Abstract
Paramyxoviruses are responsible for considerable disease burden in human and wildlife populations: measles and mumps continue to affect the health of children worldwide, while canine distemper virus causes serious morbidity and mortality in a wide range of mammalian species. Although these viruses have been studied extensively at both the epidemiological and the phylogenetic scales, little has been done to integrate these two types of data. Using a Bayesian coalescent approach, we infer the evolutionary and epidemiological dynamics of measles, mumps and canine distemper viruses. Our analysis yielded data on viral substitution rates, the time to common ancestry, and elements of their demographic history. Estimates of rates of evolutionary change were similar to those observed in other RNA viruses, ranging from 6.585 to 11.350 × 10−4 nucleotide substitutions per site, per year. Strikingly, the mean Time to the Most Recent Common Ancestor (TMRCA) was both similar and very recent among the viruses studied, ranging from only 58 to 91 years (1908 to 1943). Worldwide, the paramyxoviruses studied here have maintained a relatively constant level of genetic diversity. However, detailed heterchronous samples illustrate more complex dynamics in some epidemic populations, and the relatively low levels of genetic diversity (population size) in all three viruses is likely to reflect the population bottlenecks that follow recurrent outbreaks.
Keywords: Paramyxovirus, Coalescent, Population bottleneck, Measles virus, Mumps virus, Canine distemper virus
Introduction
Viral epidemics caused by negative-sense viruses of the family Paramyxoviridae have plagued animal populations for millennia. Three of these viruses—measles, mumps, and canine distemper virus—affect animals ranging from livestock to canines to humans (Lamb and Kolakofsky 2001; Fauquet et al. 2005) and have had a major economic and demographic impact in these species. A major disease burden is also associated with rinderpest in cattle, Newcastle disease in poultry, and phocine distemper in seals (Barrett 1994; Lamb and Kolakofsky 2001; Griffin 2001).
The first documented outbreaks of measles date to the second to fourth centuries in China and the Roman Empire (Orvell 1994), while the closest relative of measles virus—rinderpest virus—affected cattle even earlier (Carbone and Wolinsky 2001). Measles virus (genus Morbillivirus) affects nearly 30 million people annually, with a mortality of 454,000, largely in developing countries (WHO 2007). In populations with recurring multiannual epidemics, measles cases are usually clustered among children and spread through aerosol droplets (Anderson and May 1991; Grenfell and Harwood 1997; Griffin 2001). The virus causes acute systematic disease (Orvell 1994) and confers life-long immunity on those who recover (Panum 1939; Griffin 2001).
At the epidemiological level, measles epidemics display violent cyclic dynamics, with a 1- to 5-year periodicity of outbreaks in a given location (Orvell 1994). Two critical epidemiological parameters have been estimated for this important human pathogen: the reproductive number (R0) and the critical community size (CCS). R0 is the number of secondary cases that arise from each primary case in a totally susceptible population, and estimates of this parameter for measles range from 15 to 20 (Griffin 2001). CCS is the minimum number of individuals in a population required for the disease to persist in a population, and has been estimated as 250,000–500,000 for measles (Bartlett 1957; Griffin 2001). At the phylogenetic level, variability in the hemagglutinin (H) glycoprotein identifies eight major groups with 15 component genotypes, which exhibit temporal and spatial segregation (Bellini and Rota 1998; Griffin 2001).
Mumps, the viral agent of which is classified within the genus Rublavirus, was first described by Hippocrates in the fifth century BC (Rima 1994; Carbone and Wolinsky 2001) and spreads through aerosol droplets (Carbone and Wolinsky 2001). Mumps virus, like measles virus, has a very narrow host range, with humans being the only species that can actively transmit the virus and other species acting as dead-end hosts (Rima 1994). Epidemics of mumps occur in both developed and developing countries due to a lack of vaccination campaigns or vaccine failure (Carbone and Wolinsky 2001). These epidemics also display cyclical dynamics with annual outbreaks. However, the periodicity of these outbreaks can vary by as much as 2 to 7 years (Rima 1994). At the population level, R0 has been estimated as 5 (Rima 1994) and CCS = 200,000 with school terms forcing epidemic patterns (Carbone and Wolinsky 2001). Analyses of genetic variability in the hemagluttinin-neuraminidase (HN) proteins have designated 11 circulating genotypes (Orvell et al. 2002).
