Abstract
The E. coli isopentenyl diphosphate isomerase (IDI) catalyzed reaction of isopentenyl diphosphate (IPP) in D2O gives a 66% yield of dimethylallyl diphosphate labeled with deuterium at the (E)-methyl group (d-DMAPP) and a 34% yield of IPP labeled with 1-mole of deuterium at C-2 (d-IPP). This shows that the release to D2O of the initial product of the IDI-catalyzed reaction (d-DMAPP) is slower than its conversion to d-IPP. Product dissociation is therefore rate determining for isomerization of IPP with a rate constant kdis ≈ kcat = 0.08 s−1. The data provide an estimated rate constant of kas = 6 × 103 M−1 s−1 for binding of DMAPP to E. coli IDI that is similar to rate constants determined for the binding of N-protonated 2-amino ethyl diphosphate intermediate analogs to IDI from yeast [Reardon, J. E.; Abeles, R. H. Biochemistry 1986, 25, 5609–5616]. We propose that ligand binding to IDI is relatively slow because there is a significant kinetic barrier to reorganization of the initial encounter complex between enzyme, substrate and an essential Mg2+ to form the Michaelis complex where the metal cation bridges the protein and the substrate diphosphate group.
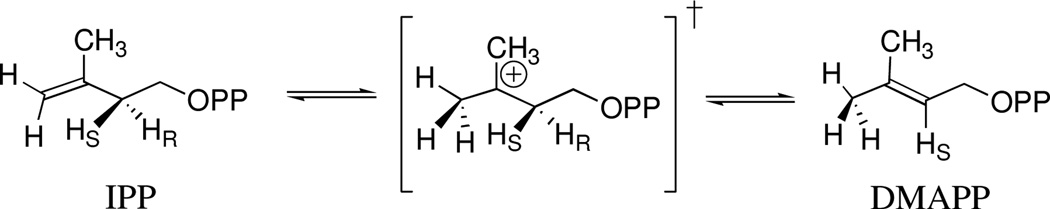
Isopentenyl diphosphate isomerase (IDI) catalyzes 1,3-isomerization of isopentenyl diphosphate (IPP) to form dimethylallyl diphosphate (DMAPP)1 through a tertiary carbocation-like transition state (Scheme 1).2,3 The central role for IPP and DMAPP in the biosynthesis of natural products,4 the deceptive simplicity of the catalyzed 1,3-proton transfer reaction, and the prominent role of tertiary carbocations in the development of our understanding of solvolysis and elimination reaction mechanisms5 have prompted penetrating mechanistic studies3,6–9 and X-ray crystallographic analyses of type 1 IDI.10–12 This work has identified several amino acid side chains essential for enzymatic activity and assigned their probable role in catalysis of the isomerization reaction.
Scheme 1.
We are interested in understanding the origin of the ≈16 kcal/mol stabilization of the tertiary carbocation-like transition state at the active site of type 1 IDI.13 It has been shown that IDI catalyzes kinetically competent transfer of deuterium from solvent D2O to the (E)-methyl group of the product DMAPP, and slow transfer of deuterium to the (Z)-methyl group of DMAPP.7 In the course of developing 1H NMR assays of the IDI-catalyzed reaction in D2O for use in studies of the reactions of truncated substrate analogs14,15 we have revisited and extended these isotope exchange experiments.
Type 1 E. coli IDI was purified from E. coli strain JM101 that contained the gene for IDI on a pARC306N expression vector. This strain, a generous gift from Prof. Dale Poulter, has been described earlier,16,17 except that the present construct contains an additional His-tag to facilitate protein purification. The protein was expressed by published procedures6,18 and purified over a Ni-Chelating Sepharose Fast Flow from Pharmacia Biotech. Enzyme-catalyzed isomerization of IPP was assayed by monitoring the appearance of the signal for the Z-methyl hydrogen of DMAPP by 1H NMR [see Supporting Information for this manuscript]. The procedures for determining a value of kcat = 0.08 s−1 for turnover of IPP are also given as Supporting Information.
