Abstract
Left-sided displacement of the abomasum (LDA) is a common disease in many dairy cattle breeds. A genome-wide screen for QTL for LDA in German Holstein (GH) cows indicated motilin (MLN) as a candidate gene on bovine chromosome 23. Genomic DNA sequence analysis of MLN revealed a total of 32 polymorphisms. All informative polymorphisms used for association analyses in a random sample of 1,136 GH cows confirmed MLN as a candidate for LDA. A single nucleotide polymorphism (FN298674:g.90T>C) located within the first non-coding exon of bovine MLN affects a NKX2-5 transcription factor binding site and showed significant associations (ORallele = 0.64; −log10Pallele = 6.8, −log10Pgenotype = 7.0) with LDA. An expression study gave evidence of a significantly decreased MLN expression in cows carrying the mutant allele (C). In individuals heterozygous or homozygous for the mutation, MLN expression was decreased by 89% relative to the wildtype. FN298674:g.90T>C may therefore play a role in bovine LDA via the motility of the abomasum. This MLN SNP appears useful to reduce the incidence of LDA in German Holstein cattle and provides a first step towards a deeper understanding of the genetics of LDA.
Introduction
Left-sided displacement of the abomasum (LDA) is a common dairy cattle disease with prevalences of about 2% in the German Holstein (GH) population [1], [2]. In the course of LDA the abomasum starts bloating and displaces from the dexter abdominal floor to the sinistral abdominal wall. LDA has to be treated surgically by fixing the abomasum into its correct position. However, even if the treatment succeeded without any incidents, a significantly reduced milk performance as well as conception problems increase the culling risk [1]–[3]. On the average, about one half of all cows affected by LDA were culled within the first year after surgery [1]. Thus, LDA increases veterinary costs for dairy farms and decreases life expectancy for dairy cows. Several QTL for LDA have already been identified in GH cows [4], among them a QTL proximal on bovine chromosome (BTA) 23. Since LDA is usually preceded by a decreased motility of the abomasum, impaired abomasal emptying, and malfunctions at the level of the intrinsic nervous system combined with impaired cholinergic muscle responses [3], the motilin (MLN) gene located proximally on BTA23 was an appropriate candidate gene. MLN encodes a small peptide hormone regulating interdigestive gastrointestinal contraction [5]. In human, MLN is mainly expressed in the stomach (UniGene, EST profile, HS.2813), which is the analog to the bovine abomasum. A reduced expression of MLN might lead to an insufficient gastrointestinal motility. This is also supported by results in human pharmacology, where medications based on MLN-agonists can remedy conditions like the diabetic gastroparesis [6], [7]. Also in cows undergoing surgical correction of LDA, the motilin agonist erythromycin increases the abomasal emptying rate by binding to motilin receptors [8].
LDA is a multifactorial disease, in which environmental effects play a role such as twin births [1], [9], housing system [1], and feeding factors [10]. However, the heritability of LDA reported in various studies ranged between 20% and 50% [1], [2] and thus is much higher than in any other common cattle disease.
The objective of this study was to investigate MLN as the most promising candidate gene for LDA on proximal BTA23. We were able to detect a significantly LDA-associated single nucleotide polymorphism (SNP) affecting a predicted transcription factor binding site of the MLN gene which also changes the expression level of this gene. Therefore, this polymorphism is useful for a genetic test to reduce the incidence of LDA in GH cows.
Results
Mutation analysis
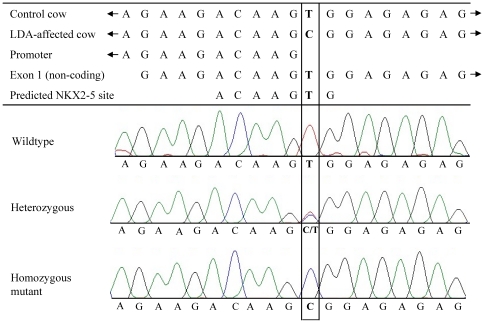
First, comparative sequencing of the complete MLN gene including all exons and introns as well as the 3′ and 5′ UTR sequences was done in four GH cows affected by LDA and four control cows of the same breed. The genomic sequence of the control cows (FN298674) was not in complete agreement with the corresponding sequence of the Bos taurus genome 4.0 (NC_007324). In comparison to the reference sequence, the first intron of MLN was shorter by 557 bp in all GH cows sequenced. Eight German Fleckvieh cattle were also analysed for this sequence variation of MLN with the result of an identical length of intron 1 as the GH cows. The putative promoter of bovine MLN extends from 73 bp to 123 bp using the genomic MLN sequence identified here (FN298674). We detected a total of 32 polymorphisms, of which 30 were SNPs and two were short tandem repeats. All polymorphisms were tested for their information content in a sample of 48 GH cows, with one half of them affected by LDA. Only eleven polymorphisms were informative (Table 1). None of the identified SNPs is located within the coding sequence of MLN, but FN298674:g.62G>A is located within the promoter and FN298674:g.90T>C within the non-coding first exon of MLN and the latter one affects a predicted NKX2-5 transcription factor binding site (Figure 1).
