Summary
Intestinal epithelial cells are exposed to both innocuous and pathogenic microbes, which need to be distinguished to mount an effective immune response. To understand the mechanisms underlying pathogen recognition, we investigated how Pseudomonas aeruginosa triggers intestinal innate immunity in Caenorhabditis elegans, a process independent of Toll-like pattern recognition receptors. We show that the P. aeruginosa translational inhibitor Exotoxin A (ToxA), which ribosylates elongation factor 2 (EF2), upregulates a significant subset of genes normally induced by P. aeruginosa. Moreover, immune pathways involving the ATF-7 and ZIP-2 transcription factors, which protect C. elegans from P. aeruginosa, are required for preventing ToxA-mediated lethality. ToxA-responsive genes are not induced by enzymatically inactive ToxA protein but can be upregulated independently of ToxA by disruption of host protein translation. Thus, C. elegans has a surveillance mechanism to recognize ToxA through its effect on protein translation rather than by direct recognition of either ToxA or ribosylated EF2.
Introduction
Intestinal epithelial cells (IECs) are continuously exposed to a complex environment populated with a vast diversity of both innocuous and pathogenic microbes. To mount an appropriate immune response, it is essential for these cells to distinguish between pathogens and commensals. It remains unclear, however, how IECs can recognize and react to virulent microbes while ignoring their innocuous counterparts.
In the current paradigm of innate immunity, pathogens are primarily detected by host pattern recognition receptors (PRRs) that recognize specific microbial features called microbe- or pathogen-associated molecular patterns (MAMPs/PAMPs) (Medzhitov, 2009). However, this model fails to address how MAMPs/PAMPs such as peptidoglycan can be used to distinguish between pathogenic and non-pathogenic bacteria. Alternate theories propose that metazoan immune responses can also be activated by indirect evidence of infection. Indeed, such a phenomenon has been observed in plants, which can recognize pathogens by the disruption of cellular homeostasis caused by pathogen-encoded virulence factors (“effector-triggered immunity”) (Jones and Dangl, 2006). Similarly, in Drosophila melanogaster, microbial virulence factors have recently been shown to initiate an immune response by modifying host proteins (Boyer et al., 2011). In mammals, the release of molecules such as DNA, uric acid, and ATP from damaged tissues can activate immune mechanisms (Kono and Rock, 2008). Moreover, concurrent inhibition of protein synthesis by secreted Legionella pneumophila effector proteins and exposure to a MAMP/PAMP upregulates immune genes in macrophages (Fontana et al., 2011). These indicators of pathogen activity have been referred to as patterns-of-pathogenesis or damage-associated molecular patterns (DAMPs) (Vance et al., 2009). In general, however, the metazoan surveillance and signaling pathways involved in these postulated mechanisms of pathogen detection remain to be discovered.
We are using the nematode Caenorhabditis elegans to investigate the mechanisms by which IECs identify pathogens. As C. elegans lack professional immune cells, IECs serve as the primary defense against ingested pathogens such as the human opportunistic pathogen Pseudomonas aeruginosa (Irazoqui et al., 2010b; Pukkila-Worley and Ausubel, 2012). The P. aeruginosa strain PA14 accumulates in the C. elegans intestinal lumen and causes host damage, upregulation of antimicrobial genes, and kills the nematode host (Irazoqui et al., 2010b; Pukkila-Worley and Ausubel, 2012). C. elegans defense against PA14 involves several parallel signaling pathways defined by PMK-1 p38 MAPK, a bZIP transcription factor called ZIP-2, and the G-protein-coupled receptor FSHR-1 (Irazoqui et al., 2010b; Pukkila-Worley and Ausubel, 2012), Interestingly, C. elegans does not encode any recognizable homologs of the NF-κB transcription factor, and its single Toll-like receptor (TLR), TOL-1, does not seem to play a key role in immunity (Irazoqui et al., 2010b; Pujol et al., 2001; Pukkila-Worley and Ausubel, 2012). In C. elegans, the P. aeruginosa-encoded signals that activate immune pathways and the corresponding PRRs, if any, are unknown.
Recent experiments show that heat-killed P. aeruginosa or virulence-attenuated mutants induce much lower levels of host gene expression than wild-type pathogen (Pukkila-Worley and Ausubel, 2012), suggesting that C. elegans immunity is triggered by DAMPs generated during a P. aeruginosa infection and/or by a process analogous to effector-triggered immunity in plants. Based on these data, we hypothesized that DAMP signals or host modifications generated by a single P. aeruginosa factor might be sufficient to trigger an immune response. However, the P. aeruginosa mutants that exhibit the most avirulent phenotypes in C. elegans correspond to global transcriptional regulators that control the activity of many downstream effector genes (Tan et al., 1999b). We therefore reasoned that it would be difficult to identify the hypothesized eliciting factor(s) by testing P. aeruginosa virulence-related mutants. Instead, we engineered a normally non-pathogenic E. coli strain to produce the P. aeruginosa-encoded AB toxin Exotoxin A (ToxA), which catalyzes the ADP-ribosylation of elongation factor 2 (EF2) by targeting the EF2 diphthamide moiety, a post-translationally modified histidine, thereby blocking protein synthesis (Yates et al., 2006).
ToxA is expressed in over 90% of clinical P. aeruginosa isolates, high toxA transcription levels are associated with severe infections (Bjorn et al., 1977; Matar et al., 2002), and toxA deficient strains are attenuated in animal virulence assays (Fogle et al., 2002; Miyazaki et al., 1995; Nicas and Iglewski, 1985; Rahme et al., 1995). Additionally, as shown in the accompanying paper, ToxA inhibits protein synthesis in C. elegans IECs during a PA14 infection (Dunbar et al., 2012). Importantly, toxins encoded by other pathogens also ribosylate EF2, including diphtheria toxin, the major virulence factor of Corynebacterium diphtheriae, and cholix toxin from Vibrio cholerae (Jorgensen et al., 2008; Yates et al., 2006). We therefore theorized that C. elegans may be able to sense P. aeruginosa and multiple other pathogens by monitoring their common molecular target (the diphthamide residue), EF2 function, or protein translation in general.
In this paper, we present evidence that C. elegans has surveillance machinery that recognizes ToxA indirectly by detecting ToxA-mediated translational inhibition. This recognition activates a potent transcriptional response in the worm that is partially dependent on known immune pathways. Moreover, the ZIP-2, PMK-1 MAPK, and FSHR-1 signaling pathways, which are necessary for resistance against P. aeruginosa, protect C. elegans from ToxA-mediated killing. In an accompanying paper, Dunbar et al. show that P. aeruginosa inhibits protein synthesis in C. elegans intestinal cells, which triggers increased levels of ZIP-2 protein and its downstream factors. Together, these studies demonstrate that the activity of a P. aeruginosa virulence factor activates a subset of C. elegans immune genes independently of either direct recognition of a bacterial-encoded molecule or effector-mediated modification of a host protein.
Results
ToxA upregulates a subset of the genes induced by P. aeruginosa
To determine if C. elegans can detect and respond to an individual P. aeruginosa factor, we used the pET100 plasmid vector to express the P. aeruginosa PA14 toxA gene in a normally non-pathogenic strain of E. coli (BL21 star). We fed these bacteria to C. elegans containing transcriptional immune reporters for irg-1 or F35E12.5, two genes robustly upregulated by P. aeruginosa PA14 (Bolz et al., 2010; Estes et al., 2010; Troemel et al., 2006). As expected, PA14 induced expression of irg-1::GFP and F35E12.5::GFP, while E. coli expressing an empty pET100 vector did not (Figures 1A, B and S1A). In contrast, E. coli expressing ToxA strongly activated irg-1::GFP (Figure 1C). However, ToxA does not fully mimic a P. aeruginosa infection as ToxA failed to upregulate the F35E12.5::GFP reporter (Figure S1A) or eight other representative P. aeruginosa-responsive genes (Figure S1B). ToxA was also unable to induce the clec-60::GFP reporter (Figure S1A), which is strongly activated by S. aureus (Irazoqui et al., 2008).
