Abstract
Endoplasmic reticulum-associated degradation (ERAD) is a process that clears the early secretory pathway of misfolded proteins. Though ERAD is of basic biological importance, the clinical importance of this pathway is emphasized by the fact that mutations that render a protein subject to the ERAD quality control pathway underlie the cause of several diseases. The yeast, Saccharomyces cerevisiae, is a valuable and frequently used model system to study biological processes, such as ERAD, as it is a relatively simple model system for which numerous biochemical and genetic tools are available. In addition, the ERAD system is highly conserved between yeast and man. In this chapter, we describe two methods for the analysis of model substrates that undergo catabolism via the ERAD pathway using S. cerevisiae. In particular, we will describe non-radioactive degradation assays and the analysis of substrate ubiquitylation in vivo with or without the use of ubiquitin overexpression systems. We also describe technical hurdles, which we have encountered in our research, and highlight remedies to overcome them.
Keywords: Yeast, ER-associated degradation, Ubiquitin, Proteasome, Cycloheximide chase
1. Introduction
Endoplasmic reticulum-associated degradation (ERAD) is a catabolic process that is of basic biological and clinical relevance. The ERAD pathway is sequential and starts with the recognition of a misfolded protein substrate, followed by substrate ubiquitylation, and then retrotranslocation from the ER for degradation by the 26S proteasome (1). Recent work from a number of fields has led to the identification of numerous ERAD substrates, and has subsequently unraveled an increasingly complex ERAD system equipped to handle substrate diversity (2, 3). One such substrate that is of clinical importance is the cystic fibrosis transmembrane conductance regulator (CFTR). When mutated, CFTR is quantitatively degraded by ERAD in both yeast and man, and even the wild-type form is mostly destroyed by ERAD (4–7). Work in our laboratory used the yeast, Saccharomyces cerevisiae, to uncover a previously unknown and evolutionarily conserved requirement for small heat shock proteins in the degradation of misfolded CFTR (8). We also identified the Hsp90 chaperone and two Hsp40 chaperones as mediators of CFTR stability in yeast (9). Other studies outlined the requirement for specific E3 ubiquitin ligases and the Cdc48p complex, which extracts ERAD substrates from the membrane, in the degradation of CFTR in yeast (10). These findings were made possible by the fact that yeast is a tractable model system with numerous biochemical and genetic tools. A tangible advantage of yeast is the commercial availability of the deletion collection, which contains individual deletions of every non-essential yeast open reading frame. Moreover, a large number of temperature sensitive alleles in essential genes exist.
In this chapter, we outline two valuable methods for analyzing the requirements for the ERAD of two model substrates in yeast. One of these substrates is CFTR, which is an integral membrane protein, and the other is a soluble substrate, a mutated form of Carboxypeptidase Y, which normally traffics to the vacuole (11). In particular, we focus on non-radioactive degradation assays and on the detection of substrate ubiquitylation in vivo with or without the use of ubiquitin overexpression vectors. These assays can be done with a variety of strains, including temperature sensitive alleles, as described below.
2. Materials
2.1. Cycloheximide Chase Assay
Plasmids encoding ERAD substrates: a hemagglutinin-tagged form of wild-type CFTR (CFTR-HA) is expressed from the pSM1152 plasmid (2 µm, URA3, pPGKCFTR::HA) and a hemagglutinin-tagged, mutated form (denoted by the asterisk) of the vacuolar peptidase, Carboxypeptidase Y (CPY*-HA) is expressed from a pRS316 plasmid under control of its endogenous promoter (CEN, URA3, pPRC1prc1*::HA) (5, 12). The indicated yeast strains are transformed with these plasmids and grown on selective media according to established protocols (13, 14).
Synthetic complete medium lacking the nucleotide uracil is used to select for strains containing the pSM1152 and CPY*-HA plasmids. Where indicated, YPD medium (10 g yeast extract, 20 g peptone, 20 dextrose, per liter of water) is used.
Cycloheximide (Microbial source, Sigma): 5 mg/ml in double-distilled water. Filter sterilize and store at −20°C in 1-ml aliquots.
MG132 (26S proteasome inhibitor): 25 mM in dimethyl sulfoxide. Store at −20°C in 100-µl aliquots.
0.5 M sodium azide (see Note 1).
Liquid nitrogen in an insulated Dewar.
