Abstract
Mieap, a p53-inducible protein, controls mitochondrial integrity by inducing the accumulation of lysosomal proteins within mitochondria. This phenomenon is designated MALM, for Mieap-induced accumulation of lysosome-like organelles within mitochondria. To identify this novel Mieap-interacting protein(s), we performed a two-dimensional image-converted analysis of liquid chromatography and mass spectrometry (2DICAL) on the proteins immunoprecipitated by an anti-Mieap antibody. We indentified 14-3-3γ as one of the proteins that was included in the Mieap-binding protein complex when MALM was induced. The interaction between Mieap and 14-3-3γ was confirmed on the exogenous and endogenous proteins. Interestingly, 14-3-3γ was localized within mitochondria when MALM occurred. A 14-3-3γ deficiency did not affect the accumulation of Mieap and lysosomal proteins within mitochondria, but dramatically inhibited the elimination of oxidized mitochondrial proteins. These results suggest that 14-3-3γ plays a critical role in eliminating oxidized mitochondrial proteins during the MALM process by interacting with Mieap within mitochondria.
Mitochondria are the sites of oxidative phosphorylation for cellular energy, a process that inevitably generates reactive oxygen species (ROS) as byproducts1,2. Therefore, mitochondria are a major source of ROS, and they are consequently highly susceptible to ROS damage. Damaged mitochondria produce much higher levels of ROS than do intact mitochondria. This increase may be due to abnormal electron transfer by dysfunctional respiratory chain proteins, impaired ATP production by dysfunctional ATP synthase proteins, and/or decreased NADH supply caused by dysfunctional TCA cycle proteins. These ROS also oxidize mitochondrial proteins, including the core proteins of energy production, leading to a vicious cycle and an accumulation of unhealthy mitochondria3,4. Furthermore, the ROS generated by unhealthy mitochondria oxidize and damage intracellular DNA, RNA, lipids, and proteins, thereby leading to a variety of cellular dysfunctions, including degenerative diseases, cancer, and aging5,6. Therefore, efficiently eliminating oxidized mitochondrial proteins and preventing mitochondrial ROS generation are critical for mitochondrial quality control.
Mieap, a p53-inducible protein, was originally identified as a key regulator of a novel mitochondrial repair system7; this phenomenon, which is designated MALM (for Mieap-induced accumulation of lysosome-like organelles within mitochondria), is critically different from canonical autophagy7. In this mechanism, Mieap induces an accumulation of intramitochondrial lysosomal proteins to eliminate oxidized mitochondrial proteins in response to mitochondrial damage7. This process leads to a decrease in ROS generation and an increase in mitochondrial ATP synthesis activity7. Therefore, this function likely mediates the repair of unhealthy mitochondria. Alternatively, another mechanism has been designated MIV, for Mieap-induced vacuole8. When MALM is inhibited, Mieap induces a vacuole-like structure known as the MIV. The MIV engulfs damaged mitochondria and fuses with lysosomes, leading to degradation of the unhealthy mitochondria. The function of the MIV is likely to act as a type of mitochondrial autophagy. Therefore, Mieap controls mitochondrial quality by repairing or eliminating unhealthy mitochondria via MALM or MIV generation, respectively7,8. Inactivating p53 or Mieap severely impairs both MALM and MIV, leading to an accumulation of unhealthy mitochondria7,8. Although Mieap-mediated mitochondrial quality control appears to be critical for a variety of diseases and biological responses, a large part of the mechanism still remains to be elucidated.
Although an accumulation of lysosomal proteins within the intramitochondrial space is evident from considerable data obtained in previous studies7, we are unaware of any molecules proven to be related to the processes of MALM-mediated mitochondrial repair, including recognizing unhealthy mitochondria, translocating lysosomal proteins into mitochondria, and degrading oxidized mitochondrial proteins. Despite the complexity of the MALM mechanisms, few molecules have been identified as MALM related. Therefore, a comprehensive identification of MALM-related molecules is required for elucidating the MALM regulatory mechanisms. Consequently, we sought to identify novel candidate MALM-related proteins by analyzing the cellular polypeptides that bind to Mieap under MALM-induced conditions.
To achieve this aim, we used two-dimensional image-converted analyses of liquid chromatography (LC) and mass spectrometry (MS) (2DICAL) to examine immunoprecipitates and identify Mieap-interacting proteins. 2DICAL is a labeling-free, MS-based quantitative proteomics platform9. In 2DICAL, large peptide data sets are defined as peaks in a single two-dimensional image with m/z values along the x-axis and retention time (RT) along the y-axis. The peaks derived from different LC-MS runs align accurately with high sensitivity and reproducibility, allowing quantitative comparisons and successful implementations of experiments with large numbers of samples10,11,12,13,14. This property of 2DICAL enables us to determine quantitative differences in immunoprecipitates between samples. Using this method, it is possible to select the proteins of interest that are significantly different between two or more immunoprecipitates. We designated this method as “immunoprecipitation-2DICAL” (IP-2DICAL).
Here, we describe identifying 14-3-3γ, a member of the 14-3-3 family of proteins, as a Mieap-interacting protein by IP-2DICAL. We demonstrated that 14-3-3γ is translocated from the cytoplasm to an intramitochondrial space during MALM. Furthermore, we showed that intramitochondrial 14-3-3γ is involved in degrading oxidized mitochondrial proteins.
Results
Identification of 14-3-3γ as a novel Mieap-interacting protein by IP-2DICAL
To clarify the MALM mechanism, we attempted to identify Mieap-interacting proteins. For this purpose, we developed the IP-2DICAL strategy depicted in Supplementary Figure S1. The immunoprecipitates were prepared from MALM-induced A549 cells using an anti-Mieap antibody and normal rabbit globulin (rIgG) as a control. Western blotting with the Mieap antibody indicated that endogenous Mieap was clearly immunoprecipitated in the Mieap-immunoprecipitated samples (Fig. 1a, lane Mieap & IP) but not in the control rIgG-immunoprecipitated samples (Fig. 1a, lane rIgG & IP). Mixtures of the proteins eluted from the immunoprecipitates were trypsinized, and the MS spectral data obtained were analyzed by 2DICAL. A total of 33,723 independent MS peaks within the 350–1,600-m/z mass range with an LC RT of 10–110 min were determined to be comparable between the samples (Fig. 1b).
Figure 1. Identifying 14-3-3γ as a Mieap-binding protein by IP-2DICAL.