While canine distemper virus (CDV) shares some similarities with measles virus, including its taxonomic identification within the genus Morbillivirus, it seems to have emerged more recently, with the first case described in 1905 (Griffin 2001). CDV also has a broader host range and can infect many mammalian species, including members of the Felidae (cats), Canidae (dogs, foxes, etc.), Mustelidae (weasles), and Proconidae (raccoons) (Barrett 1994; Griffin 2001). Although a vaccine was developed for domestic dogs in 1950, limited use means that CDV remains prevalent in many populations (Barrett 1994). CDV also displays a high degree of genetic diversity that rivals, or exceeds, that of measles virus (Bolt et al. 1997); again, genetic diversity is greatest in the hemagluttinin (H) protein and displays spatial differentiation (Mochizuki et al. 1999).
Together, the measles, mumps, and canine distemper viruses are among some of the most thoroughly studied viruses at the epidemiological scale, and all three viruses exhibit major population bottlenecks due to their cyclic epidemiological dynamics. However, little work has been done to determine how their epidemiological dynamics affect their phylogenetic patterns as inferred from gene sequence data. By performing a Bayesian coalescent analysis using serially sampled gene sequence data (Drummond et al. 2002, 2006), we are able to estimate key demographic and evolutionary parameters, and gain important insights into the evolution and epidemiology of these viral pathogens.
Methods
Data Sets
All viral sequences were downloaded from GenBank and manually aligned using the Se-Al program (Rambaut 1996). Alignment files for all data sets are available from the authors on request.
For the analysis of measles virus, both the H and the N (nucleoprotein) genes were utilized. H gene sequences were collected from localities worldwide, notably Africa. Once vaccine strains and those associated with the persistent disease manifestation subacute sclerosing panencephalitis (SSPE) were removed, as these are expected to exhibit different evolutionary dynamics (Woelk et al. 2002), 215 taxa remained with an alignment length of 1851 bp. Due to computational constraints associated with the coalescent analysis of large numbers of sequences, 120 taxa were randomly selected from this larger data set. To ensure that this subsampling introduced no bias into the analysis, it was performed an additional five times independently and all coalescent analyses were run on these five subsampled data sets. Similarly, we compiled a global sample of measles virus N gene sequences. Again, vaccine strains and those associated with SSPE were removed, which resulted in a final data set of 107 taxa, 1575 bp. The mumps data set comprised HN sequences collected globally, although with a particular emphasis on sequences from Asia. Once the vaccine strains were removed, 27 taxa remain for analysis, 1746 bp in length. Finally, in the case of CDV, H gene sequences were sampled from Japan, the United States, and Western Europe. Vaccine strains and passaged strains were removed from the data set, resulting in 35 taxa of 1949 bp.
Evolutionary Analysis
Rates of nucleotide substitution per site, the Time to the Most Recent Common Ancestor (TMRCA), and key aspects of demographic history, particularly changes in genetic diversity (an indicator of population size under strictly neutral evolution) quantified as Neτ, where Ne is the effective number of infections and τ is the infection-to-infection generation time, were estimated using a Bayesian Markov chain Monte Carlo (MCMC) method available in the BEAST package (Drummond et al. 2002; Drummond and Rambaut 2003). This method analyzes the distribution of branch lengths among viruses isolated at different times (year of collection) among millions of sampled trees. A variety of models of demographic history was investigated—constant population size, exponential population growth, logistic population growth, and expansion population growth—incorporating both relaxed (uncorrelated exponential) and constant molecular clocks (Drummond et al. 2006). For each data set, the best-fit model of nucleotide substitution was determined using MODEL-TEST (Posada and Crandall 1998). In the case of the two measles virus data sets and the mumps HN gene, the favored model was a close relative of the most general GTR + I + Γ4 model. For the CDV H gene a simpler GTR + I model was preferred. All models were compared using Akaike’s Information Criterion (AIC). To infer demographic histories we depicted the changing profile of Neτ through time in a Bayesian skyline plot (Drummond et al. 2005). In all cases statistical uncertainty in parameter values across the sampled trees is given by the 95% highest probability density (HPD) values. For all models, chains were run until convergence was achieved (as assessed using the TRACER program; http://www.evolve.zoo.ox.ac.uk/software.html?id = tracer). The BEAST analysis was also used to infer a maximum a posteriori (MAP) tree for each data set, in which tip times correspond to the year of sampling.