Ice-cold double-distilled water freshly supplemented with 1 mM phenylmethanesulfonylflouride (PMSF), 1 µg/ml leupeptin, and 0.5 µg/ml pepstatin A.
2 M NaOH, 1 M β-mercaptoethanol solution. Prepare fresh.
50% Trichloroacetic acid (TCA) (see Note 2).
Ice-cold acetone.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) sample buffer: 80 mM Tris–HCl, pH 8.0, 8 mM ethylenediaminetetracetic acid (EDTA), 3.5% SDS, 15% glycerol, 0.08% Tris base, 0.01% bromophenol blue containing freshly added dithiothreitol (DTT, final conc. 100 mM).
Kontes handheld pestle motor with Kontes disposable microcentrifuge tube pestles.
Ponceau S Red solution.
Tris-Buffered Saline Tween-20 (TBST): 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.1% Tween-20.
Blotto: 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.1% Tween-20, 3% non-fat dry milk, 10 mM sodium azide (see Note 1).
Antibodies: Anti-HA (12CA5) (Roche), anti-Sec61p (rabbit) (15), anti-mouse and anti-rabbit IgG horseradish peroxidase-conjugated secondary antibodies (GE Healthcare).
Nitrocellulose (0.2-µm pore size).
SuperSignal West Pico chemiluminescent substrate (Pierce).
2.2. In Vivo Ubiquitylation Assay
-
Plasmid encoding wild-type CFTR-HA, pSM1152, (2 µm, URA3, pPGK CFTR::HA).
Strains are transformed and grown on selective medium according to established protocols (13, 14).
Synthetic complete medium lacking the nucleotide uracil is used to select for strains containing the pSM1152 and CPY*-HA plasmids. Where indicated, YPD medium (see Subheading 2.1) is used.
0.5 M sodium azide (see Note 1).
Pure Cellulose Chromatography Paper (0.35 mm).
Borosilicate glass culture tubes (5 ml) (Fisher).
Pyrex borosilicate glass Pasteur pipette (9 in.).
Acid-washed glass beads (106 µm) (Sigma).
KNB1: 20 mM HEPES-NaOH, pH 7.4, 50 mM KOAc, 2 mM EDTA, 0.1 M sorbitol, 1 mM DTT (added fresh before use). Store at 4°C and add 1 µg/ml leupeptin, 1 mM PMSF, 0.5 µg/ml pepstatin, and 10 mM N-ethylmaleimide (NEM) immediately before use.
KNB2: 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 2% Triton X-100. Store at 4°C and add 1 µg/ml leupeptin, 1 mM PMSF, 0.5 µg/ml pepstatin, and 10 mM NEM immediately before use.
KNB3: 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1.25% SDS. Store at room temperature and add 1 µg/ml leupeptin, 1 mM PMSF, 0.5 µg/ml pepstatin, and 10 mM NEM immediately before use.
Buffer 88: 20 mM HEPES-NaOH, pH 6.8, 150 mM KOAc, 250 mM sorbitol, 5 mM MgOAc. Store at 4°C and add 1 µg/ml leupeptin, 1 mM PMSF, 0.5 µg/ml pepstatin, and 10 mM NEM immediately before use.
IP wash buffer: 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.2% SDS, 5 mM EDTA. Store at 4°C and add 1 µg/ml leupeptin, 1 mM PMSF, 0.5 µg/ml pepstatin, and 10 mM NEM immediately before use.
SDS–PAGE sample buffer: 80 mM Tris–HCl, pH 8.0, 8 mM EDTA, 3.5% SDS, 15% glycerol, 0.08% Tris base, 0.01% bromophenol blue containing freshly added DTT (final conc. 100 mM) (see Note 3).
Antibodies: Anti-ubiquitin (P4D1 (Santa Cruz), anti-HA (12CA5) (Roche), and anti-mouse IgG horseradish peroxidase-conjugated secondary antibody (GE Healthcare).
Anti-HA antibody-conjugated agarose (Roche).
Protein-A sepharose (GE Healthcare) resuspended in KNB2 to give a 50% (v / v) slurry.
Nitrocellulose (0.2-µm pore size).
SuperSignal West Femto chemiluminescent substrate (Pierce).
Blotto: 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.1% Tween-20, 3% non-fat dry milk, 10 mM sodium azide (see Note 1).