(a) Preparation of the samples for identifying endogenous Mieap-interacting proteins by IP-2DICAL. The A549 cells were γ-irradiated, and 40 h after this ionizing radiation (IR), the cell lysates were immunoprecipitated with anti-Mieap antibody (Mieap) or normal rabbit globulin (rIgG), as indicated. The immunoprecipitates were blotted with anti-Mieap antibody. Pre: total cell lysate before IP, Post: total cell lysate after IP, and IP: immunoprecipitated proteins. (b) A two-dimensional display of all (>33,000) the MS peaks in a representative sample prepared as in (a), with the m/z-value (350–1,600 m/z) along the horizontal (x) axis and the LC RT (10–110 min) along the vertical (y) axis. (c) The peak at 540 m/z and 34.9 min displayed in various combinations of axes. The immunoprecipitates generated by the anti-Mieap antibody are indicated in red and the immunoprecipitates from the control rIgG are indicated in blue. Upper left, the m/z and intensity axes, with indicators of the isotopic mass (light blue line and dot). Lower left, a gray-scale intensity pattern of the RT (x axis) and sample (y axis). Upper right, the sample and intensity axes (left) and a box-and-whisker diagram of the immunoprecipitates generated using anti-Mieap antibody and rIgG (right). Lower right, the m/z and RT axes with high (upper) and low (lower) intensities are indicated by a red dot. (d) The interaction of endogenous 14-3-3γ and Mieap proteins. The irradiated A549 cell lysates were generated as in (a) and were subjected to immunoprecipitation with the anti-Mieap antibody (Mieap). Immunoprecipitates generated using rIgG were employed as a control (rIgG). The immunoblot analysis was performed using the anti-14-3-3γ and anti-Mieap antibodies. Lysate: total cell lysate and IP: immunoprecipitated proteins. (e) The interaction of exogenous 14-3-3γ and Mieap proteins. The HCT116 cells were co-transfected with FLAG-tagged 14-3-3γ and HA-tagged Mieap α or β. The transfected cells were γ-irradiated, and 72 h after IR, the cell lysates were immunoprecipitated with anti-FLAG or anti-HA antibodies. Immunoblot analysis was carried out with the anti-FLAG or anti-HA antibodies. IB: immunoblotting, IP: immunoprecipitation, and lysate: total cell lysate.
We found 3,201 peaks for which the average intensity of the duplicates exhibited a statistically significant increase in the Mieap immunoprecipitates (>2-fold difference in intensity, P < 0.01 [T-test]). The MS/MS spectra of the 3,201 peaks were matched against the contents of the SwissProt database using the Mascot software package (version 2.2.1) (Matrix Science, London, UK), and dozens of proteins were identified as Mieap-interacting protein candidates with significance values below 0.05 (data not shown). In these candidates, we identified two MS/MS spectrum peaks at 540 m/z (34.9 min) and 822 m/z (44.3 min) that matched the YLAEVATGEK and NVTELNEPLSNEER sequences, respectively, of 14-3-3γ (NP_036611) with the highest Mascot scores (Supplementary Fig. S2).
The 2DICAL reports for various two-dimensional views of these peaks revealed significant differences between the Mieap- and rIgG-immunoprecipitated samples (Fig. 1c and Supplementary Fig. S3). Although the 14-3-3 isoforms are known to be highly homologous proteins with approximately 50% amino acid identity, two peptide sequences could be identified as being derived from 14-3-3γ (Supplementary Fig. S4). Because 14-3-3 proteins are major regulators of various proteins that are involved in diverse cellular processes15, we decided to focus on 14-3-3γ's interaction with Mieap among the Mieap-interacting protein candidates.
To confirm the interaction of Mieap and 14-3-3γ, endogenous Mieap was immunoprecipitated from the MALM-induced A549 cells. Western blotting with anti-14-3-3γ antibody indicated that 14-3-3γ was coprecipitated with endogenous Mieap under physiological conditions, while no 14-3-3γ was coprecipitated with control rIgG (Fig. 1d).
Endogenous Mieap has two alternative splicing variants, termed Mieap α and Mieap β, although the functional differences between endogenous Mieap α and β have yet to be elucidated [7, 8]. To determine whether Mieap α, Mieap β, or both variants interact with 14-3-3γ, HCT116 cells (in which endogenous Mieap expression is completely suppressed by methylation of the Mieap promoter7) were transfected with these constructs, and coimmunoprecipitation assays were performed. Our results indicated that similar amounts of 14-3-3γ were co-precipitated with both Mieap α and Mieap β (Fig. 1e). Reciprocal immunoprecipitation experiments also confirmed that Mieap α and Mieap β can be co-immunoprecipitated with 14-3-3γ (Fig. 1e). These results demonstrated that Mieap is associated with 14-3-3γ during the process of MALM, suggesting that 14-3-3γ may play a role in Mieap function.
Mapping of the interaction domains of Mieap and 14-3-3γ
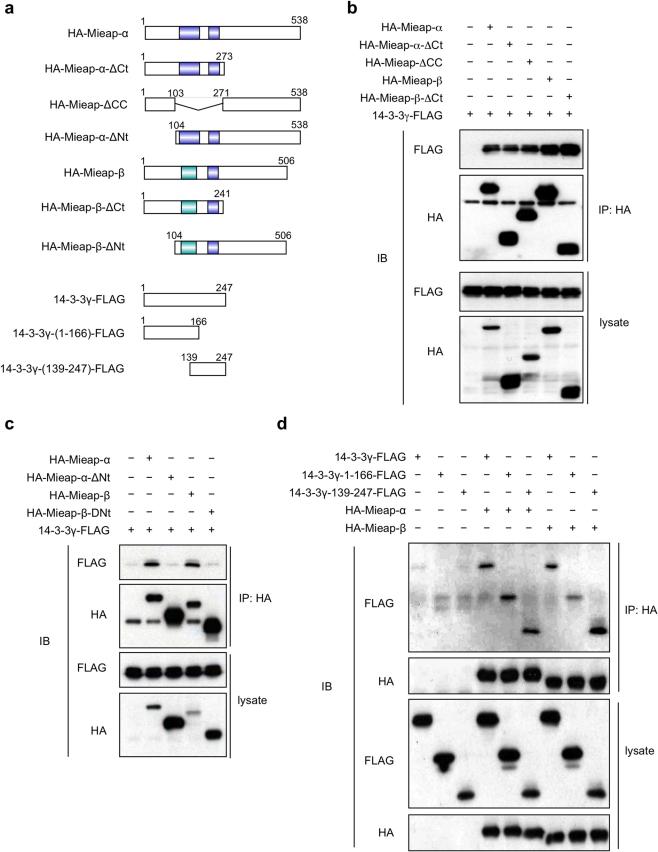
Several deletion mutants of Mieap α and β were constructed to determine the region responsible for their interaction with 14-3-3γ (Fig. 2a). Our results indicated that the N-terminal regions of Mieap α and β (ΔCt: α 1–273 and β 1–241), were sufficient for interaction with 14-3-3γ (Fig. 2b). We further confirmed that the coiled-coil domain-deleted Mieap mutant (ΔCC) also associates with 14-3-3γ at a level similar to that of the full α and full β Mieap proteins, suggesting that 14-3-3γ interacts with Mieap via its 1–103 amino acid region (Fig. 2b). To confirm this hypothesis, we constructed both Mieap α and β mutants by deleting amino acids 1–103 and designated them ΔNt (α 104–538 and β 104–506) (Fig. 2a). As expected, these deletion mutants could not associate with 14-3-3γ (Fig. 2c). Our results indicated that the 1–103 Mieap fragment contains the 14-3-3γ-binding site.
Figure 2. Mapping of the domains responsible for the interaction between Mieap and 14-3-3γ.