Table 1. All identified polymorphisms with their location within motilin (MLN).
| SNP/microsatellite | I | Location of polymorphism in MLN | Primer F (5′>3′) | Primer R (5′>3′) | AT | PS |
| FN298674:g.62G>A | + | promoter | CAGGAATCCAGACTCCTCAC | CCGGCTCTTTTGTTAACTT | 60 | 521 |
| FN298674:g.90T>C | + | exon 1 | CAGGAATCCAGACTCCTCAC | CCGGCTCTTTTGTTAACTT | 60 | 521 |
| FN298674:g.551(AG)45 | + | intron 1 | GCTCCGGGAAGTTAACAAAA | GAGCTGCTGACTTTGCCTTT * | 59 | 241 |
| FN298674:g.658A>G | intron 1 | GCTCCGGGAAGTTAACAAAA | ACATTTAATTCACCCATAAGAATCAA | 59 | 403 | |
| FN298674:g.684G>A | intron 1 | GCTCCGGGAAGTTAACAAAA | ACATTTAATTCACCCATAAGAATCAA | 59 | 403 | |
| FN298674:g.707G>A | intron 1 | GCTCCGGGAAGTTAACAAAA | ACATTTAATTCACCCATAAGAATCAA | 59 | 403 | |
| FN298674:g.869T>A | intron 1 | GAAGTTAACAAAAGAGCCGG | ACCCTCCCTTTGGAGAGAAT | 60 | 822 | |
| FN298674:g.1052C>T | intron 1 | GAAGTTAACAAAAGAGCCGG | ACCCTCCCTTTGGAGAGAAT | 60 | 822 | |
| FN298674:g.1719A>T | intron 1 | GCAGCCTTGGAAGATTCTCT | CATCCCATAAGTGGGTGTGT | 60 | 591 | |
| FN298674:g.1891insG | + | intron 1 | GACACACCCACTTATGGGAT | TCCCAGCTCCAGCAAGTCTA * | 60 | 155 |
| FN298674:g.2037T>C | intron 1 | GACACACCCACTTATGGGAT | TTGTTGGCCCTTCCCTTTG | 58 | 514 | |
| FN298674:g.2045C>G | + | intron 1 | GACACACCCACTTATGGGAT | TTGTTGGCCCTTCCCTTTG | 58 | 514 |
| FN298674:g.2451T>A | intron 1 | GCTCCTGCTTCATTAGCTC | GTGTGTCTGACCTCTCACC | 58 | 779 | |
| FN298674:g.2525G>T | intron 1 | GCTCCTGCTTCATTAGCTC | GTGTGTCTGACCTCTCACC | 58 | 779 | |
| FN298674:g.2929G>A | + | intron 2 | GCTCCTGCTTCATTAGCTC | GTGTGTCTGACCTCTCACC | 58 | 779 |
| FN298674:g.2985C>T | + | intron 2 | GCTCCTGCTTCATTAGCTC | GTGTGTCTGACCTCTCACC | 58 | 779 |
| FN298674:g.3537(T)23 | intron 2 | CCCTGGGCCTTTTACCTTAG * | TTGCCATTTCCTTTTCCAAC | 60 | 130 | |
| FN298674:g.4189C>T | intron 2 | GAGACTGAGGCTCCAACAGG | CTGGCAATTACGAGGAGGAA | 60 | 503 | |
| FN298674:g.4360G>C | + | intron 2 | GAGACTGAGGCTCCAACAGG | CTGGCAATTACGAGGAGGAA | 60 | 503 |
| FN298674:g.4942insT | + | intron 2 | AATCCGAGGACGATGTGAAG * | GCCCTTTGGGGTTTCTATTC | 60 | 128 |
| FN298674:g.5224G>T | intron 3 | GCAGCGAGAGGGTGCTTG | AGCTCGTCACCTGAGTGTC | 58 | 795 | |
| FN298674:g.5596T>G | intron 3 | CACAGAATGTGCTGCGAACT | AATGGATCTCATGGCTCTGG | 60 | 460 | |
| FN298674:g.6197G>A | intron 3 | GCCCTTCTCCCTCAGCTACT | CCAATTTCCACAGGAGCAGT | 60 | 479 | |
| FN298674:g.6689C>T | + | intron 4 | ACGCTGCCTTATTTCACAGC | ATGGATCTCTGCAGGAGGAG | 60 | 496 |
| FN298674:g.6728G>A | + | intron 4 | CTGGCAGGTTTCAGGCTCT | ATGGATCTCTGCAGGAGGAG | 58 | 672 |
| FN298674:g.6920C>T | intron 4 | CTGGCAGGTTTCAGGCTCT | ATGGATCTCTGCAGGAGGAG | 58 | 672 | |
| FN298674:g.6984C>T | intron 4 | CTGGCAGGTTTCAGGCTCT | ATGGATCTCTGCAGGAGGAG | 58 | 672 | |
| FN298674:g.7002T>A | intron 4 | TCCGAGCGGAGTCTAATCAC | CTGCCTCCAGCTTTCTGTGT | 60 | 533 | |
| FN298674:g.7153C>T | intron 4 | TCCGAGCGGAGTCTAATCAC | CTGCCTCCAGCTTTCTGTGT | 60 | 533 | |
| FN298674:g.7403delGAC | intron 4 | CAGCCTGAGTCACTGGTAC | AGCTCAGAAGGGTCAGCAGATC * | 60 | 245 | |
| FN298674:g.7408delA | intron 4 | CAGCCTGAGTCACTGGTAC | AGCTCAGAAGGGTCAGCAGATC * | 60 | 245 | |
| FN298674:g.7410TCA>CTG | intron 4 | CAGCCTGAGTCACTGGTAC | ATTTATGGCCAAACCCTCTC | 58 | 528 |
IRD labeled.
Informative (I) polymorphisms are indicated by a plus sign. The primer pairs used are given with their annealing temperatures (AT) and product sizes (PS).
Figure 1. Mutation within the bovine motilin (MLN) gene associated with LDA.
Sequence of MLN surrounding the FN298674:g.90T>C SNP for the wildtype (control cow) and the mutated allele (LDA-affected cow) as well as the 3′ end of the predicted MLN promoter, the 5′ start sequence of the non-coding first exon of MLN and the NKX2-5 transcription factor binding site are shown. Furthermore, chromatograms of this section are given for each one of the homozygous wildtype, heterozygous and homozygous mutant individuals.