3. Methods
3.1. Cycloheximide Chase Analysis
3.1.1. Cycloheximide Chase
Inoculate a single colony of cells transformed with either the CFTR-HA (pSM1152) or CPY*-HA vector into 25 ml of selective medium and then grow the cells overnight with shaking (200 rpm) at 26–30°C until log-phase (OD600 = 0.6–1.5) is reached (see Note 4).
Harvest 5.0 OD600 in a 15-ml falcon tube by centrifugation at 1000 × g for 5 min in a room temperature clinical centrifuge (see Note 5).
Resuspend the cells in YPD to a final concentration of 1.0 OD600/ml. Incubate at 26–30°C with shaking (200 rpm) for 5 min. This is done to allow the cells to recover from the effects of glucose starvation induced by centrifugation. For use of chemicals, such as MG132, and/or temperature-sensitive strains, see Note 6.
Agitate briefly using a Vortex mixer, then take 1 ml as the zero time point. Add the aliquot to a 1.5-ml microcentrifuge tube containing 30 µl of 0.5 M sodium azide (prepared in step 1, see Note 4). The final concentration of sodium azide is ~20 mM.
Pellet the cells at 15,000 × g for 1 min in a 4°C table-top microcentrifuge, aspirate the supernatant using a vacuum aspirator, and snap-freeze the pellet in liquid nitrogen. Frozen cell pellets should be stored at −80°C.
To the rest of the yeast culture, add cycloheximide to a final concentration of 200 µg/ml, agitate briefly on a Vortex mixer, and incubate the culture at 40°C for CFTR-HA or 30°C for CPY*-HA in a shaking water bath with agitation (150–200 rpm) (see Note 7).
Repeat steps 4 and 5 at additional desired time points (see Notes 7 and 8). Store all samples at −80°C until processing.
3.1.2. Sample Processing by TCA Precipitation (See Note 9)
Thaw the samples at 4°C or on ice (see Note 10).
Add 1 ml of double-distilled water supplemented with the indicated protease inhibitors (see Subheading 2.1) and resuspend the pellet.
Add 150 µl of 2 M NaOH, 1 M β-mercaptoethanol to each sample, agitate briefly on a Vortex mixer, and incubate at 4°C or on ice for 15 min.
Add 130 µl of 50% TCA, agitate briefly on a Vortex mixer, and incubate at 4°C or on ice for 15 min.
Pellet the precipitated proteins in a 4°C microcentrifuge at 15,000 × g for 10 min.
Aspirate and discard the supernatant using a vacuum aspirator and add 0.5 ml of ice-cold acetone.
Agitate briefly using a Vortex mixer and centrifuge in a 4°C microcentrifuge at 15,000 × g for 2 min.
Aspirate and discard the supernatant using a vacuum aspirator and allow the pellet to air-dry for 5 min.
Add 100 µl of SDS–PAGE sample buffer to the pellet and let sit at room temperature for 5 min.
Vigorously grind the pellet for 10 s with a Kontes handheld pestle motor with Kontes disposable microcentrifuge tube pestle to resuspend the pellet. The SDS–PAGE sample buffer should appear cloudy, and no white clumps of precipitate should be visible.
3.1.3. Gel Analysis
Heat samples at 37°C for 20 min for CFTR-HA or 75–100°C for 5 min for CPY*-HA containing samples (see Note 11).
Centrifuge samples at 15,000 × g for 1 min in a room temperature microcentrifuge prior to gel loading (see Note 12).
Load 10 µl of each sample on an 8.25 cm × 8.25 cm 10% denaturing polyacrylamide gel and electrophorese the gels at 120 V (constant voltage).
Transfer the proteins onto nitrocellulose overnight at 12 V (constant voltage) (see Note 13).
Remove the nitrocellulose membrane and wash once with double-distilled water to remove any adherent gel pieces. Incubate briefly with Ponceau S Red to check the quality of the transfer, decant the Ponceau S Red and then destain with three 5-min washes with TBST.
Block the membranes with Blotto for 30 min with gentle rocking, and then incubate the membranes with anti-HA (1:5,000) or anti-Sec61p (1:5,000) antibodies diluted in Blotto for 3 h or overnight with gentle rocking.
Wash the membranes three times for 10 min each with TBST.
Incubate with horseradish peroxidase-conjugated secondary antibodies (1:5,000) diluted in TBST for 1–2 h.
Wash the membranes three times for 10 min each with TBST.
Membranes are developed with Pierce SuperSignal West Pico chemiluminescent substrate and images are analyzed using ImageJ software (16)(see Note 14).