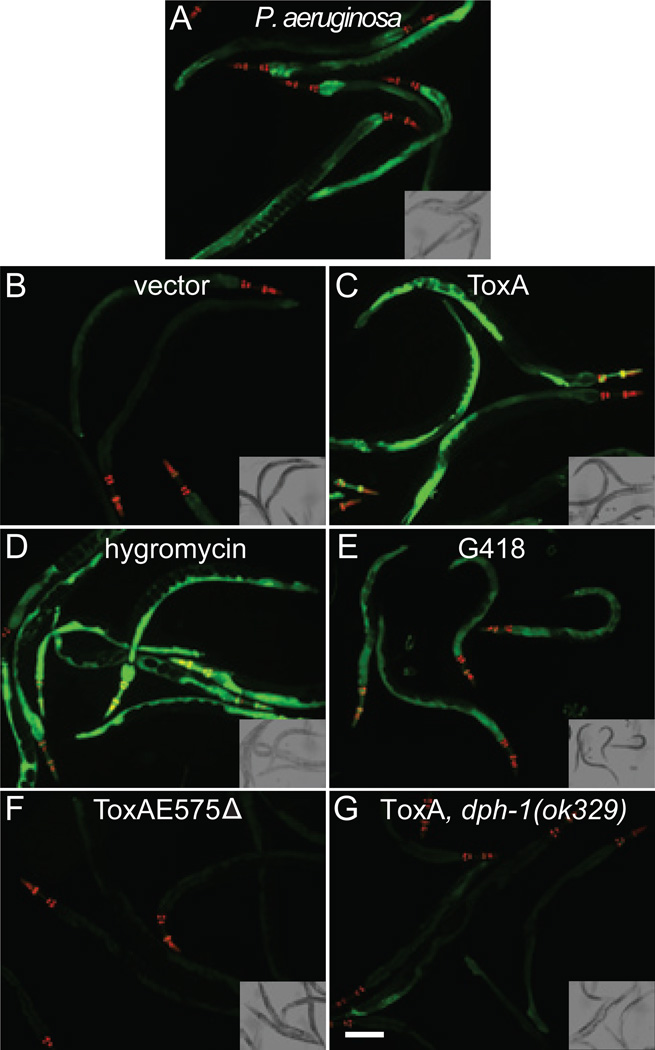
Figure 1. The immune reporter irg-1::GFP is activated by ToxA and translational inhibitors but not by inactive ToxA protein.
A C. elegans strain containing the irg-1::GFP reporter was exposed to P. aeruginosa PA14 (A); E. coli expressing either an empty expression vector (B) or ToxA (C); the translational elongation inhibitors hygromycin (D) or G418 (E); or ToxA in conditions where it causes less cellular damage because it is missing an essential catalytic residue (ToxAE575Δ; F) or because C. elegans lacks its diphthamide target (dph-1 mutant; G). All animals were exposed to the indicated condition for 24 hours starting at the L4 stage. Red pharyngeal expression is due to the co-injection marker myo-2::mCherry and confirms the presence of the transgene. All images were taken at the same time using the same camera settings. Scale bar represents 100 μm. Insets are the corresponding bright field image. See also Figure S1.
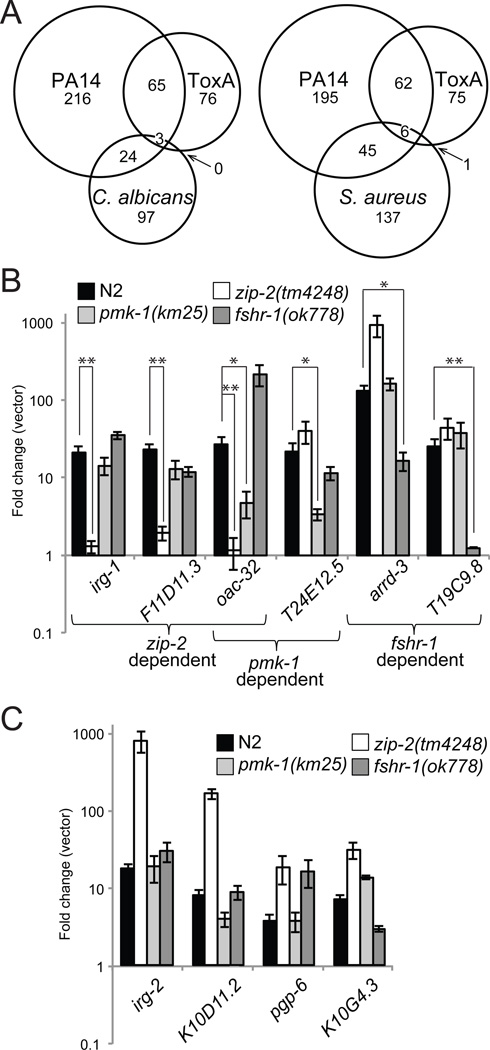
To determine whether ToxA activates other P. aeruginosa-responsive genes in addition to irg-1, we performed genome-wide transcriptional profiling analyses of wild-type animals exposed to E. coli expressing either ToxA or an empty pET100 vector. Using Affymetrix GeneChips, we identified 174 genes whose expression changed at least two-fold when exposed to ToxA (144 upregulated and 30 downregulated; see Supplemental Table 1). Based on previous Affymetrix profiling experiments, 68 of these 144 upregulated genes are also induced during the first four hours of a P. aeruginosa infection (p<1x10−13; Figure 2A) and 10 of the remaining 76 ToxA-activated genes respond to P. aeruginosa at a later time point (Troemel et al., 2006).
Figure 2. ToxA induces a subset of P. aeruginosa-induced genes through several signaling pathways.
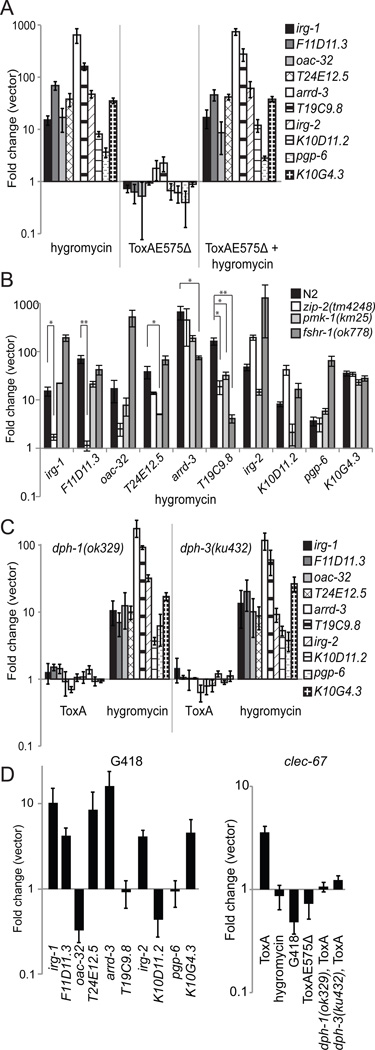
(A) Venn diagrams comparing the overlaps in genes activated by ToxA, Gram-negative P. aeruginosa PA14 (Troemel et al., 2006), and either the yeast C. albicans (Pukkila-Worley et al., 2011) or Gram-positive S. aureus (Irazoqui et al., 2010a). All microarrays were conducted with the Affymetrix GeneChip platform using animals infected at the L4/young adult stage and collected after 24 hours (ToxA), 8 hours (S. aureus), or 4 hours (PA14, C. albicans). See also Figure S2. (B and C) qRT-PCR analysis in different mutant backgrounds of genes normally activated by ToxA. ** Genes with ≥10-fold lower induction to ToxA as compared to wild-type N2 animals with p<0.05. * Genes with ≥5-fold lower induction to ToxA as compared to N2 with p<0.05. Results shown are an average of 6 (N2) or 3 (pmk-1, zip-2, fshr-1) biological replicates. Error bars represent SEM. P values determined with a 2-tailed unpaired t-test.
To further investigate the specificity of the ToxA response, we used publically available transcriptional profiling data sets to determine how many ToxA-activated genes are upregulated by other C. elegans pathogens (Irazoqui et al., 2010a; Pukkila-Worley et al., 2011). We found that ToxA only activates three of the 124 genes induced by infection with the fungal pathogen Candida albicans and seven of the 189 genes induced by infection with S. aureus (neither overlap is significant, p>0.1 for both comparisons) (Figure 2A).