Association analyses
All informative polymorphisms were tested for association with LDA using 601 affected and 535 unaffected GH dairy cows. Seven SNPs (FN298674:g.62G>A, FN298674:g.90T>C, FN298674:g.1891insG, FN298674:g.2045C>G, FN298674:g.4942insT, FN298674:g.6689C>T, FN298674:g.6728G>A; Figure 2) showed significant allelic and genotypic associations with LDA (Table 2). FN298674:g.6689C>T and FN298674:g.6728G>A were in strong linkage disequilibrium (LD) of r2 = 1.0 and FN298674:g.90T>C and FN298674:g.1891insG showed an LD of r2 = 0.8 (Figure S1). The odds ratio (OR) was highest for FN298674:g.1891insG, followed by FN298674:g.90T>C, FN298674:g.2045C>G, and FN298674:g.4942insT (Table 2). For FN298674:g.1891insG, OR was at 1.71 per allele and at 2.82 among homozygotes and the −log10P-values reached a value of 9.3. The ORs for FN298674:g.90T>C were at 0.64 and 0.4, respectively, with −log10P-values up to 7.0. For FN298674:g.90T>C, genotype frequencies for all GH cows analysed were at 0.254 (C/C), 0.509 (C/T), and 0.237 (T/T) and for FN298674:g.1891insG at 0.272 (WT/WT), 0.473 (WT/INS) and 0.255 (INS/INS). The overall genotype effect of FN298674:g.90T>C and FN298674:g.1891insG on the LDA frequency showed significant F-values (9.4 and 8.8, respectively) and the contrasts of the least-square means of the LDA frequencies among the homozygous mutant versus the heterozygous genotype or homozygous wildtype were significant. Of all cows carrying the homozygously mutated genotype, 67.3% and 68.1% were affected by LDA. Of the cows homozygous for the wildtype allele, 44.0% and 42.6% were affected by LDA and of the heterozygous individuals, 49.6% and 51.6% were affected (Table 3). Of all 601 LDA-affected animals, only 0.198 and 0.218 were homozygous for the wildtype allele.
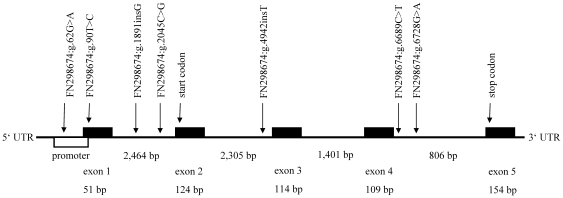
Figure 2. Structure of the motilin (MLN) gene and positions of seven SNPs associated with LDA.
The figure shows the five exons and the promoter region of the bovine MLN. Arrows indicate the positions of the seven polymorphisms associated with LDA in German Holstein cows. The size of the exons and introns is given in base pairs. The positions of the start and stop codons are indicated.
Table 2. Association study for LDA using SNPs within motilin for 1,136 German Holstein cows.
| SNP | MAF allele | MAF | OR per allele (95% CI) | OR among homozygotes | PIC | HET | P | ||||
| Cases | Controls | gt | allele | hom | strat | ||||||
| (n = 601) | (n = 535) | (95% CI) | (−log10) | (−log10) | (−log10) | (−log10) | |||||
| FN298674:g.62G>A | A | 0.49 | 0.39 | 1.47 (1.23–1.72) | 2.27 (3.22–1.59) | 0.37 | 0.47 | 4.8 | 5.1 | 5.4 | 3.3 |
| FN298674:g.90T>C | T | 0.44 | 0.55 | 0.64 (0.54–0.76) | 0.40 (0.28–0.56) | 0.37 | 0.48 | 7.0 | 6.8 | 6.8 | 3.8 |
| FN298674:g.1891insG | insG | 0.55 | 0.42 | 1.71 (1.44–2.02) | 2.82 (2.01–3.95) | 0.37 | 0.46 | 8.2 | 9.3 | 9.0 | 3.7 |
| FN298674:g.2045C>G | G | 0.52 | 0.40 | 1.62 (1.36–1.93) | 2.44 (1.74–3.42) | 0.37 | 0.40 | 6.6 | 7.1 | 6.7 | 3.2 |
| FN298674:g.4942insT | wt | 0.35 | 0.47 | 0.61 (0.51–0.72) | 0.41 (0.29–0.6) | 0.35 | 0.41 | 7.0 | 7.4 | 5.8 | 2.9 |
| FN298674:g.6689C>T | T | 0.28 | 0.40 | 0.57 (0.45–0.72) | 0.36 (0.22–0.57) | 0.32 | 0.34 | 4.3 | 5.6 | 5.0 | 1.6 |
| FN298674:g.6728G>A | A | 0.28 | 0.40 | 0.57 (0.45–0.72) | 0.36 (0.22–0.57) | 0.32 | 0.34 | 4.3 | 5.6 | 5.0 | 1.6 |
For each SNP, minor allele frequencies (MAF), the MAF allele, odds ratios (OR) with their 95% confidence intervals (CI), polymorphism information content (PIC), and heterozygosity (HET) are given. Furthermore, −log10P-values (P) are given for genotype (gt), allele, among homozygotes (hom) and for genotype after correcting the data for stratification (strat).