Quantified data are typically presented such that the signal at the zero time point is set to 100% and the subsequent time points are represented as a relative percentage to the zero time point (see Figs. 1 and 2).
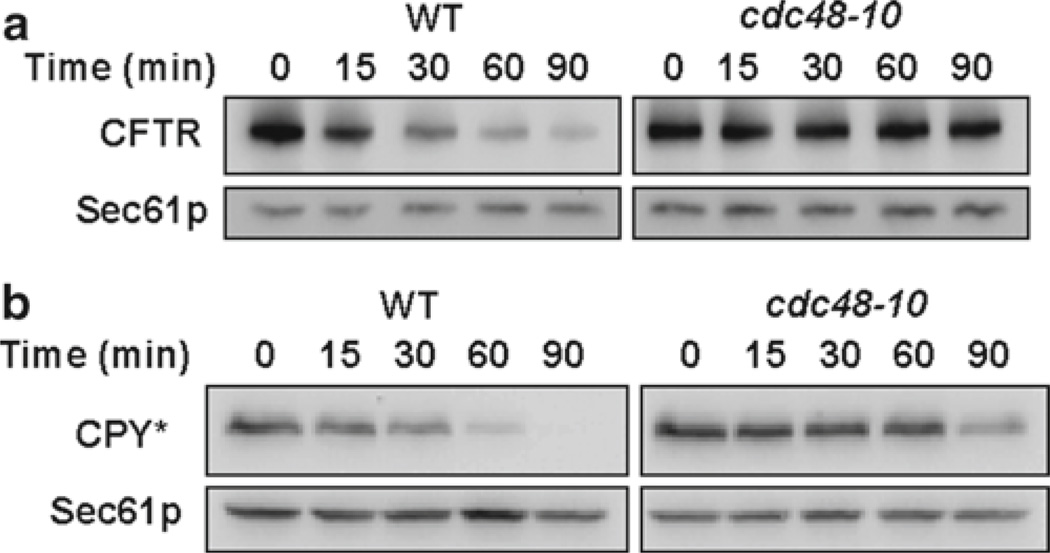
Fig. 1.
The cycloheximide chase analysis of CFTR-HA (a) and CPY*HA (b) was performed as described in Subheading 3.1 using a strain that contains a thermo-sensitive allele in the CDC48 gene (cdc48-10) and in an isogenic wild type. Western blots were probed with anti-HA antibody to detect substrate and anti-Sec61p antibody was used as a loading control
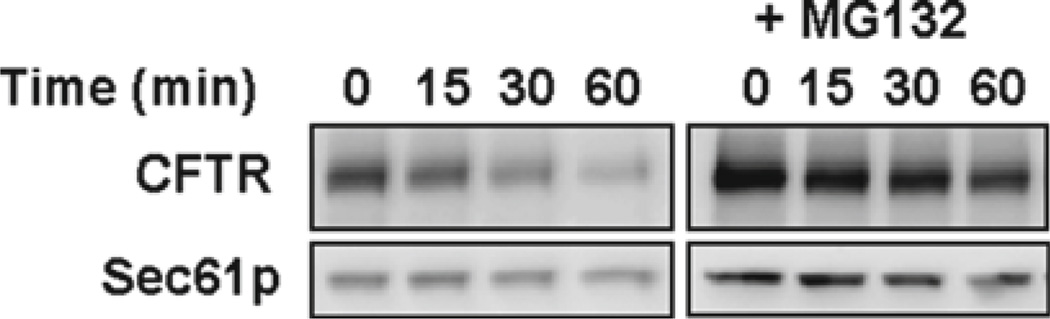
Fig. 2.
The cycloheximide chase analysis of CFTR-HA was performed as described in Subheading 3.1 using a strain lacking the multidrug resistance pump, PDR5. Cells were incubated with MG132 for 1 h prior to the cycloheximide chase (see Note 6). Western blots were probed with anti-HA antibody to detect the CFTR-HA substrate and with anti-Sec61p antibody as a loading control.
3.2. Analysis of CFTR Substrate Ubiquitylation In Vivo
3.2.1. Preparation of a Crude Membrane Fraction
Inoculate a single colony of cells transformed with the CFTR-HA (pSM1152) expression vector into 100 ml of selective medium. Incubate the culture overnight at 26–30°C with shaking (~200 rpm) until log-phase (OD600 = 0.6–1.5) is achieved (see Notes 4). If using the myc-ubiquitin overexpression system, please see Note 15.