We next tested whether ToxA acts as a general intestinal stressor by determining the number of ToxA-responsive genes that are also triggered by the heavy metal cadmium or the pore-forming toxin Cry5B (Huffman et al., 2004). Whereas ToxA induces the expression of a significant number of these stress-response genes (46 of 453 cadmium-responsive genes and 39 of 385 Cry5B-responsive genes), the majority of these “overlapping” genes (74% and 79% respectively) are also upregulated by P. aeruginosa (Figure S2). Taken together, these data show that ToxA activates a particular subset of the genes induced when C. elegans is infected with P. aeruginosa PA14, but does not trigger broad immune or stress responses.
Since ToxA upregulates a subset of P. aeruginosa-responsive genes, we investigated whether the same immune pathways are required for both ToxA- and P. aeruginosa-mediated gene induction. Previous genome-wide profiling experiments identified genes whose induced or basal expression requires the ZIP-2 or PMK-1 MAPK pathways, respectively (Estes et al., 2010; Troemel et al., 2006). We found that ToxA upregulated 19 of the 25 genes specified by Estes et al. that are activated downstream of ZIP-2 (p<5x10−4) but only five of the 101 genes identified by Troemel et al. that require the PMK-1 p38 MAPK for basal expression (p>0.05). This PMK-1 microarray, however, did not identify genes that require PMK-1 for infection-mediated upregulation and some of the genes whose induction requires PMK-1 do not require PMK-1 for basal expression (Troemel et al., 2006). To investigate the possibility that additional ToxA-responsive genes are PMK-1-dependent and to extend our analysis, we selected a representative panel of highly-upregulated, ToxA-responsive genes to assay by qRT-PCR in different mutant backgrounds. We selected 11 genes, eight of which (with the exception of arrd-3, T19C9.8, and dur-1) have been reported to be upregulated by P. aeruginosa, and verified that 10 of these genes were induced as expected in wild-type animals exposed to ToxA (Figures 2B and C; data not shown for dur-1). This verification rate is consistent with prior microarray analyses of pathogen response genes (Irazoqui et al., 2010a; Pukkila-Worley et al., 2011). qRT-PCR analysis in different mutant strains showed that irg-1 and two previously uncharacterized genes (F11D11.3 and oac-32) required ZIP-2, two genes (oac-32 and T24E12.5) were partially dependent on PMK-1, and two genes (arrd-3 and T19C9.8) were at least partially dependent on FSHR-1 for ToxA-mediated activation (Figure 2B). The four remaining genes (irg-2, K10D11.2, pgp-6, and K10G4.3) were induced in all three mutants tested (Figure 2C). Interestingly, ZIP-2 was required to activate irg-2 in response to P. aeruginosa (Estes et al., 2010) but not in response to ToxA, although we did observe that the basal expression of irg-2 was >30-fold lower in zip-2(tm4248) mutants compared to wild-type worms.
We additionally determined whether two conserved pathways that are involved in both general stress and immune responses are similarly involved in ToxA-dependent gene activation. Specifically, we tested the potential involvement of the Nrf1/2/3 ortholog SKN-1 (van der Hoeven et al., 2011) and the insulin signaling pathway mediated by the FOXO transcription factor DAF-16 (Garsin et al., 2003). Unlike the immune pathways above, ToxA-induced expression of all 10 genes was unaffected following skn-1 RNAi or in daf-16(mgDF47) mutants (not shown). Moreover, exposure to ToxA was not sufficient to cause nuclear translocation of a DAF-16::GFP translational fusion (not shown)(Henderson and Johnson, 2001).
Immune pathways are required for ToxA resistance
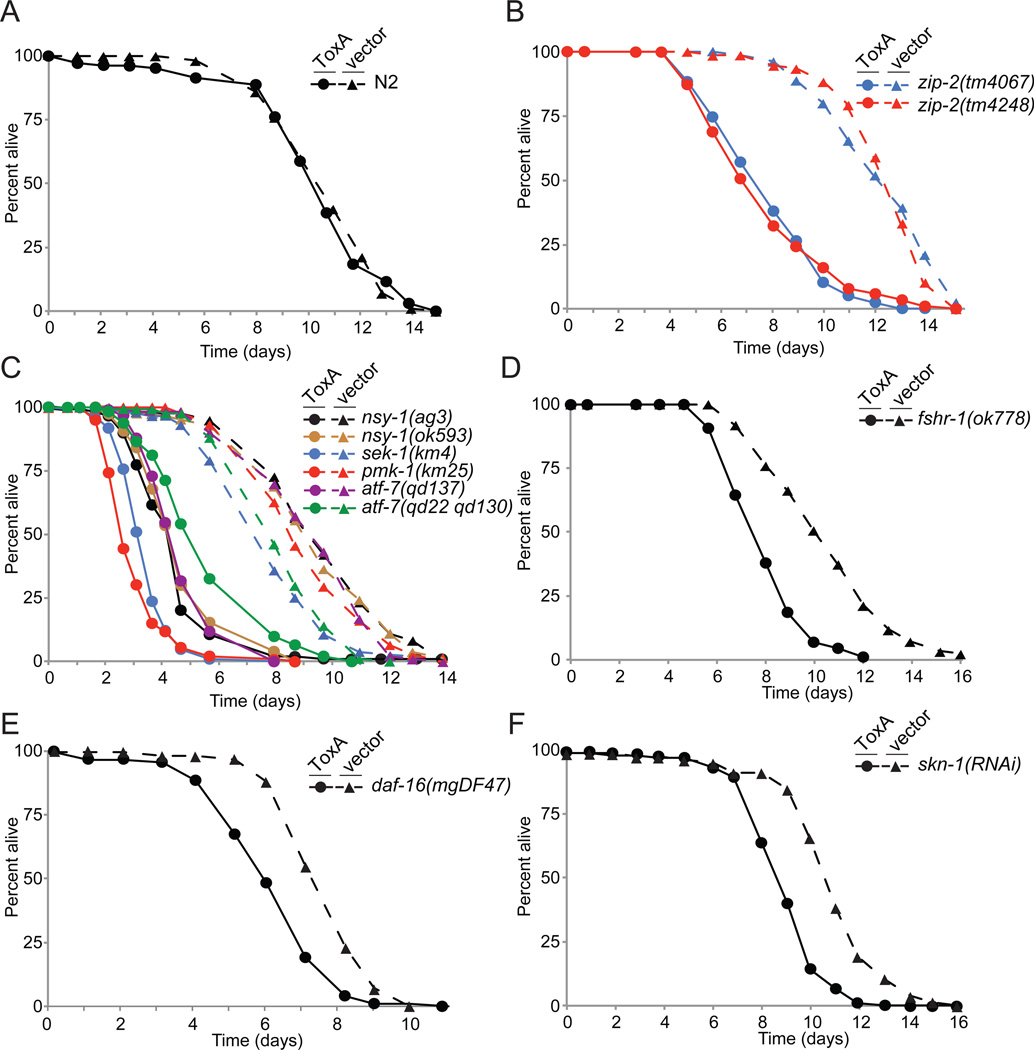
Since ToxA is a potent cytotoxin that activates C. elegans immune genes, we were surprised to observe no difference in the lifespan of wild-type worms fed E. coli expressing either ToxA or the empty vector control (p=0.8; Figure 3A), suggesting that the wild-type immune response may counteract its toxic effects. To test this possibility, we asked whether immunocompromised worms are similarly unaffected by ToxA. Since ZIP-2 acts upstream of many ToxA-responsive genes (Figure 2), we exposed zip-2(tm4067) or zip-2(tm4248) mutant worms to ToxA-expressing E. coli and discovered that, unlike wild-type animals, zip-2 mutants had significantly shorter lifespans when fed ToxA as compared to the vector control (p<5x10−9 for both alleles; Figure 3B).
Figure 3. Multiple pathways contribute to ToxA resistance.
Lifespan of wild-type N2 (A), zip-2 mutant (B), p38 MAP kinase pathway mutant (C), fshr-1 mutant (D), daf-16 mutant (E), or skn-1 RNAi (F) worms fed E. coli expressing either ToxA or an empty expression vector starting at the L4 stage. Circles represent animals fed ToxA and triangles indicate vector control food. See also Figure S3.
