Abstract
To investigate how high temperature affects viral transcription, the absolute amounts of mRNA for six bacteriophage φX174 genes were compared at 37°C and 42°C using Q-PCR. At 37°C, mRNA levels for all genes were consistent with previous studies, but at 42°C mRNA levels for four genes were significantly different from levels at 37°C. Transcript levels were higher for genes B and D; the promoter before gene B appears to be up-regulated at high temperature. Levels for genes F and G were reduced at high temperature, possibly due to increased efficiency of the transcription termination signal immediately upstream of gene F. These functional changes in φX174 gene regulation at high temperature have not been described previously. Studies of phage evolution at high temperatures indicate that this difference in transcript levels is subject to adaptation.
Introduction
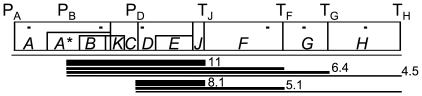
During the latter half of the twentieth century, the bacteriophage φX174 was used as a model organism for the study of many molecular processes, including DNA replication, viral infection, capsid assembly and genome organization [1], [2]. In addition, φX174 gene expression was well-studied in its host Escherichia coli at 37°C. The results of these studies indicate that φX174 recruits host-encoded trans-acting proteins and encodes a cis-regulatory network composed of three promoters and four terminators (Figure 1). The three promoters, PA, PB, and PD, are found upstream of genes A, B and D, respectively, and are bound by host sigma factors and RNA polymerase to initiate transcription [3], [4], [5]. All genes transcribed from PD are also found on transcripts starting from PB. Four terminators, TJ, TF, TG and TH, are located downstream of genes J, F, G and H, respectively, and are involved in rho-independent termination of transcription [6], [7], [8]. The terminators are not stringent, so a fraction of RNA polymerase complexes continue through these termination sites to make longer transcripts. Hayashi and colleagues found that transcript decay rates vary [9]; transcripts that begin at PB or PD and end at TJ are more stable than those ending after genes F, G and H (Figure 1). Disruption of the terminator structure of TJ accelerates the rate of mRNA degradation of the shorter transcripts [7].
Figure 1. Genetic map of φX174 based on refs. 1 and 2.
Note that the map is linearized for clarity; φX174 has a 5,386 nt circular genome. P indicates promoter; T indicates terminator. A to K indicate genes. Short bars on the genomic map indicate approximate location of Q-PCR primers for each of six genes. Below the map, bars indicate the transcripts found at 37°C, and numbers indicate transcript half-life in minutes [9].
Gene transcription in E. coli is affected by temperature due to both enzyme kinetics and the heat shock response [10], [11], [12]. Because φX174 transcription relies heavily on the host, changes that affect host transcription will also affect the phage. However, the effects of temperature on differential φX174 gene expression have not been rigorously determined. Here we demonstrate that the pattern of φX174 gene transcription is different at 42°C, providing new information about cis-regulatory regions of this well-studied phage.
Results and Discussion
Using Q-PCR, the absolute mRNA levels of six genes were compared between 37°C or 42°C infections. There was considerable variation in the total amount of phage mRNA among replicates due to differences in the number of infected cells (total phage mRNA ranged from 0.03 to 1 pg/µl; between these extremes, the average was ∼0.5 pg/µl). Each replicate experiment was done at both temperatures, and the variability between temperatures within a replicate was quite low. To account for between-replicate variation in infection rate, statistical tests were conducted on the proportion that each gene contributed to the total phage mRNA within a sample. Note from Figure 1 that each primer amplifies cDNA from multiple transcripts of different lengths, and Q-PCR does not distinguish among these transcripts. Thus the primers for gene D amplify all transcripts and the F primer set amplifies only those transcripts that read through TJ.