Add 2 ml of 0.5 M sodium azide to the 100 ml of culture and harvest the cells by centrifugation in a clinical centrifuge at 4°C at 1000× g for 5 min (see Note 16).
Decant the supernatant and resuspend the pellet in 50 ml of ice-cold water supplemented with 10 mM sodium azide. Centrifuge the mixture as in step 2.
Decant the water and resuspend the cells in 10 ml of ice-cold KNB1. Centrifuge the mixture as in step 2.
Resuspend the cells in 200 µl ice-cold KNB1 and transfer the mixture to a pre-chilled borosilicate tube. Keep the tube on ice.
Add glass beads to 3/4 volume and disrupt the cells by agitation on a Vortex mixer ten times for 30 s, leaving the mixture on ice for 30 s in between each pulse.
Collect the lysate using a Pasteur pipette that has been pulled to a fine point and transfer the extract to a 1.5-ml microcentrifuge tube (see Note 17)
Wash the glass beads with an additional 200 µl of Buffer 88, agitate briefly on a Vortex mixer, and pool with the lysate from step 7.
Pellet unbroken cells and large organelles by centrifugation at 1000 × g for 5 min in a refrigerated microcentrifuge.
Remove the supernatant without disturbing the pellet and transfer the solution to a new 1.5-ml microcentrifuge tube.
Centrifuge the supernatant again at 1000×g for 5 min in a refrigerated microcentrifuge.
Remove the supernatant and transfer to another 1.5-ml microcentrifuge tube.
Separate the crude membrane from cytosol fractions by centrifuging the supernatant fractions in steps 10 and 12 at 15,000 × g for 20 min at 4°C (see Note 18).
Aspirate and dispose of the supernatant using a vacuum aspirator and resuspend the crude membrane pellet with 1 ml of Buffer 88.
Centrifuge the resuspended membranes by centrifuging at 15,000 × g for 10 min.
Aspirate and dispose of the supernatant and resuspend the washed pellet in 1 ml of Buffer 88. Repeat the centrifugation again as described in step 15.
Thoroughly resuspend the crude membranes in 50 µl of Buffer 88 by pipetting up and down at least 30 times. The membranes should be homogenous and lack aggregates.
Add 5 µl of the membrane suspension to 995 µl of 2% SDS (1/200 dilution) and measure the absorbance of the mixture at 280 nm. The ideal absorbance of this solution is 0.2, which when multiplied by the dilution factor of 200 gives a value of 40 and corresponds to approximately 11 mg/ml of protein (see Note 19).
Prepare 23-µl aliquots of membranes and snap-freeze in a Dewar of liquid nitrogen. Store the aliquoted crude membrane fractions at −80°C.
3.2.2. Immunoprecipitation
Thaw membranes on ice and then gently mix 20 µl with 80 µl of room temperature KNB3. Mix gently by slowly pipetting up and down.
Incubate at 37°C for 20 min.
Add 400 µl of ice-cold KNB2 and agitate briefly on a Vortex mixer.
Remove the insoluble material by centrifugation at 15,000 × g for 5 min at 4°C.
Transfer the clarified supernatant to a fresh 1.5-ml microcentrifuge tube.
Preclear the extract by adding 20 µl of Protein A-sepharose (see Subheading 2.2) and rotating mixture at 4°C for 1–2 h.
Pellet the nonspecifically bound proteins in a microcentrifuge at 15,000 × g for 1 min at 4°C.
Transfer the cleared supernatant to a fresh 1.5-ml microcentrifuge tube.
Add 25 µl of anti-HA antibody-conjugated agarose and rotate the immunoprecipitation overnight at 4°C (see Note 20).
Pellet the immunoprecipitated material at 15,000 × g for 1 min in a microcentrifuge at 4°C.
Aspirate the supernatant with a vacuum aspirator, making sure to remove as much of the supernatant as possible (see Note 21).
Wash the immunoprecipitate three times with 600 µl of IP Wash Buffer. Repeat steps 10 and 11 in between each wash.
Elute immunoprecipitated CFTR-HA by adding 30 µl of SDS–PAGE sample buffer and heating at 37°C for 20 min. This mixture can be stored at −20°C until use. If stored at −20°C, incubate the mixture again at 37°C for 20 min before gel analysis.
