Summary
The combined activity of three transcription factors can reprogram adult cells into induced pluripotent stem (iPS) cells. However, the transgenic methods used to deliver reprogramming factors have raised concerns regarding the future utility of the resulting stem cells. These uncertainties could be overcome if each transgenic factor were replaced with a small molecule that either directly activated its expression from the somatic genome or in some way compensated for its activity. To this end, we have used high-content chemical screening to identify small molecules that can replace Sox2 in reprogramming. We show that one of these molecules functions in reprogramming by inhibiting Tgf-β signaling in a stable and trapped intermediate cell type that forms during the process. We find that this inhibition promotes the completion of reprogramming through induction of the transcription factor Nanog.
Introduction
Retroviral transduction with three genes: Sox2, Oct4, and Klf4, can directly reprogram somatic cells to a pluripotent stem cell state (Okita et al., 2007; Takahashi et al., 2007b). Unfortunately, the resulting induced pluripotent stem (iPS) cells are suboptimal for applications in transplantation medicine and disease modeling because both the viral vectors used for gene transfer and the reprogramming factors they encode are oncogenic (Hacein-Bey-Abina et al., 2003; Nakagawa et al., 2008; Thrasher, 2007).
One potential solution is to identify small molecules that can efficiently reprogram cells, producing unmodified iPS cell lines better suited for downstream applications. Identification of such compounds would allow reprogramming that would not be impeded by the laborious nature of protein transduction or the safety concerns surrounding transgenic approaches (Kaji et al., 2009; Kim, 2009; Okita et al., 2008).
Several small molecules that catalyze reprogramming have already been described. Compounds that alter chromatin structure, including the DNA methyltransferase inhibitor 5-aza-cytidine (AZA) and the histone deacetylase (HDAC) inhibitor valproic acid (VPA), can increase reprogramming efficiency and even reduce the number of factors required for reprogramming (Huangfu et al., 2008a; Huangfu et al., 2008b; Mikkelsen et al., 2008; Shi et al., 2008b). Treatment with these inhibitors presumably lowers the barrier to activation of endogenous pluripotency-associated genes. However, Oct4 and Sox2 not only activate genes required for pluripotency, they also function to repress genes promoting differentiation. It is therefore unlikely that this class of small molecules would be sufficient to completely replace the transgenic factors. As a result, there remains a need to identify novel small molecules that can function in reprogramming.
Here, we report the discovery of compounds that can replace the central reprogramming factor Sox2. We demonstrate that one of these chemicals specifically acts by inhibiting Tgf-β signaling. Interestingly, this compound does not act by inducing Sox2 expression in the target fibroblasts. Instead, we show that it enables reprogramming through the induction of Nanog transcription in a stable, partially reprogrammed cell type that accumulates in the absence of Sox2.
A Screen for Chemical Mediators of Reprogramming
To identify small molecules that function in reprogramming, we transduced fibroblasts with viral vectors encoding Oct4, Klf4, and cMyc and then screened for compounds that allowed for reprogramming in the absence of Sox2. We favored this approach because it was unbiased with respect to the mechanism by which a given chemical could function and would not only deliver chemical compounds with translational utility but also provide novel insights into the mechanisms controlling reprogramming.
Activation of an Oct4∷ GFP reporter gene in colonies with an ES cell morphology has been shown to be a stringent assay for reprogramming (Meissner et al., 2007). In mouse embryonic stem (mES) cell culture medium supplemented with VPA, retroviral transduction of 7500 Oct4∷GFP transgenic mouse embryonic fibroblasts (MEFs) with Oct4, Klf4, cMyc, and Sox2 (Boiani et al., 2004) routinely generated 100-200 GFP+ colonies (Figure 1A). In contrast, we observed no GFP+ colonies when Sox2 was omitted (Figure 1A). We used this robust difference to identify small molecules that can replace Sox2.
Figure 1. Identification of Small Molecules That Replace of Sox2.