Table 3. Least-squares means (LSM) with their corresponding standard errors (SE) of the frequency of LDA on the underlying logit scale for the genotypes of the SNPs FN298674:g.90T>C and FN298674:g.1891insG and the contrasts among the LSMs with their test statistics and P-values.
| SNP | Genotype or contrast | LDA-affected cows (%) | LSM ± SE or contrast among LSMs | t-value | P-value |
| FN298674:g.90T>C | T/T | 44.0 | −0.319±0.176 | ||
| C/T | 49.6 | −0.287±0.149 | |||
| C/C | 67.3 | 0.393±0.183 | |||
| C/C vs. C/T | 0.680±0.166 | 4.09 | <0.0001 | ||
| C/C vs. T/T | 0.712±0.199 | 3.57 | 0.0004 | ||
| C/T vs. T/T | 0.032±0.167 | 0.19 | 0.8 | ||
| FN298674:g.1891insG | WT/WT | 42.6 | −0.366±0.169 | ||
| WT/INS | 51.6 | −0.206±0.149 | |||
| INS/INS | 68.1 | 0.393±0.181 | |||
| WT/WT vs. WT/INS | −0.161±0.162 | −1.00 | 0.3 | ||
| WT/WT vs. INS/INS | −0.759±0.193 | −3.93 | <0.0001 | ||
| WT/INS vs. INS/INS | −0.598±0.168 | −3.56 | 0.0004 |
Genotypic means for the frequency of LDA were estimated through LSMs in a mixed logit model correcting for data stratification.
In order to correct for the data structure, we employed a logit model including a random sire effect besides the fixed genotypic SNP effects. This association analysis showed the most significant −log10P-values for FN298674:g.90T>C and FN298674:g.1891insG (3.8 and 3.7). The −log10P-values of all other polymorphisms were distinctly smaller. In addition, we performed association analyses using a random additive genetic animal effect through a numerator relationship matrix and a heritability of h2 = 0.3 for LDA estimated in the same population [1], [2]. The most significant associations with LDA were found for FN298674:g.90T>C and FN298674:g.1891insG. The FN298674:g.90T>C SNP showed the highest association with LDA. The SNPs FN298674:g.6689C>T and FN298674:g.6728G>A were no longer significantly associated with LDA. Therefore, we considered FN298674:g.90T>C and FN298674:g.1891insG as the most influential SNPs of MLN for developing LDA. The phenotypic variance explained after correction for the sire effect was at 3.1% for FN298674:g.90T>C and at 3.9% for both SNPs, FN298674:g.90T>C and FN298674:g.1891insG.
Haplotype analysis
The most common haplotypes for FN298674:g.90T>C and FN298674:g.1891insG comprising 95% of all cows were C-INS and T-WT. The haplotype including both mutant alleles (C-INS) was present in 55.1% of the LDA-affected and in 39.0% of the unaffected cows (Table S1). The haplotype trait analysis using these SNPs gave a χ2 value of 119.4 (p<0.0001) and χ2 values of 14.0 (p = 0.0002) to 60.8 (p<0.0001) for the distribution of the four single haplotypes among LDA-affected and control cows. Inclusion of further MLN polymorphisms did not improve the haplotype analysis.
Linkage analysis
We employed a multipoint non-parametric linkage analysis to re-assess linkage for MLN-associated markers in the data set previously used for the genome-wide search for LDA-QTL [4]. The chromosome-wide linkage analysis for 30 markers on BTA23 and all 14 paternal half-sib families showed a peak with a Zmean of 2.55 (P-value = 0.005) at the SNP FN298674:g.90T>C within MLN (Table S2, Figure S2).
Distribution of LDA-associated alleles in German Fleckvieh
The most significantly LDA-associated polymorphisms, FN298674:g.90T>C and FN298674:g.1891insG, were tested in 148 German Fleckvieh cattle. German Fleckvieh was used as a reference breed as this breed is known for its very low incidence of LDA. Genotypes of FN298674:g.90T>C were 0.468 T/T, 0.371 C/T and 0.161 C/C. In comparison to GH cows, German Fleckvieh showed a shift to the wildtype allele T and the homozygous wildtype genotype T/T. For FN298674:g.1891insG, the genotypic distribution was 0.591 without insertion (WT/WT), 0.295 heterozygous (WT/INS) and 0.114 homozygously inserted (INS/INS). Allele frequencies for the mutant alleles were 0.347 (FN298674:g.90T>C) and 0.262 (FN298674:g.1891insG).
Expression study
A qRT-PCR of cDNA gave evidence of a MLN expression in abomasal mucosa tissue. To test whether the FN298674:g.90T>C mutation might influence the MLN expression, samples of abomasal mucosa tissue from 55 previously genotyped GH cows were taken and analysed using qRT-PCR. Of these individuals, 26 were homozygous for the wildtype allele, 20 were heterozygous, and nine were homozygous for the mutant allele. The mean value of the ΔCT of the homozygous wildtype cows was used as calibrator [11]. The relative expression levels of MLN were decreased by 89% in cows being heterozygous or homozygous for the mutant allele C of the polymorphism FN298674:g.90T>C in comparison to cows homozygous for the wildtype allele T (Table 4). Among cows carrying the mutant allele C homozygously or heterozygously, the expression levels were identical. The error probability for the wildtype genotype versus the heterozygous or homozygous mutated genotype was at a P-value of 0.03 with a corresponding F-value of 4.8 (Table 4).
Table 4. Results of the expression analysis for FN298674:g.90T>C within motilin (MLN).
| Relative expression levels per genotype | T/T vs. C/T and C/C | ||
| T/T (n = 27) | C/T (n = 20) and C/C (n = 8) | F-value | P-value |
| 1.00±0.26 | 0.11±0.27 | 4.8 | 0.03 |
The mean relative expression levels of MLN with their standard errors are given for the wildtype genotype T/T, the heterozygously (C/T) and the homozygously mutated genotype (C/C) standardized on the mean of the genotype T/T. Furthermore, the F-values and P-values for the differences in mean relative expression levels among the genotype T/T versus the genotypes C/T and C/C as well C/T versus C/C are given. RPL4 was used as housekeeping gene.