3.2.3. Gel Analysis
If the precipitate from section 3.2.2, step 13, is stored at −20°C, heat the immunoprecipitated material at 37°C for 20 min as described, or otherwise proceed to step 2.
Pellet the resin at 15,000 × g for 1 min at room temperature.
Load 12 µl of the supernatant onto an 8.25 cm × 8.25 cm 7.5% denaturing polyacrylamide gel and electrophorese the gel at 110 V (constant voltage) until the dye front reaches the very bottom. Run duplicate gels: one will be used to detect ubiquitylated CFTR-HA and the other will be used to detect unmodified CFTR-HA.
Transfer the proteins onto a nitrocellulose membrane overnight at 12 V (see Note 14).
Remove the nitrocellulose membranes and wash once with double-distilled water to remove any residual polyacrylamide gel. Trim the nitrocellulose with a pair of scissors so there is not an excessive amount of extra, unused nitrocellulose. Store the nitrocellulose membrane in TBST until use.
Boil ~1.5 L of water in a 2-L beaker on a hot plate (see Note 22).
Cut four pieces of Pure Cellulose Chromatography Paper to be ~2 cm larger than the blot on each side, pre-moisten the chromatography paper with double-distilled water, and sandwich the blot between two sheets of chromatography paper on each side of the blot. Staple the blot closed, being sure to only staple the edge of the chromatography paper.
Place the sandwiched blot in the boiling water for 20–40 min.
Remove the beaker from the heat and let the water cool to the point where the beaker is warm to the touch before removing the sandwiched blot.
Immediately rinse the membrane with double-distilled water.
Block the membranes with Blotto for 30 min with gentle rocking. Discard the blocking agent and then incubate one membrane with anti-ubiquitin (1:1,000) and the other with anti-HA (1:5,000) antibody in Blotto for 3–16 h at 4°C with gently rocking.
Wash the membranes three times for 10 min each with TBST.
Incubate with horseradish peroxidase-conjugated anti-mouse secondary antibody (1:5,000) diluted in TBST for 1–2 h.
Wash the membranes three times for 10 min each with TBST.
Anti-ubiquitin blot is developed with the Pierce SuperSignal West Femto substrate kit and the anti-HA blot is developed with the Pierce SuperSignal West Pico kit (see Fig. 3). Images are analyzed using ImageJ software (16) (see Note 14).
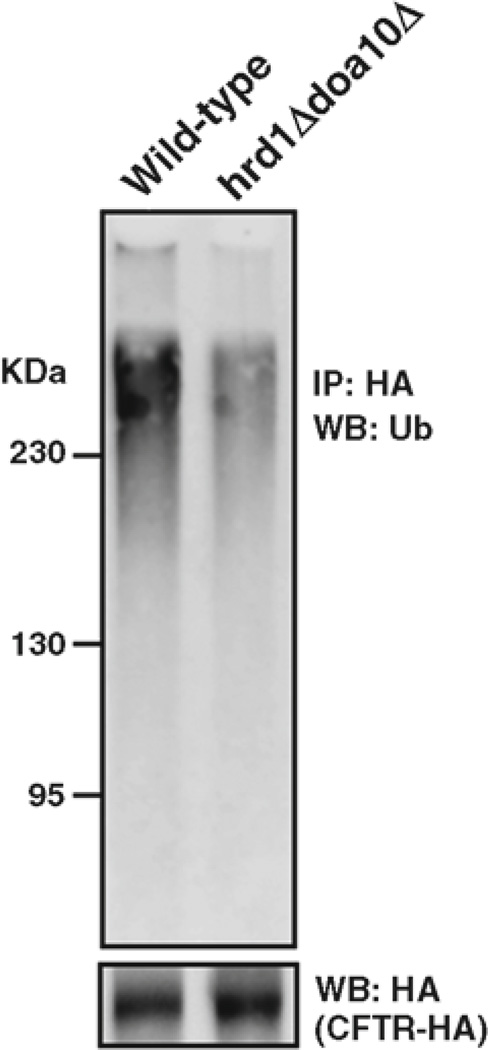
Fig. 3.
The in vivo ubiquitylation assay was performed as described in section 3.2. CFTR-HA was immunoprecipitated with anti-HA-conjugated agarose from a strain lacking both the Hrd1p and Doa10p E3 ubiquitin ligases and an isogenic wild-type strain. The immunoprecipitated CFTR-HA was resolved on a 10% denaturing polyacrylamide gel and probed with anti-ubiquitin and anti-HA antibodies.
