Abstract
Blast-induced traumatic brain injury (TBI) is of significant concern in soldiers returning from the current conflicts in Iraq and Afghanistan. Incidents of TBI have increased significantly in the current conflicts compared to previous wars, and a majority of these injuries are caused by improvised explosive devices. Currently, no specific technique or biomarker is available for diagnosing TBI when no obvious clinical symptoms are present. MicroRNAs are small RNA (∼ 22nts) molecules that are expressed endogenously and play an important role in regulating gene expression. MicroRNAs have emerged as novel serum diagnostic biomarkers for various diseases. In this study, we studied the effect of blast overpressure injury on the microRNA signatures in the serum of rats. Rats were exposed to three serial 120-kPa blast overpressure exposures through a shockwave tube. Blood and cerebrospinal fluid were collected at various time points after injury, and microRNA modulation was analyzed using real-time PCR. Five microRNAs were significantly modulated in the serum samples of these animals at three time points post-injury. Further, we also found that the levels of microRNA let-7i are also elevated in cerebrospinal fluid post-blast wave exposure. The presence of microRNA in both serum and cerebrospinal fluid immediately after injury makes microRNA let-7i an ideal candidate for further studies of biomarkers in TBI.
Key words: biomarker, blast overpressure injury, microRNA, serum, traumatic brain injury
Introduction
Traumatic brain injury (TBI) is defined as a blow or jolt to the head or a penetrating head injury resulting in the disruption of brain function (Kennedy et al., 2007). TBI has been referred to as a signature injury of the wars in Iraq and Afghanistan due to a significant increase in cases of TBI among service personnel, as well as the civilians involved in these conflicts (Risdall and Menon, 2011). Blasts resulting from improvised explosive devices account for more than 60% of war-related TBI (Ling et al., 2009). As a result of the increasing incidence of blast injury, TBI is responsible for significant mortality and long-lasting morbidity in injured soldiers and veterans returning from war zones. Blast-induced TBI may occur through several mechanisms, including the primary blast overpressure wave itself, as well as objects propelled by the explosion (secondary injuries), collision with other objects upon acceleration caused by the blast wave (tertiary injuries), or a combination of these (Elder and Cristian, 2009). An exposure to the primary blast wave results in contusions in the brain, and coup-contrecoup injuries that may result in subdural hematoma, cerebral hemorrhage, edema, and diffuse axonal injury as a result of stress wave propagation inside the brain (Leung et al., 2008). Mild or moderate brain injuries from primary blast waves often go undiagnosed and untreated due to the lack of visible symptoms and lack of availability of diagnostic markers, so immediate attention is primarily given to those with more severe visible injuries (Belanger et al., 2005; Lew, 2005). Though not visible, the primary injury caused by the blast wave leads to severe pathological and neurological sequelae in the brain (Kocsis and Tessler, 2009).
Many different imaging techniques are used to diagnose TBI. Computed tomography (CT) is the imaging method of choice to determine the severity of TBI. CT imaging provides information about the extent of focal injury, but is not capable of detecting diffuse neuronal damage due to low sensitivity and specificity (Kovesdi et al., 2010). Magnetic resonance imaging (MRI), on the other hand, has overcome the limitations of CT, but its use in diagnosing TBI has been limited because of the need for an MRI machine, the time required for scanning, and high cost (Petarakis et al., 2000). Apart from imaging techniques to diagnose TBI, many different serum proteins have been studied as biomarkers for TBI. The most well-known candidates include S100β, glial fibrillary acidic protein (GFAP), and the most recently studied ubiquitin C-terminal hydrolase-1 (UCH-L1; Svetlov et al., 2009). Though many promising protein biomarkers have been tested, none of them have been successfully validated in diagnosing TBI induced by primary blast waves.
MicroRNAs (miRNA) are a class of small (19–28nt) endogenous RNA molecules that regulate gene expression at the post-transcriptional level, either by translational repression or mRNA degradation. MiRNA binds to the complementary sequences in the mRNA and blocks its translation and accelerates mRNA decay (Brown and Naldini, 2009). MiRNAs in serum are highly stable, are resistant to repeated freeze-thaw cycles and enzymatic degradation, and can survive extreme pH conditions. Due to these properties, miRNAs have recently emerged as novel biomarkers for many diseases (Schöler et al., 2010). Further, miRNAs have been implicated as biomarkers in various tissue and organ injuries, including liver, muscle, and spinal cord (Latereza et al., 2009). Recently, miRNA has also been shown to be a biomarker in cases of human TBI. Plasma miRNAs were studied in human cases with severe to mild TBI. In severe injuries, miR-16, miR-92a, and miR-765 were significantly upregulated. However, in cases of mild TBI, miR-16 and miR-92a were significantly downregulated, and no difference was seen in the levels of miR-765 in comparison to controls. Further, the diagnostic accuracy of these markers was better for severe TBI than for mild TBI (Redell et al., 2010).
In this study, we used a rat model to study the effect of blast overpressure (BOP) injury on the expression of serum miRNAs. An air-driven shock tube was used to induce BOP. Animals were exposed to multiple BOP insults of 120 kPa to induce TBI. This intensity of BOP is shown to cause concussion and mild TBI which closely resembles clinical symptoms of a blast injury. Previously, it has been shown that 120-kPa BOP results in increases in oxidative stress, the activation of microglial cells, and blood–brain barrier (BBB) disruption in the brain (Readnower et al., 2010). In this study, we found modulation of miRNAs in the serum and cerebrospinal fluid (CSF) post-BOP injury in animals that were exposed to multiple blasts at different time intervals.