(a) The Mieap and 14-3-3γ deletion mutants. The numbers of amino acid residues are indicated. (b and c) Mieap interacts with 14-3-3γ via the N-terminal region of Mieap. The HCT116 cells were co-transfected with FLAG-tagged 14-3-3γ and various HA-tagged Mieap mutants and were γ-irradiated. Seventy-two hours after the IR, the cell lysates were immunoprecipitated with anti-HA antibody. An immunoblot analysis was performed using the anti-FLAG or anti-HA antibody. α: Mieap α, β: Mieap β, IP: immunoprecipitation, lysate: total cell lysate, and IB: immunoblotting. (d) 14-3-3γ interacts with Mieap β via the PB domain of 14-3-3γ. The HCT116 cells were co-transfected with HA-tagged Mieap and various FLAG-tagged 14-3-3γ mutants and were γ-irradiated. Seventy-two hours after the IR, the cell lysates were immunoprecipitated with anti-HA antibody. An immunoblot analysis was performed using the anti-FLAG or anti-HA antibodies. IP: immunoprecipitation, lysate: total cell lysate, and IB: immunoblotting.
To determine which region of 14-3-3γ is responsible for the interaction with Mieap, two 14-3-3γ deletion mutants were constructed next: the protein-binding (PB) domain-deficient 14-3-3γ (14-3-3γ 1–166) and the dimerization (DM) domain-deficient 14-3-3γ (14-3-3γ 139–247) mutants (Fig. 2a)16. As shown in Fig. 2d, 14-3-3γ 139–247 was able to bind to both Mieap α and β proteins. However, 14-3-3γ 1–166 displayed impaired binding with Mieap β but was able to bind to Mieap α. Consistent with the prior reports, the PB domain of 14-3-3γ likely plays a critical role in the Mieap β interaction, but both the PB and DM domains of 14-3-3γ can be independently sufficient for interaction with Mieap α.
Translocation of 14-3-3γ from the cytoplasm to the inside of mitochondria
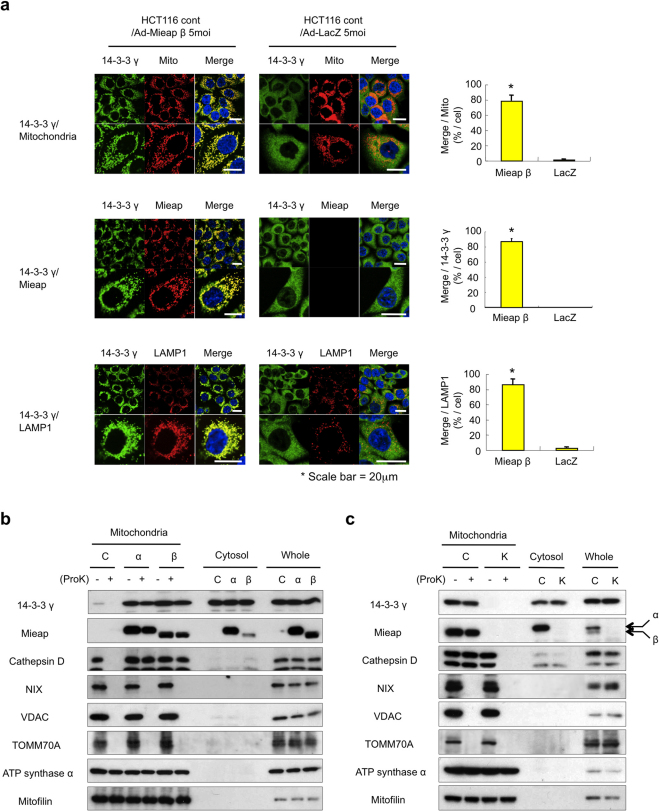
To clarify the subcellular location where 14-3-3γ interacts with Mieap, we performed immunofluorescence (IF) experiments with the anti-14-3-3γ antibody using Mieap-methylated HCT116 cells (for exogenous Mieap) and Mieap-unmethylated A549 cells (for endogenous Mieap). To induce MALM, the HCT116 cells were infected with Ad-Mieap β (Mieap β expressing adenovirus) at a multiplicity of infection (MOI) of 5 and the A549 cells were γ-irradiated at 10 Gy, as has been previously described7,8. The IF experiments were performed three days after the infection or ionizing radiation (IR).
As shown in Fig. 3a, 14-3-3γ was mainly localized in the cytoplasm of the Ad-LacZ-infected HCT116 cells. Once MALM was induced in the cells by infection with Ad-Mieap β, however, 14-3-3γ clearly colocalized with the DsRed-mito, Mieap, and LAMP1 in the MALM-induced cells (Fig. 3a), implying that 14-3-3γ is translocated from the cytoplasm to the mitochondria and interacts with Mieap in the mitochondria when MALM occurs; similar results with endogenous Mieap protein were obtained using IR-treated A549 cells (Supplementary Fig. S5).
Figure 3. Translocation of 14-3-3γ from the cytoplasm to the inside of mitochondria.
(a) The mitochondrial localization of 14-3-3γ during MALM. The Ad-LacZ and Ad-Mieap-infected HCT116 cells were subjected to IF analysis on day 3 after the IR. The 14-3-3γ protein was stained with anti-14-3-3γ antibody (14-3-3γ:green). The Mieap protein was stained with anti-Mieap antibody (Mieap: red). The lysosomes were stained with anti-LAMP1 antibody (LAMP1: red). The mitochondria are indicated by the DsRed-Mito protein signal (Mito: red). The yellow area indicates overlapping between 14-3-3γ and mitochondria, Mieap, or lysosomes. A quantitative analysis of the yellow, red, or green area was performed using 300–400 cells. The average values of the yellow to red or green ratios (merged/mitochondria, 14-3-3γ, or lysosomes; yellow bar graph) are shown with error bars indicating 1 SD. P < 0.01 (*) was considered statistically significant. Scale bar = 20 μm. (b and c) The intramitochondrial localization of 14-3-3γ during MALM. The mitochondria were fractionated from the Ad-LacZ- and Ad-Mieap-infected HCT116 cells (a) or the control and Mieap-KD A549 cells (b) on day 3 after the IR. The mitochondrial fraction was subjected to a proteinase K protection assay. (−): no treatment, (+): proteinase K treatment, Mieap: Ad-Mieap-infected HCT116 cells, LacZ: Ad-LacZ-infected HCT116 cells, IR: ionizing radiation, Mitochondria: mitochondrial fraction, Cytosol: cytoplasmic fraction, and Whole: whole cell lysate. (b) C: HCT116 cells infected with Ad-LacZ, α: HCT116 cells infected with Ad-Mieap α, β: HCT116 cells infected with Ad-Mieap β. (c) C: A549 control cells, K: A549 Mieap-KD cells, α: endogenous Mieap α protein, and β: endogenous Mieap β protein. Full-length blots are presented in Supplementary Figure S8.
To obtain a more detailed localization of 14-3-3γ in mitochondria, we performed proteinase K protection assays with mitochondrial fractions prepared from MALM-induced and non-MALM HCT116 and A549 cells. In this experiment, the mitochondria-rich fraction was incubated with proteinase K, a general protease that degrades all proteins in the fraction, which do not present within mitochondria. If a protein is localized within mitochondria, the protein is protected from proteinase K digestion and can be detected by Western blot analysis following incubation with proteinase K.