Acknowledgments
This work was supported by grant GM75061 and by grant DK79307 (“The Pittsburgh Center for Kidney Research”) from the National Institutes of Health.
Footnotes
Notes
Sodium azide is highly toxic. Gloves, goggles, and protective clothing should be worn during the preparation, handling, and use of sodium azide.
Trichloroacetic acid is highly corrosive. Gloves, goggles, and protective clothing should be worn during the preparation, handling, and use of trichloroacetic acid.
SDS–PAGE sample buffer can be supplemented with 1 µg/ml leupeptin, 1 mM PMSF, 0.5 µg/ml pepstatin, and 10 mM NEM immediately before use. Supplementing the SDS–PAGE sample buffer does not appear to affect sample electrophoresis.
This step is not trivial as some strains, in general, grow poorly and especially when expressing an ERAD substrate. In cases where cells are growing at different rates, we simply perform the cycloheximide chase experiment with cells that are in log-phase, and then perform the experiment with the slower growing cells in a parallel experiment when they too reach log-phase. Alternatively, we sometimes start 5 ml cultures and grow to an OD600 of >1.0. We then dilute the cells to an OD600 of 0.1–0.15 in the morning and allow them to grow to mid-log-phase. This is not always possible with some ERAD substrates (17). From our experience, it is not the linear phase of growth that is slow, but the lag phase is elongated.
For convenience, one can set a shaking water bath to the appropriate temperature of 40°C for CFTR-HA or 30°C for CPY*-HA and allow the water bath to equilibrate overnight. We normally also prepare and label microcentrifuge tubes at this time. Finally, it is convenient to pre-dispense 30 µl of 0.5 M sodium azide into each microcentrifuge tube and store the tubes at 4°C until use.
We normally take enough cells to give 1.0 OD600 per desired time point, plus one extra OD to account for evaporation and possible pipetting error.
For some substrates, such as ENaC, the centrifugation step leads to a rapid and unexplained loss of signal (17). This is possibly an effect of increased cell density and/or glucose deprivation. Nevertheless, the outcome with CFTR-HA or CPY*-HA cycloheximide chases is unaffected by harvesting the cells by centrifugation.
At this point, if one wants to examine whether or not degradation of a particular substrate is dependent on the 26S proteasome, one would incubate the cells with MG132 to a final concentration of 25–50 µM for an additional h at 26–30°C with shaking. For MG132 to be effective, strains lacking the multidrug exporter, PDR5, or deleted for ERG6 must be used (18, 19).
Aside from MG132, other drugs and/or chemicals can be added and incubated for the desired amount of time prior to step 4. For instance, a 15 min incubation with 20 µM cadmium was used to stabilize Pca1p prior to the cycloheximide chase assay (20). For temperature sensitive strains, this is also the point when cells can be shifted to non-permissive temperatures. For example, the cdc48-3 strain is shifted to a non-permissive temperature of 38°C for 4 h with shaking prior to use (21). One caveat is that cells may still grow during excessively long incubations with drugs, chemicals, or at non-permissive temperature, and this might necessitate checking the OD600 of the cells prior to performing the experiment. Alternatively, the compounds may be toxic and long incubations may give rise to secondary, non-specific effects.
An unexpected effect of cycloheximide exposure is the depletion of free ubiquitin, because ubiquitin has an estimated half-life of approximately 2 h (22). To minimize the impact of ubiquitin depletion on substrate clearance during the cycloheximide chase assay, we normally perform our cycloheximide chases experiments to a maximum of 120 min. This might be particularly relevant to strains that have defect in ubiquitin turnover, e.g., DOA1 / UFD3 mutants.
For CFTR-HA and CPY*-HA, we normally take 0, 30, 60, and 90-min time points. For substrates with unknown half-lives, it is ideal to allow enough time to reach the half-life of the substrate in wild-type cells, but less than 2 h (see Note 7).
We have used the simplified protein extraction method of Kushnirov (23) with equivalent results for CFTR-HA. For radioactive pulse-chase analysis, please see Chapter 35 by Wolf and colleagues in this volume.
Directly thawing samples from the −80°C freezer often results in a build-up of pressure as the air expands, causing the caps of the tubes to “pop” open. In order to avoid this and the potential loss of sample we place take the tubes from the −80°C and place them at −20°C for 10 min prior to thawing at 4°C.