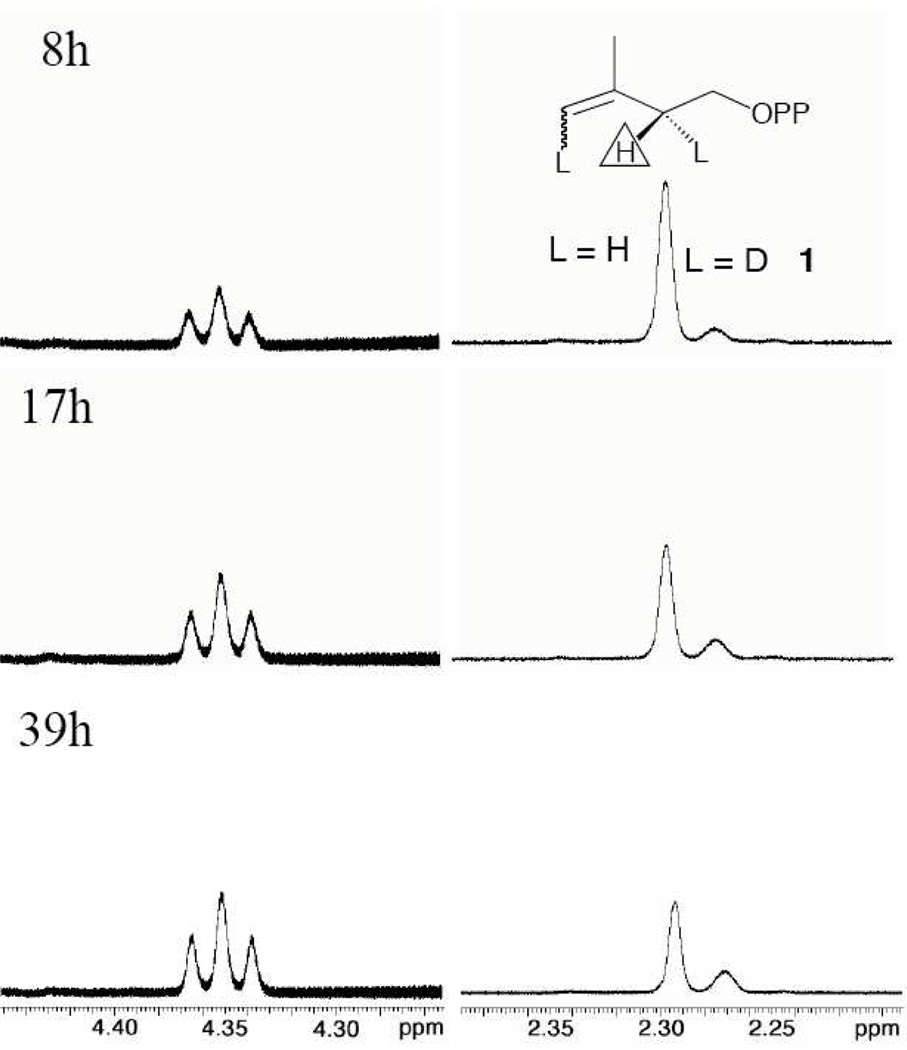
1H NMR analyses [see Supporting Information] of the products of IDI-catalyzed reactions of IPP in D2O showed at early reaction times a group of poorly resolved signals shifted up-field from the triplet for the 2-CH2- hydrogen of IPP. Homonuclear decoupling of the signals for the 1-CH2- and 2-CH2-hydrogen of IPP was accomplished by irradiation at 3.96 ppm, the resonance frequency for 1-CH2-. Decoupling causes the triplet for the 2-CH2- hydrogen to collapse to a singlet at 2.295 ppm, and a simplification of the upfield signals, which now appear as a broad singlet at 2.272 ppm. Figure 1 shows partial 1H NMR spectra obtained during the reaction of 21 mM IPP catalyzed by 3.5 µM E. coli IDI at pD 7.4 (100 mM phosphate buffer and 0.20 M KCl). The α-D at -CHD- groups of glycine and its derivatives perturbs the chemical shift of the remaining –H by ca 0.2 ppm upfield from the signal for the parent -CH2- hydrogen.19,20 We therefore assign the signal at 2.272 ppm to the 2-CHD- of 1. The apparent triplet [the coupling constants of 6.5 Hz for the spitting by the C-2 hydrogen and α-phosphorus are similar] for the C-1 methylene hydrogen of DMAPP appears at 4.35 ppm.
Figure 1.
Partial 1H-NMR spectra of the products of IDI-catalyzed isomerization of IPP in D2O, with homonuclear decoupling of the signals for the 1-CH2- and 2-CH2- of IPP by irradiation at the resonance frequency for 1-CH2- hydrogen. The triplet at 4.35 ppm and the apparent singlets at 2.295 and 2.272 are the signals for the C-1 hydrogen of DMAPP and the C-2 hydrogen of IPP and 1, respectively. The experimental details for these 1H NMR analyses are given in the Supporting Information.
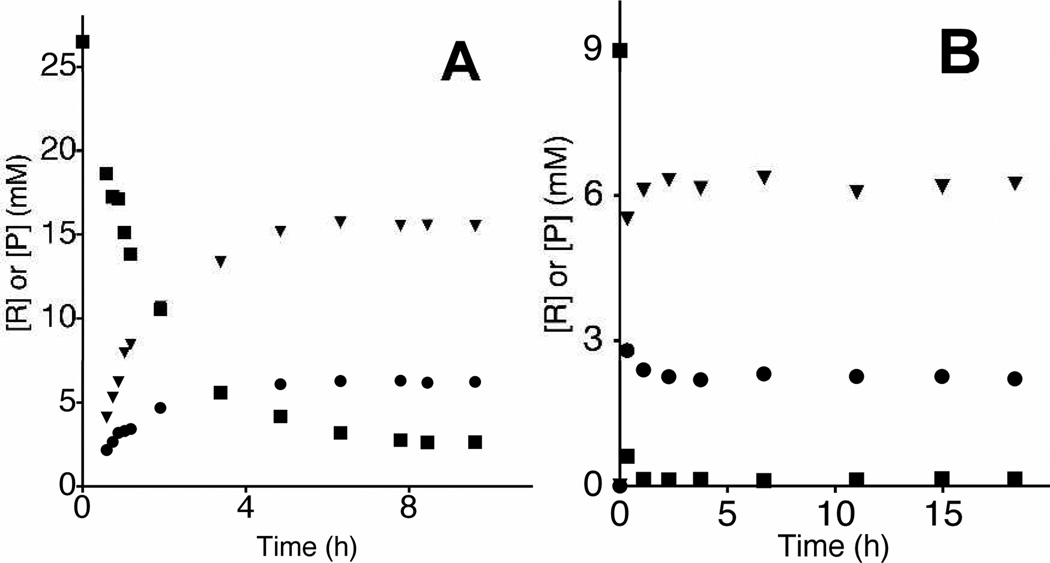
Figure 2A shows the time course for isomerization of 26.5 mM IPP catalyzed by 28 µM E. coli IDI at pD 7.4 (20 mM imidazole), 400 mM NaCl, 10 mM MgCl2 and 0.5mM DTT. The decrease in [IPP] (■) was determined by monitoring the peak at 2.295 ppm for the 2-CH2- hydrogen; and, the increase in [DMAPP]T (▲) and in [1] (●) were determined by monitoring the triplet at 4.35 for the C-1 methylene hydrogen of DMAPP and the singlet at 2.272 for the 2-CHD- hydrogen, respectively (Figure 1). Figure 2B shows the time course for the disappearance of IPP and the formation of 1 and DMAPP for isomerization of 9 mM IPP catalyzed by 71 µM E. coli IDI at pD 7.4 (20 mM imidazole), 400 mM NaCl, 10 mM MgCl2 and 0.5mM DTT. In this experiment the IPP is nearly completely consumed during the first 20 minutes of the reaction.
Figure 2.
Time courses for the IDI-catalyzed reactions of IPP (■) to form DMAPPT (▼) and 1 (●). DMAPP formed at early reaction times contains one atom of –D (d-DMAPP, Scheme 3), but we observe the incorporation of multiple atoms of –D as DMAPP and IPP achieve chemical equilibrium.7 (A) The isomerization of 26.5 mM IPP catalyzed by 28µM E. coli IDI at pD 7.4 (20 mM imidazole), 400 mM NaCl, 10 mM MgCl2 and 0.5 mM DTT. (B) The isomerization of 9 mM IPP catalyzed by 71 µM E. coli IDI at pD 7.4 (20 mM imidazole), 400 mM NaCl, 10 mM MgCl2 and 0.5 mM DTT. The procedures for these experiments are given in the Supporting Information.
