Abstract
Background
Numerous studies have investigated association of OGG1 Ser326Cys polymorphism with lung cancer susceptibility; however, the findings are inconsistent. Therefore, we performed a meta-analysis based on 27 publications encompass 9663 cases and 11348 controls to comprehensively evaluate such associations.
Methods
We searched publications from MEDLINE and EMBASE which were assessing the associations between OGG1 Ser326Cys polymorphism and lung cancer risk. We calculated pooled odds ratio (OR) and 95% confidence interval (CI) by using either fixed-effects or random-effects model. We used genotype based mRNA expression data from HapMap for SNP rs1052133 in normal cell lines among 270 subjects with four different ethnicities.
Results
The results showed that individuals carrying the Cys/Cys genotype did not have significantly increased risk for lung cancer (OR = 1.15, 95% CI = 0.98–1.36) when compared with the Ser/Ser genotype; similarly, no significant association was found in recessive, dominant or heterozygous co-dominant model (Ser/Cys vs. Cys/Cys). However, markedly increased risks were found in relatively large sample size (Ser/Ser vs. Cys/Cys: OR = 1.29, 95% CI = 1.13–1.48, and recessive model: OR = 1.19, 95% CI = 1.07–1.32). As to histological types, we found the Cys/Cys was associated with adenocarcinoma risk (Ser/Ser vs. Cys/Cys: OR = 1.32, 95% CI = 1.12–1.56; Ser/Cys vs. Cys/Cys: OR = 1.19, 95% CI = 1.04–1.37, and recessive model OR = 1.23, 95% CI = 1.08–1.40). No significant difference of OGG1 mRNA expression was found among genotypes between different ethnicities.
Conclusions
Despite some limitations, this meta-analysis established solid statistical evidence for an association between the OGG1 Cys/Cys genotype and lung cancer risk, particularly for studies with large sample size and adenocarcinoma, but this association warrants additional validation in larger and well designed studies.
Introduction
Cancer is recognized as the leading cause of death in economically developed countries and the second leading cause of death in developing countries. It has been estimated that approximately 12.7 million cancer cases and 7.6 million cancer deaths have been occurred in 2008. Lung cancer was the most commonly diagnosed type of cancer as well as the leading cause of cancer death in males in 2008. Globally, lung cancer accounts for 13% (1.6 million) of the total cases and 18% (1.4 million) of the deaths [1]. Cigarette smoking is the well known risk factor for lung cancer, which accounts for 80% of the worldwide lung cancer burden in males and at least 50% of the burden in females [2]. Tobacco smoke contains multiple carcinogens that are known to chemically modify of genomic DNA [3] and further lead to genetic mutations [4].
DNA repair genes play a crucial role in maintaining the stability and integrity of genomic DNA. In humans, more than 130 genes are involved in the five major DNA repair pathways, one of which is base excision repair (BER) pathway [5]. The BER pathway repairs lesions involving modifications to the DNA bases, including lesions generated by reactive oxygen species. The specificity of BER is supported by DNA glycosylases, which have precise substrate specificities. In mammalian cells there are four major DNA glycosylases including oxoguanine DNA glycosylase (OGG1), which primarily recognizes 8-oxodG but also active on other oxidized purines [6].
The 8-oxoguanine DNA glycosylase (OGG1) gene, located at chromosome 3p26.2, encodes the enzyme responsible for the excision of 8-oxoguanine, a mutagenic base byproduct which occurs as a result of exposure to reactive oxygen species. It catalyzes the cleavage of the glycosylic bond between the modified base and the sugar moiety, leaving an abasic apurinic/apyrimidinic site in DNA; the resulting site is then incised, followed by completing of repair with successive actions of a phosphodiesterase, a DNA polymerase and a DNA ligase [7], [8]. The OGG1 is highly polymorphic, and a number of single nucleotide polymorphisms (SNPs) have been identified [9]–[12], with at least 231 reported SNPs in the gene region (http://www.ncbi.nlm.nih.gov/projects/SNP). However, only few of these reported SNPs are potentially functional and been studied for their associations with cancer susceptibility. For OGG1, there are twenty five SNPs that reportedly change amino acid of the protein but only Ser326Cys (rs1052133) was extensively investigated for its association with cancer risk, in particular for lung cancer. Because the results from these studies were inconsistent [9], [13]–[25], we performed a meta-analysis of the published reports to further evaluate the association of OGG1 Ser326Cys SNPs with the risk of lung cancer.
Materials and Methods
Identification and eligibility of relevant studies
Studies included in this meta-analysis were to meet the following criteria: (a) evaluating the association between OGG1 Ser326Cys and cancer risk, (b) using a case-control design, (c) providing sufficient information to estimate odds ratios (ORs) and their 95% confidence intervals (CIs).