We next asked whether, as with the ZIP-2 pathway, other immune pathways that defend C. elegans from P. aeruginosa similarly prevent ToxA lethality. We found that the p38 MAPK mutant pmk-1(km25) was also sensitive to ToxA and died at a faster rate than zip-2 mutants (p<5x10−9; Figure 3C). PMK-1 acts directly downstream of the MAPKKK NSY-1 and MAPKK SEK-1 and upstream of the ATF-7 transcription factor (Pukkila-Worley and Ausubel, 2012). Similarly to pmk-1 mutants, nsy-1, sek-1, and atf-7 mutants were susceptible to ToxA (p<5x10−9 for all mutants; Figure 3C). We also determined that loss of the FSHR-1 pathway resulted in ToxA-sensitivity, although the effect was less pronounced than for the zip-2 and pmk-1 mutants (p<1x10−6; Figure 3D). Finally, we tested whether the stress response pathways mediated by DAF-16 and SKN-1 were required for ToxA resistance and found that loss of either protein conferred a slight susceptibility to ToxA (Figures 3E and F). Animals lacking both pmk-1 and zip-2, pmk-1 and fshr-1, pmk-1 and daf-16, or pmk-1 and skn-1 did not show faster ToxA-dependent lethality than single pmk-1(km25) mutants (not shown), potentially indicating that the genes regulated by pmk-1 are epistatic to those dependent on other immune pathways. Alternatively, the critical ToxA-defense genes may be regulated by multiple pathways or this E. coli ToxA killing assay is not sensitive enough to detect subtle differences in ToxA susceptibility.
The inability of immunocompromised worms to resist ToxA as effectively as wild-type worms suggests that ToxA may be responsible for the increased lethality of these immune pathway mutants during a P. aeruginosa infection. To test this hypothesis, we compared the virulence of wild-type P. aeruginosa PA14 and a toxA mutant towards either ToxA-resistant N2 animals or the extremely ToxA-sensitive pmk-1(km25) mutant. Although ToxA is important for the inhibition of C. elegans intestinal translation during a P. aeruginosa infection (Dunbar et al., 2012), we found that wild-type PA14 and the PA14 toxA mutant were indistinguishable in their ability to kill either N2 or pmk-1(km25) animals (p>0.1 for both C. elegans strains; Figure S3). While our laboratory previously reported that the PA14 toxA mutant exhibited a modest delay in killing wild-type N2 worms (Tan et al., 1999a), we speculate that this apparent discrepancy with our current results is due to minor methodological differences. Regardless, loss of ToxA does not severely hinder P. aeruginosa from killing even a ToxA-sensitive C. elegans strain, suggesting that P. aeruginosa encodes multiple, redundant virulence determinants that together contribute to killing the worms.
C. elegans responds to translational inhibition
We considered three general mechanisms by which ToxA could elicit an immune response. The ToxA protein could be directly recognized as a MAMP/PAMP, EF2-ribosylation by ToxA could be sensed directly, or ToxA could be indirectly detected by surveying for the inhibition of protein synthesis, the consequence of which might include the generation of DAMPs. To distinguish amongst these possibilities, we first tested whether the irg-1::GFP reporter could be activated by a ToxA protein that is defective in EF2 ribosylation (MAMP/PAMP-like mechanism) or could be activated by disrupting protein synthesis independently of ToxA (DAMP/effector-like mechanism).
To mimic a ToxA-mediated translational block, we used the aminoglycoside antibiotics hygromycin B (hereafter referred to as “hygromycin”) and G418, both of which bind to the eukaryotic ribosome and prevent elongation of the polypeptide chain (Eustice and Wilhelm, 1984). While prolonged exposure to these drugs resulted in nematode lethality (Figure S4A), at early times in the treatment, both antibiotics triggered the upregulation of irg-1::GFP (Figures 1D, E).
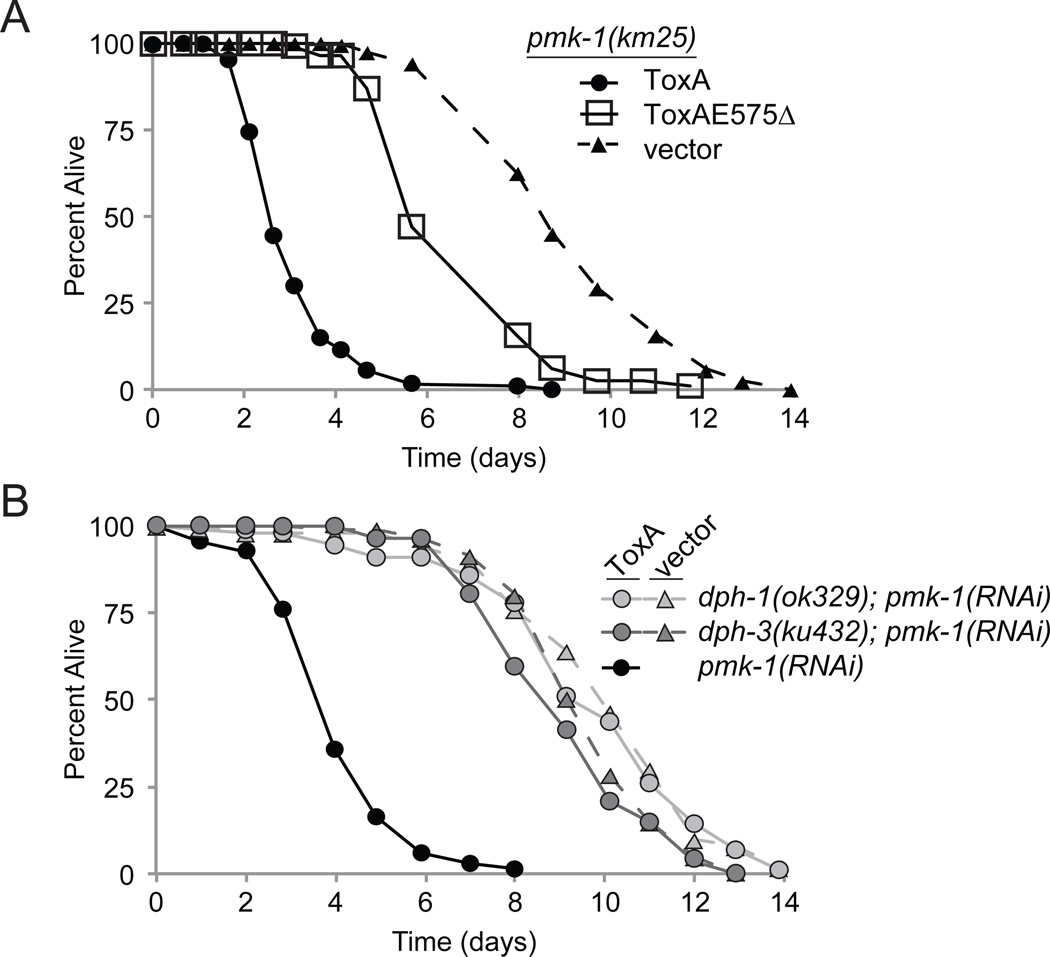
We next tested whether irg-1::GFP is upregulated by a catalytically-inactive ToxA protein that does not inhibit host translation. For this experiment, we constructed a ToxAE575Δ mutant, which is missing a key catalytic residue directly involved in the ADP-ribosylation of EF2 but not required for the stability or cellular trafficking of ToxA (Killeen and Collier, 1992; Lukac et al., 1988; Pastrana et al., 2008). We determined by Western blotting analysis that both wild-type ToxA and ToxAE575Δ mutant proteins were expressed by E. coli at comparable levels (see Methods). To confirm that the ToxAE575Δ mutant is attenuated in C. elegans, we assayed its effect on pmk-1(km25) mutants. As expected, E. coli expressing ToxAE575Δ did not rapidly kill these animals like E. coli expressing wild-type ToxA. It is worth noting that the ToxAE575Δ mutant may contain residual catalytic activity, as exposure to it slightly shortened the nematode lifespan as compared to the vector control (Figure 4A). In a complementary set of experiments, we inhibited the function of ToxA by preventing the formation of its diphthamide target. We confirmed that C. elegans mutants of the diphthamide synthesis genes dph-1 and dph-3 were insensitive to wild-type toxin by feeding them E. coli expressing ToxA after knocking down pmk-1 with RNAi (p>0.1 for both mutants; Figure 4B). To ensure that dph-1 and dph-3 do not have background RNAi mutations making them resistant to pmk-1 knockdown, we confirmed that both strains responded like wild-type animals to RNAi targeting lin-29 and nhr-23 (not shown) (Zhang et al., 2011). Thus, by using these two methods, we could expose animals to ToxA without affecting host translation. When E. coli expressing ToxAE575Δ was fed to wild-type C. elegans or E. coli expressing wild-type ToxA was fed to the C. elegans dph-1 mutant, the irg-1::GFP reporter was not induced, demonstrating that the presence of ToxA protein alone was not sufficient to trigger the full C. elegans response (Figures 1F, G). Additional data supporting this conclusion is presented below.