Discussion
This study demonstrated that the non-coding MLN transcription factor binding site mutation FN298674:g.90T>C is significantly associated with MLN expression levels and the incidence of LDA in GH cows. Furthermore, this polymorphism showed a significant linkage with LDA in GH cows from the same population. The MLN gene encodes the 22 amino-acid peptide motilin, a hormone which plays a role in stimulating gastrointestinal motility. Motilin is considered as an endocrine regulator and initiator of phase III of the migrating motor complex, the interdigestive peristaltic reflex in the gastrointestinal tract [5], [12]. This hormone therefore causes gastric emptying in periods of fasting and its release ceases with the ingestion of food. Plasma levels of motilin increase cyclically every 90–120 minutes within the interdigestive periods, which evoke short time intervals of five to ten minutes of strong peristaltic contractions initiating from the stomach [5]. In a study using the house musk screw as animal model for motilin studies, the contractile responses induced by motilin were dose-dependent [13]. There are also therapeutical approaches in using the positive effect of motilin agonists on gastric contractions concerning the treatment of diabetic gastroparesis [6], [7] and altered motilin plasma levels are seen in some human diseases [14]. This study shows that MLN plays a role in the development of bovine LDA, a disease known to be initiated among other things by a reduced peristalsis of the gastrointestinal tract [3]. The reduced food-intake often seen in dairy cows after calving necessitates a high interdigestive peristalsis, which is a function of MLN. Naturally, its influence on the development of LDA is a fractional amount, because the disease is known to be genetically complex. After correcting the data for stratification, the polymorphism FN298674:g.90T>C within bovine MLN showed the highest association with LDA. This SNP affects a NKX2-5 transcription factor binding site. The effect of NKX2-5 on gene expression has been previously described [15] and SNPs within NKX2-5 binding sites have been reported to be functional for complex genetic diseases as systemic lupus erythematosus, rheumatoid arthritis, and graves disease in man as the mutations lead to a reduced expression of the respective gene [16]. Therefore, FN298674:g.90T>C is presumably the reason for the reduced expression of MLN in GH cows carrying a C allele at this position. Interestingly, while the association study showed the highest associations for the homozygous mutant genotype with LDA, the expression of MLN was reduced in cows with at least one copy of the mutated allele. This indicates a dominant effect of FN298674:g.90T>C. These results might be explained by the motilin-level regulation process. Motilin controls the gastrointestinal peristalsis reflex to provide gastric emptying during the interdigestive phases [5], [12]. Plasma levels of motilin increase cyclically within these phases and evoke short time intervals of strong peristaltic contractions [5]. Therefore, though cows heterozygous for the FN298674:g.90T>C SNP show a considerably reduced basis expression of MLN, they might still be able to raise their motilin level more frequently or higher during the digestive periods where it is required and therefore reach a threshold to prevent LDA. Individuals homozygous for the mutated allele cannot raise their motilin level much beyond basis expression. Therefore, the gastric motility in heterozygous cows might be sufficient to prevent LDA.
Though the homozygously mutated genotype of FN298674:g.90T>C significantly increases the risk of a cow to be affected by LDA and is the most likely causal SNP due to its effect on a transcription factor binding site, FN298674:g.1891insG within the first intron of MLN also showed a highly significant association with LDA. As FN298674:g.1891insG is in strong linkage disequilibrium (r2 = 0.8) with FN298674:g.90T>C, its high association with LDA might be explained by this fact. However, the insertion mutation FN298674:g.1891insG might also contribute to the negative effect on MLN expression since an effect on a further regulatory domain can not be excluded.
To reveal the genotypic distribution of the SNPs FN298674:g.90T>C and FN298674:g.1891insG in a cattle breed with a low incidence of LDA [17], these SNPs were tested in German Fleckvieh. In German Fleckvieh, the wildtype allele was prevalent with a frequency of 0.653 at FN298674:g.90T>C and of 0.738 at FN298674:g.1891insG and the homozygous mutant genotypes for these polymorphisms showed an even lower frequency (0.161 and 0.114, respectively) than in GH cows unaffected by LDA (0.176 and 0.170, respectively). This corroborates the impact of MLN as a causal gene for LDA.
GH are high-performance dairy cows and due to their energy adapted-feeding and the cattle specific anatomic peculiarities they might be more susceptible for diseases caused by gastric motility decrease than other species. However, alongside cattle and human [18], gastric dilatation and displacement have also been reported in other species like dogs [19], cats [20], pigs [21], guinea pigs [22], and horses [23]. Therefore, this study might be of interest in the research of gastric motility disorders in these species.
In conclusion, we identified a SNP (FN298674:g.90T>C) which affects a predicted NKX2-5 binding site within the first non-coding exon of the bovine MLN gene. This polymorphism was shown to be correlated with a significantly lower expression of MLN and a significant increase of LDA incidence in GH cows. This is the first polymorphism showing a functional association with LDA and it can be used to test for LDA-susceptibility in this breed. MLN might be a useful candidate for the genetic analysis of gastric motility disorders in further species.
Materials and Methods
Ethics statement
All animal work has been conducted according to the national and international guidelines for animal welfare. All blood-sampling of LDA-affected cows was done in university clinics for cattle along with diagnostic protocols prior to surgery. The blood samples of unaffected cows were taken by veterinarians on farms. The project has been notified to the Lower Saxony state veterinary office, Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Oldenburg, Germany, and registered as a notifiable experiment with the number 33.9-42502-05-08A541.