Heating CFTR-HA-containing samples at temperatures higher than 37°C can result in CFTR-HA aggregation (24). The HA-reactive signal is then found trapped in the well of an SDS–PAGE, or at the top of the separating phase of the gel.
After heating the samples, the vast majority of cell debris remains in the insoluble pellet. From our experience, quantitation of samples without centrifugation versus with centrifugation gives similar quantitation results for both CFTR-HA and CPY*-HA. One noticeable difference is the quality of the band; samples that were centrifuged give sharper bands.
We have also transferred blots at 35 V for 2–2.5 h.
We use a Kodak Image Station 440cf equipped with a Charge-Coupled Device camera to capture images. It is essential to avoid chemiluminescent signal saturation.
In some cases, we have introduced the copper-inducible myc-ubiquitin construct, YEp105 (25) to increase the signal in select strains. Incubating cells with 100 µM of CuSO4 for 3 h gives an even greater signal.
At this point in time, it is a good idea to chill borosilicate tubes on ice. Pasteur pipettes can also be pulled to a fine point using the flame of a Bunsen burner.
An alternative to using flame-pulled Pasteur pipettes is to use gel loading tips. We, however, prefer Pasteur pipettes because the glass is rigid enough to pierce through the slurry of glass beads to reach extract at the bottom of the borosilicate tube.
Unexpectedly, using excessively high forces (e.g., 150,000 × g) to separate membranes from cytosol leads to inconsistent CFTR ubiquitylation results.
If the absorbance is lower than 0.2, we readjust the volume by first centrifuging the crude membrane fraction at 15,000 × g for 10 min, and then pipetting off the required amount of supernatant. We then resuspend the membranes as done in section 3.2.1, step 17. If the absorbance value is greater than 0.2, we simply add the appropriate amount of Buffer 88 and mix thoroughly as described in section 3.2.1, step 17.
Immunoprecipitations can also be performed for as few as 3 h.
The appearance of the HA-agarose beads turns from light gray to white when the supernatant is completely removed.
To save time, it is quicker to microwave the 1.5 L of water for ~10 min before placing it on a preheated hot plate.
References
- 1.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brodsky JL. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation) Biochem J. 2007;404:353–363. doi: 10.1042/BJ20061890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Nijbroek G, Sullivan ML, et al. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward CL, Kopito RR. Intracellular turn-over of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- 7.Jensen TJ, Loo MA, Pind S, et al. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 8.Ahner A, Nakatsukasa K, Zhang H, et al. Small heat-shock proteins select deltaF508-CFTR for endoplasmic reticulum-associated degradation. Mol Biol Cell. 2007;18:806–814. doi: 10.1091/mbc.E06-05-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Youker RT, Walsh P, Beilharz T, et al. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis trans-membrane conductance regulator degradation in yeast. Mol Biol Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gnann A, Riordan JR, Wolf DH. Cystic fibrosis transmembrane conductance regulator degradation depends on the lectins Htm1p/EDEM and the Cdc48 protein complex in yeast. Mol Biol Cell. 2004;15:4125–4135. doi: 10.1091/mbc.E04-01-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- 12.Bhamidipati A, Denic V, Quan EM, et al. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 13.Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser C, Michaelis S, Mitchell A, et al. Methods in yeast genetics : a Cold Spring Harbor Laboratory course manual. 1994 ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 15.Stirling CJ, Rothblatt J, Hosobuchi M, et al. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 17.Buck TM, Kolb AR, Boyd CR, et al. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol Biol Cell. 2010;21:1047–1058. doi: 10.1091/mbc.E09-11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming JA, Lightcap ES, Sadis S, et al. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc Natl Acad Sci USA. 2002;99:1461–1466. doi: 10.1073/pnas.032516399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee DH, Goldberg AL. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J Biol Chem. 1996;271:27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- 20.Adle DJ, Lee J. Expressional control of a cadmium-transporting P1B-type ATPase by a metal sensing degradation signal. J Biol Chem. 2008;283:31460–31468. doi: 10.1074/jbc.M806054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakatsukasa K, Huyer G, Michaelis S, et al. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 24.Glozman R, Okiyoneda T, Mulvihill CM, et al. N-glycans are direct determinants of CFTR folding and stability in secretory and endocytic membrane traffi. J Cell Biol. 2009;184:847–862. doi: 10.1083/jcb.200808124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochstrasser M, Ellison MJ, Chau V, et al. The short-lived MAT alpha 2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]