Gene and site-specific selection pressures for all four data sets were measured as the ratio of nonsynonymous (dN) to synonymous substitutions (dS) per site (dN/dS), estimated using the single likelihood ancestor counting (SLAC) and random effects likelihood (REL) maximum likelihood methods available at the Datamonkey facility (Kosakovsky et al. 2005). Because of the large number of sequences in the measles virus data sets, our analyses in this case were restricted to the SLAC method. In all cases we utilized the general reversible substitution (GTR) model with input neighbor-joining trees.
Results
Evolutionary Dynamics of the Paramyxoviruses
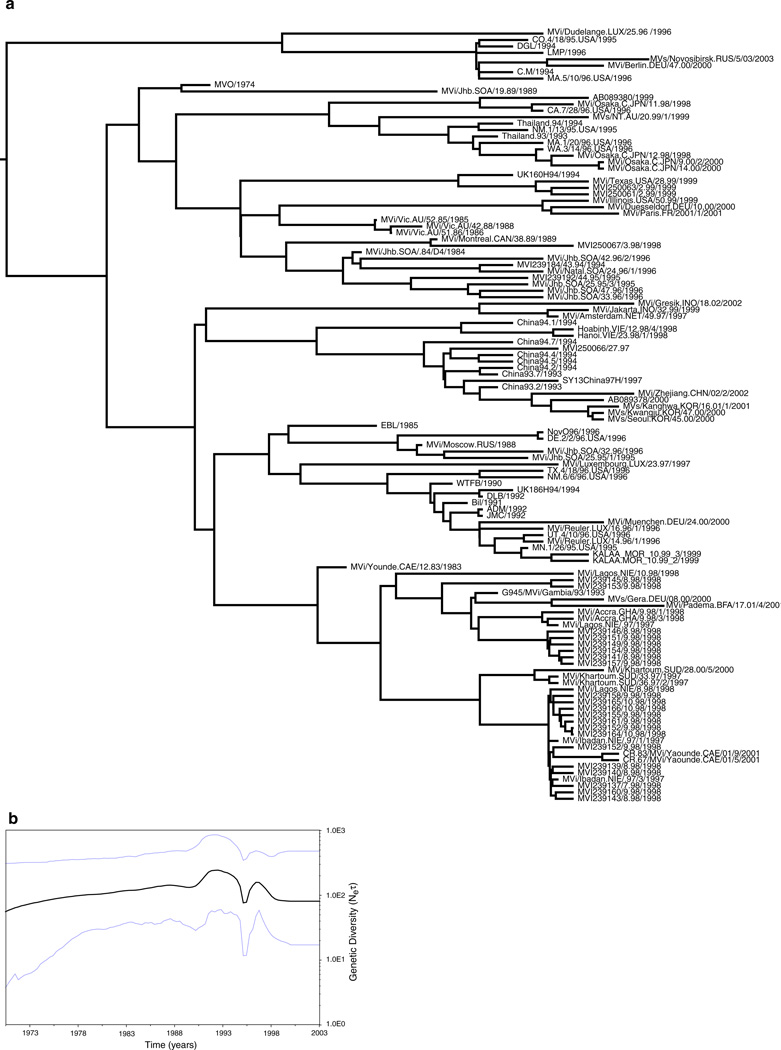
Maximum a posteriori (MAP) trees were inferred for each paramyxovirus data set and are shown in Figs. 1a–4a (equivalent maximum likelihood trees are available from the authors on request). In each case the temporal structure in the data is reflected in the diversity of tip times, which provide a means of estimating evolutionary dynamics.
Fig. 1.
a Maximum a posteriori (MAP) tree of the H gene of measles virus. Tip times reflect the year of sampling (which is provided by the final four digits in the isolate name) and correspond to the time scale provided in b. b Bayesian skyline plot of the changing levels of genetic diversity (Neτ) of the measles virus H gene sampled between 1965 and 2003. The bold line represents the median estimate, while the light lines depict the 95% highest probability density values
Fig. 4.
a Maximum a posteriori (MAP) tree of the H gene of canine distemper virus. b Bayesian skyline plot of the CDV H gene sampled between 1982 and 2001
Rates of evolutionary change, measured as the number of nucleotide substitutions per site, per year (subs/site/year), were estimated using a Bayesian coalescent method (Table 1). In all cases a relaxed (uncorrelated exponential) molecular clock was a better fit to the data than a strict molecular clock, and the best-fit demographic model was either constant population size or exponential population growth (see below), although parameter estimates were consistent among models. Notably, similar parameter values were estimated for all three viruses and with overlapping HPD values in all cases (Table 1). Measles virus was found to have a mean substitution rate of 6.585 × 10−4 for the H gene and 8.693 × 10−4 for the N gene; the HN gene of the mumps virus exhibited a substitution rate of 9.168 × 10−4 subs/site/year, while the H gene of CDV had the highest mean substitution rate, at 11.350 × 10−4 subs/site/year. In the case of the measles H gene, similar rates were found in all subsampled data sets (Table 1) indicating that sampling did not introduced major biases into these analyses. These rates are slightly higher than those previously estimated for the paramyxoviruses (Jenkins et al. 2002), although with overlapping sampling errors, which most likely reflect the use of strict molecular clocks in earlier analyses and relaxed molecular clocks in the current study.
Table 1.
Bayesian estimates of substitution and demographic parameters in the paramyxoviruses studied here
| Virus | Gene | No. of sequences |
Date range of sequences |
Molecular clock |
Demographic model |
Substitution rate, 10−4 subs/site/yr (95% HPD) |
TMRCA, years (95% HPD) |
TMRCA date (95% HPD) |
|---|---|---|---|---|---|---|---|---|
| Measles | H | 120 | 1974–2003 | Relaxed | Exponential growth | 6.585 (4.792–8.306) [6.341–8.154]a |
60 (36–90) [57–63]a |
1943 (1913–1967) [1940–1946]a |
| Measles | N | 107 | 1977–2003 | Relaxed | Constant | 8.693 (5.886–11.270) | 77 (37–140) | 1926 (1863–1966) |
| Mumps | HN | 27 | 1945–1999 | Relaxed | Constant | 9.168 (4.832–14.170) | 91 (58–149) | 1908 (1850–1941) |
| Canine distemper | H | 35 | 1982–2001 | Relaxed | Constant | 11.65 (5.438–18.050) | 58 (27–107) | 1943 (1894–1974) |
Note. HPD, highest probability density
Range of mean values in five additional subsampled data sets of 120 sequences given in brackets
Using the same coalescent approach we also estimated the TMRCA (i.e., the age of the sampled genetic diversity) of each virus (Table 1). For the measles virus H gene, the mean TMRCA was ~60 years before the most recent sequence, collected in 2003, which equates to approximately 1943. The measles N gene analysis yielded a similar result: the mean TMRCA was ~77 years before the most recent sequence, isolated in 2003. The mean TMRCA in the HN gene of mumps virus was ~91 years before 1999, dating the MRCA to ~1908, although with wide HPD values, reflecting the small sample size. Finally, for CDV, the mean TMRCA was ~58 years before the most recent sequence, sampled in 2001, suggesting that the ancestor of these sequences existed close to 1943. Despite differences in sample size, all analyses point to a similar time frame for the age of the current genetic diversity in the measles, mumps, and canine distemper viruses, equating to the first half of the 20th century.
Population Demography
The serially sampled coalescent method available in the BEAST program also allows the inference of the modes and rates for population growth. However, because all three viruses undergo cyclic epidemiological dynamics, and coalescent methods that deal, quantitatively, with such dynamics are currently unavailable, it is not possible to accurately estimate population growth rates in these circumstances. We therefore chose to depict population dynamics using a Bayesian skyline plot which provides a piecewise graphical depiction of changes in genetic diversity (population size) through time (Drummond et al. 2005). Ecologically, measles epidemics occur in multiannual cycles; hints of this periodicity can be seen in the Bayesian skyline plots for this virus. First, in the case of the H gene (Fig. 1b) a peak in population size occurs in ~1991 (although the HPD interval is wide), followed by a population bottleneck in ~1995. This peak in population size is also apparent in the five additional subsampled H gene data sets, although the size of the bottleneck varies in magnitude (not shown; available from the authors on request). An identical demographic signal is apparent in the Bayesian skyline plot of the N gene of measles virus (Fig. 2b), with a sharp population decline clearly depicted. Notably, the timing of the population decline depicted in both genes corresponds with the advent of WHO vaccination campaigns in six southern African countries—Botswana, Malawi, Namibia, South Africa, Swaziland, and Zimbabwe (Uzicanin et al. 2002; WHO 1999)—and African sequences constitute a major component of our measles data sets. Although vaccination campaigns may cause a decrease in cases due to depletion of susceptible individuals, additional epidemics can follow when susceptible individuals are replenished through births or immigration. Given a more intensive sampling regime, in which multiple viruses are collected each year from individual localities, it may be possible to recover the complex cyclic behavior of measles virus.
Fig. 2.
a Maximum a posteriori (MAP) tree of the N gene of measles virus. b Bayesian skyline plot of the measles virus N gene sampled between 1962 and 2003
In the case of mumps virus, the small sample size precludes a detailed analysis of population dynamics, although the Bayesian skyline plot reveals that Neτ is consistently low (see below) (Fig. 3b). Similar constraints imposed by sample size also apply to CDV, the population size of which appears to be relatively constant in the Bayesian skyline plot and with a low Neτ (Fig. 4b). Strikingly, in all the viruses studied here our estimates of genetic diversity, measured as Neτ, are between 10 and 100 and, hence, considerably lower than those previously recorded in chronic infections such as HIV (Robbins et al. 2003) and hepatitis C (Pybus et al. 2001). Such consistently low levels of genetic diversity suggest that recurrent population bottlenecks, a characteristic of paramyxovirus population dynamics, are regularly purging, and hence limiting, genetic variation.
Fig. 3.
a Maximum a posteriori (MAP) tree of the HN gene of mumps virus. b Bayesian skyline plot of the mumps virus HN gene sampled between 1950 and 2000
Selection Pressures
To obtain a measure of the selection pressures acting on the three paramyxoviruses studied here, we computed the relative numbers of nonsynonymous (dN) to synonymous (dS) substitutions per site (ratio dN/dS) using two likelihood-based methods (SLAC and REL [Kosakovsky Pond and Frost 2005]). This analysis revealed little evidence for positive selection, and an abundance of negatively selected sites (Table 2). Overall, across all the sequences analyzed, only four sites were found to be positively selected at a significance value of p < 0.1: one site each for the measlesH and N genes using SLAC and two for the mumps HN gene under REL. No sites were found to be selected in the CDV H gene. No positively selected sites in any virus were detected using the more stringent significance value of p < 0.05.
Table 2.
Summary of selection pressures acting in the paramyxoviruses studied
| Data set | Mean dN/dS | SLAC no. positively selected sites (codon position)a |
SLAC no. negatively selected sitesa |
REL no. positively selected sites (codon position)a |
|---|---|---|---|---|
| Measles H | 0.232 | 1 (476) | 82 | NA |
| Measles N | 0.155 | 1 (329) | 95 | NA |
| Mumps HN | 0.143 | 0 | 49 | 2 (353, 402) |
| Canine distemper N | 0.274 | 0 | 21 | 0 |
p < 0.1 (no positively selected sites were detected at p < 0.05)
Discussion
Our results indicate that the nucleotide substitution rates of measles, mumps, and canine distemper viruses resemble those of many other RNA viruses (Hanada et al. 2004; Jenkins et al. 2002). Measles virus has traditionally been thought to be a relatively conserved RNA virus, since it confers life-long immunity on its hosts and those in the vaccinated class (Panum 1939) and because some earlier studies revealed low levels of both antigenic and genetic variation (Rota et al. 1992). These observations led to suggestions that measles—and its relatives within in the Paramyxoviridae—evolve anomalously slowly. However, our analyses indicate that all three paramyxoviruses studied here have substitution rates in the range of 10−3 to 10−4 subs/site/year, typical of the rapid mutation and replication dynamics that define RNA viruses (Domingo and Holland 1997). It is therefore important to note that high rates of molecular evolutionary change do not necessarily translate into high levels of antigenic variation.
Striking, the TMRCA of all three viruses was surprisingly recent and always within the last century. In principle, such shallow genetic diversity can be attributed to one of four causes: the erroneous estimation of substitution rates and divergence times, recent cross-species transmission, a large-scale and global population bottle-neck which purged preexisting genetic diversity, or neutral evolution within a small effective population. The first of these four theories—estimation error—seems unlikely, as substitution rates of between 10−3 and 10−4 subs/site/year have been estimated previously for the paramyxoviruses (Woelk et al. 2001, 2002; Jenkins et al. 2002; Hanada et al. 2004) and for RNA viruses in general. Similarly, we regard recent cross-species transmission as unlikely since measles-like disease has been documented in human populations for millennia, associated with the start of human urbanization (Orvell 1994; Carbone and Wolinsky 2001; Rima 2004).