Figure 4. Mutating ToxA or removing the C. elegans diphthamide modification inhibits ToxA-mediated lethality.
(A) Lifespan comparison of pmk-1(km25) mutants fed either wild-type ToxA, the catalytic mutant ToxAE575Δ, or vector control food. (B) Lifespan comparison between wild-type N2 and diphthamide synthesis mutants when fed ToxA following pmk-1 RNAi. All assays were started with L4 stage animals.
To determine whether detecting ToxA’s inhibition of protein synthesis can induce the complete ToxA-response described in Figure 2A, we carried out transcriptional profiling of wild-type animals exposed to hygromycin. We identified 1297 genes whose expression was altered at least two-fold by the inhibitor (861 induced and 436 repressed; see Supplementary Table 2). Strikingly, of the 144 genes activated by ToxA, 136 (94%) were also upregulated by hygromycin in the absence of ToxA (6.3 genes expected by chance; Figure S4B). For those genes activated by both ToxA and hygromycin, hygromycin induced an average 8-fold stronger response than ToxA under the conditions of these transcriptional profiling analyses. The larger effect of hygromycin on both the number of genes impacted and the magnitude of their response suggests that C. elegans translation was more effectively disrupted by the hygromycin treatment. Alternatively, hygromycin could have multiple cellular targets and thus trigger a broad response involving many non-specific immune and stress genes. To help distinguish these possibilities, we determined which functional classes were overrepresented among the hygromycin-induced genes using DAVID enrichment analysis (Huang et al., 2009). Similar to ToxA, hygromycin preferentially upregulated genes related to transcriptional regulation and innate immunity (Figure S4C). Hygromycin also activated genes associated with protein kinase cascades, Ras-GTPases, and proteins containing F-box domains, all of which have been previously directly or indirectly implicated in C. elegans host-pathogen defenses (Kim et al., 2002; Lundquist, 2006; Thomas, 2006). We additionally identified enriched gene classes by analyzing our results in the context of gene mountains, which are clusters of co-regulated genes as determined by meta-array analysis (Kim et al., 2001). Intestinal and antibacterial genes were disproportionally upregulated following exposure to either hygromycin or ToxA (mountain 8 enriched 3- and 3.5-fold compared to the genome respectively, p<0.01). In contrast, hygromycin but not ToxA upregulated neuronal genes (mountains 1 and 6 each 2-fold enriched, p<0.01), suggesting that hygromycin may affect a wider variety of targets than ToxA.
We further analyzed the specificity of hygromycin by determining the percent of hygromycin-induced genes that are also triggered by the abiotic stresses and pathogens described above. While hygromycin activated 42% of P. aeruginosa-responsive genes, it only upregulated 25% and 23% of C. albicans and S. aureus–responsive genes respectively, which was similar to the overlap between these pathogens and P. aeruginosa (21% and 27% respectively). Furthermore, many of the genes induced by both hygromycin and either C. albicans or S. aureus were also upregulated by P. aeruginosa, suggesting that these genes are not specific for the response to a single type of pathogen (Figure S4D). Finally, while hygromycin induced a greater percentage of stress genes (35% of cadmium- and 40% of Cry5B-induced genes) than P. aeruginosa (22% of cadmium- and 23% of Cry5B-induced genes), many of the genes upregulated by both hygromycin and either cadmium or Cry5B were also upregulated by P. aeruginosa (Figure S4E). Based on these analyses, we conclude that hygromycin activates the same general gene classes as ToxA and does not broadly activate immune response genes or a large subset of stress response genes. Instead, like ToxA, hygromycin primarily upregulates a subset of the genes specifically activated in response to infection by P. aeruginosa PA14. The extensive overlap between genes induced by ToxA and hygromycin suggests that ToxA-responsive genes are triggered by the enzymatic function (translational inhibition) of ToxA.
To confirm our transcriptional profiling results and further rule out the alternative hypothesis that recognition of ToxA protein per se functions as an immune trigger, we analyzed the 10 genes in Figure 2, which are activated by ToxA and are predicted by microarray analysis to be upregulated by hygromycin. Specifically, we carried out qRT-PCR analysis under conditions that distinguish direct and indirect recognition mechanisms. Consistent with the profiling data, exposure to hygromycin was sufficient to induce the expression of all 10 genes (Figure 5A). Moreover, these gene activations largely required the same immune pathways as the ToxA response (Figure 5B). In contrast, none of these 10 genes were upregulated by E. coli expressing the catalytic mutant ToxAE575Δ (Figure 5A). We also investigated whether catalytically inactive ToxAE575Δ could amplify an established immune response by acting as a secondary trigger, but we found that simultaneous exposure to hygromycin and E. coli expressing ToxAE575Δ did not significantly enhance the level of activation compared to treatment with hygromycin alone (p>0.2 for each gene; Figure 5A). In addition, none of the 10 genes were upregulated by E. coli expressing wild-type ToxA in either of the diphthamide mutants (Figure 5C). In contrast, the diphthamide mutants responded to hygromycin by activating these 10 genes, confirming that they were still capable of recognizing and reacting to translational inhibition. Finally, we tested whether G418, a different ribosomal inhibitor, would also trigger expression of these ToxA-activated genes. When exposed to G418, six of the 10 genes were upregulated, indicating that their response was not specific to hygromycin (Figure 5D). Exposure to G418 did not result in C. elegans lethality as quickly as for hygromycin (Figure S4A), suggesting that activation of the four uninduced genes may require a higher, hygromycin-like, level of protein inhibition.
Figure 5. ToxA genes are upregulated by translational inhibition but not by catalytically inactive ToxA protein.
(A-E) qRT-PCR analysis of ToxA-responsive genes exposed to the indicated condition. N2 animals were tested unless otherwise noted. As shown in B, 4 genes (irg-1, F11D11.3, T24E12.5, and arrd-3) required the same pathways to respond to both hygromycin and ToxA (compare to Figure 2B). Hygromycin-induced expression of oac-32 was reduced in zip-2 and pmk-1 mutants (7 and 2 fold respectively), but this reduction was not statistically significant (p>0.05). Both hygromycin and ToxA-induced expression of T19C9.8 required fshr-1 but only hygromycin-mediated activation additionally required zip-2 and pmk-1. None of these gene inductions depended on skn-1 but daf-16 was necessary for the upregulation of T19C9.8 in response to hygromycin [11-fold reduction in daf-16(mgDF47) mutants, p<0.05]. However, as with ToxA, exposure to hygromycin was not sufficient to cause strong nuclear translation of DAF-16 (not shown). ** Genes with ≥ 10-fold lower induction to hygromycin as compared to wild-type N2 animals with p<0.05. * Genes with ≥5-fold lower induction to hygromycin as compared to N2 with p<0.05. Results shown are an average of 6 (A, B and E N2 hygromycin; D ToxA) or 3 (all remaining conditions) biological replicates. Error bars represent SEM. P values determined with a 2-tailed unpaired t-test. See also Figure S4.