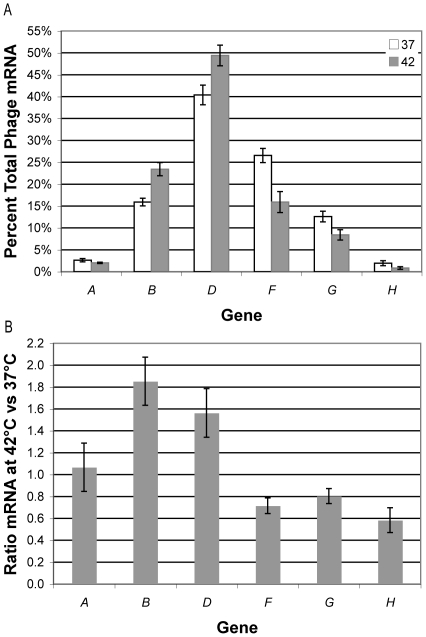
Qualitatively, our results at 37°C (Figure 2A) were consistent with the previously published results shown in Figure 1 [1], [2]. Transcript from PA is low due in large part to mRNA instability [9]. There is more mRNA for gene D than gene B due to PD, and there is successively less mRNA for the genes downstream of gene J due to the four terminators. There were important differences between the results at 37°C and those at 42°C (Figure 2A). The mRNA levels were greater for genes B and D at 42°C than at 37°C (df = 1, p-value<0.005), but were less for genes F and G at 42°C than at 37°C (df = 1, p-value<0.05). Differences in mRNA levels for genes A and H were not significantly different between temperatures, although there appears to be a two fold reduction in transcript for gene H at the higher temperature. The transcript levels for genes A and H were at the lower limit of detection for our Q-PCR assay, leading to greater variability in their estimates and lower statistical power.
Figure 2. Transcript levels of six φX174 genes measured 4 min post ejection.
A. Proportion (%) each gene contributed to the total phage mRNA in a sample averaged over six samples for growth at 37°C and 42°C. Error bars are ±1 standard error. B. Ratio of the absolute quantity of each gene at 42°C to 37°C averaged over six samples. Error bars are ±1 standard error.
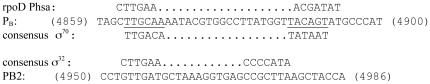
The amount of mRNA for genes B and D were greater at 42°C than at 37°C, suggesting that at least one promoter behaves in a manner similar to a heat shock promoter (Figure 2B) [10]. The absolute amount of mRNA for gene B is on average 85% greater at 42°C than at 37°C. The increase in the absolute amount of gene D mRNA (56%) at 42°C can be accounted for by the increase in transcription from PB. Our data reveal that PB responds to high temperature by up-regulating gene expression. This response is like that of a heat shock promoter, which is regulated by the heat shock sigma factor, σ32. A sequence with similarity to the −10 and −35 σ32 binding sites of the heat shock promoter of the rpoD gene overlaps the σ70 binding site identified in vitro for PB (Figure 3) [3], [13], [14]. These sequence motifs are completely conserved among 16 phages of the phiX clade sampled from nature [15]. There is also another sequence downstream of PB that is similar to the consensus σ32 binding site and is also conserved within the phiX clade (Figure 3). These two potential σ32 binding sites have low similarity to the consensus σ32 site, coinciding with the weak increase in transcription at higher temperature. The apparent importance of PB at high temperature has not been reported previously.
Figure 3. Sequence alignment of sigma factor binding sites.
Top: Alignment of the experimentally-determined binding sites for PB [3] (underlined) with the σ32 binding sites from the E. coli rpoD promoter and with the canonical σ70 binding sites [13], [30]. Bottom: Potential heat shock promoter (PB2) downstream of PB aligned with the consensus σ32 binding sites [14]. Numbers indicate start and end position of the φX174 sequence, and a dot indicates spacing to maintain alignment.
The increase in transcript levels for genes B and D appear to be biologically as well as statistically significant. Previous studies in which φX174 was evolved in chemostats to investigate adaptation of the phage to high temperature resulted in adaptive substitutions in PD [16], [17], [18]. Our previous work on these substitutions indicate that they down-regulate gene expression at both 37°C and 42°C, virtually eliminating the bottom set of transcripts in Figure 1 [19]. Phage differing from the wild type at any one of these adaptive substitutions have greater doublings per hour than wild type in batch cultures growing at 42°C and can outcompete the wild type phage in a chemostat at 42°C but not at 37°C. Thus there is strong selection to decrease the levels of transcripts for genes after PD, indicating that the increase in transcription at 42°C impacts the biology of the phage.