Animals
We used EDTA-blood samples of a total of 1,136 GH cows. Of these animals, 601 cows had undergone a surgery because of LDA at the clinics for cattle of the veterinary universities Hannover or Giessen, Germany. The remaining 535 GH cows were unaffected by LDA. This sample was balanced by paternal grandsires. Each grandsire had to have at least one descendant affected by LDA and one unaffected descendant. The data set included 72 grandsires. Furthermore, EDTA-blood samples of 148 German Fleckvieh individuals with diverse ancestry were included into this study. Genomic DNA from EDTA-blood was extracted using the QIAamp 96 Spin Blood Kit (Qiagen, Hilden, Germany). In addition, samples from the abomasal mucosa tissue were taken from 55 GH cows at the slaughterhouse (Schlachthof Hannover and Schlachthof Giessen, Germany). All these animals were fasting prior to sampling. The samples were taken under equal conditions directly after slaughtering.
Sequencing and genotyping
The complete MLN gene was sequenced and scanned for polymorphisms in each four LDA-affected and unaffected GH cows. To ensure that the four control cows would not develop a LDA later in life, only cows with an age of at least nine years and without ever showing signs of LDA in their lifes were chosen for this first scan for polymorphisms. Sequence data were analysed using the Sequencher 4.9 software (GeneCodes, Ann Arbor, MI). PCR primers used for sequencing of MLN and for genotyping are given in Table 1 and Table S3. Sequencing of PCR products was performed on a MegaBACE 1,000 capillary sequencer (GE Healthcare, Freiburg, Germany). After this, each polymorphism identified was tested in 24 LDA-affected cows and 24 controls and, if it was informative, genotyped in a total of 1,136 GH cows. Markers were regarded as informative, if their PIC and heterozygosity were at 0.25 or higher. Genotyping of SNPs was performed using enzymatic digestion or sequencing (Table S4). For the genotyping of microsatellites, insertions, and deletions, each one primer was IRD-labeled and the PCR products were size-fractionated by gel electrophoresis on automated sequencers (LI-COR 4300, Lincoln, NE, USA) using 6% polyacrylamide denaturing gels (Table S4).
Expression analysis
For the analysis of MLN expression, RNA was isolated from abomasal mucosa tissue samples of 55 different cows using the NucleoSpin®RNA II kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions. The reverse transcription into cDNA was performed using 200 U SuperScript III Reverse Transcriptase (Invitrogen, Karlsruhe, Germany). A single PCR assay was used for quantification of the MLN gene transcript (GenBank NM_173938.2) using a forward primer situated on the boundary of exon 2 and exon 3 (5′- TGC AGG AAA AGG AGA GGT ACA AG -3′) and a reverse primer within exon 4 (5′- GTT CAT CCT CAT TCC AAT TTC CA -3′) amplifying an 155-bp product with a FAM-labeled TaqMan minor groove binding (MGB) probe (Applied Biosystems, Darmstadt, Germany) located at the boundary of exon 3 and 4 (5′-CAG GAA GTT ATC AAG CTG A-3′). Bovine RPL4 was used as a housekeeping gene. We used a forward primer within exon 2 (5′-TTG CGC AAA AAC AAC AGA CA-3′) and a reverse primer in exon 3 (5′-GCC GGT ACC CCA AGA CTC A-3′) amplifying a 81-bp product in combination with a VIC-labeled TaqMan MGB probe (Applied Biosystems) spanning the boundary of exon 2 and 3 (5′-TAG CAG GTC ATC AAA CC-3′) according to the bovine reference mRNA sequence (GenBank NM_001014894). The quantitative real-time (qRT)-PCR was carried out using an ABI7300 sequence detection system (Applied Biosystems). The MLN transcript specific expression was normalised by the bovine RPL4 expression level (ΔCT), and the relative expression level was calculated by the ΔΔCT method using the average ΔCT of the homozygous wildtype samples as calibrator [11]. All assays were performed in duplicates.
Statistical analyses
To test for association with LDA, case-control analyses based on χ 2-tests for genotypes and alleles were performed using SAS/Genetics, version 9.3 (Statistical Analysis System, Cary, NC, USA, 2011). The ALLELE and HAPLOTYPE procedures of SAS/Genetics were used for estimation of allele and haplotype frequencies, Hardy-Weinberg equilibrium and linkage disequilibrium among marker alleles. Logistic regression analysis was performed using the GLIMMIX procedure of SAS. The GLIMMIX procedure fits statistical models to data with correlations or nonconstant variability and where the response is not necessarily normally distributed and was therefore also used with our data. An animal model analysis was parameterized using a numerator relationship matrix for each individual and a heritability of 0.3 for LDA. This heritability was chosen as a mean value from former studies for LDA in GH cows where heritabilities of 0.2 to 0.5 were estimated [1], [2]. Odds ratios (OR) and P-values were obtained from the CASECONTROL and GLIMMIX procedure. Promoter prediction was carried out by Promoter Prediction software (http://www.fruitfly.org). To detect the impact of intronic polymorphisms and to search for transcription factor binding sites, the TFSEARCH software (http://www.cbrc.jp/research/db/TFSEARCH.html) was used. Statistical calculation of pairwise linkage disequilibrium (LD) was performed using HAPLOVIEW 4.0 (http://www.broad.mit.edu/mpg/haploview/). Statistical evaluation of MLN expression results was performed using standard procedures for comparing means with GLM (general linear model) of SAS, version 9.3.