We searched the electronic literature MEDLINE and EMBASE databases for all relevant articles using the search terms: “OGG1, HMMH, MUTM, OGH1 or hOGG1”, “variant or variation or polymorphism” and “lung cancer” (last search was updated on Nov 30, 2011). All eligible studies were retrieved, and their bibliographies were manually checked for other relevant publications. Review articles and bibliographies of other relevant studies identified were hand-searched as well to search for additional eligible studies. Only published studies with full-text articles in English were included. If more than one article was published using the same patient population, only the latest or the largest study would be used in this meta-analysis. Two authors (Wei-Xun Duan and Rui-Xi Hua) independently assessed the articles for compliance with the inclusion criteria, and any disagreement was resolved by discussions till consensus was reached. In addition, investigations departure from Hardy-Weinberg equilibrium (HWE) was excluded from the final analysis.
Data extraction
The following information was collected from each study: first author's surname, year of publication, ethnicity of the study population, cancer types, histological types, source used for controls, total number of cases and controls, genotype methods and numbers of cases and controls with the Ser/Ser, Ser/Cys, and Cys/Cys genotypes for OGG1. For those studies that included subjects of different ethnic groups, genotypes data were extracted separately for each of ethnic groups, categorized as Caucasians, Asians, Africans or Mixed which contained more than one ethnic group.
Genotype and gene expression correlation analysis
The data on OGG1 Ser326Cys (rs1052133C>G) genotype and transcript (mRNA) expression levels were available by SNPexp online tool (http://app3.titan.uio.no/biotools/help.php?app=snpexp) [26]. The genotyping data for OGG1 were derived from the HapMap phase II release 23 data set consisting of 3.96 million SNP genotypes from 270 individuals from four populations (CEU: 90 Utah residents with ancestry from northern and western Europe; CHB: 45 unrelated Han Chinese in Beijing; JPT: 45 unrelated Japanese in Tokyo; YRI: 90 Yoruba in Ibadan, Nigeria) [27]. The transcript (mRNA) expression data were detected by using genome-wide expression arrays (47294 transcripts) from EBV-transformed lymphoblastoid cell lines from the same 270 individuals [28].
Statistical methods
The strength of association between OGG1 Ser326Cys and lung cancer risk was assessed by calculating ORs with the corresponding 95% CIs. For OGG1 Ser326Cys, the pooled ORs were also performed for additive (Ser/Ser vs. Cys/Cys and Ser/Cys vs. Cys/Cys), recessive model (Ser/Ser+Ser/Cys vs. Cys/Cys), and dominant model (Ser/Ser vs. Ser/Cys+Cys/Cys). The homogeneity assumption was verified by Chi square-based Q-test. If the studies were found to be homogeneous (with P>0.10 for the Q test), the pooled OR estimate of all studies would be calculated by the fixed-effects model (the Mantel–Haenszel method) [29]. If homogeneity could not be assumed, a random-effects model (the DerSimonian and Laird method) would be used [30]. Subgroup analyses were performed by cancer type, ethnicity, study design and sample size. To verify the potential publication bias, a standard error of log (OR) for each study was plotted against its log (OR). Funnel plot asymmetry was assessed by Egger's linear regression test [31]. To assess the effect of individual studies on the overall risk of cancers, sensitivity analyses were performed by excluding each study individually and recalculating the ORs and the 95% CIs. The mRNA expression levels between two strata were assessed by using Student's t test. The transcript expression level trends by genotypes were evaluated by using General linear model. This meta-analysis was performed by using the software STATA version 11.0 (Stata Corporation, College Station, TX) and SAS software (version 9.1; SAS Institute, Cary, NC). All the P values were two-sided, and a P<0.05 was considered statistically significant.
Results
Study characteristics
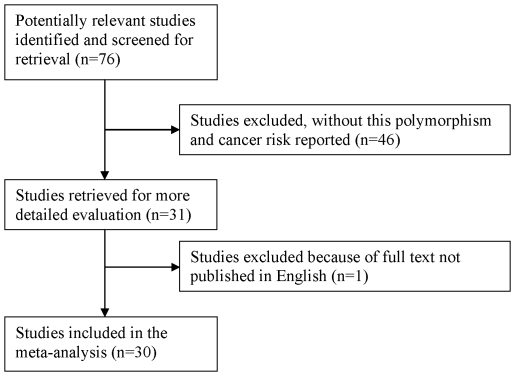
As shown in Figure 1, a total of 76 published records were retrieved, of which 45 were excluded after the abstract was found to be irrelevant, and one paper was excluded since they published in Korean [32]. A total of 30 case-control studies met the inclusion criteria [9], [13]–[25], [33] and were included in the meta-analysis (Table 1). The study by Bonner et al. [33] was excluded in the final analysis because they used the same samples as a previous article [34]. The distribution of genotypes for the OGG1 polymorphism in the controls of all studies was consistent with that expected from the HWE, except for three studies [23], [35], [36]. Chang et al. [19] evaluate the differences in genetic contribution to lung cancer risk in Latinos and African-American ethnic groups, so this study was separated in two. Overall, 27 studies with 9663 cases and 11348 controls investigating the OGG1 Ser326Cys SNP were included in this meta-analysis. The study of Klinchid et al. [20] was included only in the calculation of the dominant model, because the genotype distribution was not presented in sufficient detail. Of the 27 studies, sample sizes ranged from 45 to 2155, in which eight studies focused on non-small cell lung cancer (NSCLC) and nineteen on mixed lung cancers. There were eleven studies on Caucasians, twelve studies on Asians, two studies on Africans and two on mixed ethnicity. Almost all of the cases were histologically confirmed. Controls were mainly matched for sex and age. Of all the studies, twelve were population-based, fifteen were hospital-based; three studies with sample size less than 100, nineteen studies with sample size between 100 to 500, and five studies with sample size more than 500. For available histological types, six studies with small cell lung cancer (SCLC), seven with squamous cell carcinoma (SCC), ten with adenocarcinoma (ADC), one with large cell carcinoma (LCC), six with NSCLC thus histological type details not available, and twelve with lung cancer but details not available that were considered as mixed.