The absolute amounts of mRNA for genes F and G were on average 28% and 20% less, respectively, at 42°C than at 37°C (Figure 2B). Although this is a relatively small decrease between the two temperatures, it represents a large decrease in the total amount of mRNA that is transcribed from PB and PD at 42°C. Whereas 68% of transcripts read through TJ at 37°C, only 34% of transcripts read through at 42°C (Figure 2A). This substantial drop in the absolute amount of gene F transcript relative to gene D may be explained by the increased termination efficiency of TJ at high temperature. The termination efficiency of rho-independent transcription terminators is determined by competition between the rates of elongation and termination [12]. At increasing temperature, mRNA synthesis increases linearly due to enzyme kinetics [11]. Counteracting the increased elongation rate, the ternary complex including DNA, mRNA and RNA polymerase may dissociate faster at higher temperature when disrupted by a strong terminator. This effect on translation efficiency is not seen for TF and TG possibly due to the weak structural stability of their hairpin structures. Computationally determined hairpin scores for each terminator, TJ, TF, TG and TH, are −9.5, −2.6, −2.1 and −13.2, respectively, where smaller values indicate more stable hairpin structures [20]. Transcription terminators both terminate transcription and stabilize the upstream transcript [21]. The lower structural stability of TF and TG is also suggested by the shorter half-lives of transcripts that end at TF and TG than those that end at TJ (Figure 1). Our results illustrate the increased efficiency of the rho-independent transcription terminator TJ at high temperature.
Visualization of the B, D, F, G and H proteins on SDS gels show that these differences in mRNA levels are not reflected by differences in protein levels (data not shown). mRNA and protein levels are not always correlated in bacteria, and this is particularly true at increased temperatures where mRNA secondary structure and subsequent accessibility of ribosome binding sites may change [22], [23], [24]. We speculate that the transcript level differences at high temperature may be necessary to maintain the ratio of proteins involved in capsid assembly as has been found in phages T4 and lambda [25], [26]. In particular, protein B is stabilized when bound to the coat protein, otherwise, it degrades rapidly [27]. The amount of protein B seen in the gels therefore may be determined by the amount of F protein. Similarity in the ratio of proteins at the two temperatures suggests that adaptive changes that down-regulate transcription may affect some other aspect of phage biology, possibly genome replication rate. Understanding these two possibilities is the direction of our current research program.
Although the biology of φX174 is well-characterized, the functions of its regulatory regions at high temperature have not been fully described. Comparing gene expression at 37°C and 42°C shows that growth at high temperature alters transcription in two ways. Transcript levels were up-regulated from PB as if it were a heat shock promoter and down-regulated for several genes possibly due to increased efficiency of TJ. These results indicate that there is still more to learn about the regulation of gene expression of φX174 and its related microvirid phage.
Materials and Methods
To obtain RNA, replicate cultures were grown at either 37°C or 42°C using our previously developed protocol [19]. Briefly, wild-type φX174 (GenBank no. AF176034) infections were synchronized by incubating 1.5×108 E. coli C cells with 1.5×107 phages in 1.0 mL of phage Luria-Bertani medium [18] containing 2.0 mM CaCl2 at 15°C for 1 hr. Unattached phage were removed by centrifuging at 13000 rpm for 5 minutes and discarding the supernatant. The pellet was resuspended in 1 mL cold medium and added to 9 mL medium pre-warmed to 37°C or 42°C. Samples were taken at 4 min, which is less than the half-life of most transcripts [9]; and added to RNAprotect™ Bacteria Reagent (Qiagen Cat. No. 76506). Total RNA was purified following the RNeasy Mini Kit (Qiagen Cat. No. 74104). RNase-Free DNase Set (Qiagen Cat. No. 79254) was used for on-column DNA digestion. The E. coli C strain and the wild type φX174 phage were kindly provided by HA Wichman, University of Idaho [17].
Q-PCR was used to determine the absolute amount of mRNA for each of the six genes. Compared with the method used by Hayashi [28], Q-PCR is more sensitive and accurate, although it does not provide information about individual transcripts. cDNA was synthesized from each mRNA sample, and mRNA levels were detected for genes A, B, D, F, G and H using the protocol described previously and the primer sets listed in Table 1 [19]. Additionally, solutions of purified phage DNA at known concentrations (0.002, 0.0002, 2E-05, 2E-06 ng/µL) were used to determine a standard curve relating absolute mRNA quantity to the cycle threshold (the number of cycles required for the fluorescent signal to cross a threshold value). This standard curve was used to estimate the total amount of mRNA in each sample for each gene.
Table 1. Oligonucleotide primers used in Q-PCR of φX174 genes.
| Primer Name* | Location** | Sequence (5′ to 3′) |
| pA_F | 4188–4212 | GGTGATATGTATGTTGACGGCCATA |
| pA_R | 4309–4333 | GGGCGGTGGTCTATAGTGTTATTAA |
| pA_M | 4216–4232 | ACGAACGTCAGAAGCAG |
| pB_F | 5337–5360 | CTCAAATTTATGCGCGCTTCGATA |
| pB_R | 5377–5397 | TTCTGCGTCATGGAAGCGATA |
| pB_M | 27–42 | CCAACCTGCAGAGTTT |
| pD_F | 400–424 | TTACTGAACAATCCGTACGTTTCCA |
| pD_R | 457–466 | ACGGCAGAAGCCTGAATGAG |
| pD_M | 427–443 | CCGCTTTGGCCTCTATT |
| pF_F | 1778–1801 | CAGTTTTCTGGTCGTGTTCAACAG |
| pF_R | 1839–1863 | GCAAGAGTAAACATAGTGCCATGCT |
| pF_M | 1820–1836 | CCGCGTTTCTTTGTTCC |
| pG_F | 2611–2635 | GTTTCTGTTGGTGCTGATATTGCTT |
| pG_R | 2662–2685 | AGAAGACTCAAAGCGAACCAAACA |
| pG_M | 2642–2656 | CCGACCCTAAATTTT |
| pH_F | 3318–3333 | GCTTGGGAGCGTGCTG |
| pH_R | 3467–3484 | CGTGAAGTCGCCGACTGA |
| pH_M | 3451–3466 | ATGCCAGCAATCTCTT |
Amplified gene designated by letter before underscore; F and R after underscore indicate forward and reverse amplification primers; M designates labeled probe.
Numbering based upon GenBank accession AF176034.
To account for between-replicate variation in infection rate, statistical tests were conducted on the proportion that each gene contributed to the total phage mRNA within a sample. A standard variance-stabilizing transformation (arcsin(√p)) was used on the proportions prior to performing the statistical tests [29]. A linear model testing the effect of gene, temperature and the gene-by-temperature interaction on the transformed proportions indicated that gene (F = 268.25, df = 5, p-value<0.0001) and gene-by-temperature interaction (F = 10.37, df = 5, p-value<0.0001) were significant. Thus, each gene was tested separately for the difference in expression between the two temperatures, and p-values were adjusted for multiple tests on the same data using a Bonferroni correction.
Acknowledgments
We thank Dr. Holly Wichman for providing the wild type phage strain and advice, and Dr. Bentley Fane, our reviewers and the University of Idaho phage group for helpful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by National Institutes of Health (www.nih.gov) grant NCRR P20RR16448. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hayashi M, Aoyama A, Richardson DL, Hayashi MN. Biology of the Bacteriophage phiX174. In: Calendar R, editor. The Bacteriophages. New York: Plenum Press; 1988. pp. 1–71. [Google Scholar]
- 2.Fane BA, Brentlinger KL, Burch AD, Chen M, Hafenstein S, et al. øX174 et al. The Microviridae. In: Calendar R, editor. The Bacteriophages. 2nd ed. Oxford Press; 2006. pp. 129–145. [Google Scholar]
- 3.Arquint M, Spencer JH. Mapping the B and D gene promoters of bacteriophage S13 by footprinting and exonuclease III analysis. J Virol. 1987;61:3694–3700. doi: 10.1128/jvi.61.12.3694-3700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arquint M, Spencer JH. Mapping of RNA polymerase binding sites in the H/A gene region of bacteriophage S13 by footprinting and exonuclease III analysis. Biochim Biophys Acta. 1994;1218:339–345. doi: 10.1016/0167-4781(94)90186-4. [DOI] [PubMed] [Google Scholar]
- 5.Sorensen SE, Barrett JM, Wong AK, Spencer JH. Identification of the in vivo promoters of bacteriophages S13 and phi X174 and measurement of their relative activities. Biochem Cell Biol. 1998;76:625–636. doi: 10.1139/bcb-76-4-625. [DOI] [PubMed] [Google Scholar]
- 6.Hayashi MN, Hayashi M, Imai M. Bacteriophage phi X174-specific mRNA synthesis in cells deficient in termination factor rho activity. J Virol. 1981;38:198–207. doi: 10.1128/jvi.38.1.198-207.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi MN, Hayashi M, Muller UR. Role for the J-F intercistronic region of bacteriophages phi X174 and G4 in stability of mRNA. J Virol. 1983;48:186–196. doi: 10.1128/jvi.48.1.186-196.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi MN, Yaghmai R, McConnell M, Hayashi M. mRNA stabilizing signals encoded in the genome of the bacteriophage phi X174. Mol Gen Genet. 1989;216:364–371. doi: 10.1007/BF00334377. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi MN, Hayashi M. Stability of bacteriophage phi X174-specific mRNA in vivo. J Virol. 1981;37:506–510. doi: 10.1128/jvi.37.1.506-510.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neidhardt FC, VanBogelen RA, Vaughn V. The genetics and regulation of heat-shock proteins. Annu Rev Genet. 1984;18:295–329. doi: 10.1146/annurev.ge.18.120184.001455. [DOI] [PubMed] [Google Scholar]