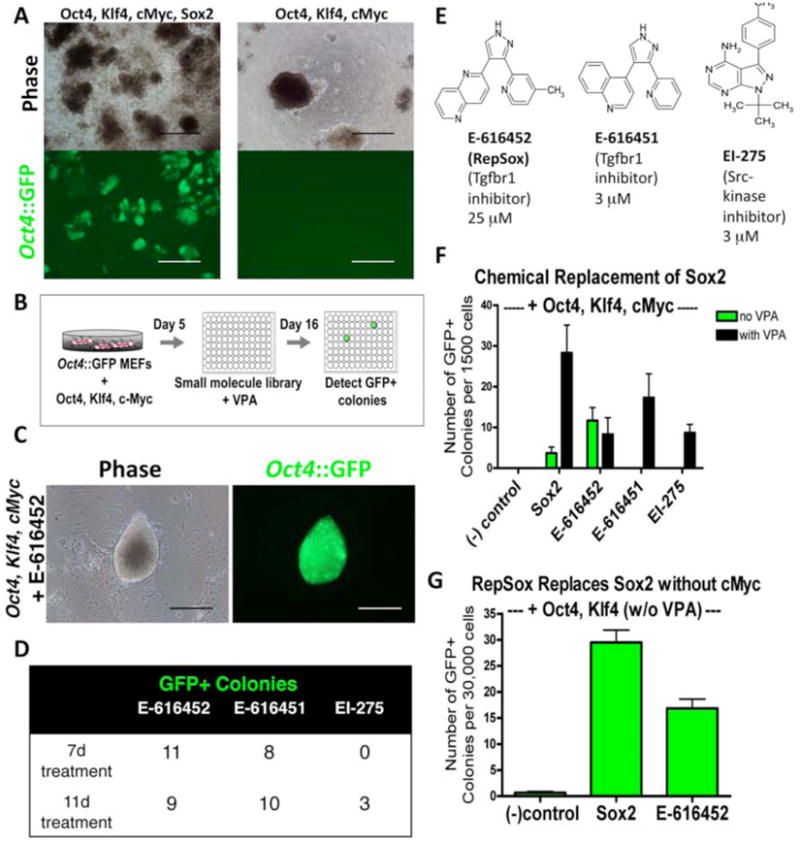
(A) Oct4∷GFP+ colonies form readily in Oct4, Klf4, cMyc, and Sox2-infected MEF cultures and do not form in Oct4, Klf4, and cMyc-infected MEF cultures. Scale bars = 500 μm.
(B) Overview of chemical screen for replacement of Sox2.
(C) A P0 colony from Oct4, Klf4, and cMyc-infected MEFs + RepSox that displays a mES-like morphology and is Oct4∷GFP+. Scale bars = 200 μm.
(D) Number of Oct4∷GFP+ colonies detected for each hit in the primary screen after transduction of Oct4, Klf4, and cMyc and VPA treatment.
(E) Chemical structures of E-616452, E-616451, and EI-275, with the optimal concentrations for reprogramming listed.
(F) Quantification of small molecule replacement of Sox2 in Oct4, Klf4, and cMyc-infected MEFs with and without VPA treatment.
(G) Sox2 replacement by RepSox is not dependent on cMyc (no VPA treatment).
To facilitate the identification of cellular targets and signaling pathways affected by any compounds we discovered, we utilized a library of molecules with known pharmacological targets. We transduced Oct4∷GFP MEFs with Oct4, Klf4, and cMyc, and then plated 2000 cells per well in 96-well format. To each well we added one of 200 distinct compounds for 7-11 days, while also treating with 2 mM VPA for the first 7 days (Figure 1B). It was our hope that this approach would allow us to identify both compounds that required chromatin remodeling to induce reprogramming (Huangfu et al., 2008a) and compounds that did not. After 16 days, we scored each well for the presence of GFP+ colonies with a mES-like morphology (Figure 1C) and identified 3 independent hit compounds (Figure 1D). Two of these compounds were distinct Transforming Growth Factor-β Receptor 1 (Tgfbr1) kinase inhibitors (E-616452 and E-616451 (Figure 1E) (Gellibert et al., 2004)), while the third was a Src-family kinase inhibitor (EI-275 (Figure 1E) (Hanke et al., 1996)).
Efficient Small Molecule Replacement of Sox2
Next, we optimized the effective concentration for each hit molecule (Figure S1) and quantified the efficiency at which it synergized with VPA to replace Sox2. When 1500 MEFs were transduced with only Oct4, Klf4, and cMyc and then treated with VPA, we did not observe GFP+ colonies (Figure 1F). However, the addition of E-616452 (25 μM), E-616451 (3 μM), or EI-275 (3 μM), led to the formation of GFP+ colonies with an ES cell morphology at a rate that was comparable to transduction with Sox2 (Figure 1F).
Since the three compounds were identified in the presence of VPA, we next determined whether these molecules were dependent on this HDAC inhibitor for their reprogramming activities. We found that E-616451 and EI-275 could not induce the appearance of GFP+ colonies in the absence of VPA (Figure 1F), while E-616452 could do so and at a rate that was similar to a positive control transduced with the Sox2 retrovirus (Figure 1F).
Although cMyc does increase the efficiency of reprogramming, it is not required for the generation of iPS cells (Nakagawa et al., 2008). Since the elimination of cMyc is an important step towards reducing the risk of tumor formation, we tested whether E-616452 could function in the absence of this oncogene. When added to MEFs transduced with only Oct4 and Klf4, E-616452 induced the formation of GFP+ colonies with an efficiency similar to viral Sox2 (Figure 1G).