To induce MALM, the HCT116 cells were infected with Ad-Mieap α or Ad-Mieap β at an MOI of 5. As previously reported, the Mieap and cathepsin D proteins in the mitochondrial fraction from the MALM-induced cells (Fig. 3b, lanes α and β) were protected from proteinase K digestion in parallel with the two intramitochondrial proteins (ATP synthase α and mitofilin), while the cathepsin D protein was degraded by proteinase K, similar to the three mitochondrial outer membrane proteins (NIX, VDAC, and TOMM70A) in the non-MALM cells (Fig. 3b, lane C). Interestingly, the 14-3-3γ levels in the mitochondrial fraction were markedly increased in the MALM-induced cells, while little 14-3-3γ was detected in the mitochondrial fraction prepared from the MALM non-induced cells (Fig. 3b, lanes C, α, and β). Surprisingly, the 14-3-3γ protein in the mitochondrial fraction prepared from the MALM-induced HCT116 cells was resistant to proteinase K digestion (Fig. 3b, lanes C, α, and β). This result indicated that the 14-3-3γ protein is present within mitochondria. A similar result was obtained from the IR-treated A549 cells, in which endogenous Mieap was activated by IR and MALM was induced by endogenous Mieap (Fig. 3c). Taken together, these results suggest that 14-3-3γ is translocated from the cytoplasm to the inside of mitochondria through the interaction with Mieap during the process of MALM.
Fig. 3c also indicates that the endogenous Mieap β protein was preferentially localized to mitochondria and translocated into the inside of mitochondria during MALM, while the Mieap α protein was preferentially localized to the cytoplasm (Fig. 3c). The data suggests that Mieap β may play a specific functional role in MALM under physiological conditions.
Potential role of 14-3-3γ in the elimination of oxidized mitochondrial proteins
To further explore the role of 14-3-3γ in Mieap-regulated mitochondrial quality control, we established 14-3-3γ knockdown (14-3-3γ KD) cells using the HCT116 and A549 cells. We confirmed that 14-3-3γ expression was severely inhibited in the 14-3-3γ KD HCT116 and A549 cells (Supplementary Fig. S6).
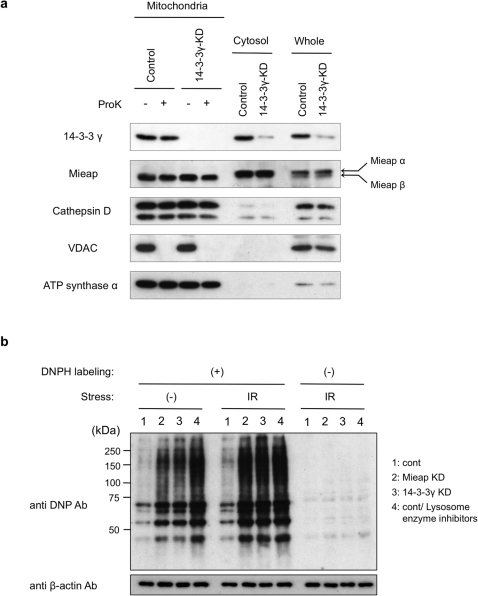
Using these cell lines, we first performed a proteinase K protection assay to examine whether 14-3-3γ deficiency affects the translocation of Mieap and lysosomal proteins to the inside of mitochondria during MALM. As shown in Fig. 4a, when MALM was induced by endogenous Mieap, we clearly observed the accumulation of the Mieap and cathepsin D proteins within mitochondria in the 14-3-3γ KD A549 cells, implying that 14-3-3γ is not involved in regulating mitochondrial translocation during MALM.
Figure 4. 14-3-3γ plays a role in degrading oxidized proteins.
(a) 14-3-3γ does not affect the accumulation of Mieap and lysosomal proteins within mitochondria. The mitochondria were fractionated from the control and 14-3-3γ-KD A549 cells on day 3 after the IR. The mitochondrial fraction was subjected to a proteinase K protection assay. ProK: Proteinase K, (−): no treatment, (+): proteinase K treatment, Control: A549 control cells, 14-3-3γ-KD: A549 14-3-3γ-KD cells, Mitochondria: mitochondrial fraction, Cytosol: cytoplasmic fraction, Whole: whole cell lysate, Mieap α: endogenous Mieap α protein, and Mieap β: endogenous Mieap β protein. (b) 14-3-3γ deficiency causes the accumulation of oxidized proteins. The total cell lysates isolated from the control (1: cont), Mieap-KD (2: Mieap KD), and 14-3-3γ-KD (3: 14-3-3γ KD) A549 cells on day 3 after the IR. To evaluate the role of lysosomes, lysosome enzyme inhibitors were added to the A549 control cells on day 2 after the IR (4: cont/lysosome enzyme inhibitors). Then, the cell lysates were labeled by DNPH and subjected to Western blot analysis with anti-DNP antibody to detect the carbonyl-oxidized proteins. β-actin was used as a loading control. Full-length blots are presented in Supplementary Figure S8.
Because MALM degrades oxidized mitochondrial proteins7, we next examined the role of 14-3-3γ in this process. First, we examined the oxidative modification of the carbonyl groups in the protein side chains. Interestingly, oxidized proteins accumulated in the Mieap KD and the 14-3-3γ KD A549 cells, even in the absence of stress (Fig. 4b). After the IR, the accumulation of oxidized proteins was enhanced in both the Mieap KD and 14-3-3γ KD A549 cells, but not in the control A549 cells. These results clearly suggest that 14-3-3γ is involved in degrading oxidized proteins by MALM.
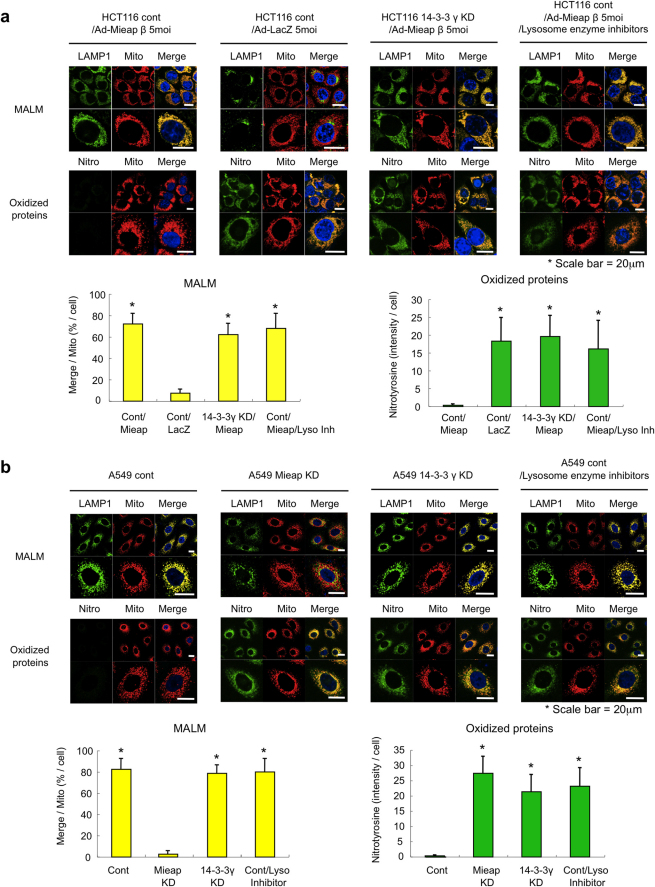
Using an antibody against nitrotyrosine, another oxidative stress marker, we next tried to confirm the role of 14-3-3γ in degrading oxidized mitochondrial proteins by carrying out IF experiments. Consistent with our previous study7, the accumulation of oxidized proteins in mitochondria was observed in the non-MALM cells (Fig. 5a, HCT116 cont/Ad-LacZ 5moi) after the IR, while the accumulation of oxidized mitochondrial proteins was significantly inhibited in the MALM-induced cells (Fig. 5a, HCT116 cont/Ad-Mieap β 5moi).