A more plausable explanation for the recent dates of common ancestry is a large-scale and geographically extensive population bottleneck (as opposed to local cyclical dynamics), either neutral or selectively determined, which would have purged most standing genetic variation. Although possible, it seems unlikely that such bottlenecks would occur in all the paramyxoviruses studied here on roughly the same time scale. Further, host immune selection, mediated by either antibody or T-cell responses, is only sporadically observed in measles virus and is not associated with global selective sweeps (Woelk et al. 2001). Indeed, we found only weak evidence for positive selection in the sequence data analyzed here.
Finally, and perhaps most likely of all, the recent age of the three viruses may simply be a fundamental property of neutral evolutionary dynamics. Under a haploid Wright-Fisher model, the expected mean TMRCA is 2Ne generations (Ewens 2004). Although there are 30 million measles cases per year (WHO 2007), the number of cases in the troughs between epidemic peaks is evidently much lower. Indeed, estimates for case numbers during epidemic troughs, which provide a more realistic measure of effective population size, can be as low as 5000 individuals (Grenfell, pers. comm.). Under these parameters, and assuming an epidemic generation time of 9–13 days the mean TMRCA of measles virus would be 90,000–130,000 days, or ~250–300 years—with a large variance. It is therefore possible that a history of neutral genetic drift alone is sufficient to cause the shallow genetic diversity observed in the paramxyoviruses studied here, particularly given their complex cyclical dynamics, which will reduce effective population sizes.
Acknowledgments
We thank Rubing Chen for assistance with the BEAST analyses, Bryan Grenfell for advice on measles demography and statistics, and two anonymous reviewers for useful comments. Laura Pomeroy was supported by the National Science Foundation, under the NSF Graduate Teaching Fellowship in K-12 Education (DGE-0338240). This work was also supported by NIH Grant GM080533-01.
Contributor Information
Laura W. Pomeroy, Email: LPomeroy@psu.edu, Center for Infectious Disease Dynamics, Department of Biology, The Pennsylvania State University, 501 ASI Building, University Park, PA 16802, USA.
Ottar N. Bjørnstad, Center for Infectious Disease Dynamics, Department of Biology, The Pennsylvania State University, 501 ASI Building, University Park, PA 16802, USA Fogarty International Center, National Institutes of Health, Bethesda, MD 20892, USA; Department of Entomology, The Pennsylvania State University, University Park, PA 16802, USA.
Edward C. Holmes, Center for Infectious Disease Dynamics, Department of Biology, The Pennsylvania State University, 501 ASI Building, University Park, PA 16802, USA Fogarty International Center, National Institutes of Health, Bethesda, MD 20892, USA.
References
- Anderson RM, May RM. Infectious diseases of humans: dynamics and control. Oxford: Oxford University Press; 1991. [Google Scholar]
- Barrett T. Rinderpest and distemper viruses. In: Webster RG, Granoff A, editors. Encyclopedia of virology. New York: Academic Press; 1994. pp. 1260–1269. [Google Scholar]
- Bartlett MS. Measles periodicity and community size. J R Stat Soc A. 1957;120:48–70. [Google Scholar]
- Bellini WJ, Rota PA. Genetic diversity of wild-type measles viruses: implications for global measles elimination programs. Emerg Infect Dis. 1998;4:29–35. doi: 10.3201/eid0401.980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt G, Jensen TD, Gottschalck E, Arctander P, Appel MJ, Buckland R, Blixenkrone-Moller M. Genetic diversity of the attachment (H) protein gene of current field isolates of canine distemper virus. J Gen Virol. 1997;78:367–372. doi: 10.1099/0022-1317-78-2-367. [DOI] [PubMed] [Google Scholar]
- Carbone KM, Wolinsky JS. Mumps virus. In: Fields BN, Knipe DM, Howley PM, editors. Virology. Philadelphia, PA: Lippincott Williams & Wilkins; 2001. pp. 1381–1400. [Google Scholar]