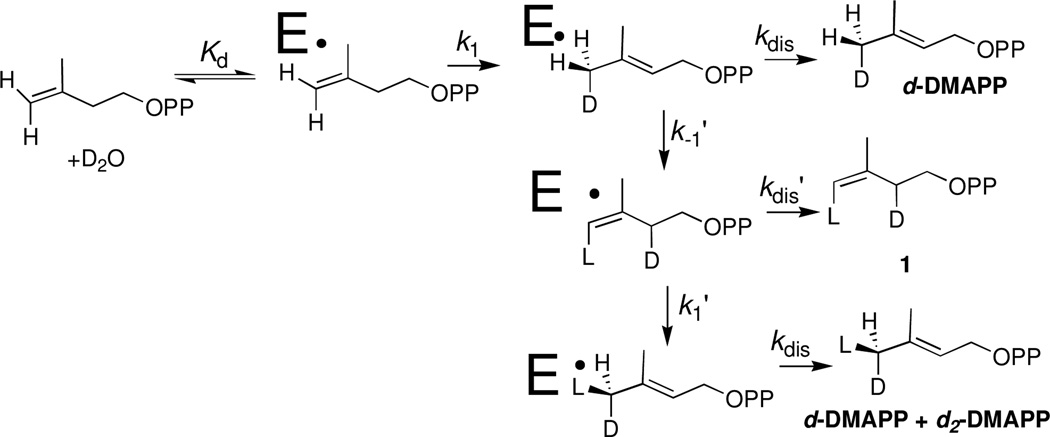
The data in Figures 2A and 2B are inconsistent with the sequential reaction shown in Scheme 2, where d-DMAPP is released to D2O and then converted to 1 in a second independent reaction. The requirement for product release will result in a lag in the formation of 1 as d-DMAPP accumulates, but this lag is not observed (Figure 2A). Instead, the product ratio [1]T/[DMAPP]T = 0.50 is constant over the first three time points (0.9 h) and decreases to Keq = [IPP]T/[DMAPP]T = 0.36 as total DMAPP and IPP achieve chemical equilibrium. This equilibrium constant is in fair agreement with the value of Keq = 0.45 determined in earlier work.7 We also observe at later reaction times the extensive washout of -H from the -CH3 groups of DMAPP and IPP reported in the same earlier study.7
Scheme 2.
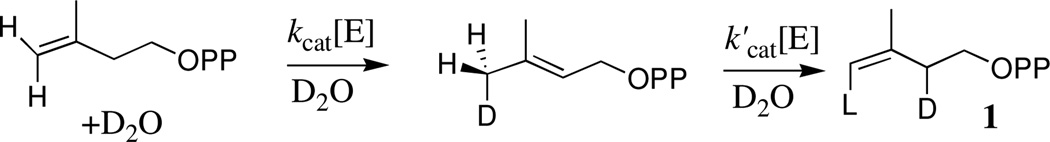
Figure 2A shows that d-DMAPP and 1 form at the same reaction velocity during the first 0.9 h of the reaction. This requires that the initial enzyme-catalyzed reaction of IPP give d-DMAPP and 1 as products, so that the release of d-DMAPP to D2O is slower than its conversion to 1 (k−1' > kdis, Scheme 3). The initial product ratio [1]T/[DMAPP]T = 0.50 is determined by the relative velocity for formation of d-DMAPP by k1, kdis and for formation of 1 by k1, k−1', kdis' (Scheme 3). At longer reaction times the signals for a poorly resolved broad pentet for the (Z)–CHD2 group of d2-DMAPP were detected. We are uncertain of the yield (if any) for the formation of this product at early reaction times by k1, k−1', k1', kdis (Scheme 3).
Scheme 3.
The initial product ratio [1]T/[DMAPP]T = 0.50 from Figure 2B is the same as determined for Figure 2A. The very high enzyme concentration used in this experiment results in the rapid consumption of IPP. We were surprised to observe (Figure 2B) that the concentration of 1 at early reaction times is larger than the concentration of 1 once chemical equilibrium with DMAPP is established! The IDI-catalyzed conversion of IPP to 1 is therefore faster than the formation of an equilibrium mixture of [IPP]T and [DMAPP]T, because of the rapid washout of the C-2 hydrogen from IPP. This result is inconsistent with the sequential reaction shown in Scheme 2. It may be rationalized by the fast enzyme-catalyzed conversion of enzyme bound IPP to 1 through E•d-DMAPP (Scheme 3) provided the concentration ratio [E•1]/[E•d-DMAPP] during steady state turnover is larger than the ratio of the concentration of the free species at chemical equilibrium in D2O
We conclude that conversion of E•d-DMAPP to 1 is faster than release of d-DMAPP to D2O (k−1' > kdis, Scheme 3). If product release is slower than the chemical steps at the enzyme active site, then it is rate determining for turnover of IPP, so that kdis ≈ kcat = 0.08 s−1 at 25 °C. It is not unusual for product release to limit the rate for enzyme turnover. For example, the release of glyceraldehyde 3-phosphate is rate determining for TIM-catalyzed isomerization of dihydroxyacetone phosphate.21 However, product release from TIM is fast (ca 4000 s−1) and rate determining only because the chemical isomerization of bound substrate is even faster.21 By contrast, E. coli IDI is a sluggish enzyme with kcat = 0.08 s−1, and the enzyme-product complex has the surprisingly long lifetime of ca 10s.