Figure 5. Potential role of 14-3-3γ in eliminating oxidized proteins within mitochondria.
(a and b) An IF analysis of MALM and oxidized proteins. The Ad-Mieap-β- or Ad-LacZ-infected control and Ad-Mieap-β-infected 14-3-3γ KD HCT116 cells (a) or the control, Mieap-KD, and 14-3-3γ KD A549 cells (b) were γ-irradiated. Three days after the IR, an IF analysis was performed using anti-LAMP1 antibody (LAMP1) to detect MALM (MALM) or using anti-nitrotyrosine antibody (Nitrotyrosine) to detect nitrotyrosine-oxidized proteins (Oxidized proteins). To evaluate the role of lysosomes, lysosome enzyme inhibitors were added to the A549 control and Ad-Mieap-β-infected HCT116 control cells on day 2 after the IR. The mitochondria are indicated by the DsRed-mito protein signal (Mito). Representative images are shown (upper panel of a or b). A quantitative analysis of MALM or nitrotyrosine intensity was performed using 300-400 cells. The average intensities of the MALM or nitrotyrosine-oxidized proteins per cell are shown with error bars indicating 1 standard deviation (SD; lower panel). P < 0.01 (*) was considered statistically significant. Scale bar = 20 μm.
We confirmed that the degradation of oxidized mitochondrial proteins, but not MALM, is severely inhibited by lysosomal enzyme inhibitors (Fig. 5a, HCT116 cont/Ad-Mieap β 5moi/lysosomal enzyme inhibitors) and NH4Cl (Supplementary Fig. S7a), which neutralizes the acidic environment of lysosomes, suggesting that lysosomal enzymes play a critical role in removing oxidized mitochondrial proteins.
Similarly, although the accumulation of Mieap and lysosomal proteins within mitochondria was not inhibited, we detected the accumulation of oxidized mitochondrial proteins in the MALM-induced 14-3-3γ KD cells (HCT116 14-3-3γ KD/Ad-Mieap β 5moi) (Fig. 5a). We also confirmed similar results in the IR-treated A549 14-3-3γ KD cells, implying a similar role for 14-3-3γ in degrading oxidized mitochondrial proteins by endogenous Mieap protein (Fig. 5b and Supplementary Fig. S7b). These results, taken together, suggest that 14-3-3γ is involved in degrading oxidized mitochondrial proteins by MALM.
Discussion
Immunoprecipitation is a major method used to screen the binding partners of target proteins. In general, the immunoprecipitates are separated by SDS-PAGE and visualized by silver staining. Then, selected visualized protein bands are excised and subjected to in-gel digestion with trypsin. The resulting peptides are analyzed by MS analysis. Although silver staining is the most sensitive nonradioactive detection method and is widely used for screening protein-protein interactions, it has some limitations. For example, visualizing particular proteins that are less sensitive to silver is difficult when using silver staining17,18. In addition, quantitative comparisons of large numbers of silver-stained proteins are difficult.
By contrast, 2DICAL can overcome these limitations because it is highly sensitive and reproducible; 60,000–160,000 peptides can be readily detected in a 1-h LC run and accurately quantified without labeling9,10,11,12,13,14. Therefore, using 2DICAL to analyze immunoprecipitates may become a valuable tool for screening protein-protein interactions. In the present study, we demonstrated the utility of a novel protein-protein interaction analysis system, IP-2DICAL, in which two or more comparable trypsinized immunoprecipitates are directly subjected to 2DICAL analysis. Indeed, we detected dozens of Mieap-interacting protein candidates from the endogenous Mieap immunoprecipitates using IP-2DICAL (data not shown); among them, we confirmed that 14-3-3γ interacts with Mieap in MALM-induced cells.
The 14-3-3 proteins constitute a family of acidic dimeric proteins that are highly conserved from yeast to mammals19. Crystal structure analyses of the 14-3-3 protein have revealed the existence of a binding groove in each 14-3-3 monomer, and cocrystallization of 14-3-3 with short synthetic phosphopeptides has revealed the ability of each monomer to independently bind a phosphopeptide, indicating that DM-deficient 14-3-3 mutants can bind target proteins with the same efficiency as wild-type 14-3-320,21,22,23. Consistent with these reports, the DM-deficient 14-3-3γ, but not the PB-deficient 14-3-3γ, was able to bind to the N-terminal region of Mieap β (Figure 2). However, both the DM-deficient and PB-deficient 14-3-3γ mutants were able to bind to Mieap α. Therefore, we speculate that the PB-deficient 14-3-3γ mutants may interact with Mieap α by hetero-dimerization with the endogenous 14-3-3γ protein; if so, 14-3-3γ dimerization may be required for 14-3-3γ to interact with Mieap α. However, further examination is absolutely needed.
In many, but not all, cases, 14-3-3 proteins bind to phosphorylated proteins24. A sequence analysis by Scansite (http://scansite.mit.edu/)25 predicted the existence of three putative 14-3-3-binding sites on Mieap α: Ser268, Ser274, and Ser507. Whether these sites are phosphorylated under physiological conditions remains to be elucidated. In addition, given that these putative 14-3-3-binding sites are not located in the 14-3-3-binding region of Mieap, these sites may not directly affect the interaction of 14-3-3γ with Mieap, even if they are phosphorylated under physiological conditions.
The MALM phenomenon is a novel mitochondrial quality control system that raises many interesting biological questions7. The first question relates to how unhealthy mitochondria are recognized by Mieap. We have previously reported on several experiments that addressed this question and demonstrated that mitochondrial ROS and NIX are essential for MALM induction8. Recently, we found that BNIP3 also mediates MALM26. We assumed that excess mitochondrial ROS production from unhealthy mitochondria modifies NIX, BNIP3, and/or Mieap, and that the oxidized NIX or BNIP3 at the outer mitochondrial membrane functions as a marker for unhealthy mitochondria, which are the target of MALM. Indeed, Mieap interacts with NIX and BNIP3 in a ROS-dependent manner, and this interaction is required for MALM induction8. Because 14-3-3 proteins can stimulate protein-protein interactions, e.g., Raf-1 and Bcr interactions27, we first speculated that 14-3-3γ plays an essential role in recognizing unhealthy mitochondria by Mieap through stimulating Mieap-NIX interactions. However, the accumulation of Mieap and lysosomal proteins in unhealthy mitochondria was observed in the 14-3-3γ-deficient cells in the IF experiment, suggesting that 14-3-3γ is not required for this process.