Figure 1. Flow chart of Included Studies.
Table 1. Characteristics of studies included in the meta-analysis.
| Surname | Year | Country | Ethnicity | Cancer type | Cases/controls | Control source | Genotype method | MAF | H-W |
| Khono | 1998 | Japan | Asian | Mixed | 45/42 | PB | PCR-SSCP | 0.40 | 0.939 |
| Sugimura | 1999 | Japan | Mixed | Mixed | 241/197 | HB | PCR-SSCP | 0.41 | 0.082 |
| Wikman | 2000 | Germany | Caucasian | NSCLC | 105/105 | HB | PCR-RFLP | 0.22 | 0.067 |
| Ito | 2002 | Japan | Asian | ADC | 138/240 | HB | PCR-CTPP | 0.47 | 0.837 |
| Le | 2002 | USA | Mixed | Mixed | 298/405 | PB | PCR-RFLP | 0.35 | 0.350 |
| Sunaga | 2002 | Japan | Asian | ADC | 198/152 | HB | PCR-RFLP | 0.45 | 0.126 |
| Lan | 2004 | China | Asian | Mixed | 118/109 | PB | SNP500 | 0.33 | 0.232 |
| Park | 2004 | USA | Caucasian | Mixed | 179/350 | HB | PCR-RFLP | 0.15 | 0.857 |
| Vogel | 2004 | Denmark | Caucasian | Mixed | 256/269 | PB | real-time PCR | 0.24 | 0.237 |
| Hung | 2005 | European | Caucasian | SCC, ADC | 2155/2163 | HB | TaqMan | 0.20 | 0.215 |
| Khono | 2006 | Japan | Asian | ADC | 1097/394 | HB | pyrosequencing | 0.45 | 0.627 |
| Loft | 2006 | Denmark | Caucasian | Mixed | 251/261 | PB | real-time PCR | 0.24 | 0.200 |
| Sorensen | 2006 | Denmark | Caucasian | Mixed | 431/796 | PB | real-time PCR | 0.22 | 0.258 |
| Matullo | 2006 | European | Caucasian | Mixed | 116/1094 | PB | DHPLC | 0.22 | 0.901 |
| De Ruyck | 2007 | Belgium | Caucasian | Mixed | 110/110 | HB | PCR-RFLP | 0.25 | 0.176 |
| Hatt | 2008 | Denmark | Caucasian | Mixed | 158/164 | PB | TaqMan | 0.25 | 0.536 |
| Karahalil | 2008 | Turkey | Caucasian | Mixed | 165/250 | PB | PCR-RFLP | 0.33 | 0.546 |
| Chang | 2009 | Taiwan | Asian | Mixed | 1096/997 | HB | MassARRAY | 0.60 | 0.741 |
| Miyaishi | 2009 | Japan | Asian | Mixed | 108/121 | HB | PCR-RFLP | 0.45 | 0.271 |
| Okasaka | 2009 | Japan | Asian | Mixed | 515/1030 | PB | TaqMan | 0.49 | 0.070 |
| Chang | 2009 | USA | African | Mixed | 112/296 | PB | Illumina | 0.32 | 0.691 |
| Chang | 2009 | USA | African | Mixed | 254/280 | PB | Illumina | 0.15 | 0.521 |
| Klinchid | 2009 | Thailand | Asian | NSCLC | 76/75 | HB | diASA-AMP | ||
| Janik | 2011 | Poland | Caucasian | NSCLC | 88/79 | HB | PCR-MSSCP | 0.15 | 0.542 |
| Kohno | 2011 | Japan | Asian | SCC | 377/325 | HB | pyrosequencing | 0.45 | 0.704 |
| Li | 2011 | China | Asian | Mixed | 395/443 | HB | PCR-CTPP | 0.62 | 0.329 |
| Qian | 2011 | China | Asian | NSCLC | 581/601 | HB | TaqMan | 0.55 | 0.592 |
| Liang | 2005 | China | Asian | SCC, ADC | 227/227 | HB | diASA-AMP | 0.61 | 0.043 |
| Zienolddiny | 2006 | Norway | Caucasian | NSCLC | 326/386 | PB | APEX | 0.35 | 0.000 |
| Liu | 2010 | Taiwan | Asian | Mixed | 358/716 | HB | PCR-RFLP | 0.64 | 0.004 |
HB, Hospital based; PB, Population based; SCC, Squamous Cell Carcinoma; NSCLC, non-small cell lung cancer; ADC, Adenocarcinoma; PCR-SSCP, PCR-single strand conformation polymorphism; PCR-RFLP, PCR-restriction fragment length polymorphisms; PCR-CTPP, polymerase chain reaction with confronting two-pair primers; diASA-AMP, di-allele-specific amplification with artificially modified primers; DHPLC, denaturing high performance liquid chromatography.
Meta-analysis results
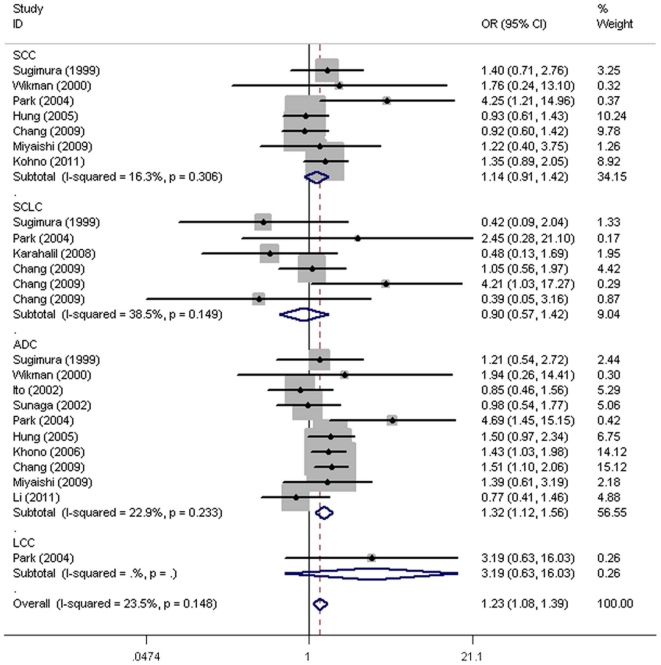
The overall results suggested there was no significant association between OGG1 Ser326Cys and risk of lung cancer (Ser/Ser vs. Cys/Cys: OR = 1.15, 95% CI = 0.98–1.36; Ser/Cys vs. Cys/Cys: OR = 1.09, 95% CI = 0.95–1.25), recessive (OR = 1.11, 95% CI = 0.97–1.28) or dominant model (OR = 1.09, 95% CI = 0.98–1.21) (Table 2). In the subgroup analysis by sample size, a statistically significant association was found for studies with large sample size (Ser/Ser vs. Cys/Cys: OR = 1.29, 95% CI = 1.13–1.48; Ser/Ser+Ser/Cys vs. Cys/Cys: OR = 1.19, 95% CI = 1.07–1.32), and a relative small sample size (<100) for dominant model (OR = 1.79, 95% CI = 1.08–2.98) which just included three studies. Further stratification by cancer type, ethnicity, and source of controls all yielded no statistically significant estimates. In the histological type subgroup analysis with more details, the OGG1 Ser326Cys allele was significantly associated with risk of ADC (Ser/Ser vs. Cys/Cys: OR = 1.32, 95% CI = 1.12–1.56; Ser/Cys vs. Cys/Cys: OR = 1.19, 95% CI = 1.04–1.37) and recessive model (OR = 1.23, 95% CI = 1.08–1.40), but not with cancers of the SCC, SCLC and LCC (Table 3, Figure 2).
Table 2. Meta-analysis of the association between OGG1 Ser326Cys polymorphism and lung cancer risk.
| Variables | No. of studies | Homozygous | P het a | Heterozygous | P het a | Recessive | P het a | Dominant | P het a |
| Ser/Ser vs. Cys/Cys | Ser/Cys vs. Cys/Cys | (Ser/Ser+Ser/Cys) vs. Cys/Cys | Ser/Ser vs. (Ser/Cys+Cys/Cys) | ||||||
| All | 27 | 1.15 (0.98–1.36) | 0.001 | 1.09 (0.95–1.25) | 0.010 | 1.11 (0.97–1.28) | 0.002 | 1.09 (0.98–1.21) | <0.001 |
| Cancer type | |||||||||
| NSCLC | 8 | 1.29 (0.99–1.69) | 0.118 | 1.14 (0.85–1.55) | 0.054 | 1.18 (0.90–1.54) | 0.029 | 1.20 (0.99–1.45) | 0.125 |
| Mixed | 19 | 1.09 (0.89–1.34) | 0.002 | 1.08 (0.92–1.26) | 0.016 | 1.09 (0.92–1.29) | 0.006 | 1.05 (0.93–1.19) | 0.001 |
| Ethnicity | |||||||||
| Caucasian | 11 | 1.12 (0.78–1.62) | 0.014 | 1.08 (0.79–1.47) | 0.107 | 1.11 (0.79–1.55) | 0.028 | 1.05 (0.88–1.25) | 0.001 |
| Asian | 12 | 1.15 (0.92–1.43) | 0.004 | 1.04 (0.89–1.22) | 0.030 | 1.07 (0.91–1.27) | 0.007 | 1.15 (0.98–1.35) | 0.018 |
| African | 2 | 1.00 (0.54–1.84) | 0.802 | 0.98 (0.53–1.82) | 0.423 | 1.01 (0.56–1.83) | 0.653 | 1.12 (0.82–1.52) | 0.279 |
| Mixed | 2 | 1.48 (0.96–2.27) | 0.224 | 1.73 (1.23–2.43) | 0.368 | 1.61 (1.14–2.28) | 0.284 | 1.01 (0.79–1.29) | 0.334 |
| Source of controls | |||||||||
| Population | 12 | 1.15 (0.94–1.40) | 0.335 | 1.09 (0.88–1.36) | 0.221 | 1.14 (0.94–1.38) | 0.273 | 1.08 (0.97–1.19) | 0.617 |
| Hospital | 15 | 1.20 (0.94–1.54) | <0.001 | 1.11 (0.92–1.33) | 0.005 | 1.13 (0.93–1.38) | <0.001 | 1.10 (0.93–1.31) | <0.001 |
| Sample size | |||||||||
| <100 | 3 | 4.46 (0.29–68.84) | 0.021 | 3.88 (0.41–36.47) | 0.059 | 4.33 (0.33–57.22) | 0.024 | 1.79 (1.08–2.98) | 0.302 |
| 100–500 | 19 | 1.05 (0.83–1.34) | 0.002 | 1.01 (0.82–1.25) | 0.010 | 1.04 (0.84–1.29) | 0.002 | 1.04 (0.91–1.19) | 0.002 |
| >500 | 5 | 1.29 (1.13–1.48) | 0.920 | 1.05 (0.81–1.37) | 0.842 | 1.19 (1.07–1.32) | 0.953 | 1.12 (0.96–1.31) | 0.046 |
P value of the Q-test for heterogeneity test.
Table 3. Meta-analysis of the association between OGG1 Ser326Cys polymorphism and lung cancer with definite histological types.
| Variables | No. of studies | Homozygous | P het a | Heterozygous | P het a | Recessive | P het a | Dominant | P het a |
| Ser/Ser vs. Cys/Cys | Ser/Cys vs. Cys/Cys | (Ser/Ser+Ser/Cys) vs. Cys/Cys | Ser/Ser vs. (Ser/Cys+Cys/Cys) | ||||||
| Histological types | |||||||||
| SCLC | 6 | 0.90 (0.57–1.42) | 0.149 | 0.98 (0.67–1.44) | 0.696 | 0.97 (0.68–1.39) | 0.357 | 1.07 (0.67–1.72) | 0.014 |
| SCC | 7 | 1.14 (0.91–1.42) | 0.306 | 1.16 (0.96–1.41) | 0.163 | 1.14 (0.95–1.38) | 0.143 | 1.04 (0.84–1.28) | 0.101 |
| ADC | 10 | 1.32 (1.12–1.56) | 0.233 | 1.19 (1.04–1.37) | 0.186 | 1.23 (1.08–1.40) | 0.248 | 1.13 (0.93–1.38) | 0.014 |
| LCC | 1 | 3.19 (0.63–16.03) | - | 2.18 (0.41–11.69) | - | 2.85 (0.58–14.03) | - | 1.61 (0.76–3.42) | - |
| NSCLC | 6 | 1.34 (1.03–1.75) | 0.065 | 1.11 (0.89–1.39) | 0.155 | 1.18 (0.95–1.46) | 0.086 | 1.25 (0.97–1.61) | 0.147 |
| Mixed | 12 | 1.24 (1.02–1.49) | 0.364 | 1.20 (1.00–1.43) | 0.320 | 1.24 (1.05–1.46) | 0.307 | 1.08 (0.96–1.22) | 0.371 |
SCLC, Small cell lung cancer; SCC, Squamous Cell Carcinoma; ADC, Adenocarcinoma; LCC, Large cell carcinoma; NSCLC, non-small cell lung cancer.
P value of the Q-test for heterogeneity test.
Figure 2. ORs of lung cancer with definite histological types associated with the OGG1 Ser/Ser genotype compared with the Cys/Cys genotype.
For each study, the estimates of OR and its 95% CI are plotted with a box and a horizontal line. ⋄, pooled ORs and its 95% CIs.
The mRNA expression by genotypes
The mRNA expression level of OGG1 by the genotypes of four ethnicities is shown in Table 4. We did not find any mRNA expression difference between different genotypes among the four different ethnicities. Thus, mRNA expression level was a little higher though no significant difference for rs1052133G variant allele was found in Asian populations. No trend of transcript expression levels by genotypes was found for OGG1.
Table 4. OGG1 mRNA expression by the genotypes of SNPs, using data from the HapMapa.
| Population | Genotypes | No. | Mean ± SD | P b | P trend c |
| CHB | CC | 11 | 7.84±0.20 | 0.267 | |
| CG | 23 | 7.94±0.26 | 0.280 | ||
| GG | 11 | 7.96±0.29 | 0.264 | ||
| CG/GG | 34 | 7.95±0.26 | 0.234 | ||
| JPT d | CC | 8 | 7.67±0.18 | 0.125 | |
| CG | 26 | 7.80±0.18 | 0.084 | ||
| GG | 10 | 7.81±0.16 | 0.102 | ||
| CG/GG | 36 | 7.80±0.17 | 0.057 | ||
| CEU d | CC | 55 | 7.87±0.31 | 0.283 | |
| CG | 26 | 7.89±0.28 | 0.781 | ||
| GG | 6 | 7.63±0.18 | 0.082 | ||
| CG/GG | 32 | 7.84±0.28 | 0.689 | ||
| YRI d | CC | 65 | 7.80±0.23 | 0.753 | |
| CG | 20 | 7.77±0.20 | 0.568 | ||
| GG | 3 | 7.83±0.20 | 0.869 | ||
| CG/GG | 23 | 7.78±0.19 | 0.637 | ||
| Asian d | CC | 19 | 7.77±0.21 | 0.106 | |
| CG | 49 | 7.86±0.23 | 0.117 | ||
| GG | 21 | 7.89±0.24 | 0.102 | ||
| CG/GG | 70 | 7.87±0.23 | 0.083 | ||
| All d | CC | 139 | 7.82±0.26 | 0.620 | |
| CG | 95 | 7.85±0.24 | 0.420 | ||
| GG | 30 | 7.83±0.24 | 0.883 | ||
| CG/GG | 125 | 7.85±0.24 | 0.468 |
Genotyping data and mRNA expression levels for OGG1 Ser326Cys by genotypes obtained from the HapMap phase II release 23 data from EBV-transformed lymphoblastoid cell lines from 270 individuals. CHB: 45 unrelated Han Chinese in Beijing; JPT: 45 unrelated Japanese in Tokyo; CEU: 90 Utah residents with ancestry from northern and western Europe; YRI: 90 Yoruba in Ibadan, Nigeria.
Two-side Student's t test within the stratum.
P values for the trend test of OGG1 mRNA expression among 3 genotypes for each SNP from a general linear model.
There were missing data because genotyping data were not available.
Publication bias
No publication bias was detected for OGG1 Ser326Cys (the Egger's test, Ser/Ser vs. Cys/Cys: P = 0.944, Ser/Cys vs. Cys/Cys: P = 0.987, recessive model: P = 0.892, dominant model: P = 0.217).
Discussion
It is well recognized that individual susceptibility to cancer varies, even after exposure to the same environment. Therefore, it has been suggested that genetic variation, such as SNPs of genes is involved in carcinogenesis. We conducted a meta-analysis of published studies to evaluate the association between the OGG1 Ser326Cys polymorphism and lung cancer risk because no such up-to-date meta-analysis including histological types has been published to date. We found no statistical evidence of an overall effect of the Ser326Cys polymorphism on lung cancer risk in either recessive or dominant effect models. Compared with the Ser/Ser genotype, the variant Cys/Cys genotype was not significantly associated with overall lung cancer risk in all subjects from 27 eligible studies included in the analysis. In a previous meta-analysis with 17 studies consist of 6375 cases and 6406 controls, significantly increased risks were found among Asian subjects in a dominant model, and lung cancer risk associated with the OGG1 Cys/Cys genotype was significantly increased in population-based studies [9]. However, these were not found in ours, may be attributed to a larger sample size.
Previous studies demonstrated that genetic variation in OGG1 affects cancer susceptibility; the frequency of the OGG1 326Cys allele was found to be significantly higher in patients when compared with controls [22], [24], [25], [37], [38]; however, this association was not replicated by other studies [9], [13]–[17]. Overall, we did not found that individuals carrying the Cys/Cys genotype had significantly increased risk of lung cancer when compared with the Ser/Ser genotype, no significant association with lung cancer risk was found in dominant model, recessive model and heterozygous co-dominant model (Ser/Cys vs. Cys/Cys). However, markedly increased risks were found in relatively large sample size, and this hinted us that in the future studies, only the studies with large sample size would be reliable. In the histological type subgroup analysis, the OGG1 Ser326Cys allele was significantly associated with risk of ADC, but not with cancers of the SCC, SCLC and LCC. These may be attributed to the tumor specificity.
Mambo et al. [39] analyzed the expression of hOGG1 mRNA in 18 lung cancer and three normal cell lines and found hOGG1 was over expressed in most cell lines, 2/18 (11.1%) showed a lower hOGG1 mRNA and protein expression (∼80% decrease) relative to normal cell lines, indicating 8-Hydroxyguanine repair defects in certain lung cancers. When we compared the mRNA expression levels of OGG1 Ser326Cys by the genotypes of four different ethnicities, no difference or trend was found. Lung cancer is known to be a complex and multifactorial disease, gene-gene and gene-environment interactions both contribute greatly to the occurrence of this disease, a single nucleotide variation may be insufficient to alter the OGG1 mRNA expression especially the ones in the coding regions which just lead to amino acid alteration.
Though we included the latest data, there are several limitations in this meta-analysis must also be considered. First, lack of the original data of lung cancer histological types limited our further evaluation of histological types and genotypes interactions. Second, lack of the original data limited our further evaluation of potential gene-gene and gene-environment interactions. Third, lack of information on disease status, genotypes, and well-documented smoking status may also influence the results. Fourth, most of the studies except for five [17], [21], [22], [40], [41] had a relatively small sample sizes (<500 cases and controls). Finally, the studies included in the analysis have used more than ten different genotyping methods that had different quality control issues.
In conclusion, this meta-analysis found that the OGG1 326Cys/Cys genotype was not associated with significantly increased risk of lung cancer. However, given the relatively limited lung cancer histological types and sample size, Cys/Cys was associated with adenocarcinoma risk. However, further studies are warranted to validate the association between the OGG1 Ser326Cys polymorphism and lung cancer risk with larger sample size and more detailed histological types.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by grants from the National Natural Science Foundation of China (NO. 81102687, NO. 81070198). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ezzati M, Lopez AD. Estimates of global mortality attributable to smoking in 2000. Lancet. 2003;362:847–852. doi: 10.1016/S0140-6736(03)14338-3. [DOI] [PubMed] [Google Scholar]
- 3.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 4.Livneh Z. DNA damage control by novel DNA polymerases: translesion replication and mutagenesis. J Biol Chem. 2001;276:25639–25642. doi: 10.1074/jbc.R100019200. [DOI] [PubMed] [Google Scholar]
- 5.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 6.Scharer OD, Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 7.Dianov GL, Souza-Pinto N, Nyaga SG, Thybo T, Stevnsner T, et al. Base excision repair in nuclear and mitochondrial DNA. Prog Nucleic Acid Res Mol Biol. 2001;68:285–297. doi: 10.1016/s0079-6603(01)68107-8. [DOI] [PubMed] [Google Scholar]
- 8.Karahalil B, Hogue BA, de Souza-Pinto NC, Bohr VA. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J. 2002;16:1895–1902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Hao X, Zhang W, Wei Q, Chen K. The hOGG1 Ser326Cys polymorphism and lung cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17:1739–1745. doi: 10.1158/1055-9965.EPI-08-0001. [DOI] [PubMed] [Google Scholar]
- 10.Chevillard S, Radicella JP, Levalois C, Lebeau J, Poupon MF, et al. Mutations in OGG1, a gene involved in the repair of oxidative DNA damage, are found in human lung and kidney tumours. Oncogene. 1998;16:3083–3086. doi: 10.1038/sj.onc.1202096. [DOI] [PubMed] [Google Scholar]
- 11.Nohmi T, Kim SR, Yamada M. Modulation of oxidative mutagenesis and carcinogenesis by polymorphic forms of human DNA repair enzymes. Mutat Res. 2005;591:60–73. doi: 10.1016/j.mrfmmm.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 12.Hill JW, Evans MK. A novel R229Q OGG1 polymorphism results in a thermolabile enzyme that sensitizes KG-1 leukemia cells to DNA damaging agents. Cancer Detect Prev. 2007;31:237–243. doi: 10.1016/j.cdp.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karahalil B, Emerce E, Kocer B, Han S, Alkis N, et al. The association of OGG1 Ser326Cys polymorphism and urinary 8-OHdG levels with lung cancer susceptibility: a hospital-based case-control study in Turkey. Arh Hig Rada Toksikol. 2008;59:241–250. doi: 10.2478/10004-1254-59-2008-1924. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Guan W, Li MX, Zhong ZY, Qian CY, et al. Genetic polymorphism of DNA base-excision repair genes (APE1, OGG1 and XRCC1) and their correlation with risk of lung cancer in a Chinese population. Arch Med Res. 2011;42:226–234. doi: 10.1016/j.arcmed.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Matullo G, Dunning AM, Guarrera S, Baynes C, Polidoro S, et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27:997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- 16.Hatt L, Loft S, Risom L, Moller P, Sorensen M, et al. OGG1 expression and OGG1 Ser326Cys polymorphism and risk of lung cancer in a prospective study. Mutat Res. 2008;639:45–54. doi: 10.1016/j.mrfmmm.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Qian B, Zhang H, Zhang L, Zhou X, Yu H, et al. Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer. 2011;73:138–146. doi: 10.1016/j.lungcan.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 18.Miyaishi A, Osawa K, Osawa Y, Inoue N, Yoshida K, et al. MUTYH Gln324His gene polymorphism and genetic susceptibility for lung cancer in a Japanese population. J Exp Clin Cancer Res. 2009;28:10. doi: 10.1186/1756-9966-28-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, et al. Base excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African-Americans. Carcinogenesis. 2009;30:78–87. doi: 10.1093/carcin/bgn261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klinchid J, Chewaskulyoung B, Saeteng S, Lertprasertsuke N, Kasinrerk W, et al. Effect of combined genetic polymorphisms on lung cancer risk in northern Thai women. Cancer Genet Cytogenet. 2009;195:143–149. doi: 10.1016/j.cancergencyto.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Okasaka T, Matsuo K, Suzuki T, Ito H, Hosono S, et al. hOGG1 Ser326Cys polymorphism and risk of lung cancer by histological type. J Hum Genet. 2009;54:739–745. doi: 10.1038/jhg.2009.108. [DOI] [PubMed] [Google Scholar]
- 22.Chang CH, Hsiao CF, Chang GC, Tsai YH, Chen YM, et al. Interactive effect of cigarette smoking with human 8-oxoguanine DNA N-glycosylase 1 (hOGG1) polymorphisms on the risk of lung cancer: a case-control study in Taiwan. Am J Epidemiol. 2009;170:695–702. doi: 10.1093/aje/kwp019. [DOI] [PubMed] [Google Scholar]
- 23.Liu CJ, Hsia TC, Tsai RY, Sun SS, Wang CH, et al. The joint effect of hOGG1 single nucleotide polymorphism and smoking habit on lung cancer in Taiwan. Anticancer Res. 2010;30:4141–4145. [PubMed] [Google Scholar]
- 24.Janik J, Swoboda M, Janowska B, Ciesla JM, Gackowski D, et al. 8-Oxoguanine incision activity is impaired in lung tissues of NSCLC patients with the polymorphism of OGG1 and XRCC1 genes. Mutat Res. 2011;709–710:21–31. doi: 10.1016/j.mrfmmm.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Kohno T, Kunitoh H, Mimaki S, Shiraishi K, Kuchiba A, et al. Contribution of the TP53, OGG1, CHRNA3, and HLA-DQA1 genes to the risk for lung squamous cell carcinoma. J Thorac Oncol. 2011;6:813–817. doi: 10.1097/JTO.0b013e3181ee80ef. [DOI] [PubMed] [Google Scholar]
- 26.Holm K, Melum E, Franke A, Karlsen TH. SNPexp - A web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC Bioinformatics. 2010;11:600. doi: 10.1186/1471-2105-11-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 28.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 30.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 31.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CH, Lee KY, Choe KH, Hong YC, Noh SI, et al. [Effects of oxidative DNA damage and genetic polymorphism of the glutathione peroxidase 1 (GPX1) and 8-oxoguanine glycosylase 1 (hOGG1) on lung cancer]. J Prev Med Public Health. 2006;39:130–134. [PubMed] [Google Scholar]
- 33.Bonner MR, Rothman N, Mumford JL, He X, Shen M, et al. Green tea consumption, genetic susceptibility, PAH-rich smoky coal, and the risk of lung cancer. Mutat Res. 2005;582:53–60. doi: 10.1016/j.mrgentox.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Lan Q, Mumford JL, Shen M, Demarini DM, Bonner MR, et al. Oxidative damage-related genes AKR1C3 and OGG1 modulate risks for lung cancer due to exposure to PAH-rich coal combustion emissions. Carcinogenesis. 2004;25:2177–2181. doi: 10.1093/carcin/bgh240. [DOI] [PubMed] [Google Scholar]
- 35.Liang G, Pu Y, Yin L. Rapid detection of single nucleotide polymorphisms related with lung cancer susceptibility of Chinese population. Cancer Lett. 2005;223:265–274. doi: 10.1016/j.canlet.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 36.Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 37.Le Marchand L, Donlon T, Lum-Jones A, Seifried A, Wilkens LR. Association of the hOGG1 Ser326Cys polymorphism with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:409–412. [PubMed] [Google Scholar]
- 38.Park J, Chen L, Tockman MS, Elahi A, Lazarus P. The human 8-oxoguanine DNA N-glycosylase 1 (hOGG1) DNA repair enzyme and its association with lung cancer risk. Pharmacogenetics. 2004;14:103–109. doi: 10.1097/00008571-200402000-00004. [DOI] [PubMed] [Google Scholar]
- 39.Mambo E, Chatterjee A, de Souza-Pinto NC, Mayard S, Hogue BA, et al. Oxidized guanine lesions and hOgg1 activity in lung cancer. Oncogene. 2005;24:4496–4508. doi: 10.1038/sj.onc.1208669. [DOI] [PubMed] [Google Scholar]
- 40.Hung RJ, Brennan P, Canzian F, Szeszenia-Dabrowska N, Zaridze D, et al. Large-scale investigation of base excision repair genetic polymorphisms and lung cancer risk in a multicenter study. J Natl Cancer Inst. 2005;97:567–576. doi: 10.1093/jnci/dji101. [DOI] [PubMed] [Google Scholar]
- 41.Kohno T, Kunitoh H, Toyama K, Yamamoto S, Kuchiba A, et al. Association of the OGG1-Ser326Cys polymorphism with lung adenocarcinoma risk. Cancer Sci. 2006;97:724–728. doi: 10.1111/j.1349-7006.2006.00240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]