Methods
Animals and injury
Adult, male Spargue-Dawley rats (250–300 g) were used for this study. Six animals for each experimental and control group were used. Care was taken to prevent secondary and tertiary blast injuries by anesthetizing the animals with isoflurane and placing them in a holder as previously described (Chavko et al., 2006). The animals were kept in the end of the expansion chamber of an air-driven shock tube (2.5-ft compression chamber connected to a 15-ft expansion chamber) with the right side ipsilateral to the direction of the BOP. Two injury groups were used. The short interval injury group (SII), in which animals were exposed to three serial BOP of 120 kPa at an interval of 2 h. The other group is the long interval injury group (LII), for which BOP of 120 kPa was given at an interval of 24 h. From each of these groups, serum and CSF samples were collected at 3 h and 24 h after the last BOP exposure. Blood and CSF samples were collected from the control animals at the same time points sample collection was done for the injured animals.
RNA isolation
Total RNA was isolated from serum and CSF samples combining protocols of Trizol LS reagent (Invitrogen, Inc., Carlsbad, CA), and the mirVana miRNA isolation kit (Ambion Inc., Streetsvillle, Missisauga, Canada). Initially 2 volumes of Trizol LS were added to the samples along with 1 volume of chloroform. After centrifugation, the aqueous layer was collected and mixed with 1.25 volumes of absolute ethanol. It was then loaded onto a cartridge provided in the mirVana miRNA isolation kit and total RNA was isolated according to the manufacturer's protocol. RNA quantitation was done using a Nanodrop 2000 spectrophotometer (Thermo Scientific Inc. Pittsburgh, PA).
cDNA synthesis
Reverse transcription (RT) was performed with a TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). The RT reaction mixture contained megaplex pools of stem-loop RT primers (10×) 0.8 μL, 100 mM dNTPs (with dTTP) 0.2 μL, multiscribe reverse transcriptase (50 U/μL) 1.5 μL, 10× RT buffer 0.8 μL, MgCl2 (25 mM) 0.1 μL, RNAse inhibitor (20 U/μL) 0.1 μL, total RNA template (20 ng/μL) 3 μL, and nuclease free water to a final volume of 7.5 μL. The RT reaction was carried out according to the manufacturer's recommendation in a Thermal Cycler TC-3000G (Techne Corp., Minneapolis, MN).
cDNA pre-amplification
The cDNA (2.5 μL) was pre-amplified using megaplex pre-amplification master mix (2×) 12.5 μL, megaplex preamp primers (10×) 2.5 μL, and nuclease free water to a final volume of 25 μL. The pre-amplification reaction was carried out using a Thermal Cycler TC-3000G according to the manufacturer's recommended thermal cycling conditions. Following the pre-amplification, 75 μL of 0.1× TE (pH 8.0) was added to each amplified RT product.
Taqman Low Density array real-time PCR
From the amplified RT product, 6 μL was added to 450 μL of 2× TaqMan Universal PCR Master mix, no AmpErase UNG (Applied Biosystems), and nuclease free water to a final volume of 900 μL. Then 100 μL PCR reaction mix was loaded into each row of the 384-well TaqMan Low Density Rodent MicroRNA array (TLDA). The PCR reaction was carried out at default thermal cycling conditions in an AB7900 Real Time HT machine (Applied Biosystems). High-throughput profiling of 821 miRNAs was carried out using a TaqMan® Rodent microRNA Array Set v2.0 (Applied Biosystems). Real-time PCR data were analyzed using Statminer® (Integromics Inc., Madison, WI). Data were filtered for Ct values <35, and the expressions were normalized to a computationally determined endogenous control which was computed by using three endogenous control miRNAs: mammU6, U87, and Y1.
Specific miRNA real time assay
Twenty nanograms of total RNA isolated from BOP-exposed rat serum and CSF samples were subjected to RT using mmu-let-7i miRNA and endogenous control MammU6-specific primers per the manufacturer's protocol (Applied Biosystems). Briefly, RT was conducted in a final volume of 15 μL, which contained 1.5 μL of 10× RT buffer, 0.19 μL of RNase inhibitor, 0.15 μL of dNTPs with dTTP, 1 μL of multiscribe reverse transcriptase, 3 μL of miRNA-specific stem-loop RT primer, 20 ng of total RNA, and water. RT was carried out on a TC-3000G Thermal Cycler (Techne) at 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. Thereafter, real-time qPCR was performed using a TaqMan MicroRNA assay kit (Applied Biosystems) to quantitate individual miRNAs. In brief, 1.33 μL of 1:10 diluted RT product was combined with 10 μL of TaqMan gene expression master mix, 1 μL of Taqman miRNA assay mix, and water to make a final volume of 20 μL. qPCR was carried out on an ABI 7900 HT qRT PCR instrument (Applied Biosystems) at 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec, and 60°C for 1 min. All qPCR reactions were performed in triplicate. Fold change was calculated using the comparative Ct method.
Bioinformatic analysis
For predicting the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of mRNAs that are targeted by miRNAs altered by BOP, we used a DNA intelligent analysis (DIANA) miRPath algorithm, combined with the miRNA target prediction web tools of DIANA microT 4.0, TargetScan 5, and PicTar (Lewis et al., 2005; Papadopoulos et al., 2009). Network analyses of modulated miRNAs were done using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood, City, CA).
Results
MiRNA expression profile in serum samples of rats exposed to repeated BOP
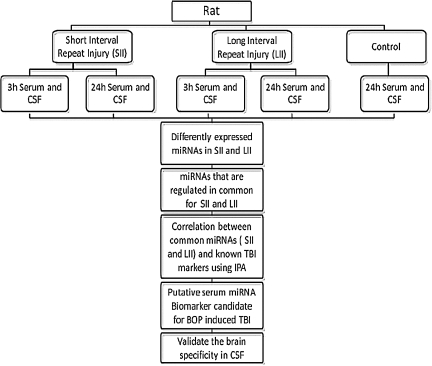
The objective of this study was to identify serum miRNAs as biomarkers of BOP-induced TBI in war fighters. On the battlefield military personnel may be exposed to multiple blasts, which can happen in a short interval of time or over a span of days. An ideal biomarker should be one that has the sensitivity and specificity to predict injury in these scenarios. To address this, in our study we included two injury groups, SII and LII, as shown in Figure 1.
FIG. 1.
Flow chart showing the experimental design of blast overpressure experiments in the rat (CSF, cerebrospinal fluid; IPA, Ingenuity Pathway Analysis).
Serum samples from these groups were analyzed for their miRNA modulation in response to BOP injury. In the case of SII, a total of 123 and 75 miRNAs were dysregulated in samples collected at 3 h and 24 h post-BOP injury, respectively. Among these, 33 miRNAs were common in both of the subgroups. In the case of LII, 17 and 19 miRNAs were dysregulated in serum samples collected 3 h and 24 h after the last BOP injury, respectively (Supplementary Table 1; see online supplementary material at http://www.liebertonline.com). This difference in the number of miRNAs modulated between SII and LII may be attributable to the fact that in the case of LII, the time between the two injuries was 24 h, and this may help in recovery from the trauma. Previously, in a rat model it was shown that increasing the time from 15 min to 4 h between three successive 26-psi blast exposures decreased the mortality rate from 87% to 27%. If the time interval was increased to 1 day, the mortality dropped further to 7% (Stuhmiller et al., 1991). This suggests that increasing the time interval between repetitive blasts allows a physiological recovery time, which may explain the reduced miRNA modulation in the LII group observed in this study. Moreover, many miRNAs were found to be modulated in specific injury groups, whereas others were modulated in multiple injury groups. From a biomarker perspective, miRNAs that show modulation in various injury groups will be the best candidates for a BOP injury biomarker. In our study, we found a total of 47 miRNAs modulated in two or more groups with a significance level of p<0.05.
Further, five candidate miRNAs were selected for analysis, which includes miR-let-7i, miR-122, miR-340-5p, miR-200b*, and miR-874, since these miRNAs were modulated in the serum of three injury groups. No miRNA was found to be significantly modulated in all the injury groups (Table 1). MiR-let-7i, miR-122, and miR-340-5p were upregulated in both SII and LII, whereas miR-874 was downregulated. Mir-200b was upregulated in SII, but was found to be downregulated in one injury subgroup of LII. An important role of these miRNAs in various pathologic conditions of the central nervous system (CNS) has been reported (Supplementary Table 2; see online supplementary material at http://www.liebertonline.com). Among these miRNAs, miR-122 and let-7i were shown to be upregulated in liver and brain, respectively, upon exposure to an RDX blast, whereas miR-200b was downregulated in brain tissue (Zhang and Pan, 2009). Further, members of the let-7 miRNA family are highly enriched in brain, specifically in the hippocampus and frontal cortex (Bak et al., 2008; Lagos-Quintana et al., 2002). These studies suggest that among all the modulated miRNAs in serum post-BOP injury, miR-let-7i may be a good candidate biomarker for BOP injury, since it is upregulated in three injury groups and is highly enriched in brain.
Table 1.
MiRNAs Modulated in Serum of Animals Exposed to BOP Injury in Both SII and LII Groups
| |
|
Fold change (Log 10) |
|
|
Fold change (Log10) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
Blast 2h interval×3 |
Blast 24h interval×3 |
|
|
Blast 2h interval×3 |
Blast 24h interval×3 |
||||
| S# | miRBase (mature) | 3 h serum | 24 h serum | 3 h serum | 24 hserum | S# | miRBase (mature) | 3 h serum | 24 h serum | 3 h serum | 24 h serum |
| 1 | mmu-let-7c-1* | 0.978 | −1.669 | 25 | mmu-miR-31* | 1.797 | 1.485 | ||||
| 2 | mmu-let-7i | 1.560 | 1.881 | 1.574 | 26 | mmu-miR-340-5p | 1.543 | 1.273 | 1.069 | ||
| 3 | mmu-miR-1 | −1.696 | −1.236 | 27 | mmu-miR-34c* | 1.069 | 1.397 | ||||
| 4 | mmu-miR-101a* | 1.861 | 1.128 | 28 | mmu-miR-350 | 1.525 | 1.623 | ||||
| 5 | mmu-miR-101b | 3.077 | 1.492 | 29 | mmu-miR-425* | 2.347 | 1.588 | ||||
| 6 | mmu-miR-106b* | 3.178 | 1.487 | 30 | mmu-miR-429 | 1.317 | −1.822 | ||||
| 7 | mmu-miR-122 | 1.602 | 1.449 | 1.054 | 31 | mmu-miR-449a | 1.821 | 1.055 | |||
| 8 | mmu-miR-130a | 1.677 | 1.698 | 32 | mmu-miR-449c | 2.333 | 1.096 | ||||
| 9 | mmu-miR-140 | 1.161 | 1.132 | 33 | mmu-miR-450a-5p | 2.592 | 2.241 | ||||
| 10 | mmu-miR-142-5p | 1.311 | −1.030 | 34 | mmu-miR-455* | 2.304 | 1.446 | ||||
| 11 | mmu-miR-148a | 1.453 | 1.700 | 35 | mmu-miR-542-3p | 1.283 | 1.126 | ||||
| 12 | mmu-miR-17* | 1.603 | 1.307 | 36 | mmu-miR-652 | 1.148 | 2.317 | ||||
| 13 | mmu-miR-185 | 1.021 | 1.259 | 37 | mmu-miR-744* | 1.638 | 1.536 | ||||
| 14 | mmu-miR-192 | 1.243 | 1.607 | 38 | mmu-miR-7a | 1.554 | 1.366 | ||||
| 15 | mmu-miR-193* | 1.700 | 1.606 | 39 | mmu-miR-802 | 2.113 | 1.493 | ||||
| 16 | mmu-miR-194 | 1.794 | –1.253 | 40 | mmu-miR-872 | 1.620 | 3.198 | ||||
| 17 | mmu-miR-19a | 1.024 | 1.352 | 41 | mmu-miR-93* | 0.882 | 1.274 | ||||
| 18 | mmu-miR-200b* | 1.411 | 1.235 | –1.810 | 42 | rno-let-7e* | 1.989 | 1.245 | |||
| 19 | mmu-miR-20b* | 1.965 | 1.225 | 43 | rno-miR-219-1-3p | 1.077 | 1.571 | ||||
| 20 | mmu-miR-21* | 1.649 | 1.097 | 44 | rno-miR-363* | 1.802 | 1.967 | ||||
| 21 | mmu-miR-26b* | 1.124 | 1.233 | 45 | rno-miR-505 | 1.671 | 1.840 | ||||
| 22 | mmu-miR-29c* | 1.327 | −1.337 | 46 | rno-miR-532-5p | 0.927 | 1.101 | ||||
| 23 | mmu-miR-301a | 1.015 | 1.243 | 47 | mmu-miR-874 | −1.676 | −1.423 | −1.914 | |||
| 24 | mmu-miR-301b | 1.048 | 1.325 | ||||||||
The data were normalized using endogeneous control by using Statminer software (p<0.05).
Highlighted MiRNAs are modulated in three injury groups.
BOP, blast overpressure; miRNA, microRNA; SII, short interval injury; LII, long interval injury.
Validation of miR-let-7i expression
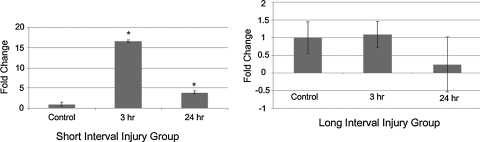
Most of the published studies to quantify miRNAs for the purpose of biomarker development involve the use of real-time PCR. However, due to low serum concentrations of miRNAs, a pre-amplification step is performed, which can potentially introduce bias in later quantitation (Schöler et al., 2010). Hence, we validated miR-let-7i in the serum of animals exposed to short-interval BOP with independent Taqman miRNA assays in which pre-amplification of the cDNA is not required. We found that miR-let-7i expression was significantly upregulated in SII in both the subgroups. In the case of LII, no significant change in the expression of miR-let-7i was observed. Overall these results correlate with the miRNA profiling done using TLDA cards (Fig. 2).
FIG. 2.
Validation of miR-let 7i miRNA in the short interval (SII) and long interval (LII) groups. The levels of miRNA were normalized by the level of MammU6 endogenous control RNA, and all reactions were performed in triplicate. (A) Expression levels of let-7i were significantly upregulated in SII. (B) No significant upregulation of let-7i was observed in both subgroups of LII (*p<0.05).
MiR-Let-7i expression in CSF
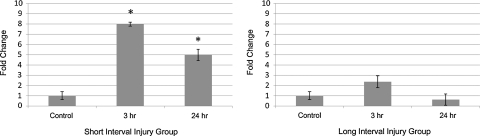
Earlier, it was suggested that a biomarker of brain injury is often detected in serum as well as CSF (Papa et al., 2010), therefore we also analyzed the expression of miR-let-7i in the CSF of the animals exposed to BOP injury. CSF samples from the same group of animals were analyzed using a specific real-time miRNA assay for miR-let-7i. Overexpression of miR-let-7i in CSF samples in the SII group of more than eightfold and fivefold were observed for the 3 h and 24 h groups, respectively (p<0.05), whereas in the LII group only the 3 h injury group showed a twofold increase, which failed to reach significance. The group with injuries 24 h apart, and samples collected 24 h after the last BOP exposure, also did not show any significant increase over the control samples (Fig. 3). This pattern correlates with the expression of miR-let-7i in the serum of these animals, which further supports our hypothesis that increased expression of miR-let-7i in serum and CSF is of CNS origin.
FIG. 3.
Expression of miR-let-7i in both the short interval (SII) and long interval (LII) groups in cerebrospinal fluid (CSF) of rats exposed to blast overpressure (BOP). (A) A significant increase in the expression of miR-let-7i was observed in the SII groups post-BOP exposure. (B) No significant difference was observed in the expression of miR-let-7i in the LII groups (*p<0.05).
Functional pathway analysis of modulated miRNAs
Even though only five miRNAs were found to be modulated in both the SII and LII groups out of a total of 47, the remaining miRNAs may also play important roles in the pathology of BOP injury. Biological pathways are the major functional units that are involved in physiological and pathological processes. A single miRNA can act on several targets, and a single mRNA can be targeted by multiple miRNAs. To determine the role of the modulated miRNAs after BOP injury in different biological pathways, we performed DIANA mirPath analysis of 47 modulated miRNAs (Table 1). Three miRNA prediction web tools, DIANA microT 4.0, TargetScan 5, and PicTar, were used by this program, which identified axon guidance and Wnt signaling as the two topmost KEGG pathways (Table 2). Previously, neuropathology of the brains of the animals exposed to 120-kPa blasts was studied. Diffuse axonal injury was observed, including mechanical shearing of axons, damage to the cytoskeleton, interruption of axoplasmic flow, and calcium influx into axons, resulting in interruption of traffic along these interconnecting pathways. Further, BBB breakdown and oxidative stress were also observed (Readnower et al., 2010; Young, 2010). Wnt signaling has been implicated in the regulation of the BBB, along with the regulation of β-catenin, claudin, occludins, and other tight junction proteins in the endothelial cells that form the blood vessels (Polakis, 2008). Together, the results of the pathway analysis and the evidence from the previous studies indicate that miRNAs modulated in the serum after BOP injury may play an important role in neurological pathways.
Table 2.
Functional Pathway Analysis Using DIANA mirPath Software
| S# | KEGG Pathway-Target Scan | No. of Genes (Union) | -In (p-value) Union | KEGG Pathway microT4 | No. of Genes (Union) | -In (p-value) Union | KEGG Pathway PicTar | No. of Genes (Union) | -In (p-value) Union | KEGG Pathway-Target Scan-PicTar-microT4 | No. of Genes (Union) | -In (p-value) Union |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Axon guidance | 63 | 29.59 | Axon guidance | 67 | 38.55 | Axon guidance | 44 | 24.8 | Axon guidance | 85 | 44.34 |
| 2 | Focal adhesion | 84 | 28.9 | MAPK signaling pathway | 106 | 33.93 | MAPK signaling pathway | 69 | 21.6 | Wnt signaling pathway | 86 | 33.73 |
| 3 | MAPK signaling pathway | 99 | 23.59 | Focal adhesion | 84 | 30.96 | Wnt signaling pathway | 45 | 20.33 | Focal adhesion | 105 | 31.86 |
| 4 | Glioma | 35 | 21.89 | Renal cell carcinoma | 40 | 27.8 | Colorectal cancer | 27 | 12.72 | MAPK signaling pathway | 128 | 29.14 |
| 5 | TGF-β signaling pathway | 43 | 19.15 | Glioma | 36 | 25.13 | Melanogenesis | 29 | 11.46 | Colorectal cancer | 54 | 24.96 |
| 6 | Chronic myeloid leukemia | 38 | 18.55 | Regulation of actin cytoskeleton | 84 | 24.11 | TGF-β signaling pathway | 27 | 11.4 | Regulation of actin cytoskeleton | 106 | 24.86 |
| 7 | Renal cell carcinoma | 35 | 17.16 | Wnt signaling pathway | 63 | 23.1 | mTOR signaling pathway | 18 | 10.1 | Melanogenesis | 58 | 22.01 |
| 8 | Wnt signaling pathway | 59 | 16.67 | Chronic myeloid leukemia | 39 | 21.39 | Insulin signaling pathway | 35 | 9.14 | ErbB signaling pathway | 52 | 21.07 |
| 9 | Adherens junction | 36 | 16.47 | Colorectal cancer | 42 | 20.99 | Renal cell carcinoma | 21 | 8.77 | Glioma | 41 | 20.46 |
| 10 | Regulation of actin cytoskeleton | 78 | 16.13 | ErbB signaling pathway | 41 | 18.79 | ErbB signaling pathway | 24 | 8.26 | Chronic myeloid leukemia | 47 | 20.37 |
For all the significantly modulated microRNAs (miRNAs), the DNA intelligent analysis (DIANA) mirPath algorithm combined with miRNA target prediction web tools of DIANA microT 4.0, TargetScan 5, and PicTar was performed. The results represented here indicate the top predicted biological functions of modulated miRNAs.
KEGG, Kyoto Encyclopedia of Genes and Genomes.
MiR-Let-7i may regulate important TBI-related proteins
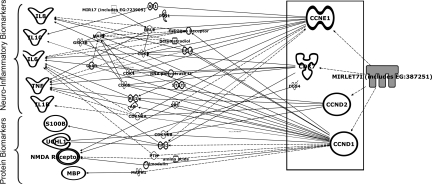
The DIANA mirPath analysis for all the modulated miRNAs indicated their role in neurological processes. We further specifically studied miR-let-7i for its role in the regulation of neurological pathways. We used IPA network analysis software and made a network using all of the important TBI-related proteins and correlated it with miR-let-7i gene targets. The analysis showed that miR-let-7i may regulate many proteins and inflammatory cytokines, including S100β and UCH-L1, which are currently proposed as candidate protein biomarkers for TBI through intermediate downstream molecules (Fig. 4).
FIG. 4.
Functional interaction networks of known TBI-related protein biomarkers and inflammatory molecules predicted to be regulated targets of miRNA let-7i miRNA as predicted by the Ingenuity Pathway Analysis program (NMDA, N-methyl-D-aspartate; UCHL-1, ubiquitin C-terminal hydrolase-1; TNF-α, tumor necrosis factor-α; IL-1β, interleukin-1β; IL-6, interleukin-6; IL-8, interleukin-8).
Discussion
Biomarker development often uses two approaches: “top-down” and “bottom-up” (Dash et al., 2010). Most commonly a top-down approach has been used for biomarker discovery. In this method, a biomarker, commonly a protein, will be identified in the injured brain, and then its presence in peripheral body fluids is evaluated. Depending on its level of expression, its utility as a diagnostic marker is determined, which might be lessened if its expression is too low. In this study, we have followed a bottom-up approach, in which a biomarker in the peripheral fluid is first determined, and then is correlated with the pathology of brain injury. A collective signature of these biomarkers may help in diagnosing a condition such as brain injury.
Serum-based biomarkers are valuable for the early diagnosis of TBI due to the non-invasive method of sample collection. Many serum proteins have been studied for developing a diagnostic biomarker for TBI. S100β is a small dimeric calcium-binding protein that has been extensively studied as a TBI biomarker. It is abundantly expressed in glial cells of the CNS, and is detected in serum following TBI (Geyer et al., 2009). Another promising candidate is GFAP, a filamentous protein found only in the astroglial cytoskeleton. Although GFAP has shown to be a good biomarker for severe TBI, no significant changes have been detected in GFAP expression in cases of mild TBI (Pelinka et al., 2004). Recently, UCH-L1, a neuronal marker, has been identified as a biomarker for severe TBI. Elevated levels of UCH-L1 were detected in serum and CSF samples in patients with severe TBI (Papa et al., 2010). High levels of UCH-L1 were detected in serum and CSF of rats exposed to severe BOP injury (Svetlov et al., 2010). Despite the numerous studies on the development of protein-based biomarkers for TBI, none of the candidates have been successfully used in the clinic to diagnose patients.
Recently, miRNAs have emerged as novel diagnostic biomarkers for various diseases, including neurodegenerative disorders like Alzheimer's disease, Parkinson's disease, Huntington's disease, and schizophrenia (Barbato et al., 2009; Lukiw et al., 2008). Further, miRNAs have also been implicated as circulating biomarkers in tissue injury, including liver, muscle, and brain (Laterza et al., 2009).
The systemic effects of the level of blast (∼120 kPa) used in the present study were previously characterized and were shown to produce acute reflex suppression in rats, which is consistent with concussion and mild TBI, and closely resembles the clinical manifestations of blast-induced TBI (Bruns and Jagoda, 2009; Jones et al., 2007). After exposure to 120-kPa BOP, a substantial reactive astrocytosis was observed in the cerebellum and hippocampus as early as 3 h post-blast (Cullen et al., 2011). Widespread increased expression of GFAP and inflammation, including microglial activation and neutrophil infiltration, was observed in the cerebrum, hippocampus, and cortex region (Cullen et al., 2011; Readnower et al., 2010). In our experiments, we used two different groups for comparing the short-term and long-term effects of multiple BOP injury. We found more miRNAs modulated in the SII groups than in the LII groups. This can be correlated with the recent study by Readnower and associates (2010), who demonstrated that the maximum BOP injury severity is visible immediately after injury, and that the animals show signs of recovery by the end of third day. However, we did not observe any increase in the number and extent of miRNA modulation between 3 and 24 h post-injury.
Another important factor to keep in mind when studying biomarkers for TBI is whether the biomolecule is modulated directly in response to the injury, or is a secondary effect of the injury. A promising biomarker candidate will be one that is not only detected for an extended period of time after injury, but also appears immediately after the injury. Keeping these criteria in mind, we analyzed the data and selected five miRNAs to study (miR-let-7i, miR-122, miR-200b*, miR-340-5p, and miR-874). We searched the literature for the expression and role of these miRNAs and found that only miR-let-7i is highly enriched in brain and is overexpressed upon blast exposure. Mir-122 is also upregulated following blast, but has been shown to be liver-specific. Based on these observations it appeared that increased expression of miR-let-7i was a direct effect of injury to the brain, and therefore it may be a good candidate for further studies. The next step was to see if its expression was increased in CSF of the animals, since most of the neuronal markers that are detected in serum are also suggested to be present in CSF (Papa et al., 2010). The increased expression of miR-let-7i in the CSF samples correlated with that of serum, which further indicated that expression of miR-let-7i was caused from brain injury.
To further validate this hypothesis, we selected the existing TBI biomarker candidates, such as protein and neuroinflammatory cytokine biomarkers (Kövesdi et al., 2010; Svetlov et al., 2009), and the five aberrant miRNAs in rat serum that are expressed in both SII and LII BOP exposures. These were added to “my pathway workflow” in Ingenuity Pathway Analysis to build customized pathways. Among the five aberrant miRNAs, only miR-let-7i showed involvement in the regulatory pathways of several proteins and neuroinflammatory cytokine biomarkers.
Recently, Redell and colleagues (2010) have shown miRNA modulation in a patient with TBI. Five miRNAs, miR-16, miR-26a, miR-92a, miR-638, and miR-765, were modulated in severe TBI patient samples. In our study, we did not observe significant modulation of these miRNAs in any of the injury groups. This may be because the type of injury was different in our study in comparison to the patient study, in which there was no BOP exposure. This suggests that more studies are required to understand whether a single miRNA signature can be used as a biomarker for different types of TBI.
Some molecular biomarkers are released very early after brain injury, while others may not appear until 24–48 h after the injury. The biomarkers that are released very early after TBI will be the most relevant to the early diagnosis, particularly for the field diagnosis of mild and moderate TBI. In this study, we have for the first time shown modulation of miRNAs in serum and CSF in response to BOP injury. Specifically, miR-let-7i appeared at elevated levels in the serum as early as 3 h post-injury, and was upregulated in the SII groups. Moreover, we found elevated levels of miR-let-7i in the CSF samples in the SII groups, whereas a modest upregulation was also observed in the LII groups. Together, these indicate that miR-let-7i can be used as a biomarker for the early detection of blast-induced TBI. Validation of its role may lead to new therapeutic interventions for TBI involving miR-let-7i.
To our knowledge this is the first study to report miR-let-7i as a biomarker for blast-induced TBI. We are currently investigating the miRNA expression pattern in the brains of animals with BOP injury, which will help in correlating serum and CSF miRNA expression with the pathology of BOP injury. We are also planning to investigate if miR-let-7i is also upregulated in a weight-drop model and a CCI model of TBI. These studies will help to establish the specificity of miR-let-7i for blast-induced TBI and other models of TBI.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Defense Medical Research and Development Program. The authors are also thankful to Dr. Anuj Sharma for critically reviewing the manuscript, and Dr. Deepti Parashar for her help in real-time PCR assays.
Author Disclosure Statement
The opinions expressed here are those of the authors and should not be construed as official or reflecting the views of the Uniformed Services University of Health Sciences, Bethesda, Maryland; the Naval Medical Research Center, Silver Spring, Maryland; and the Birla Institute of Technology and Science, Pilani, Rajasthan, India. No competing financial interests exist.
References
- Bak M. Silahtaroglu A. Moller M. Christensen M. Rath M.F. Skryabin B. Tommerup N. Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato C. Ruberti F. Cogoni C. Searching for MIND: microRNAs in neurodegenerative diseases. J. Biomed. Biotechnol. 2009;87:1313. doi: 10.1155/2009/871313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger H.G. Scott S.G. Scholten J. Curtiss G. Vanderploeg R.D. Utility of mechanism-of-injury-based assessment and treatment: Blast Injury Program case illustration. J. Rehabil. Res. Dev. 2005;42:403–412. doi: 10.1682/jrrd.2004.08.0095. [DOI] [PubMed] [Google Scholar]
- Brown B.D. Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat. Rev. Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- Bruns J.J., Jr. Jagoda A.S. Mild traumatic brain injury. Mt. Sinai J. Med. 2009;76:129–137. doi: 10.1002/msj.20101. [DOI] [PubMed] [Google Scholar]
- Chavko M. Prusaczyk W.K. McCarron R.M. Lung injury and recovery after exposure to blast overpressure. J. Trauma. 2006;61:933–942. doi: 10.1097/01.ta.0000233742.75450.47. [DOI] [PubMed] [Google Scholar]
- Cullen D.K. Browne K.D. Xu Y. Adeeb S. Wolf J.A. McCarron R.M. Yang S. Chavko M. Smith D.H. Blast-induced change in photonic crystals corresponds with brain pathology. J. Neurotrauma. 2011;28:2307–2318. doi: 10.1089/neu.2011.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash P.K. Zhao J. Hergenroeder G. Moore A.N. Biomarkers for the diagnosis, prognosis, and evaluation of treatment efficacy for traumatic brain injury. Neurotherapeutics. 2010;7:100–114. doi: 10.1016/j.nurt.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder G.A. Cristian A. Blast-related mild traumatic brain injury: mechanisms of injury and impact on clinical care. Mt. Sinai J. Med. 2009;76:111–118. doi: 10.1002/msj.20098. [DOI] [PubMed] [Google Scholar]
- Geyer C. Ulrich A. Grafe G. Stach B. Till H. Diagnostic value of S100B and neuron-specific enolase in mild pediatric traumatic brain injury. J. Neurosurg. Pediatr. 2009;4:339–344. doi: 10.3171/2009.5.PEDS08481. [DOI] [PubMed] [Google Scholar]
- Jones E. Fear N.T. Wessely S. Shell shock and mild traumatic brain injury: a historical review. Am. J. Psychiatry. 2007;164:1641–1645. doi: 10.1176/appi.ajp.2007.07071180. [DOI] [PubMed] [Google Scholar]
- Kennedy J.E. Jaffee M.S. Leskin G.A. Stokes J.W. Leal F.O. Fitzpatrick P.J. Posttraumatic stress disorder and posttraumatic stress disorder-like symptoms and mild traumatic brain injury. J. Rehabil. Res. Dev. 2007;44:895–920. doi: 10.1682/jrrd.2006.12.0166. [DOI] [PubMed] [Google Scholar]
- Kocsis J.D. Tessler A. Pathology of blast-related brain injury. J. Rehabil. Res. Dev. 2009;46:667–672. doi: 10.1682/jrrd.2008.08.0100. [DOI] [PubMed] [Google Scholar]
- Kövesdi E. Luckl J. Bukovics P. Farkas O. Pal J. Czeiter E. Szellar D. Doczi T. Komoly S. Buki A. Update on protein biomarkers in traumatic brain injury with emphasis on clinical use in adults and pediatrics. Acta Neurochir. (Wien.) 2010;152:1–17. doi: 10.1007/s00701-009-0463-6. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M. Rauhut R. Yalcin A. Meyer J. Lendeckel W. Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Laterza O.F. Lim L. Garrett-Engele P.W. Vlasakova K. Muniappa N. Tanaka W.K. Johnson J.M. Sina J.F. Fare T.L. Sistare F.D. Glaab W.E. Plasma microRNAs as sensitive and specific biomarkers of tissue injury. Clin. Chem. 2009;55:1977–1983. doi: 10.1373/clinchem.2009.131797. [DOI] [PubMed] [Google Scholar]
- Leung L.Y. VandeVord P.J. Dal Cengio A.L. Bir C. Yang K.H. King A.I. Blast related neurotrauma: a review of cellular injury. Mol. Cell Biomech. 2008;5:155–168. [PubMed] [Google Scholar]
- Lew H.L. Rehabilitation needs of an increasing population of patients: Traumatic brain injury, polytrauma, and blast-related injuries. J. Rehabil. Res. Dev. 2005;42:xiii–xvi. [PubMed] [Google Scholar]
- Lewis B.P. Burge C.B. Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Ling G. Bandak F. Armonda R. Grant G. Ecklund J. Explosive blast neurotrauma. J. Neurotrauma. 2009;26:815–825. doi: 10.1089/neu.2007.0484. [DOI] [PubMed] [Google Scholar]
- Lukiw W.J. Zhao Y. Cui J.G. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J. Biol. Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa L. Akinyi L. Liu M.C. Pineda J.A. Tepas J.J., 3rd. Oli M.W. Zheng W. Robinson G. Robicsek S.A. Gabrielli A. Heaton S.C. Hannay H.J. Demery J.A. Brophy G.M. Layon J. Robertson C.S. Hayes R.L. Wang K.K. Ubiquitin C-terminal hydrolase is a novel biomarker in humans for severe traumatic brain injury. Crit. Care Med. 2010;38:138–144. doi: 10.1097/CCM.0b013e3181b788ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos G.L. Alexiou P. Maragkakis M. Reczko M. Hatzigeorgiou A.G. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–1993. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- Paterakis K. Karantanas A.H. Komnos A. Volikas Z. Outcome of patients with diffuse axonal injury: the significance and prognostic value of MRI in the acute phase. J. Trauma. 2000;49:1071–1075. doi: 10.1097/00005373-200012000-00016. [DOI] [PubMed] [Google Scholar]
- Pelinka L.E. Kroepfl A. Schmidhammer R. Krenn M. Buchinger W. Redl H. Raabe A. Glial fibrillary acidic protein in serum after traumatic brain injury and multiple trauma. J. Trauma. 2004;57:1006–1012. doi: 10.1097/01.ta.0000108998.48026.c3. [DOI] [PubMed] [Google Scholar]
- Polakis P. Formation of the blood-brain barrier: Wnt signaling seals the deal. J. Cell Biol. 2008;183:371–373. doi: 10.1083/jcb.200810040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Readnower R.D. Chavko M. Adeeb S. Conroy M.D. Pauly J.R. McCarron R.M. Sullivan P.G. Increase in blood-brain barrier permeability, oxidative stress, and activated microglia in a rat model of blast-induced traumatic brain injury. J. Neurosci. Res. 2010;88:3530–3539. doi: 10.1002/jnr.22510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redell J.B. Moore A.N. Ward N.H., 3rd. Hergenroeder G.W. Dash P.K. Human traumatic brain injury alters plasma microRNA levels. J. Neurotrauma. 2010;27:2147–2156. doi: 10.1089/neu.2010.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risdall J.E. Menon D.K. Traumatic brain injury. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:241–250. doi: 10.1098/rstb.2010.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler N. Langer C. Dohner H. Buske C. Kuchenbauer F. Serum microRNAs as a novel class of biomarkers: a comprehensive review of the literature. Exp. Hematol. 2010;38:1126–1130. doi: 10.1016/j.exphem.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Stuhmiller J.H. Phillips Y.Y. Richmond D.R. The physics and mechanisms of primary blast injury. In: Zajtchuk R., editor; Quick C.M., editor. Conventional Warfare: Ballistic Blast and Burn Injuries. Washington, DC: TMM Publications; 1991. pp. 241–270. [Google Scholar]
- Svetlov S.I. Larner S.F. Kirk D.R. Atkinson J. Hayes R.L. Wang K.K. Biomarkers of blast-induced neurotrauma: profiling molecular and cellular mechanisms of blast brain injury. J. Neurotrauma. 2009;26:913–921. doi: 10.1089/neu.2008.0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlov S.I. Prima V. Kirk D.R. Gutierrez H. Curley K.C. Hayes R.L. Wang K.K. Morphologic and biochemical characterization of brain injury in a model of controlled blast overpressure exposure. J. Trauma. 2010;69:795–804. doi: 10.1097/TA.0b013e3181bbd885. [DOI] [PubMed] [Google Scholar]
- Young G.B. Traumatic brain injury: the continued quest for early prognostic determination. Crit. Care Med. 2010;38:325–326. doi: 10.1097/CCM.0b013e3181bfeb89. [DOI] [PubMed] [Google Scholar]
- Zhang B. Pan X. RDX induces aberrant expression of microRNAs in mouse brain and liver. Environ. Health Perspect. 2009;117:231–240. doi: 10.1289/ehp.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.