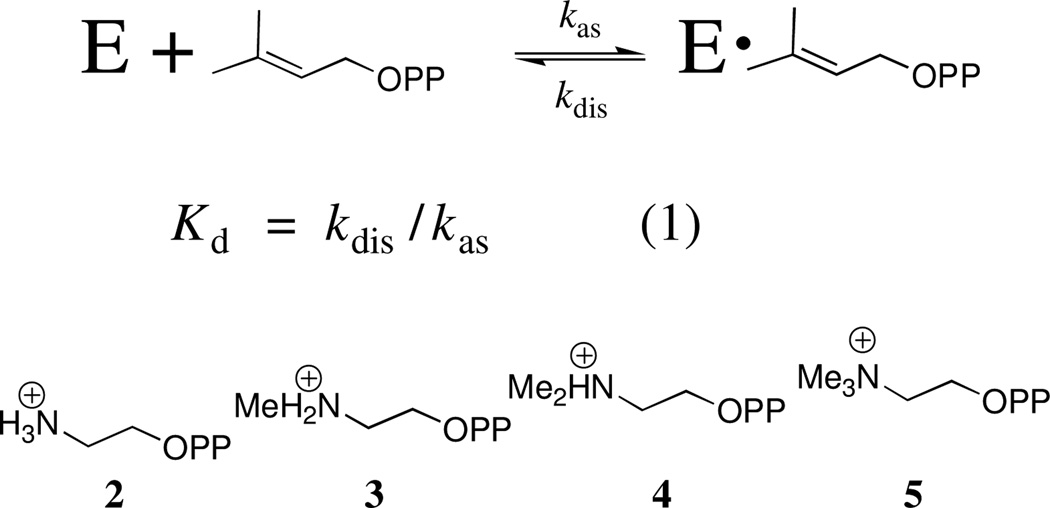
The value of Kd = 14 µM for inhibition of IDI-catalyzed isomerization of IPP by the product DMAPP17 may be combined with kdis ≈ kcat ≈ 0.08 s−1 for release of DMAPP to give kas ≈ 6 × 103 M−1 s−1 for ligand binding (eq 1). By comparison, the second-order rate constants kas for association of tight-binding inhibitors 2 – 5 (Ki is in the nanomolar to picomolar range) to IDI from yeast range from 105 M−1 s−1 for 2 to 103 M−1 s−1 for 5.2 We conclude that this slow ligand binding is a property of both the substrate and of tight-binding cationic inhibitors. The high-affinity inhibitors are released exceedingly slowly to water.2,3 For example, the value of kdis < 5 × 10−7 s−1 for dissociation of 4 corresponds to a > 16 day halftime for breakdown of the protein-inhibitor complex.2
Scheme 4.
There is likely a single mechanism-based explanation for the slow binding of both reactant DMAPP and inhibitors 2 – 5 to IDI. IDI requires two metal dications for activity, and accepts a variety of dications at each of the two metal ion binding sites.6 One metal ion forms a distorted octahedral complex at a binding site that is partly composed of N-terminal amino acid side chain ligands that are disordered at the metal-free enzyme.12 The second metal cation bridges the coordinated diphosphate group of substrate with the IDI main-chain amide carbonyl oxygen of Cys-67 and the carboxylate side chain of Glu-87.12 The slow binding of DMAPP and 2 – 5 to IDI shows that most encounters between the ligand and IDI are nonproductive. We suggest that the slow ligand binding reflects the substantial kinetic barrier to reorganization of the initial ligand•Mg2+•enzyme encounter complex. This might include the barrier to reorganization of the side chains of Cys-67 and Glu-87 so that they are positioned to coordinate to the metal dication.
Supplementary Material
ACKNOWLEDGMENT
We thank Prof. Dale Poulter for providing us with the E. coli strain JM101 that contained the gene for IDI on an expression vector.
Funding Sources
This work was generously supported by Grant GM39754 from the National Institutes of Health.