The second question relates to how the cytoplasmic MALM-related proteins, including Mieap and the components of lysosome-like organelles or lysosomal proteins, are translocated to the inside of mitochondria without destroying the mitochondrial structure. Therefore, we speculated that 14-3-3γ may mediate the translocation of these proteins and lysosome-like organelles into the intramitochondrial space. However, the accumulation of Mieap and lysosomal proteins within mitochondria was not inhibited in the 14-3-3γ-deficient cells in the proteinase K protection assay, suggesting that 14-3-3γ is not required for translocating Mieap and lysosomal proteins from the cytosol to the inside of mitochondria. Interestingly, we found that 14-3-3γ, which is a major cytosolic protein, is also translocated from the cytosol to the inside of mitochondria during MALM. Notably, a similar observation on the localization of 14-3-3 to the mitochondrial matrix has been previously reported in plant cells28. These findings suggest that mitochondria have an undefined pore or channel that facilitates importing cytosolic proteins that do not contain mitochondrial-targeting presequences and do not normally localize to mitochondria.
The third question relates to how oxidized mitochondrial proteins are degraded by Mieap-regulated lysosome-like organelles or lysosomal proteins within mitochondria. As shown in this study, the degradation of oxidized mitochondrial proteins by MALM is inhibited by lysosomal enzyme inhibitors and by neutralizing the acidic environment of the mitochondria with NH4Cl (Figure 4B); these results support our speculation that lysosomes and lysosomal enzymes are involved in this process7. Interestingly, the degradation of oxidized mitochondrial proteins was dramatically inhibited in the 14-3-3γ-deficient cells, despite the accumulation of lysosomal proteins within mitochondria. This result suggests that 14-3-3γ is involved in degrading oxidized mitochondrial proteins.
The roles of the 14-3-3 proteins have been classified into three modes of actions: (1) directed conformational changes to the target protein; (2) masking of a specific region of the target protein; and (3) colocalization of two proteins (scaffolding)29. In the case of MALM, we speculate that 14-3-3γ may act as a scaffold to anchor proteins within close proximity of one another. Within mitochondria, 14-3-3γ may regulate the import of oxidized mitochondrial proteins into lysosome-like organelles by mediating the interaction between Mieap and oxidized proteins. Further investigation on this point is needed.
MALM raises many important questions about well-established concepts in cell biology. Identifying 14-3-3γ as a Mieap-interacting protein will facilitate the elucidation of the MALM process. In addition, our finding that 14-3-3γ localizes to the intramitochondrial space suggests that we should rethink the importing system for mitochondria-localized proteins. The clarification of the exact role of 14-3-3γ and its import mechanism in MALM is a challenge for future studies.
Methods
Cell lines
The following human cancer cell lines were purchased from the American Type Culture Collection: HCT116 (colorectal adenocarcinomas) and A549 (lung cancer). Cells were cultured under the conditions recommended by their depositors.
Antibodies
The anti-Mieap antibody was prepared as described previously7. The other primary antibodies used in this study were rabbit polycolonal anti 14-3-3γ antibody (Santa Cruz Biotechnology), mouse monoclonal anti-beta-actin antibody (clone AC-74, Sigma), mouse monoclonal anti-NIX antibody (Abnova), mouse monoclonal anti-FLAG antibody (Sigma), rat monoclonal anti-HA antibody (Roche), mouse monoclonal anti-cathepsin D antibody (Novus Biologicals), mouse monoclonal anti-VDAC/Porin antibody (Abcam), rabbit polyclonal anti-TOMM70A antibody (Novus Biologicals), mouse monoclonal anti-ATP synthase alpha antibody (Invitrogen), mouse monoclonal anti-mitofilin antibody (Calbiochem), and mouse monoclonal anti-LAMP1 antibody (BD Pharmingen).
DNA-damaging treatments
Cells were seeded 12 h before treatment and were 60–70% confluent at the time of the treatment. To induce MALM, the cells were γ-irradiated at 10 Gy or 60 Gy using a 60Co source.
Immunoprecipitation
Immunoprecipitation was carried out at 0–4°C essentially as described8. Briefly, 2×106 A549 or 5×106 HCT116 cells were lysed on ice for 10 min in 500 μl of ice-cold buffer A [20 mM Tris-HCl (pH 7.4), 120 mM NaCl, 50 mM β-glycerophosphate, 5 mM NaF, and protease inhibitor (Roche)] containing 1% NP40. After centrifugation at 20,600×g for 20 min, indicated antibody were added to the cleared supernatant and incubated with rotation for 1 hr. Then protein A-sepharose (Invitrogen) or protein G-sepharose (Invitrogen) was added and the incubation continued for 1 hr. Captured immunoprecipitates were washed five times with buffer A. For 2DICAL analysis, 2×107 A549 cells were subjected to immunoprecipitation.
IP-2DICAL analysis
The immunoprecipitants were released from the sepharose by boiling them in 1% sodium deoxycholate (SDC). Then, the eluted immunoprecipitants were trypsinized in buffer B [1 M urea and 50 mM NH4HCO3]. The digested solution was acidified with 0.8% formic acid (final concentration) and centrifuged at 20,600×g for 5 min. The supernatant was collected and equal volume of ethyl acetate was added. The mixture was shaken for 1 min, then centrifuged at 20,600×g for 5 min to obtain aqueous and organic phases. The aqueous phase was collected and was dried with a SpeedVac concentrator (Thermo Electron, Holbrook, NY). The dried peptides were dissolved in 0.1% formic acid.
LC-MS
LC-MS and data acquisition were performed as reported previously9,10,11,12,13,14. Briefly, the peptide samples were separated with a linear gradient from 0 to 80% acetonitrile in 0.1% formic acid at a flow rate of 200 nl/min for 60 min using the splitless nano-flow HPLC systems (NanoFrontier nLC; Hitachi High-Technologies, Tokyo, Japan)9,10,11,12,13,14. MS spectra were acquired every second in duplicate with Synapt HDMS (Waters, Milford, MA) and QSTARXL (ABSciex, Foster City, CA) for identification of endogenous Mieap-interacting proteins.
Peak Alignment Across Multiple LC-MS
MS peaks were detected, normalized, and quantified using the in-house 2DICAL software package, as described previously9,10,11,12,13,14.
Protein Identification
MS/MS spectra were acquired from preparative LC. LC-MS data were aligned and targeted MS/MS was performed. The MS/MS data were analyzed with Mascot software (Matrix Sciences, Londn, UK).
Sequence Analysis
The amino acid sequence identity and similarity were analyzed by using ClustalW.