- 11.Ryals J, Little R, Bremer H. Temperature dependence of RNA synthesis parameters in Escherichia coli. J Bacteriol. 1982;151:879–887. doi: 10.1128/jb.151.2.879-887.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson KS, von Hippel PH. Stability of Escherichia coli transcription complexes near an intrinsic terminator. J Mol Biol. 1994;244:36–51. doi: 10.1006/jmbi.1994.1702. [DOI] [PubMed] [Google Scholar]
- 13.Harley CB, Reynolds RP. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wade JT, Roa DC, Grainger DC, Hurd D, Busby SJ, et al. Extensive functional overlap between sigma factors in Escherichia coli. Nat Struct Mol Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- 15.Rokyta DR, Burch CL, Caudle SB, Wichman HA. Horizontal gene transfer and the evolution of microvirid coliphage genomes. J Bacteriol. 2006;188:1134–1142. doi: 10.1128/JB.188.3.1134-1142.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ. Different trajectories of parallel evolution during viral adaptation. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- 17.Bull JJ, Badgett MR, Wichman HA, Huelsenbeck JP, Hillis DM, et al. Exceptional convergent evolution in a virus. Genetics. 1997;147:1497–1507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crill WD, Wichman HA, Bull JJ. Evolutionary reversals during viral adaptation to alternating hosts. Genetics. 2000;154:27–37. doi: 10.1093/genetics/154.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown CJ, Zhao L, Evans KJ, Ally D, Stancik AD. Positive selection at high temperature reduces gene transcription in the bacteriophage φX174. BMC Evol Biol. 2010;10:378. doi: 10.1186/1471-2148-10-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. Prediction of transcription terminators in bacterial genomes. J Mol Biol. 2000;301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- 21.Abe H, Aiba H. Differential contributions of two elements of rho-independent terminator to transcription termination and mRNA stabilization. Biochimie. 1996;78:1035–1042. doi: 10.1016/s0300-9084(97)86727-2. [DOI] [PubMed] [Google Scholar]
- 22.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 23.de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci U S A. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geissmann T, Marzi S, Romby P. The role of mRNA structure in translational control in bacteria. RNA Biol. 2009;6:153–160. doi: 10.4161/rna.6.2.8047. [DOI] [PubMed] [Google Scholar]
- 25.Floor E. Interaction of morphogenetic genes of bacteriophage T4. J Mol Biol. 1970;47:293–306. doi: 10.1016/0022-2836(70)90303-7. [DOI] [PubMed] [Google Scholar]
- 26.Sternberg N. Properties of a mutant of Escherichia coli defective in bacteriophage lambda head formation (groE). II. The propagation of phage lambda. J Mol Biol. 1973;76:25–44. doi: 10.1016/0022-2836(73)90079-x. [DOI] [PubMed] [Google Scholar]
- 27.Bernal RA, Hafenstein S, Olson NH, Bowman VD, Chipman PR, et al. Structural studies of bacteriophage alpha3 assembly. J Mol Biol. 2003;325:11–24. doi: 10.1016/s0022-2836(02)01201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi M, Fujimura FK, Hayashi M. Mapping of in vivo messenger RNAs for bacteriophage phiX-174. Proc Natl Acad Sci U S A. 1976;73:3519–3523. doi: 10.1073/pnas.73.10.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ott RL. An Introduction to Statistical Methods and Data Analysis. Belmont, CA: Duxbury Press; 1993. [Google Scholar]
- 30.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]