Linkage Analysis
In addition to the association analysis, a multipoint non-parametric linkage analysis was conducted. For this purpose, 14 previously described paternal half-sib families segregating for LDA and containing a total of 360 animals were employed [4]. These families can be grouped into five grandsire families. We have taken the same microsatellites on BTA23 as in the previous study. In addition, we included the SNPs FN298674:g.90T>C and FN298674:g.1891insG within MLN as well as five new microsatellite markers located on the proximal end of BTA23. Positions of all markers were determined using a BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis against the UMD_3.1 bovine genome assembly. The markers DIK5319 and BM47 could not be placed or were located on a different chromosome and therefore excluded from this study. A total of 30 markers on BTA23 were used. PCR was conducted according to a standard protocol. One primer of each pair was endlabeled with fluorescent IRD700 or IRD800. Primers and positions of the five newly developed microsatellites are given in Table S5. For the analysis of the microsatellite genotypes, the PCR products were size-fractionated by gel electrophoresis on an automated sequencer (LI-COR 4300, Lincoln, NE, USA) using 6% polyacrylamide denaturing gels. Multipoint non-parametric linkage analyses based on haplotypes identical-by-descent (IBD) for a half-sib design were employed [24], [25]. The statistical analyses were performed using MERLIN, version 1.1.2 [26].
Supporting Information
Linkage disequilibrium (LD) among the seven LDA-associated SNPs in German Holstein cows. The pairwise r2-values are shown for each SNP pair. The red square between the markers FN298674:g.6689C>T and FN298674:g.6728G>A indicates complete LD.
(TIF)
The Zmean score profile with corresponding chromosome-wide P-values for LDA in German Holstein cattle for the bovine chromosome (BTA) 23. The horizontal line indicates the threshold of chromosome-wide significance (P-value = 0.05).
(TIF)
Haplotype analysis of the polymorphisms FN298674:g.90T>C and FN298674:g.1891insG. The haplotype frequencies as well as their standard errors, χ2-values and P-values are given. The test for marker-trait association gave a χ2 value of 119.4 (p<0.0001).
(DOC)
Zmeans and LOD-scores with their P-values of the multipoint non-parametric linkage analysis for BTA23 including the SNPs FN298674:g.90T>C and FN298674:g.1891insG within MLN . For the analysis, 14 paternal half-sib families comprising 360 individuals were used.
(DOC)
Additional pairs of primers used for sequencing motilin . For each pair of primers, the amplified region, annealing temperature (AT) and product size (PS) is given.
(DOC)
Methods used for genotyping of informative SNPs. All SNPs genotyped using RFLPs (restriction fragment length polymorphisms) or IRD (infrared dye) labeled gel electrophoresis are given. Not specified markers were genotyped by sequencing.
(DOC)
Description of the five new microsatellites used for the linkage analysis. The positions (Pos) determined on the bovine UMD_3.1 assembly, primer sequences, repeat motif, heterozygosity (Het), polymorphism information content (PIC), number of alleles (A), annealing temperature (AT) and product sizes (PS) are given.
(DOC)
Acknowledgments
We thank all members of the staff of the Clinic for Cattle, University for Veterinary Medicine Hannover, Germany, and of the Clinic for Ruminants and Swine, Faculty for Veterinary Medicine, Justus-Liebig-University Giessen, Germany, who participated in collecting samples. We also wish to thank the breeding organizations and all breeders for the samples and the pedigree information of their cattle.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The project and SM were supported by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG), Bonn (MO1691/1-2 and 2-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wolf V, Hamann H, Scholz H, Distl O. [Influences on the development of abomasal displacement in German Holstein cows]. Dtsch tierärztl Wschr. 2001;108:403–408. [PubMed] [Google Scholar]
- 2.Hamann H, Wolf V, Scholz H, Distl O. Relationships between lactational incidence of displaced abomasum and milk production traits in German Holstein cows. J Vet Med A. 2004;51:203–208. doi: 10.1111/j.1439-0442.2004.00626.x. [DOI] [PubMed] [Google Scholar]
- 3.Geishauser T, Reiche D, Schemann M. In vitro motility disorders associated with left-sided displaced abomasum in dairy cows. Neurogastroenterol Motil. 1998;10:395–401. doi: 10.1046/j.1365-2982.1998.00119.x. [DOI] [PubMed] [Google Scholar]
- 4.Mömke S, Scholz H, Doll K, Rehage J, Distl O. Mapping quantitative trait loci for milk performance traits in German Holstein cows affected by left-sided displaced abomasum (LDA). J Dairy Sci. 2008;91:4383–4392. doi: 10.3168/jds.2008-1260. [DOI] [PubMed] [Google Scholar]
- 5.Poitras P, Peeters TL. Motilin. Curr Opin Endocrinol Diabetes Obes. 2008;15:54–57. doi: 10.1097/MED.0b013e3282f370af. [DOI] [PubMed] [Google Scholar]
- 6.McCallum RW, Cynshi O Investigative Team. Clinical trial, effect of mitemcinal (a motilin agonist) on gastric emptying in patients with gastroparesis - a randomized, multicentre, placebo-controlled study. Aliment Pharmacol Ther. 2007;15:1121–1130. doi: 10.1111/j.1365-2036.2007.03461.x. [DOI] [PubMed] [Google Scholar]
