been characterized in detail, RTT109 was previously linked tothe DDR through genome-wide studies systematically identifying mutants required for resistance to genotoxic agents (2, 3). To further characterize Rtt109p, we generated a deletion mutant and examined its sensitivities to a wider range of DNA damaging agents. We found that rtt109Δ mutants were hypersensitive to agents that generate replication stress (Fig. 1A upper panel). We also observed hypersensitivity of rtt109Δ cells to continuous growth on plates containing phleomycin, an ionizing radiation (IR) mimetic that induces DNA double-strand breaks (DSBs). However, rtt109Δ cells were not markedly hypersensitive to acute IR treatment (Fig. 1A). Chronic DSB induction by phleomycin impinges on S-phase repair pathways, whereas most cells in an asynchronous culture subjected to acute IR treatment arrest cell cycle progression and repair the DNA damage in G2 (4).Together, these results therefore suggest a role for Rtt109p in DNA damage tolerance during S-phase.
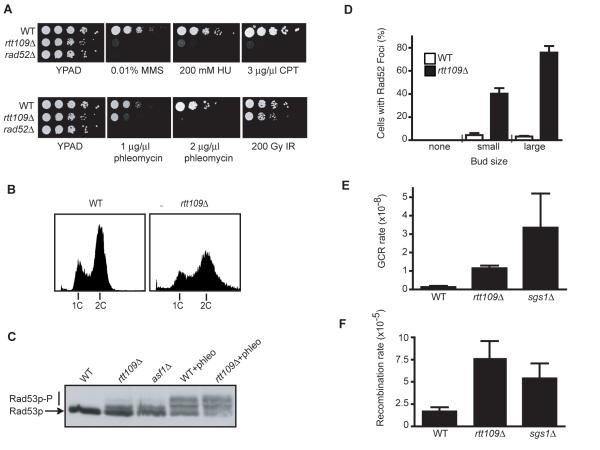
Figure 1. rtt109Δ cells display hypersensitivity to DNA damaging agents, DNA damage checkpoint activation and genomic instability.
(A) Serial dilutions (ten-fold) of the indicated mutants were spotted on YPAD alone or on YPAD containing the indicated drug (B) The DNA content of asynchronous cultures of wild-type (WT) and rtt109Δ strains were determined by flow cytometric analysis. (C) Electrophoretic mobility of Rad53p was analyzed in extracts from asynchronous cultures of wild-type (WT), rtt109Δ and asf1Δ cells, and WT and rtt109Δ cells treated with phleomycin for 1 h. (D) Cell cycle distribution of WT and rtt109Δ cells with Rad52-YFP foci as determined by bud size. The average and standard deviation of two independent experiments are shown (E) GCR frequency was measured for the indicated strains (7). The average and standard deviation of three fluctuation tests are shown. (F) Recombination frequencies were measured in the indicated strains with a direct repeat recombination assay (8); the average and standard deviation of three fluctuation tests are shown.
In addition to displaying DNA damage hypersensitivity, we observed that rtt109Δ cells were slow growing. Flow cytometric analysisof an asynchronous rtt109Δ culture revealed a high proportion of cells in the G2-M stage of the cell cycle (Fig.1B); and furthermore, the DNA content profile for rtt109Δ cells was much broader than wild-type, indicating continued cell growth despite slowed cell cycle progression. Cells deleted for both RTT109 and MEC1 (which encodes the central DNA damage checkpoint kinase) had a budding index equivalent to that of the wild-type strain (Fig. S1), indicating that the altered cell cycle profile of rtt109Δ cells reflects activation of the DNA damage checkpoint. This was confirmed by the presence of partially phosphorylated DDR effector kinase Rad53p in rtt109Δ cells, indicative of chronic checkpoint activation (Fig. 1C).
Quantification of DSBs can be used to assess the presence of DNA damage in yeast cells (5, 6). Strikingly, we found that Rad52-YFP foci – markers of DSBs – occur at much higher frequencies in rtt109Δ cells than in wild-type cells (Fig. 1D). Very few foci were seen in wild-type cells and unbudded rtt109Δ cells, whereas they occured in around 40% and 75% of small-budded and large budded rtt109Δ cells, corresponding to S and G2 cells, respectively. These data therefore indicate increased spontaneously arising DNA damage in cells lacking Rtt109p, and suggest that such damage largely arises during S-phase. To examine the effects of this DNA damage on genome stability, we measured rates of gross chromosomal rearrangements (GCRs) (7). Thus, we found that RTT109 deletion increased GCR rates ~9-fold compared to wild-type cells (Fig. 1E). As a further test for genome instability in rtt109Δ cells, we used a system to measure spontaneous recombination between tandem direct repeats (8). This revealed that RTT109 deletion yields a moderate hyper-recombination phenotype, similar to that exhibited by an sgs1Δ hyper-recombination mutant (Fig. 1F). These data indicate that rtt109Δ mutants display increased genomic instability, possibly as a consequence of spontaneously arising DNA damage, and furthermore suggest that this reflects defects in responding to and/or repairing DNA damage arising during S-phase.
We noted that the phenotypesof rtt109Δ strains were similar to those reported for strains lacking the histone chaperone Asf1p (9, 10), and that large-scale genetic network analyses had revealed the genetic interaction profile of RTT109 to be highly similar to that of ASF1 (11, 12) suggesting they might act within the same pathway. Indeed, we found that the rtt109Δ asf1Δ double mutant was no more sensitive to HU or MMS than either single mutant (Fig. 2A). Although Asf1p stimulates histone deposition by the CAF-1 chromatin assembly complex in vitro (13), it also acts in a distinct and/or partially overlapping role in providing resistance to DNA damaging agents (14). To address whether Rtt109p also acts synergistically with CAF-1, we combined the RTT109 deletion with a disruption of CAC1, which encodes a CAF-1 subunit. As for asf1Δ cac1Δ cells, rtt109Δ cac1Δ cells were more sensitive to HU or MMS than the single mutant strains. Furthermore, rtt109Δ asf1Δ cac1Δ cells were no more sensitive than rtt109Δ cac1Δ cells (Fig 2A). From these data, we thus conclude that Rtt109p acts in the same pathway as Asf1p in providing resistance to DNA damaging agents.
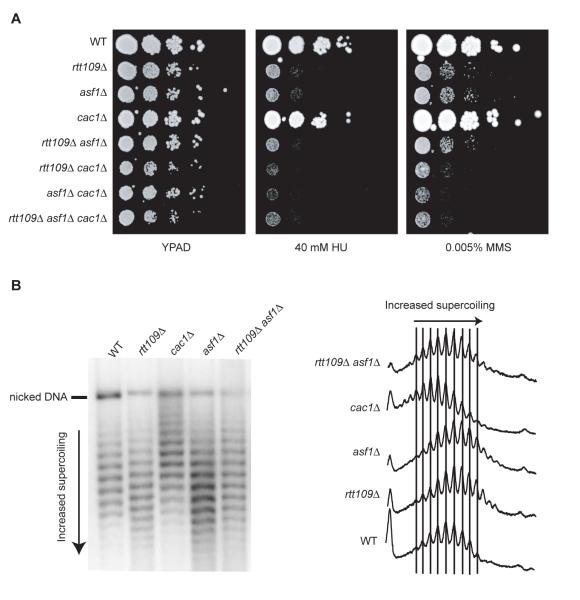
Figure 2. Effects of RTT109, ASF1 and CAC1 disruption on DNA damage sensitivity and 2μ plasmid supercoiling.
(A) Serial dilutions (ten-fold) of the indicated strains were plated on YPAD or YPAD containing HU or MMS. (B) DNA isolated from the indicated strains was electrophoresed on an agarose gel containing chloroquine. Superhelical density of the 2μ plasmid was analyzed by southern blotting and hybridization with a radioactively-labelled probe (left panel). Topoisomers were quantified by densitometric tracing (right panel).
Given the reported role of Asf1p in promoting nucleosome assembly (10, 13) we also analyzed the superhelical density of the 2μ plasmid in rtt109Δ cells. RTT109 deletion caused a shift in the distribution of 2μ topoisomers, indicating increased supercoiling compared to wild-type cells (Fig. 2B). Furthermore, a similar change in topoisomer distribution was caused by ASF1 deletion, and no further change was seen in an asf1Δ rtt109Δ double mutant strain. These results therefore indicate that Rtt109p and Asf1p act together in governing chromatin structure.
The DNA damage hypersensitivity and slow growth of asf1Δ cells are associated with loss of acetylation on histone H3 lysine 56 (H3 K56) (15), but it has hitherto been unclear why K56 acetylation is absent in asf1Δ cells or which histone acetyl-transferase (HAT) is responsible for acetylating this residue (16). Because of the epistatic relationship between Asf1p and Rtt109p, we examined histone H3 K56 acetylation in rtt109Δ cells and found an absence of detectable K56 acetylation (Fig. 3A) While either RTT109 deletion or mutation of histone H3 K56 to arginine resulted in hypersensitivity towards HU, the double mutant was no more sensitive than the single mutants (Fig. 3B). Growth curves also revealed an epistatic relationship between the two mutants (Fig. S2), indicating that the growth defect of rtt109Δ cells is due to loss of H3 K56 acetylation.
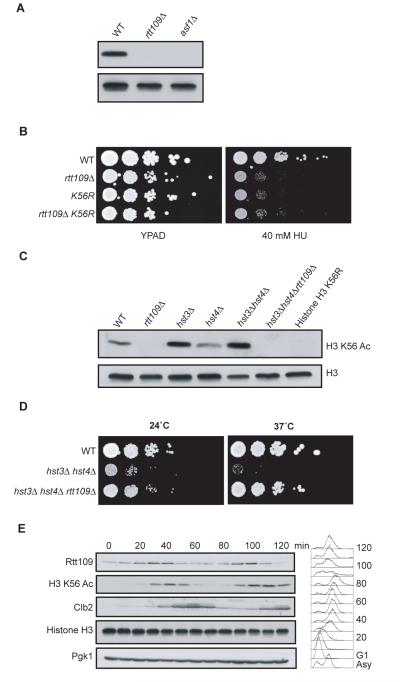
Figure 3. Rtt109p is needed for histone H3 K56 acetylation in vivo.
(A) Western blot analysis of whole cell extracts isolated from the indicated strains was performed with antibodies specific for histone H3 or histone H3 acetylated on K56 (K56 Ac). (B) Serial dilutions (ten-fold) of the indicated strains were plated on YPAD or YPAD containing HU. (C) Whole cell extracts from the indicated strains were probed with antibodies for histone H3 and histone H3 K56 Ac. (D) Serial dilutions (ten-fold) of the indicated strains were plated on YPAD at the indicated temperature. (E) A strain expressing a HA-tagged version of Rtt109p was arrested in G1 with α-factor and released. Samples were taken at the indicated times for FACS and western analysis performed, probing for the indicated proteins with Pgk1p and Histone H3 as loading controls and Clb2p as a G2-M marker.
We reasoned that Rtt109p could either facilitate K56 acetylation or prevent K56 deacetylation by the known histone H3 K56 deacetylases Hst3p/Hst4p (17, 18). Significantly, we found that, despite elevated K56 acetylation in the absence Hst3p and Hst4p, K56 acetylation was undetectable in a hst3Δ hst4Δ rtt109Δ triple mutant strain (Fig. 3C), suggesting Rtt109p acts to promote H3 K56 acetylation. Consistent with these findings, and as reported for ASF1 deletion (17), we found that RTT109 deletion suppressed the temperature sensitive growth defect of a hst3Δ hst4Δ strain (Fig. 3D). Furthermore, prompted by the fact that H3 K56 acetylation shows cell-cycle control (19), we examined Rtt109p expression during cell cycle progression. Thus, we found that Rtt109p levels peak just prior to maximal K56 acetylation (Fig. 3E), as would be expected if Rtt109p is required for generating this modification.
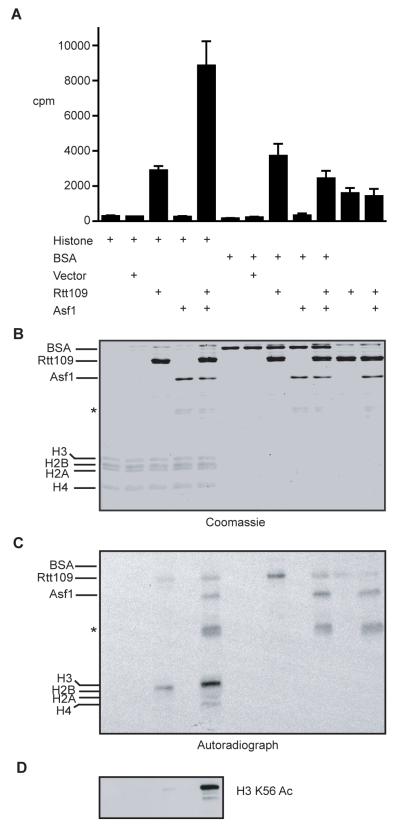
To examine whether and how Rtt109p might mediate H3 K56 acetylation, we expressed and purified recombinant Rtt109p and Asf1p. Histone acetylation assays revealed that while histone H3 acetylation took place with Rtt109p alone, this activity was enhanced in the presence of Asf1p (Fig. 4A). Autoradiographic analysis of the reactions also revealed that Rtt109p auto-acetylates and weakly acetylates Asf1p, but not bovine serum albumin (Fig. 4C). Moreover, western immunoblots of acetylation reactions probed with an antibody directed against acetylated H3 K56, revealed that Asf1p markedly stimulates the ability of Rtt109p to acetylate this site (Fig. 4D), indicating that that Asf1p governs the substrate specificity of Rtt109p. Because we have been unable to obtain evidence for a physical interaction between Asf1p and Rtt109p, we currently favour a model whereby an Asf1p-H3/H4 complex provides the optimal substrate for H3 acetylation by Rtt109p.