Plasmids
A HA-tagged full-length human Mieap α and Mieap β (DDBJ accession numbers AB465501 and AB465502) were cloned in pcDNA3.1 (Invitrogen) and designated as HA-Mieap α and HA-Mieap β, respectively. A full-length human 14-3-3γ with a 3×FLAG epitope at the C terminus, designated as 14-3-3γ-FLAG, was cloned in p3×FLAG-CMV-14 (Sigma). The deletion mutants of HA-Mieap-α-full(HA-Mieap-α-ΔCt: α1-273, HA-Mieap-α-ΔNt: α104-538) HA-Mieap-β-full (HA-Mieap-β-ΔCt: β1-241, HA-Mieap-β-ΔNt: β104-506), coiled-coil-deleted Mieap-α and β (HA-Mieap-ΔCC), and 14-3-3γ-full (14-3-3γ-(1-166)-FLAG: 14-3-3γ 1-166, 14-3-3γ-(139-247)-FLAG: 14-3-3γ 139-247) were generated by PCR. The transfection with these expression vectors was carried out by using FuGENE6 Transfection Reagent (Roche) according to the manufacture's protocol.
Immunocytochemistry
For immunocytochemistry, cells were grown on eight-well chamber slides (2×104 cells/well) at 37°C in conventional culture medium and fixed in 2% paraformaldehyde for 10 min at room temperature. Slides were incubated with 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 3 min and washed three times with PBS at room temperature. Cells were blocked with 3% bovine serum albumin (BSA) in PBS for 1 h and incubated with primary antibodies for 1 h at room temperature. After being washed three times with PBS, slides were incubated with fluorescein isothiocyanate (FITC)-conjugated or Alexa Fluor 546-conjugated secondary antibodies for 1 h at room temperature. Slides were treated with 1 µM TO-PRO-3 (Invitrogen) for 15 min to stain the nuclei and washed four times with PBS. Slides were mounted with VECTASHIELD H-1000 (Vector Laboratories) and observed under an Olympus IX70 (Olympus) inverted fluorescence microscope coupled to a Radiance 2000 laser-scanning confocal system (Bio-Rad).
Quantitative analysis of colocalization of 14-3-3γ with mitochondria, Mieap, or lysosomes
The cells were subjected to IF analysis using the anti-14-3-3γ, anti-Mieap, and anti-LAMP1 antibodies and DsRed-mito. 14-3-3γ and Mieap proteins were detected with anti-14-3-3γ antibody (green) and anti-Mieap antibody (red). Lysosomes were detected with anti-LAMP1 antibody (red), and mitochondria were indicated by the signals of DsRed-mito (red). The overlappings (yellow) of the 14-3-3γ (green) and mitochondrial (red), Mieap (red), or lysosomal signals (red) were analyzed by LuminaVision image analysis software in 300–400 cells using the optimal threshold parameters. The ratio of yellow to red or green was presented as the average of the calculated values per cell with error bars.
Subcellular fractionation and proteinase K protection assay
The experiment was performed as described previously7. HCT116 cells were infected with Ad-LacZ, Ad-Mieap α, and Ad-Mieap β at an MOI of 5, and 12 h after the infection, the cells were γ-irradiated at 60 Gy and collected 3 days after IR. The control, Mieap-KD, and 14-3-3γ-KD cells of A549 were γ-irradiated at 10 Gy, and collected 3 days after IR. The cells were suspended in cold homogenization buffer (20 mM HEPES, pH 7.4, and 250 mM sucrose) at 2×107 cells/ml and homogenized using Dounce homogenization on ice. The homogenized samples were centrifuged at 800 × g at 4°C for 10 min to pellet the nuclear fraction. The centrifugation was repeated, and the resulting supernatant was centrifuged at 10,000 × g at 4°C for 10 min to obtain a mitochondrial pellet and cytosolic supernatant. The mitochondrial pellets were centrifuged again with fresh homogenization buffer, and the pellets were suspended in 25 mM Tris-HCl, pH 7.4, and 150 mM NaCl to react with the protease. The mitochondrial suspensions were treated or not with 20 µg/ml proteinase K for 20 min on ice. Then, proteinase K was inactivated using 2 mM PMSF for 10 min on ice. The samples were dissolved in Laemmli buffer for SDS–PAGE, and western blotting was performed using primary antibodies against the indicated proteins.
Establishment of Mieap KD and NIX KD cell lines using RNA interference
We established the Mieap KD cell line using A549 cells, as described previously7. We also established 14-3-3γ KD cell lines using A549 cells. The 14-3-3γ expression was inhibited in the cell line by retroviral expression of short-hairpin RNA (shRNAs; 14-3-3γ KD1: 5′-gatccccGGAGGGTCATCAGTAGCATTGttcaagagaCAATGCTACTGATGACCCTCCtttttggaaa-3′, 14-3-3γ KD2: 5′- gatccccACGAGGACTCCTACAAGGACttcaagagaGTCCTTGTAGGAGTCCTCGTtttttggaaa-3′, 14-3-3γ KD3: 5′- gatccccGCCACTGTCGAATGAGGAAttcaagagaTTCCTCATTCGACAGTGGCtttttggaaa-3′) against the 14-3-3γ sequence. We also established control cell lines by infection with a retroviral vector containing shRNA (Control) against the green fluorescent protein (GFP) sequence.
Analysis of oxidized proteins
For detection of carbonyl-oxidized proteins, the control, Mieap-KD, and 14-3-3γ-KD cells of A549 (3×105 cells/6 cm-dish) were γ-irradiated at 10Gy. To evaluate the role of lysosomes in the degradation of oxidized proteins, lysosomal enzyme inhibitors [75 µl of 2 mg/ml Leupeptin (Sigma) and 60 µl of 5 mg/ml Pepstatin A (Sigma)] were added to the culture medium (3 ml) of the A549 control cells 2 days after IR. Then, 3 days after IR, cell lysates were isolated from the cells. The supernatants were adjusted at 3 mg/ml protein extractions by Lowly-protein assay Kit (Bio-Rad). To derivatize the carbonyl groups on the protein side chains by 2,4-dinitrophenylhydrazine (DNPH), 5 µl of 12% SDS was added to 5 µl of protein extraction, and the mixture was incubated with 10 µl of 10 mM DNPH solution (OxyBlot kit, Chemicon), or 10 µl of control solution without DNPH for 15 min. After neutralization, the samples were subjected to the western blot analysis with rabbit polyclonal antibody against DNP-KLH (Invitrogen).