While hygromycin upregulated 94% of the ToxA-responsive genes, we did identify eight genes that were induced by ToxA and not hygromycin, although these genes were not highly upregulated by ToxA (all eight genes induced < 4.5 fold and four genes induced ≤ 2.5 fold). To further characterize these transcripts, we tested which conditions triggered the expression of a representative gene, clec-67 (upregulated 4.2-fold). As predicted from the microarray analysis, clec-67 was induced by ToxA-encoding E. coli but not by hygromycin or G418. Like the other genes tested in Figures 5A-C, clec-67 was not activated by E. coli expressing ToxAE575Δ or activated in dph-1(ok329) or dph-3(ku432) mutants exposed to E. coli expressing wild-type ToxA (Figure 5E), indicating that clec-67 is also not upregulated by the ToxA protein itself.
Discussion
In this study, we established that the P. aeruginosa factor ToxA is sufficient to trigger a C. elegans immune response and that signaling pathways activated by P. aeruginosa infection and mediated by PMK-1 p38 MAPK, ZIP-2, FSHR-1, DAF-16, and SKN-1 also protect the nematode from ToxA-mediated killing to varying extents. Through microarray analysis, we defined a set of C. elegans genes that respond to an inhibition of translational elongation and determined that a significant number of these genes are also activated by a P. aeruginosa infection. Although C. elegans recognizes and reacts to ToxA, this recognition does not appear to involve detection of the ToxA protein per se. Catalytically-inactive ToxA did not trigger the expression of a representative set of genes upregulated by both ToxA and hygromycin, and co-administering inactive ToxA with hygromycin did not elicit a stronger response than hygromycin alone. Similarly, C. elegans diphthamide mutants, which are insensitive to ToxA-mediated inhibition, did not activate the representative ToxA-responsive genes when exposed to ToxA but did upregulate them following hygromycin treatment. We therefore conclude that the transcriptional response to ToxA is a consequence the enzymatic activity of ToxA resulting from its ribosylation of EF2.
There are at least two mechanisms by which EF2 ribosylation could activate an immune response. First, C. elegans could have a receptor that recognizes the ribosylated diphthamide residue. This mechanism is conceptually similar to the so-called “guard hypothesis” in plant immunity which postulates that plant NB-LRR receptor proteins “guard” the targets of microbial effector proteins (Jones and Dangl, 2006). Alternatively, ToxA-mediated inhibition of protein synthesis could generate a secondary signal, analogous to the DAMP hypothesis, which in turn activates immune signaling. Our data shows that 94% of ToxA-responsive genes are induced by hygromycin, which, like ToxA, blocks elongation of growing protein chains. This finding, combined with the observation that disrupting protein synthesis through genetic or chemical approaches activates the ZIP-2 signaling pathway (Dunbar et al., 2012), strongly suggests that the C. elegans immune response is triggered by inhibiting protein synthesis rather than by recognizing ribosylated EF2. However, we have not eliminated the possibility that C. elegans may simultaneously detect and react to ToxA-mediated ribosylation. Similarly, we cannot rule out that the E. coli food source used in these experiments contain MAMPs/PAMPs that function in conjunction with ToxA to activate immunity. However, it is likely that C. elegans in their natural environment regularly encounter innocuous Gram-negative bacteria like E. coli, and so it is unclear how relevant putative MAMPs/PAMPs could be for discriminating between pathogenic and benign microbes.
While we identified several pathways that are activated by ToxA/translational inhibition, we do not know which specific signal(s) connects disrupted protein synthesis with the upregulation of immune genes. A recent report by Fontana et al. showed that inhibiting translation in macrophages with L. pneumophila virulence factors or pharmacological agents activates mammalian immunity through an NF-kB-dependent mechanism (Fontana et al., 2011). In these macrophages, blocking translation prevents the synthesis of the NF-κB inhibitor IκB, thereby enabling continuous NF-κB function. Although, as stated above, we cannot rule out the possibility that C. elegans also receives a MAMP/PAMP-like signal since they are in constant contact with a bacterial food source, it is likely that the nematode responds to translational disruption via a different mechanism since, unlike C. elegans, the macrophage response required two simultaneous inputs: a translational inhibitor and a TLR agonist (Fontana et al., 2011). The absence of an identifiable ortholog for NF-κB in C. elegans further strengthens this conclusion.
The data shown in Figure 5B demonstrated that the p38 PMK-1, ZIP-2, and FSHR-1 immune signaling pathways are involved in the C. elegans response to translational inhibition. In addition to our work, Li et al., 2011 have shown that PMK-1 phosphorylation in C. elegans increases following exposure to a translational inhibitor or RNAi of endogenous translation factors (Li et al., 2011), suggesting a mechanistic link between the identification of protein synthesis abnormalities and activation of the p38 MAPK pathway. In addition to p38 MAPK signaling, Dunbar et al. have shown that blocking translation upregulates a ZIP-2::GFP reporter through a process that shares similarities with amino acid starvation, which is regulated by inhibitory upstream ORFs (uORFs) (Dunbar et al., 2012). It is an open question whether mammalian cells have a mechanism for sensing protein synthesis inhibition similar to the one that we and Dunbar et al. have identified in C. elegans, although evidence has been mounting in recent years suggesting that mammalian innate immunity utilizes a variety of non-TLR receptors and signaling pathways that may respond to DAMP-like signals (Medzhitov, 2009).
Our observations that wild-type immuno-competent C. elegans can survive the lethal effects of ToxA and P. aeruginosa virulence is undiminished in a toxA mutant despite apparently normal levels of intestinal translation, raise the question of whether ToxA should be considered a virulence factor, at least in the context of these laboratory C. elegans infection assays. In the wild, it is likely that C. elegans is subjected to many biotic and abiotic stresses and that C. elegans individuals may often be functionally immuno-compromised and potentially susceptible to ToxA. On the other hand, if the main effect of ToxA is to rapidly upregulate immune genes and not to enhance virulence, ToxA could function analogously to a Type III effector in a plant pathogen that elicits effector-triggered immunity (ETI) in particular hosts (Jones and Dangl, 2006). However, the analogy is imperfect, since plant pathogens that elicit ETI are less virulent, whereas in these experiments, PA14 strains expressing ToxA have the same virulence towards C. elegans as a P. aeruginosa toxA mutant.
The efficacy of the ToxA response suggests that C. elegans in the wild may encounter virulent strains of P. aeruginosa or other pathogens, such as C. diphtheriae, with toxins that have similar enzymatic activities. Furthermore, protein synthesis can be disrupted by many microbially-synthesized xenobiotic compounds such as antibiotics produced by actinomycetes in the soil. Thus, the C. elegans immune response to protein synthesis inhibitors may enable the nematode to quickly respond to diverse environmental threats. In addition to mediating immune defenses, the surveillance pathway(s) that recognize translational inhibition may function more generally to protect the worms against a variety of biological and chemical onslaughts. Interestingly, RNAi knockdown of translational components can enable many beneficial phenotypes including an extended lifespan (Curran and Ruvkun, 2007; Hansen et al., 2007; Rogers et al., 2011; Wang et al., 2010), increased stress resistance (Li et al., 2011), and avoidance behaviors (Melo and Ruvkun, 2012). Exposure of C. elegans to E. coli-expressing ToxA does not increase N2 longevity, suggesting that the enhanced longevity observed following the RNAi experiments described above may be triggered by different levels of translational inhibition or by alternative DAMP-like signals that are not generated by ToxA. Alternatively, the negative effects of ToxA may outweigh its potential benefits including this increased longevity.
Our data suggest that C. elegans at least partially recognizes P. aeruginosa through the disruption of a core cellular process, but it is unknown if this is the primary mechanism of C. elegans pathogen identification, or whether it is unique for P. aeruginosa and xenobiotics that inhibit translation. In addition to the translational component knockdowns described above, RNAi against many essential C. elegans genes including those involved in molting, proteasome machinery, and mitochondrial processes can both activate immune reporters and stimulate a bacterial aversion behavior that resembles pathogen avoidance (Melo and Ruvkun, 2012). Regardless of the results reported here and those of Melo and Ruvkun, other data from our laboratory indicate that C. elegans may recognize some infectious microbes through more traditional MAMP/PAMP mechanisms. For example, C. elegans mounts an immune reaction to heat-killed avirulent S. aureus and C. albicans (Pukkila-Worley and Ausubel, 2012).
In general, the relative roles of identifying highly conserved MAMPs/PAMPs, microbial effectors, effector-modified host proteins, or host damage caused by bacterial toxins and cellular invasion remain to be determined for C. elegans as well as other metazoans. MAMP/PAMP-triggered signaling mediated by TLRs is well-established in insects and vertebrates. In addition, the NAIP family of intracellular receptors has been recently shown in mammals to directly recognize conserved components of bacterial type-III secretion systems, which are involved in the delivery of a variety of virulence-related factors directly into the host cell cytoplasm (Kofoed and Vance, 2011; Zhao et al., 2011). Moreover, as described above, Boyer et al. show that an immune response can be triggered in both insects and mammalian cells by deamidation of a host RhoGTPase mediated by the E. coli-encoded toxin CNF1, in a process that is independent of MAMP/PAMP recognition (Boyer et al., 2011), and Fontana et al. have shown that translational inhibitors can activate mammalian immune genes but only where there is concurrent activation via a PRR pathway (Fontana et al., 2011). Our data, however, suggest that the C. elegans ToxA-response differs from all of these examples because it does not appear to involve the direct recognition of a bacterial-encoded molecule or effector-mediated modification of a host protein. Instead, the ToxA-mediated activation of immunity in C. elegans only requires the disruption of an essential cellular function. Whether this involves specific DAMP signaling molecules and host receptors remains to be elucidated.
Experimental Procedures
Strains
C. elegans were maintained using standard methods (Brenner, 1974). The following C. elegans strains were used in this study: N2 (wild-type), pmk-1(km25), nsy-1(ag3), nsy-1(ok593), sek-1(km4), agIs219 atf-7(qd22 qd130), agIs219 atf-7(qd137), zip-2(tm4067), zip-2(tm4248), fshr-1(ok778), daf-16(mgDf47), dph-1(ok329), dph-3(ku432), agIs17(irg-1::GFP), agEx39(myo-2::mCherry, clec-60::GFP), acIs101[pDB09.1(F35E12.5::GFP); pRF4[rol-6(su1006)]], ldl5007[skn-1B/C::GFP+pRF4[rol-6(su1006)]], and zls356IV[daf-16::DAF-16::GFP+pRF4[rol-6(su1006)]]. ToxA proteins and the vector control were expressed in E. coli BL21 Star (DE3) (Invitrogen). All RNAi clones were obtained from the Ahringer library (Kamath and Ahringer, 2003) and confirmed by sequencing. UCBPP-PA14 toxA was obtained from the Rahme lab (Rahme et al., 1995).
Cloning
Exotoxin A was amplified from PA14 genomic DNA with the primers 5’-caccatgcacctgataccccattg and 5’-ctacttcaggtcctcgcgcg and inserted into the pET100d vector using directional TOPO cloning (Invitrogen). E575 was removed by site-directed mutagenesis with the primers 5’-ggcgggcgcctgaccattctcggc and 5’-gccgagaatggtcaggcgcccgcc. To test whether removing E575 affected ToxA expression or stability, both ToxA and ToxAE575Δ were fused to green fluorescence protein (GFP). After confirming that the GFP tag does not disrupt protein function (i.e. ToxA::GFP is still lethal to pmk-1(km25) mutants), we determined that BL21 cells express full length ToxAE575Δ::GFP at the same concentration as they express ToxA::GFP through western blot analysis with an anti-GFP antibody (ab290, abcam) and an anti-GAPDH antibody loading control (pierce-antibodies).
C. elegans lifespan/killing assays
Synchronized animals were prepared by hypochlorite treatment and L1 arrest. Synchronized L1 larvae were plated onto 6-cm NGM plates seeded with E. coli OP50 or 6-cm RNAi plates prepared as described (Kamath and Ahringer, 2003). Animals were grown to the L4 stage at 20°C and transferred to 6-well assay plates prepared as described below. All assays were conducted at 25°C and in the presence of FUDR (100 μg/ml) to inhibit progeny production. ~30 worms were transferred to each well and at least 3 replicates were performed for each condition. Animals were considered dead when they failed to respond to the touch of a platinum wire (worm pick). Animals that died by crawling up the side of the plate were censored from the analysis. Each assay was performed a minimum of 3 times and a representative experiment was included in the paper. Survival curves were compared using a log-rank test
Assay plate preparation
Luria Broth (LB) containing 100 μg/ml ampicillin was inoculated with an overnight culture of E. coli (1:20 dilution), grown for 2 hours at 37°C, induced with IPTG to a final concentration of 1 mM, and grown an additional hour. 10x concentrated bacteria was seeded onto NGM plates containing 5 mM IPTG and 100 μg/ml ampicillin and used immediately. For hygromycin and G418 experiments, plates were dried for 1 hour before a mixture of inhibitor plus water was spread evenly over the agar. Once this mixture soaked into the agar, the plates were seeded with bacteria prepared as described above. 3.13 μl of hygromycin B (50.37 mg/ml in water, Sigma) or 12.5 μl of G418 (50 mg/ml in water, Sigma) was added per ml agar. P. aeruginosa slow killing plates were prepared as described (Troemel et al., 2006).
RNA extraction and Microarray Analysis
Worms were treated as described for the killing assays with the exception that ~4000–6000 L4 worms were plated onto 10-cm assay plates. After growing on the assay plates for 24 hours, worms were washed into a 15 ml conical tube with water and rinsed once. The animals were resuspended in TRI Reagent (Molecular Research Center) and frozen at −80°C. Once thawed, RNA was extracted according to manufacturer’s protocols and purified with an RNeasy column (Qiagen). RNA samples were prepared and hybridized to Affymetrix full-genome GeneChips for C. elegans at the Partners Center for Personalized Genetic Medicine, Boston, MA, according to manufacturer’s protocols. Three biological replicates were tested for each condition. Gene expression was analyzed using GCRMA (http://www.bioconductor.org/packages/release/bioc/html/gcrma.html). Differentially regulated genes were determined as described (Kirienko and Fay, 2007) using a fold change ≥2 and a modified Wilcoxon rank test with a minimal quotient of >1.45. Area proportional Venn Diagrams were generated with BioInfoRx software (http://bioinforx.com/free/bxarrays/overlap.php). We determined the number of genes expected by chance as reported (Troemel et al., 2006).
Quantitative RT-PCR (qRT-PCR) analysis
RNA was collected as described above and reverse transcribed using random decamers and the Retroscript kit (Ambion). qRT-PCR was performed with IQ SYBR green supermix (Bio-Rad) in a CFX96 system (Bio-Rad). Primers were designed with IDT SciTools (IDT) and tested for efficiency with a dilution series of template. At least 3 biological and 2 technical replicates were measured for each gene and condition. All Ct values were normalized against the snb-1 control gene. Fold changes were calculated using the Pfaffl method and compared using through a 2-tailed unpaired t-test. Primer sequences are available upon request.
Imaging
Animals were mounted onto agar pads and paralyzed with 20 mM levamisole (Sigma). Images were acquired with a Zeiss AXIO Imager ZI microscope with a Zeiss AxioCam HRm camera and Axiovision 4.6 (Zeiss) software.
Highlights.
P. aeruginosa exotoxin A (ToxA) upregulates a subset of C. elegans immune genes
Multiple immune signaling pathways are required to mount a response to ToxA
C. elegans immune pathways prevent ToxA-mediated lethality
C. elegans responds to translational inhibition by ToxA rather than the ToxA protein
Supplementary Material
Acknowledgements
We are grateful to the Caenorhabditis Genetics Center (CGC), Min Han, Laurence Rahme, and Keith Blackwell for C. elegans and bacterial strains and to Lynda Stuart and members of the Ausubel lab for critical reading of the manuscript. We also thank Emily Troemel, Gary Ruvkun and members of the Ruvkun lab, particularly Justine Melo, Yan Qi, and Dave Shore, for helpful discussions and for sharing unpublished data. This work was supported by MBRC Tosteson Postdoctoral Fellowship Award and by NIH grants R01 AI085581 and P30 DK040561.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing interests.
References
- Bjorn MJ, Vasil ML, Sadoff JC, Iglewski BH. Incidence of exotoxin production by Pseudomonas species. Infect Immun. 1977;16:362–366. doi: 10.1128/iai.16.1.362-366.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolz DD, Tenor JL, Aballay A. A Conserved PMK-1/p38 MAPK Is Required in Caenorhabditis elegans Tissue-specific Immune Response to Yersinia pestis Infection. Journal of Biological Chemistry. 2010;285:10832–10840. doi: 10.1074/jbc.M109.091629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L, Magoc L, Dejardin S, Cappillino M, Paquette N, Hinault C, Charriere Guillaume M, Ip WKE, Fracchia S, Hennessy E, et al. Pathogen-Derived Effectors Trigger Protective Immunity via Activation of the Rac2 Enzyme and the IMD or Rip Kinase Signaling Pathway. Immunity. 2011;35:536–549. doi: 10.1016/j.immuni.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan Regulation by Evolutionarily Conserved Genes Essential for Viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar TL, Yan Z, Balla KM, Troemel ER. Pathogen infection attenuates host intestinal translation to activate bZIP immune signaling in C. elegans. Cell Host & Microbe. 2012 doi: 10.1016/j.chom.2012.02.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes KA, Dunbar TL, Powell JR, Ausubel FM, Troemel ER. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 2010;107:2153–2158. doi: 10.1073/pnas.0914643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustice DC, Wilhelm JM. Mechanisms of Action of Aminoglycoside Antibiotics in Eucaryotic Protein Synthesis. Antimicrobial Agents and Chemotherapy. 1984;26:53–60. doi: 10.1128/aac.26.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle MR, Griswold JA, Oliver JW, Hamood AN. Anti-ETA IgG Neutralizes the Effects of Pseudomonas aeruginosa Exotoxin A. Journal of Surgical Research. 2002;106:86. doi: 10.1006/jsre.2002.6433. [DOI] [PubMed] [Google Scholar]
- Fontana MF, Banga S, Barry KC, Shen X, Tan Y, Luo Z-Q, Vance RE. Secreted Bacterial Effectors That Inhibit Host Protein Synthesis Are Critical for Induction of the Innate Immune Response to Virulent Legionella pneumophila. PLoS Pathog. 2011;7:e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. Long-Lived C. elegans daf-2 Mutants Are Resistant to Bacterial Pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee S-J, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Current Biology. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Huang dw, Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huffman DL, Abrami L, Sasik R, Corbeil J, van der Goot FG, Aroian RV. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10995–11000. doi: 10.1073/pnas.0404073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui JE, Ng A, Xavier RJ, Ausubel FM. Role for b-catenin and HOX transcription factors in Caenorhabditis elegans and mammalian host epithelial-pathogen interactions. 2008;105:17469–17474. doi: 10.1073/pnas.0809527105. P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui JE, Troemel ER, Feinbaum RL, Luhachack LG, Cezairliyan BO, Ausubel FM. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010a:e1000982. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010b;10:47. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Purdy AE, Fieldhouse RJ, Kimber MS, Bartlett DH, Merrill AR. Cholix Toxin, a Novel ADP-ribosylating Factor from Vibrio cholerae. Journal of Biological Chemistry. 2008;283:10671–10678. doi: 10.1074/jbc.M710008200. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Killeen KP, Collier RJ. Conformational integrity of a recombinant toxoid of Pseudomonas aeruginosa exotoxin A containing a deletion of glutamic acid-553. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1992;1138:162. doi: 10.1016/0925-4439(92)90057-t. [DOI] [PubMed] [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan M-W, et al. A Conserved p38 MAP Kinase Pathway in Caenorhabditis elegans Innate Immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Min J, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A Gene Expression Map for Caenorhabditis elegans. Science. 2001;293:2087. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Kirienko NV, Fay DS. Transcriptome profiling of the C. elegans Rb ortholog reveals diverse developmental roles. Developmental Biology. 2007;305:674–684. doi: 10.1016/j.ydbio.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8:279. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Matilainen O, Jin C, Glover-Cutter KM, Holmberg CI, Blackwell TK. Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity. PLoS Genet. 2011;7:e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac M, Pier GB, Collier RJ. Toxoid of Pseudomonas aeruginosa exotoxin A generated by deletion of an active-site residue. Infect Immun. 1988;56:3095–3098. doi: 10.1128/iai.56.12.3095-3098.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist EA. In: Small GTPases. Wormbook, editor. The C elegans Research Community, WormBook; 2006. http://wwwwormbookorg. [Google Scholar]
- Matar GM, Ramlawi F, Hijazi N, Khneisser I, Abdelnoor AM. Transcription Levels Pseudomonas aeruginosa Exotoxin A Gene and Severity of Symptoms in Patients with Otitis Externa. Current Microbiology. 2002;45:350. doi: 10.1007/s00284-002-3703-z. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Approaching the Asymptote: 20 Years Later. Immunity. 2009;30:766. doi: 10.1016/j.immuni.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Melo JA, Ruvkun G. Inactivation of essential cellular pathways stimulates microbial avoidance behavior, drug detoxification, and pathogen defense-related responses in C. elegans. Cell. 2012 in press. [Google Scholar]
- Miyazaki S, Matsumoto T, Tateda K, Ohno A, Yamaguchi K. Role of exotoxin A in inducing severe Pseudomonas aeruginosa infections in mice. J Med Microbiol. 1995;43:169–175. doi: 10.1099/00222615-43-3-169. [DOI] [PubMed] [Google Scholar]
- Nicas TI, Iglewski BH. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can J Microbiol. 1985;31:387–392. doi: 10.1139/m85-074. [DOI] [PubMed] [Google Scholar]
- Pastrana D, Yun C, McKee M, FitzGerald D. Mammalian cell expression of an active site mutant of Pseudomonas exotoxin disrupts LRP1 maturation. Journal of Biomedical Science. 2008;15:427. doi: 10.1007/s11373-008-9245-z. [DOI] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan M-W, Ray KP, Solari R, Johnson CD, Ewbank JJ. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Current Biology. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- Pukkila-Worley R, Ausubel FM. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Current Opinion in Immunology. 2012 doi: 10.1016/j.coi.2011.10.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Ausubel FM, Mylonakis E. Candida albicans Infection of Caenorhabditis elegans Induces Antifungal Immune Defenses. PLoS Pathog. 2011;7:e1002074. doi: 10.1371/journal.ppat.1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Jing S, Tompkins RG, Ausubel FM. Common Virulence Factors for Bacterial Pathogenicity in Plants and Animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- Rogers Aric N, Chen D, McColl G, Czerwieniec G, Felkey K, Gibson Bradford W, Hubbard A, Melov S, Lithgow Gordon J, Kapahi P. Life Span Extension via eIF4G Inhibition Is Mediated by Posttranscriptional Remodeling of Stress Response Gene Expression in C. elegans. Cell Metabolism. 2011;14:55. doi: 10.1016/j.cmet.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M-W, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999a;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M-W, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proceedings of the National Academy of Sciences. 1999b;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006;16:1017–1030. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK Regulates Expression of Immune Response Genes and Contributes to Longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven R, McCallum KC, Cruz MR, Garsin DA. Ce-Duox1/BLI-3 Generated Reactive Oxygen Species Trigger Protective SKN-1 Activity via p38 MAPK Signaling during Infection in C. elegans. PLoS Pathog. 2011;7:e1002453. doi: 10.1371/journal.ppat.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of Pathogenesis: Discrimination of Pathogenic and Nonpathogenic Microbes by the Innate Immune System. Cell Host & Microbe. 2009;6:10. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Robida-Stubbs S, Tullet JMA, Rual J-Fo, Vidal M, Blackwell TK. RNAi Screening Implicates a SKN-1 Dependent Transcriptional Response in Stress Resistance and Longevity Deriving from Translation Inhibition. PLoS Genet. 2010;6:e1001048. doi: 10.1371/journal.pgen.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates SP, Jørgensen R, Andersen GR, Merrill AR. Stealth and mimicry by deadly bacterial toxins. Trends in Biochemical Sciences. 2006;31:123. doi: 10.1016/j.tibs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Gabel HW, Fischer SEJ, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 2011;108:1201–1208. doi: 10.1073/pnas.1018695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.