- Domingo E, Holland JJ. RNA virus mutations for fitness and survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST v1.0. 2003 Available at: http://www.evolve.zoo.ox.ac.uk/beast/ [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4(5):e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens WJ. Mathematical population genetics. 2nd ed. New York: Springer-Verlag; 2004. [Google Scholar]
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus taxomomy: classification and nomenclature of viruses. New York: Elsevier Academic Press; 2005. [Google Scholar]
- Grenfell B, Harwood J. (Meta)population dynamics of infectious diseases. Trend Ecol Evol. 1997;12:395–399. doi: 10.1016/s0169-5347(97)01174-9. [DOI] [PubMed] [Google Scholar]
- Griffin DE. Measles virus. In: Fields BN, Knipe DM, Howley PM, editors. Virology. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1401–1441. [Google Scholar]
- Hanada K, Suzuki Y, Gojobori T. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol Biol Evol. 2004;21:1074–1080. doi: 10.1093/molbev/msh109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 2002;54:156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Virology. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1305–1340. [Google Scholar]
- Mochizuki M, Hashimoto M, Hagiwara S, Yoshida Y, Ishiguro S. Genotypes of canine distemper virus determined by analysis of the hemagglutinin genes of recent isolates from dogs in Japan. J Clin Microbiol. 1999;37:2936–2942. doi: 10.1128/jcm.37.9.2936-2942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvell C. Measles virus. In: Webster RG, Granoff A, editors. Encyclopedia of virology. New York: Academic Press; 1994. pp. 838–847. [Google Scholar]
- Orvell C, Tecle T, Johansson B, Saito H, Samuelson A. Antigenic relationships between six genotypes of the small hydrophobic protein gene of mumps virus. J Gen Virol. 2002;83:2489–2496. doi: 10.1099/0022-1317-83-10-2489. [DOI] [PubMed] [Google Scholar]
- Panum PL. Observations made during the epidemic of measles on Faroe Islands in the year 1846. Med Class. 1939;3:829–886. [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Pybus OG, Charleston MA, Gupta S, Rambaut A, Holmes EC, Harvey PH. The epidemic behaviour of the hepatitis C virus. Science. 2001;292:2323–2325. doi: 10.1126/science.1058321. [DOI] [PubMed] [Google Scholar]
- Rambaut A. Se-Al: Sequence Alignment Editor. 1996 Available at: http://www.evolve.zoo.ox.ac.uk/ [Google Scholar]
- Rima BK. Mumps virus. In: Webster RG, Granoff A, editors. Encyclopedia of virology. New York: Academic Press; 1994. pp. 876–883. [Google Scholar]
- Robbins KE, Lemey P, Pybus OG, Jaffe HW, Youngpairoj AS, Brown TM, Salemi M, Vandamme A-M, Kalish ML. US human immunodeficiency virus type 1 epidemic: Date of origin, population history, and characterization of early strains. J Virol. 2003;77:6359–6366. doi: 10.1128/JVI.77.11.6359-6366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota JS, Hummel KB, Rota PA, Bellini WJ. Genetic variability of the glycoprotein genes of current wild-type measles isolates. Virology. 1992;188:135–142. doi: 10.1016/0042-6822(92)90742-8. [DOI] [PubMed] [Google Scholar]
- Uzicanin A, Eggers R, Webb E, Harris B, Durrheim D, Ogunbanjo G, Isaacs V, Hawkridge A, Biellik R, Strebel P. Impact of the 1996–1997 supplementary measles vaccination campaigns in South Africa. Int J Epidemiol. 2002;31:968–976. doi: 10.1093/ije/31.5.968. [DOI] [PubMed] [Google Scholar]
- WHO. Progress toward measles elimination—Southern Africa, 1996–1998. MMWR. 1999;48:585–589. [PubMed] [Google Scholar]
- WHO. Measles fact sheet. 2007 Available at: http://www.who.int/mediacentre/factsheets/fs286/en/
- Woelk CH, Jin L, Holmes EC, Brown DWG. Immune and artificial selection in the hemagglutin (H) glycoprotein of measles virus. J Gen Virol. 2001;82:2463–2474. doi: 10.1099/0022-1317-82-10-2463. [DOI] [PubMed] [Google Scholar]
- Woelk CH, Pybus OG, Jin L, Brown DWG, Holmes EC. Increased positive selection pressure in persistent (SSPE) versus acute measles virus infections. J Gen Virol. 2002;83:1419–1430. doi: 10.1099/0022-1317-83-6-1419. [DOI] [PubMed] [Google Scholar]