- 7.McCallum RW, Cynshi O Investigative Team. Efficacy of mitemcinal, a motilin agonist, on gastrointestinal symptoms in patients with symptoms suggesting diabetic gastropathy, a randomized, multi-center, placebo-controlled trial. Aliment Pharmacol Ther. 2007;26:107–116. doi: 10.1111/j.1365-2036.2007.03346.x. [DOI] [PubMed] [Google Scholar]
- 8.Wittek T, Tischer K, Körner I, Sattler T, Constable PD, et al. Effect of preoperative erythromycin or dexamethasone/vitamin C on postoperative abomasal emptying rate in dairy cows undergoing surgical correction of abomasal volvulus. Vet Surg. 2008;37:537–544. doi: 10.1111/j.1532-950X.2008.00401.x. [DOI] [PubMed] [Google Scholar]
- 9.Rohrbach BW, Cannedy AL, Freeman K, Slenning BD. Risk factors for abomasal displacement in dairy cows. J Am Vet Med Assoc. 1999;214:1660–1663. [PubMed] [Google Scholar]
- 10.Grymer J, Willeberg P, Hesselholt M. Milk production and left displaced abomasum: cause and effect relationships. Nord Vet Med. 1982;34:412–415. [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Wierup N, Björkqvist M, Weström B, Pierzynowski S, Sundler F, et al. Ghrelin and motilin are cosecreted from a prominent endocrine cell population in the small intestine. J Clin Endocrinol Metab. 2007;92:3573–3581. doi: 10.1210/jc.2006-2756. [DOI] [PubMed] [Google Scholar]
- 13.Tsutsui C, Kajihara K, Yanaka T, Sakata I, Itoh Z, et al. House musk shrew (Suncus murinus, order: Insectivora) as a new model animal for motilin study. Peptides. 2009;30:318–329. doi: 10.1016/j.peptides.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Zhang ZH, Wu SD, Su Y, Jin JZ, Fan Y, et al. Differences and significance of motilin, vasoactive intestinal peptide and gastrin in blood and gallbladder tissues of patients with gallstones. Hepatobiliary Pancreat Dis Int. 2008;7:58–64. [PubMed] [Google Scholar]
- 15.Riazi AM, Takeuchi JK, Hornberger LK, Zaidi SH, Amini F, et al. NKX2-5 regulates the expression of β-Catenin and GATA4 in ventricular myocytes. PLoS One. 2009;28:e5698. doi: 10.1371/journal.pone.0005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oishi T, Iida A, Otsubo S, Kamatani Y, Usami M, et al. A functional SNP in the NKX2.5-binding site of ITPR3 promotor is associated with susceptibility to systemic lupus erythematosus in Japanese population. J Hum Genet. 2008;53:151–162. doi: 10.1007/s10038-007-0233-3. [DOI] [PubMed] [Google Scholar]
- 17.Doll K, Sickinger M, Seeger T. New aspects in the pathogenesis of abomasal displacement. Vet J. 2009;181:90–96. doi: 10.1016/j.tvjl.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Tsai JJ, Tseng CW. Education and imaging. Gastrointestinal: gastric volvulus. J Gastroenterol Hepatol. 2009;24:1306. doi: 10.1111/j.1440-1746.2009.05956.x. [DOI] [PubMed] [Google Scholar]
- 19.Monnet E. Gastric dilatation-volvulus syndrome in dogs. Vet Clin North Am Small Anim Pract. 2003;33:987–1005. doi: 10.1016/s0195-5616(03)00059-7. [DOI] [PubMed] [Google Scholar]
- 20.Bredal WP, Eggertsdóttir AV, Austefjord O. Acute gastric dilatation in cats, a case series. Acta Vet Scand. 1996;37:445–451. doi: 10.1186/BF03548084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bethlehem M, Hilvering J. [Stomach dilatation-stomach torsion syndrome, a ‘new’ clinical picture in sows?] Tijdschr Diergeneeskd. 1987;112:1101–1105. [PubMed] [Google Scholar]
- 22.Willemse A. Gastric torsion in a guinea pig. Tijdschr Diergeneeskd. 1975;100:840–841. [PubMed] [Google Scholar]
- 23.Hudson NP, Merritt AM. Equine gastrointestinal motility research: where we are and where we need to go. Equine Vet J. 2008;40:422–428. doi: 10.2746/042516408X312932. [DOI] [PubMed] [Google Scholar]
- 24.Kong A, Cox NJ. Allele-sharing models:LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 26.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Linkage disequilibrium (LD) among the seven LDA-associated SNPs in German Holstein cows. The pairwise r2-values are shown for each SNP pair. The red square between the markers FN298674:g.6689C>T and FN298674:g.6728G>A indicates complete LD.
(TIF)
The Zmean score profile with corresponding chromosome-wide P-values for LDA in German Holstein cattle for the bovine chromosome (BTA) 23. The horizontal line indicates the threshold of chromosome-wide significance (P-value = 0.05).
(TIF)
Haplotype analysis of the polymorphisms FN298674:g.90T>C and FN298674:g.1891insG. The haplotype frequencies as well as their standard errors, χ2-values and P-values are given. The test for marker-trait association gave a χ2 value of 119.4 (p<0.0001).
(DOC)
Zmeans and LOD-scores with their P-values of the multipoint non-parametric linkage analysis for BTA23 including the SNPs FN298674:g.90T>C and FN298674:g.1891insG within MLN . For the analysis, 14 paternal half-sib families comprising 360 individuals were used.
(DOC)
Additional pairs of primers used for sequencing motilin . For each pair of primers, the amplified region, annealing temperature (AT) and product size (PS) is given.
(DOC)
Methods used for genotyping of informative SNPs. All SNPs genotyped using RFLPs (restriction fragment length polymorphisms) or IRD (infrared dye) labeled gel electrophoresis are given. Not specified markers were genotyped by sequencing.
(DOC)
Description of the five new microsatellites used for the linkage analysis. The positions (Pos) determined on the bovine UMD_3.1 assembly, primer sequences, repeat motif, heterozygosity (Het), polymorphism information content (PIC), number of alleles (A), annealing temperature (AT) and product sizes (PS) are given.
(DOC)