For detection of 3-nitrotyrosine oxidized proteins, the control, Mieap-KD, and 14-3-3γ-KD cells of A549 (2×104 cells/well in 8 well chamber) were γ-irradiated at 10Gy. The Ad-Mieap and Ad-LacZ infected cells of HCT116 (2×104 cells/well in 8 well chamber) were also γ-irradiated at 10Gy. To evaluate the role of lysosomes in the degradation of oxidized proteins, lysosomal enzyme inhibitors [7.5 µl of 2 mg/ml Leupeptin (Sigma) and 6 µl of 5 mg/ml Pepstatin A (Sigma)] or 7.5 µl of 1 mM NH4Cl was added to the culture medium (300 µl) of the Ad-Mieap infected HCT116 cells or the A549 control cells 2 days after IR. Then, 3 days after IR, or immediately (non-treatment (-)), the cells were subjected to IF experiment with rabbit polyclonal anti-nitrotyrosine antibody. The cells on 8-well chamber slides were fixed and blocked, and incubated with rabbit polyclonal antibody against nitrotyrosine (1:100, Upstate) in blocking solution for 1.5 h. After washing with PBS, the cells were incubated with FITC conjugated goat anti-rabbit IgG secondary antibody (1:300, Santa cruz) for 1 h. Nucleus and mitochondria in the cells were counterstained with TO-PRO-3 and DsRed-Mito, respectively. The nitrotyrosine signals in 300–500 cells were analyzed by confocal microscopy. The intensity of nitrotyrosine in each captured image was analyzed by LuminaVision image analysis software (Version 2.4). The total areas of nitrotyrosine in 300–400 cells were extracted using the optimal threshold parameters and calculated with LuminaVision image analysis software. The nitrotyrosine intensity was presented as the average of the calculated values per cell with error bars.
Quantitative analysis of MALM
MALM was evaluated as described previously8. In brief, the indicated cells were subjected to IF analysis using anti-LAMP1 antibody and Ad-DsRed-mito vector. Lysosomes were detected with anti-LAMP1 antibody (green), and mitochondria were indicated by the signals of DsRed-mito (red). The overlapping of the mitochondrial and lysosomal signals (yellow area) were analyzed by LuminaVision image analysis software in 300–400 cells using the optimal threshold parameters. The yellow intensity was presented as the average of the calculated values per cell with error bars.
Author Contributions
T.M., N.K., M.O. contributed equally to this work; T.M., N.K., M.O., Y.N., M.Y., H.K., R.M. and H.A. carried out the biochemical and cell biological experiments and analyzed the data; T.M., N.K., M.O., T.Y. and H.A. designed the experiments; T.M., N.K., M.O. and H.A. produced the figures; T.M. and H.A. wrote the paper. All authors discussed the results and commented on the manuscript.
Supplementary Material
Supplementary figures
Acknowledgments
We thank Izumi Hyo for technical assistance. This work is supported, in part, by grants from the Ministry of Education, Culture, Sports, Science, and Technology, the Ministry of Health, Labour, Welfare, the Japan Society for the Promotion of Science, and the National Cancer Center Research and Development Fund.
References
- Murphy M. P. How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkov A. A. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann. N Y Acad. Sci. 1147, 37–52 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban R. S., Nemoto S. & Finkel T. Mitochondria, oxidants, and aging. Cell 120, 483–495 (2005). [DOI] [PubMed] [Google Scholar]
- Choksi K. B., Boylston W. H., Rabek J. P., Widger W. R. & Papaconstantinou J. Oxidatively damaged proteins of heart mitochondrial electron transport complexes. Biochim. Biophys. Acta 1688, 95–101 (2004). [DOI] [PubMed] [Google Scholar]
- Wallance D. C. Mitochondrial diseases in man and mouse. Science 283, 1482–1488 (1999). [DOI] [PubMed] [Google Scholar]
- Wallance D. C. A mitochondrial paradigm of metabolic and degenerative disease, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet 39, 359–407 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y. et al. Possible existence of lysosome-like organella within mitochondria and its role in mitochondrial quality control. PLoS One 6, e16054 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N. et al. Mieap, a p53-inducible protein, controls mitochondrial quality by repairing or eliminating unhealthy mitochondria. PLoS One 6, e16060 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M. et al. Label-free quantitative proteomics using large peptide data sets generated by nanoflow liquid chromatography and mass spectrometry. Mol. Cell Proteomics. 5, 1338–1347 (2006). [DOI] [PubMed] [Google Scholar]
- Negishi A. et al. Large-scale quantitative clinical proteomics by label-free liquid chromatography and mass spectrometry. Cancer Sci. 100, 514–519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi A. et al. Quantitative proteomics using formalin-fixed paraffin-embedded tissues of oral squamous cell carcinoma. Cancer Sci. 100, 1605–1611 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M. et al. Prolyl 4-hydroxylation of alpha-fibrinogen: a novel protein modification revealed by plasma proteomics. J. Biol. Chem. 284, 29041–29049 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara J. et al. Identification of a predictive biomaker for hematologic toxicities of gemcitabine. J. Clin. Oncol. 27, 2261–2268 (2009). [DOI] [PubMed] [Google Scholar]
- Matsubara J. et al. Survival prediction for pancreatic cancer patients receiving gemcitabine treatment. Mol. Cell Proteomics 9, 695–704 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. K. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 19, 16–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y. H. et al. Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Mol. Biol. Cell. 14, 4721–4733 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersch-Björkman Y., Thomsson K. A., Holmén Larsson J. M., Ekerhovd E. & Hansson G. C. Large scale identification of proteins, mucins, and their O-glycosylation in the endocervical mucus during the menstrual cycle. Mol. Cell Proteomics. 6, 708–716 (2007). [DOI] [PubMed] [Google Scholar]
- Chuba P. J. & Palchaudhuri S. Requirement for cysteine in the color silver staining of proteins in polyacrylamide gels. Anal. Biochem. 156, 136–139 (1986). [DOI] [PubMed] [Google Scholar]
- van Heusden G. P. 14-3-3 Proteins: Regulators of Numerous Eukaryotic Proteins. IUBMB Life. 57, 623–629 (2005). [DOI] [PubMed] [Google Scholar]
- Gu M. & Du X. A novel ligand-binding site in the zeta-form 14-3-3 protein recognizing the platelet glycoprotein lbalpha and distinct from the c-Raf-binding site. J. Biol. Chem. 273, 33465–33471 (1998). [DOI] [PubMed] [Google Scholar]
- Ichimura T. et al. Identification of the site of interaction of the 14-3-3 protein with phosphorylated tryptophan hydroxylase. J. Biol. Chem. 270, 28515–28518 (1995). [DOI] [PubMed] [Google Scholar]
- Rittinger K. et al. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell. 4, 153–166 (1999). [DOI] [PubMed] [Google Scholar]
- Yaffe M. B. et al. The structural basis for 14-3-3, phosphopeptide binding specificity. Cell 91, 961–971 (1997). [DOI] [PubMed] [Google Scholar]
- Mhawech P. 14-3-3 proteins–an update. Cell Res. 15, 228–236 (2005). [DOI] [PubMed] [Google Scholar]
- Yaffe M. B. et al. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotechnol. 19, 348–353 (2001). [DOI] [PubMed] [Google Scholar]
- Nakamura Y. et al. BNIP3 and NIX mediate Mieap-induced accumulation of lysosomal proteins within mitochondria. PLoS One 7, e30767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braselmann S. & McCormick F. BCR and RAF form a complex in vivo via 14-3-3 proteins. EMBO J. 14, 4839–4848 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney T. D., van Walraven H. S. & de Boer A. H. 14-3-3 protein is aregulator of the mitochondrial and chloroplast ATP synthase.. Proc. Natl. Acad. Sci USA. 98, 4249–4254 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D. & Moorhead G. B. 14-3-3 proteins: A number of functions for a numbered protein. Sci. STEK 296, re10 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures