Abstract
Objective
Left ventricular hypertrophy (LVH) is a highly prevalent and robust predictor of cardiovascular morbidity and mortality. Existing studies have finely detailed mechanisms involved with its development, yet clinical translation of these findings remains unsatisfactory. We propose an alternative strategy focusing on mechanisms of LVH regression rather than its progression and hypothesize that LVH regression is associated with a distinct genomic profile
Methods
Minimally-invasive transverse arch banding and debanding (or their respective sham procedures) were performed in C57Bl6 male mice. LVH was assessed physiologically by transthoracic echocardiography, structurally by histology, and molecularly by real-time PCR. Mouse hearts were genomically analyzed with Agilent mouse 44k developmental gene chips.
Results
Compared to controls, animals banded for 28 days developed a robust hypertrophic response by heart weight/body weight ratio, histology, echocardiography, and fetal gene expression. These parameters were reversed within 1 week of debanding. Whole genome arrays on LV tissue revealed 288 genes differentially expressed during progression, 265 genes differentially expressed with regression, and only 23 genes shared by both processes. Signaling-related expression patterns were more prevalent with regression rather than the structural-related patterns associated with LVH progression. In addition, regressed hearts showed comparatively more changes in energy metabolism and protein production.
Conclusions
This study demonstrates an effective model for characterizing LVH and reveals that regression is genomically distinct from its development. Further examination of these expression profiles will broaden our understanding of LVH and provide a novel therapeutic paradigm focused on promoting regression of LVH, not just halting its progression.
Left ventricular hypertrophy (LVH) affects approximately 25% of adult Americans and is most often associated with pressure overload as seen with essential hypertension, aortic stenosis and ischemic heart disease.1 LVH is highly morbid and is a robust risk factor for cardiovascular mortality.2 Yet, despite optimal blood pressure control, treated hypertensive patients rarely achieve more than a 15–20% reduction in LV mass. Furthermore, nearly half of these patients will continue to have LVH progression and increased cardiovascular events, including death.3 Thus, there remains a great need to expand our understanding and treatment of LVH.
To date, most experimental and therapeutic approaches addressing pressure-induced LVH have been directed at its progression. Numerous studies have profiled the genomic response to aortic banding at multiple time points,4 in different chambers,5 and in different genders.6 Collectively, these types of studies have exquisitely detailed and identified mechanistic targets that may be blocked to delay or retard LVH progression,7, 8 but have not been effective in advancing therapy for existing disease. As opposed to the plethora of studies focusing on LVH development, few studies have attempted to characterize independent features of LVH regression. Friddle and colleagues first detailed genomic differences with LVH regression using nascent gene array technology following administration and withdrawal from beta-adrenergic agonists.9 Although no significant physiologic differences between groups were discerned, 8 genes unique to the drug removal were identified. Despite only 4,000 genes on the array platform, this study revealed that gene expression differences between groups may exist in the absence of detectable physiologic changes, and that array technology might be sensitive enough to highlight these differences.
We and others have demonstrated that removal of a previously placed constrictive aortic band can allow reversal of LVH.10, 11, 12 Similarly, studies of human patients with LVH that have aortic valve replacement for aortic stenosis are highly successful in causing LVH regression.13 It remains unknown, however, if LVH regression is truly a unique process and thus a target for focused study and intervention. Accordingly, we sought to determine if gene expression associated with physiologic LVH regression was not simply the reverse of those genes associated with LVH progression, but rather a distinct genomic pattern.
MATERIALS AND METHODS
Surgical Model and Experimental Design
In accordance with both an institutionally approved IACUC protocol and NIH guidelines, 10-week-old C57Bl6 male mice were randomly assigned to one of 4 groups: Sham (S), Band (B), Sham Deband (SDB), and Deband (DB). The 60 mice enrolled were partitioned as follows: 10 S, 15 B, SDB 13, and DB 21. Mice in the Sham and Band groups underwent minimally invasive transverse aortic banding as previously described.10 Briefly, animals were anesthetized using inhaled isoflurane by facemask. A midline neck incision was used to approach the anterior mediastinum. The transverse arch was identified and a constrictive band was placed and tightened to the approximate diameter of a 27 gauge needle. The only difference between sham and band groups was that in the sham animals, the constrictive band was not tightened. Adequate placement of the band was verified by evaluation of carotid Dopplers both before and after placement of the aortic band. Adequate banding was accepted when the Doppler velocity ratio doubled from right to left carotid arteries. Animals in the Deband and Sham Deband had initial procedures identical to those for the Band and Sham animals, respectively. At 4 weeks, the aortic band was removed, heretofore referred to as debanded.10 Efficiency of debanding was verified by carotid Doppler with normalization of carotid velocities. Deband and Sham Deband animals were sacrificed one week following the deband procedure for all evaluations except histology, where animals were sacrificed both at 1 and 4 weeks after debanding.
Transthoracic echocardiography
Transthoracic echocardiography was performed using the Vevo 660 High Resolution Biomicroscopy System equipped with a 30 Mhz transducer (Visual Sonics, Toronto, CA). During examination, mice were anesthetized with 1–1.5% inhaled isoflurane. Depth of anesthesia was standardized by recording images at heart rates of 480–520 bpm. Images were recorded in all animals before surgery, at 2 and 4 weeks after banding, and at 1, 2, 3, and 4 weeks after removal of aortic constriction (debanding). Two technicians, blinded to the animals’ experimental status, performed exams and measurements.
Tissue Procurement
At time of sacrifice, hearts were rapidly excised and the left ventricular apices were sectioned and flash frozen in liquid nitrogen for gene array analysis. Additional animals allocated to histological analysis were perfused with PBS followed by 10% Formalin, fixed overnight in Formalin, and then processed for histological analysis using periodic acid Schiff staining.14 Cardiomyocyte cross-sectional area was measured using ImageJ (v. 1.38j).15
RNA Preparation, MicroArray Process, and Real-Time PCR
To minimize variation between mice within a tested group and to enhance detection of variation between experimental groups, semi-pooled groups of mice were created. For each experimental group, 9 mice were selected from a group of up to 22 based on echocardiographic criteria of both progression with banding (increase in LVmass of >20%) and regression with debanding (decrease in LVmass of >10%). Within these groups, subgroups of 3 animals were randomly pooled so that 3 samples would be created for each experimental group.
LV apices were homogenized in 0.5ml of ice cold Trizol solution (Sigma) using a bead mill homogenizer (Retsch, Newtown, PA). RNA was then isolated using the standard trizol procedure with additional steps for removal of DNA and fibrous tissue. Purity of RNA was verified by using a 260/280 ratio ≥1.95. An Agilent BioAnalyzer 2100 instrument was used to verify RNA integrity for all samples. 500 ng of total RNA was labeled with Cyanine-5 CTP in a T-7 transcription reaction using the Agilent Low Input Linear RNA Amplification/Labeling System. Labeled cRNA from samples was then hybridized to Agilent mouse 44k developmental microarray slides in the presence of equimolar concentrations of Cyanine-3 CTP labeled mouse reference RNA prepared from pools of 1-day-old mouse pups. Real-time PCR was performed using Taqman primers and probes (Applied Biosystems International (ABI), Foster City, CA).
Western blot
Protein fractions were isolated in ice cold lysis buffer during dounce homogenization. Concentrations were determined using the Bradford assay. Protein fractions were denatured in loading buffer. 30 μg of each sample was then loaded into alternating lanes for gel electrophoresis. Membrane transfer was performed overnight and rabbit anti-mouse antibody was used to probe for Plk1 and Hpcal1. GAPDH was used as the loading control.
Statistical Methods
All physiologic data are presented as mean ± standard error (SE) except where noted. Real-time PCR data were log transformed prior to comparison. All comparisons of physiologic data were performed using 2-tailed, type 3 or type 1 t-tests. Microarray data (N=12 arrays) were Loess normalized and probes were filtered for features having a normalized intensity of <30 aFU in either red or green channels. Probes were removed if data was not present in at least 9 of 12 samples. Missing data points were imputed (to facilitate further statistical comparisons) using the k nearest neighbors algorithm (k=2). Samples were then standardized (μ=0, σ=1) using a custom Perl script (ActiveState Perl 5.8.1, build 807, release date 2003-11-6). Differences in expressed genes were validated using significance analysis of microarrays (SAM) with a false discovery rate of 5%.
Linear models for microarray (Limma)16 was used to model the variation in all data sets, and perform the following contrasts: Band vs. Sham, Deband vs. Sham Deband, and (Deband –Sham Deband) vs. (Band – Sham). These contrasts were executed using custom scripts written in the R statistical language and environment (“R”; Version 2.2.1, build r36812, release date 2005-12-20). Genes were identified as significant in each contrast with the criteria P ≤0.01 and mean fold change ≥ 1.2.
Unsupervised gene and array clustering was performed for each comparison: Band vs. Sham, Deband vs. Band, and Deband vs. Sham Deband. Data for each gene being compared were filtered for 3 instances of mean fold change ≥2. Data were further adjusted by median centering the genes. Average linkage hierarchical clustering using centered correlation similarity metric for both genes and arrays was performed using Gene Cluster 3.0 (Michiel de Hoon, University of Tokyo, Human Genome Center, 2006). Data were then visualized using Java TreeView (version 1.0.13).
Gene ontologies of significant genes in each contrast were identified using the NIH curated DAVID (Database for Annotation, Visualization, and Integrated Discovery) 2007.17 To facilitate interpretation of gene ontology, functional clustering was performed and results were rank ordered by DAVID using enrichment analysis, and were further organized according to the number of genes in that cluster relative to the total number of genes identified within a particular contrast. The primary advantage of the DAVID technique is its ability to take large lists of genes and identify the biological processes and functions that are most important (within the set of significant genes) to the biological phenomenon under study. Orthogonal filtering of data sets was performed using the Significance Analysis of Functional Expression (SAFE)18 using custom R scripts. In this method, also called orthogonal filtering, all genes are first organized based on their gene ontologies, or GO categories. Each ontologic category is then evaluated based on the degree of variation of individual genes within that category, regardless of the significance of individual genes. The more randomly the genes within each ontology are distributed, the less likely that that ontology is contributing to the observed differences in the experimental groups. Ontologies are then ranked based on their relative significance to the process under study. Essentially, the entire gene array is used to generate meaningful information about the process under study, rather than just a few dozen genes that are identified as significantly different. Gene ontologies with a P-value of 0.1 or less were identified in all contrasts.
RESULTS AND DISCUSSION
Regression of pressure-induced LVH
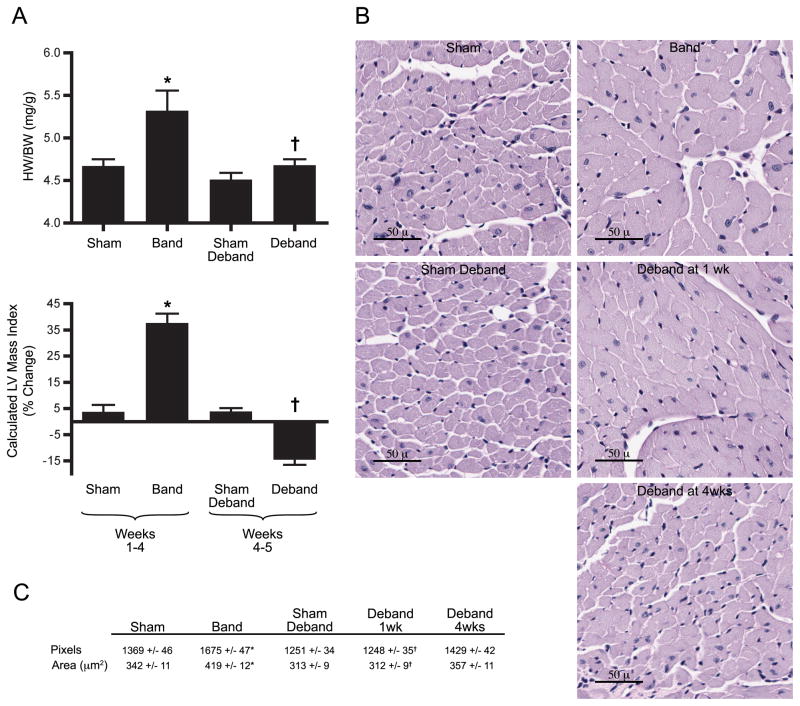
Sixty mice were used in this study: 2 died perioperatively and 2 were excluded with wound infections. Our technique of minimally-invasive banding and debanding effectively produced LVH with subsequent regression. Doppler velocity ratios between right and left carotid arteries verified creation and relief of arch obstruction as previously demonstrated.10 Grossly, heart-weight to body-weight ratios similarly increased with banding and normalized after debanding (Figure 1A). Echocardiographically, aortic constriction resulted in increased wall thickness, greater chamber dimensions, and significantly greater LV mass (Figure 1A, Supplementary Data, Table 1). One week following band removal, most parameters changed in the direction of baseline, with a favorable trend noted in fractional shortening. Histologically, cardiomyocyte cross sectional areas increased approximately 20% following banding, and declined significantly following debanding (Figure 1B, C). To determine the corresponding molecular changes, three common genetic expression markers of ventricular hypertrophy were evaluated using real-time PCR, including beta myosin heavy chain, natriuretic peptide type B, and skeletal muscle α1-actin (n=6–8). All were significantly upregulated in the banded animals and normalized in debanded mice. For example, natriuretic peptide levels increased 65.4 ± 25.4 fold with banding (sham 1.0 ± 0.2 fold, p<0.05) and decreased with debanding (2.9 ± 0.8, versus banding p<0.05). Taken together, these data comprehensibly demonstrate a reproducible model to study mechanisms independently associated with LVH regression.
Figure 1.
Physiologic and structural evidence of LVH reversal. (A) Heart weight/body weight ratios (n=11–22 per group) and serial echocardiographic measurements showing changes over depicted time period (n=11–22 per group) (abbreviations in Supplement). (B) Left ventricle cross-sections stained with PAS. (C) Calculated cardiomyocyte cross-sectional areas (n=4 per group). *p<0.05 vs. Sham, †p<0.05 vs. Band.
Identification and clustering of significant genes
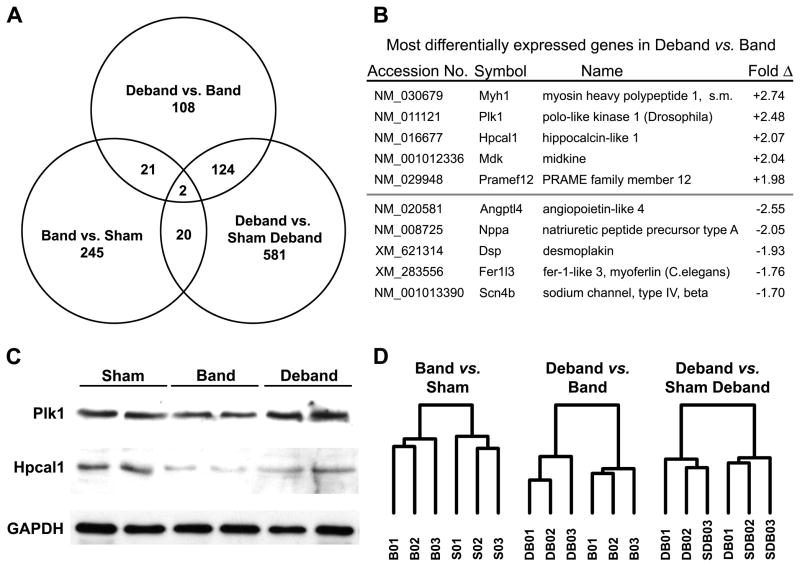
In order to minimize variation in for our array experiments, we used echocardiography to select 9 animals from a larger starting cohort of mice for each group (n=11–22). The sensitivity of the array technology demanded that we study the best representative animals for each group. Furthermore, at the time of these studies, it was cost prohibitive to do individual arrays for each animal. Nevertheless, changes in LV mass index were not significantly different between the full and array data sets. i.e. %Δ Band (total, n=17) 35.2 ± 4.3 compared to Band (array, n=9) 42.6 ± 5.0, p=0.29 and the %Δ Deband (total, n=22) −9.8 ± 1.9 compared to Deband (array, n=9) −15.1 ± 1.6, p=0.09. After preliminary filtering and standardization, a total of 14,693 distinct transcripts were identified in our heart tissue and analyzed using linear models for microarray (Limma). In the regression contrast (Deband vs. Band), we identified 255 differentially expressed genes: 92 were upregulated, and 133 were downregulated. Contrasts for LVH progression (Band vs. Sham) and for the regressed state (Deband vs. Sham Deband) identified 288 and 727 differentially expressed genes respectively (Figure 2A). During LVH progression, there were 158 upregulated and 130 downregulated genes, and in the Deband vs. Sham Deband contrast there were 244 upregulated and 483 downregulated (detailed profiles for each contrast provided in Supplemental Data, Table 2).
Figure 2.
Gene expression associated with LVH regression. (A) Venn diagram depicts different and shared genes in each experimental contrast (p<0.01 and absolute fold change >1.2). (B) Most differentially expressed genes in the regression contrast (Deband versus Band). (C) Western Blot for selected proteins identified in the Deband vs. Band comparison. (D) Unsupervised gene clustering between groups (B, band; S, sham; DB, deband; SDB, sham-deband).
Interestingly, we identified only 23 genes that were significantly differentially expressed in both LVH progression and regression. Conversely, 108 genes were distinctly associated with regression. In examining the most differentially upregulated genes in regression, there were 2 cell cycle control genes, polo-like kinase 1 (Plk1) and midkine (Mdk), a myosin peptide not previously associated with the heart (Myh1), the neural conduction protein Hpcal1, and Pramef12, a protein with homology to a melanoma associated protein (Figure 2B). Of these, only midkine has previously been associated with cardiovascular disease.19 Western blot analysis confirmed the relationship between our gene array and protein expression for Plk1 and Hpcal 1 (Figure 2C). Among the most downregulated genes in regression, only the trends in Nppa expression were consistent with our prior understanding of LVH, and changes in expression of the remaining genes were unique to this experiment.
Although we identified changes in expression of individual genes, we sought to determine if LVH regression was associated with a distinct genomic pattern. Indeed, unsupervised clustering of the samples in each comparison showed clear separation of Band and Sham groups, and of the Deband and Band groups (Figure 2D). The separation was less distinct in the Deband vs. Sham Deband comparison, with one sample from Deband and one sample from Sham Deband not clustering appropriately. This observation indicates that the expression patterns of the Deband and Sham Deband groups were less significant than might otherwise have been predicted based on individual gene differences originally identified by Limma. Taken together, the global expression pattern depicted in the unsupervised clustering suggests that differences between groups were not limited to a few hundred genes, but rather several thousand, and those differences were consistent from one group of animals to another.
Gene expression profiles associated with LVH regression
In the LVH progression analysis, gene ontology clustering of our most significant genes showed expected characteristics of hypertrophic stress, including a large focus on actin, microtubules, cytoskeletal integrity, and contractile function, with the remainder including organelle production, transport, and organization (Table 1; Supplementary data, Figure 1). Similarly designed gene array studies of LVH progression (in mice) have identified these same genomic themes.5 In the regression analysis, none of these cytoskeletal or contractile clusters were differentially expressed. Instead, there was a predominance of intra- and extra-cellular signaling themes. Further breakdown of the functional clusters involved in the regression process indicated that cell growth, morphogenesis, and cell cycle were all highly upregulated, while metabolism, and protein and nucleotide production, were downregulated. Lastly, the regressed state (DB vs. SDB) had 126 overlapping genes that demonstrated mostly activity in metabolism and in mitochondria and other organelles. This is markedly different from the patterns associated with either LVH progression or regression. This latter observation suggests that although the debanded hearts appear structurally similar to sham-debanded hearts, they remain genomically distinct.
Table 1.
Gene ontology clustering of significant genes between respective groups as assessed by DAVID 2007 and listed in order of prevalence.
| Band vs Sham | Deband vs Band | Deband vs Sham Deband |
|---|---|---|
| Organelle Production | Protein glycosylation | Cellular metabolism |
| Intracellular transport | Extracellular signaling | Protein metabolism |
| Actin/cytoskeletal organization | Glycosaminoglycan binding | Organelle organization |
| Contractile protein binding | Cell adhesion | Intracellular transport |
| Nucleotide binding | Cell cycle control | Ion binding |
We next evaluated all 14,693 expressed genes using the orthogonal filtering methodology SAFE in order to identify significant gene ontologies and gene set enrichment analysis (GSEA) categories that were relevant to the processes of both LVH progression and LVH regression. The SAFE analysis is particularly useful as it gathers information from all genes in the experiment, not merely ontologies characterized by statistically significant genes. In regression, there were 52 gene ontologies and 34 GSEA categories with p≤0.1 (Supplementary Data, Table 3). Importantly, only 5 gene ontologies were identified as common to both LVH progression and regression (Supplementary Data, Table 4). These data further distinguish LVH regression as an independent process. Taken together, the DAVID and SAFE ontologic analysis corroborate differences identified at the individual gene level and demonstrate that LVH regression is associated with a distinct thematic pattern of gene expression.
Commentary
Our results compare favorably to a recently published study that detailed gene expression by microarray in rats following transverse arch debanding.12 Although they identified 52 regression-related genes, animals that received an operation 2 weeks prior to heart procurement were compared to animals 24 hours after reoperative chest surgery to remove an inflammatory silk band. Thus, many transcripts might actually reflect differences attributed to the perioperative systemic inflammatory response. We purposefully and rigorously designed (cage strategy, circadian cycling of procedures) our study, in a minimally-invasive murine model, to avoid confounding perioperative variables. Furthermore, no quantifiable physiologic data is reported to indicate that LVH regression actually occurred. Additionally, statistical issues evaluating fold-changes between groups (differences are expressed in relation to a single sham control group, not between the contrasting groups) are magnified when the essential comparison only utilized 3 animals, as opposed to 9 animals in each of our groups. Finally, their study utilized a limited array platform with less than 10,000 transcripts, possibly limiting their scope of observation, especially when compared to the whole genome platform used in our present study.
Hypertrophy and atrophy have similarly been studied in skeletal muscle, and the processes have much in common with the progression and regression of LVH. For example, atrophy has been previously demonstrated to be an active process with a distinct pattern of gene expression, rather than simply the genomic reverse of muscle hypertrophy.20, 21 Functional genes associated with atrophy and regression involve increased protein degradation, downregulation of ATP production, decreased glycolysis, a decrease in growth protein synthesis, and altered ECM production – all categories that we identified in regression compared to LVH. Fundamentally, however, muscle atrophy typically describes progression to a disease state, while regression of LVH is a return to a normal physiologic state. In this regard, and because of the intrinsic physiologic differences between skeletal and cardiac muscle, we believe that mechanistic inferences must be limited in all but the final pathways that directly result in decreased muscle mass.
Our results must be viewed with several caveats. In this study, we have examined in detail the effects of banding at only 1 timepoint (4 weeks) and for only a discreet period of pressure unloading (1 week). We suspect that the influence of pressure-relief when the ventricle is either less (2–3 weeks of banding) or more stressed (6 weeks or more of banding) would offer different structural and genomic patterns. We thus are likely witnessing a continuum of change as the heart both negatively and positively remodels. We chose to organize the array around the 1 week timepoint because our preliminary data indicated that the effective physiologic and structural reversal of LVH was nearly complete at this time (and far-enough away from the surgical procedure), despite differences in gene expression. As demonstrated simply in the Venn diagram, structural reversal did not create a “normal heart” as assessed genomically. These observations will propel more detailed studies focusing on timing and better understanding of the reversed state. Finally, we acknowledge that this model is really a model that relieves acute pressure overload (banding). Thus, parallels to the insidious pressure-overload that occurs with typical aortic stenosis in human, and its subsequent reversal with aortic valve replacement,13, 22 can only be inferred.
In conclusion, we demonstrate that relief of pressure overload-induced LVH is not simply the reverse of LVH progression, but rather a unique process with a specific gene expression profile. Therapeutic approaches to LVH based on enhancing regression, rather than impeding progression, are rarely pursued. Elaborating mechanisms associated with both regression and the regressed state have great potential for identifying novel and clinically relevant strategies for treating patients with LVH.
Clinical Implications
Much attention is given to the study of mechanisms involved with the development of LVH and heart failure. Unfortunately, current strategies have not markedly altered the natural history of this disease. Aortic valve replacement for severe aortic stenosis (ie: removing the band) has clearly demonstrated a favorable impact in cardiac remodeling. Yet, there is a large subset of patients with LVH from pressure-overload that do not regress and have increased risk of cardiovascular morbidity and mortality. While an allure exists to jump to elaborate therapies for HF (for example, cellular replacement and stem cell injections), a great need exists to identify underlying mechanisms associated with myocardial recovery that can subsequently be targeted for sole therapy or perhaps in combination with mechanical unloading (AVR or ventricular assist). While LVH experimentation in mice is temporally quite different from humans, it is none-the-less an important first step in characterizing the genomic signature associated with cardiac remodeling, and will enhance future work in myocardial recovery therapy.
Supplementary Material
Acknowledgments
Supported by grants from the Thoracic Surgery Foundation for Research and Education (WES, CHS), American Heart Association (PCC), National Institutes of Health (CP), and American College of Surgeons (CHS)
We wish to thank Monte Willis, MD, PhD, for his critical comments; David Threadgill, PhD, for his assistance with experimental design and interpretation; and Margaret Alford Cloud for her editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levy D, Anderson KM, Savage DD, Kannel WB, Christiansen JC, Castelli WP. Echocardiographically detected left ventricular hypertrophy: prevalence and risk factors. The Framingham Heart Study. Ann Intern Med. 1988;108:7–13. doi: 10.7326/0003-4819-108-1-7. [DOI] [PubMed] [Google Scholar]
- 2.Vakili BA, Okin PM, Devereux RB. Prognostic implications of left ventricular hypertrophy. Am Heart J. 2001;141:334–41. doi: 10.1067/mhj.2001.113218. [DOI] [PubMed] [Google Scholar]
- 3.Koren MJ, Ulin RJ, Koren AT, Laragh JH, Devereux RB. Left ventricular mass change during treatment and outcome in patients with essential hypertension. Am J Hypertens. 2002;15:1021–8. doi: 10.1016/s0895-7061(02)03061-3. [DOI] [PubMed] [Google Scholar]
- 4.Zhao M, Chow A, Powers J, Fajardo G, Bernstein D. Microarray analysis of gene expression after transverse aortic constriction in mice. Physiol Genomics. 2004;19:93–105. doi: 10.1152/physiolgenomics.00040.2004. [DOI] [PubMed] [Google Scholar]
- 5.Wagner RA, Tabibiazar R, Powers J, Bernstein D, Quertermous T. Genome-wide expression profiling of a cardiac pressure overload model identifies major metabolic and signaling pathway responses. J Mol Cell Cardiol. 2004;37:1159–70. doi: 10.1016/j.yjmcc.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Weinberg EO, Mirotsou M, Gannon J, Dzau VJ, Lee RT, Pratt RE. Sex dependence and temporal dependence of the left ventricular genomic response to pressure overload. Physiol Genomics. 2003;12:113–27. doi: 10.1152/physiolgenomics.00046.2002. [DOI] [PubMed] [Google Scholar]
- 7.Caron KM, James LR, Kim HS, Knowles J, Uhlir R, Mao L, et al. Cardiac hypertrophy and sudden death in mice with a genetically clamped renin transgene. Proc Natl Acad Sci U S A. 2004;101:3106–11. doi: 10.1073/pnas.0307333101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keys JR, Greene EA, Cooper CJ, Naga Prasad SV, Rockman HA, Koch WJ. Cardiac hypertrophy and altered beta-adrenergic signaling in transgenic mice that express the amino terminus of beta-ARK1. Am J Physiol Heart Circ Physiol. 2003;285:H2201–11. doi: 10.1152/ajpheart.00112.2003. [DOI] [PubMed] [Google Scholar]
- 9.Friddle CJ, Koga T, Rubin EM, Bristow J. Expression profiling reveals distinct sets of genes altered during induction and regression of cardiac hypertrophy. Proc Natl Acad Sci. 2000;97:6745–50. doi: 10.1073/pnas.100127897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stansfield WE, Rojas M, Corn D, Willis M, Patterson C, Smyth SS, et al. Characterization of a model to independently study regression of ventricular hypertrophy. J Surg Res. 2007;142:387–93. doi: 10.1016/j.jss.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 11.Gao XM, Kiriazis H, Moore XL, Feng XH, Sheppard K, Dart A, et al. Regression of pressure overload-induced left ventricular hypertrophy in mice. Am J Physiol - Heart Circ Physiol. 2005;288:H2702–7. doi: 10.1152/ajpheart.00836.2004. [DOI] [PubMed] [Google Scholar]
- 12.Yang DK, Choi BY, Lee YH, Kim YG, Cho MC, Hong SE, et al. Gene profiling during regression of pressure overload-induced cardiac hypertrophy. Physiol Genomics. 2007;30:1–7. doi: 10.1152/physiolgenomics.00246.2006. [DOI] [PubMed] [Google Scholar]
- 13.Taniguchi K, Takahashi T, Toda K, Matsue H, Shudo Y, Shintani H, et al. Left ventricular mass: impact on left ventricular contractile function and its reversibility in patients undergoing aortic valve replacement. Eur J Cardiothorac Surg. 2007;32:588–95. doi: 10.1016/j.ejcts.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Stansfield WE, Tang RH, Moss NC, Baldwin AS, Willis MS, Selzman CH. Proteasome inhibition promotes regression of left ventricular hypertrophy. Am J Physiol Heart Circ Physiol. 2008;294:H645–50. doi: 10.1152/ajpheart.00196.2007. [DOI] [PubMed] [Google Scholar]
- 15.Abramoff M, Magelhaes P, Ram S. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 16.Smyth G. Bioinformatics and computational biology solutions using R and Bioconductor: Limma: linear models for microarray data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Statistics for Biology and Health. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 17.Dennis G, Jr, Sherman B, Hosack D, Yang J, Gao W, Lane H, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 18.Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005;21:1943–9. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- 19.Horiba M, Kadomatsu K, Yasui K, Lee JK, Takenaka H, Sumida A, et al. Midkine plays a protective role against cardiac ischemia/reperfusion injury through a reduction of apoptotic reaction. Circulation. 2006;114:1713–20. doi: 10.1161/CIRCULATIONAHA.106.632273. [DOI] [PubMed] [Google Scholar]
- 20.Glass DJ. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol. 2003;5:87–90. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- 21.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 22.Kunadian B, Vijayalakshmi K, Thornley AR, de Belder MA, Hunter S, Kendall S, et al. Meta-analysis of valve hemodynamics and left ventricular mass regression for stentless versus stented aortic valves. Ann Thorac Surg. 2007;84:73–8. doi: 10.1016/j.athoracsur.2007.02.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.