Figure 4. Rtt109p displays histone acetyltransferase activity in vitro.
(A) (3H) acetyl-CoA was incubated with histone octamers, BSA, Rtt109p or Asf1p as indicated. The 3H counts were measured and the mean and standard deviation from six independent experiments are shown. (B) Half the reaction mixture from (A) was electrophoresed on a SDS-PAGE gel and subsequently stained with Coomassie brilliant blue, or transferred onto a nitrocellulose membrane and subjected to autoradiography (C). Asterisks mark a degradation product of Asf1p. (D) Alternatively, a nitrocellulose membrane was prepared as in (C) and probed with an antibody specific for H3 K56 Ac.
Taken together, our findings reveal that S. cerevisiae Rtt109p is the predominant HAT for histone H3 K56 in vivo and that this acetylation plays a critical role(s) in conferring resistance to spontaneously-arising or experimentally-induced DNA damage or replication stress. Notably, while the putative acetyl-CoA binding sites of various previously-known acetyl-transferases display some sequence homologies with one another (20), we have not found significant homologies between these and Rtt109p. This raises the possibility that Rtt109p evolved catalytic activity independently of other known HATs, and highlights the prospect of there being further, as yet uncharacterized, acetyl-transferases that have not come to light through sequence analyses. Finally, our findings provide a mechanism for how Asf1 promotes histone H3 acetylation and thereby influences chromatin structure and genome stability.
Supplementary Material
Acknowledgments
We thank A. Aguilera, M. Lisby, R. Rothstein, D.P. Toczyski, Z. Zhang, P. D. Kaufman, T. Bartke and N. Lowndes for strains and reagents and S. Gravel, P. Huertas, A. Bannister and T. Kouzarides for skilful technical advice and critical reading of the manuscript. RD is supported by a BBSRC CASE studentship with KuDOS Pharmaceuticals and by a Cancer Research UK Programme Grant to SPJ. Research in the SPJ laboratory is made possible by core infrastructure funding from Cancer Research UK and the Wellcome Trust.
References
- 1.Scholes DT, Banerjee M, Bowen B, Curcio MJ. Genetics. 2001;159:1449. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett CB, et al. Nat. Genet. 2001;29:426. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- 3.Chang M, Bellaoui M, Boone C, Brown GW. Proc. Natl. Acad. Sci. U.S. A. 2002;99:16934. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinert TA, Hartwell LH. Science. 1988;241:317. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 5.Lisby M, Barlow JH, Burgess RC, Rothstein R. Cell. 2004;118:699. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 6.Lisby M, Rothstein R, Mortensen UH. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8276. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myung K, Chen C, Kolodner RD. Nature. 2001;411:1073. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 8.Aguilera A, Klein HL. Genetics. 1989;122:503. doi: 10.1093/genetics/122.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramey CJ, et al. Mol. Cell. Biol. 2004;24:10313. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prado F, Cortes-Ledesma F, Aguilera A. EMBO Rep. 2004;5:497. doi: 10.1038/sj.embor.7400128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan X, et al. Cell. 2006;124:1069. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 12.Tong AH, et al. Science. 2004;303:808. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 13.Tyler JK, et al. Nature. 1999;402:555. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 14.Linger J, Tyler JK. Genetics. 2005;171:1513. doi: 10.1534/genetics.105.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Recht J, et al. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6988. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozdemir A, et al. J. Biol. Chem. 2005;280:25949. doi: 10.1074/jbc.C500181200. [DOI] [PubMed] [Google Scholar]
- 17.Celic I, et al. Curr. Biol. 2006;16:1280. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 18.Maas NL, Miller KM, DeFazio LG, Toczyski DP. Mol. Cell. 2006;23:109. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Masumoto H, Hawke D, Kobayashi R, Verreault A. Nature. 2005;436:294. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 20.Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V. Cell. 1998;94:427. doi: 10.1016/s0092-8674(00)81584-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.