ABBREVIATIONS
- IPP
isopentenyl diphosphate
- DMAPP
dimethylallyl diphosphate
- IDI
isopentenyl diphosphate isomerase
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental procedures for the synthesis of IPP, for the determination of kcat for IDI-catalyzed isomerization of IPP using an 1H NMR assay, and for 1H NMR analyses of the yields of the products of IDI catalyzed isomerization of IPP in D2O. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Agranoff BW, Eggerer H, Henning U, Lynen F. J. Am. Chem. Soc. 1959;81:1254–1255. [Google Scholar]
- 2.Reardon JE, Abeles RH. Biochemistry. 1986;25:5609–5616. doi: 10.1021/bi00367a040. [DOI] [PubMed] [Google Scholar]
- 3.Muehlbacher M, Poulter CD. Biochemistry. 1988;27:7315–7328. doi: 10.1021/bi00419a021. [DOI] [PubMed] [Google Scholar]
- 4.Kuzuyama T, Seto H. Nat. Prod. Rep. 2003;20:171–183. doi: 10.1039/b109860h. [DOI] [PubMed] [Google Scholar]
- 5.Toteva MM, Richard JP. J. Am. Chem. Soc. 1996;118:11434–11445. [Google Scholar]
- 6.Lee S, Poulter CD. J. Am. Chem. Soc. 2006;128:11545–11550. doi: 10.1021/ja063073c. [DOI] [PubMed] [Google Scholar]
- 7.Street IP, Christensen DJ, Poulter CD. J. Am. Chem. Soc. 1990;112:8577–8578. [Google Scholar]
- 8.Street IP, Coffman HR, Baker JA, Poulter CD. Biochemistry. 1994;33:4212–4217. doi: 10.1021/bi00180a014. [DOI] [PubMed] [Google Scholar]
- 9.Clifford K, Cornforth JW, Mallaby R, Phillips GT. J. Chem. Soc. D. 1971:1599–1600. [Google Scholar]
- 10.Zhang C, Liu L, Xu H, Wei Z, Wang Y, Lin Y, Gong W. J. Mol. Biol. 2007;366:1437–1446. doi: 10.1016/j.jmb.2006.10.092. [DOI] [PubMed] [Google Scholar]
- 11.Durbecq V, Sainz G, Oudjama Y, Clantin B, Bompard-Gilles C, Tricot C, Caillet J, Stalon V, Droogmans L, Villeret V. EMBO J. 2001;20:1530–1537. doi: 10.1093/emboj/20.7.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wouters J, Oudjama Y, Barkley SJ, Tricot C, Stalon V, Droogmans L, Poulter CD. J. Biol. Chem. 2003;278:11903–11908. doi: 10.1074/jbc.M212823200. [DOI] [PubMed] [Google Scholar]; Wouters J, Oudjama Y, Stalon V, Droogmans L, Poulter CD. Proteins. 2004;54:216–221. doi: 10.1002/prot.10573. [DOI] [PubMed] [Google Scholar]
- 13.Toteva MM, Richard JP. Bioorg. Chem. 1997;25:239–245. [Google Scholar]
- 14.O'Donoghue AC, Amyes TL, Richard JP. Biochemistry. 2005;44:2622–2631. doi: 10.1021/bi047953k. [DOI] [PubMed] [Google Scholar]
- 15.Amyes TL, Richard JP. Biochemistry. 2007;46:5841–5854. doi: 10.1021/bi700409b. [DOI] [PMC free article] [PubMed] [Google Scholar]; Goryanova B, Amyes TL, Gerlt JA, Richard JP. J. Am. Chem. Soc. 2011;133:6545–6548. doi: 10.1021/ja201734z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Street IP, Coffman HR, Poulter CD. Tetrahedron. 1991;31:5919–5924. [Google Scholar]
- 17.Hahn FM, Hurlburt AP, Poulter CD. J. Bact. 1999;181:4499–4504. doi: 10.1128/jb.181.15.4499-4504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrigan CN, Poulter CD. J. Am. Chem. Soc. 2003;125:9008–9009. doi: 10.1021/ja0350381. [DOI] [PubMed] [Google Scholar]
- 19.Rios A, Richard JP, Amyes TL. J. Am. Chem. Soc. 2002;124:8251–8259. doi: 10.1021/ja026267a. [DOI] [PubMed] [Google Scholar]
- 20.Rios A, Amyes TL, Richard JP. J. Am. Chem. Soc. 2000;122:9373–9385. [Google Scholar]
- 21.Knowles JR, Albery W. J. Acc. Chem. Res. 1977;10:105–111. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.