Previous reports on small molecules that affect reprogramming have focused on MEFs or neural stem cells (NSCs). These cells may be reprogrammed more easily due to either their proliferative capacity or their expression of iPS factors (Huangfu et al., 2008a; Shi et al., 2008a; Shi et al., 2008b). However, it may be that chemical modulation of gene expression is cell-type specific and we therefore determined if the reprogramming compound we identified functioned in a more patient-relevant cell type. When we infected adult tail tip fibroblasts with Oct4, Klf4, and cMyc alone, we did not observe Oct4∷GFP+ colonies. However, when we added E-616452, we readily observed reprogramming (Figure S2A). The resulting Oct4∷GFP+ colonies could be expanded into cell lines that maintained homogeneous Oct4∷GFP expression and self-renewed similarly to mES and 4-factor control iPS lines (Figure S2B). Because it could efficiently replace transgenic Sox2 in the absence of VPA and cMyc, as well as in both embryonic and adult fibroblasts, we chose to further characterize E-616452 and named it RepSox, for Replacement of Sox2.
RepSox-reprogrammed Cells are iPS cells
Investigation of self-renewal capacity (Figure 2A), gene expression program, and pluripotency demonstrated that Oct4∷GFP+ cells induced by the RepSox replacement of Sox2 were bona fide iPS cells. PCR with primers specific to the Oct4, Klf4, cMyc, and Sox2 transgenes confirmed that this cell line did not harbor transgenic Sox2 (Figure S3A). Chromosomal analysis indicated it was karyotypically normal (Figure S3B).
Figure 2. RepSox-reprogrammed Cells Are Pluripotent.
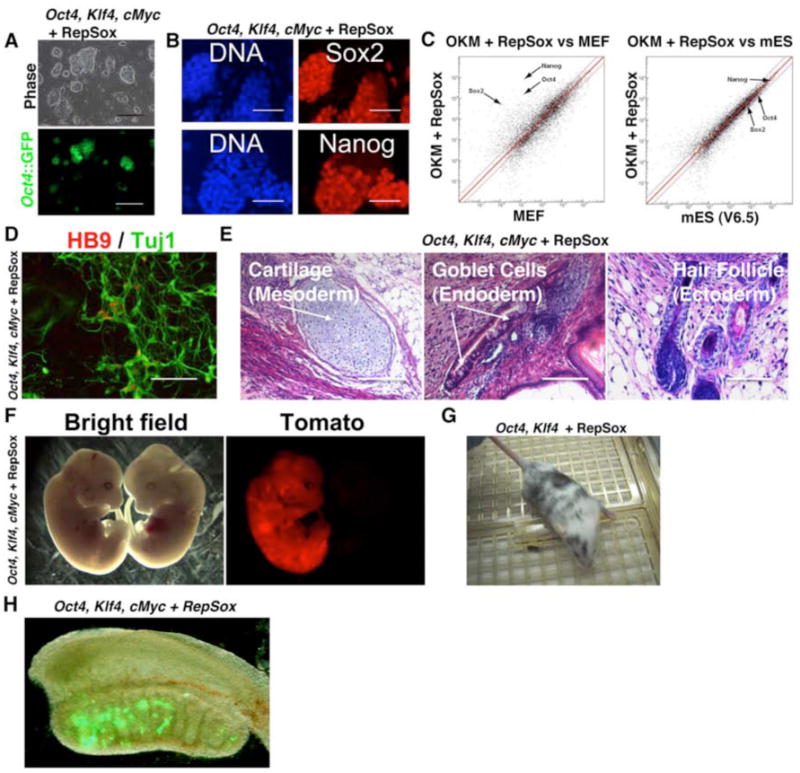
(A) An Oct4∷GFP+ iPS line that was derived from a culture of RepSox-treated Oct4, Klf4, and cMyc-infected MEFs (OKM + RepSox line 1) displays the characteristic mES-like morphology and self-renewal properties. Passage 11. Scale bars = 500 μm.
(B) Antibody staining of OKM + RepSox line 1 cells shows that they express markers of pluripotent stem cells Sox2 and Nanog. Scale bars = 100 μm.
(C) Microarray scatter plots showing that the global gene expression profile of OKM + RepSox line 1 is highly similar to that of mES line V6.5 and very different from that of somatic MEFs.
(D) Motor neurons differentiated in vitro from OKM + RepSox line 1. Scale bar = 200 μm.
(E) Teratomas containing cells of all three germ layers formed by injection of OKM + RepSox line 1 cells into nude mice.
(F) E12.5 chimeric mouse embryo (left, vs. non-chimeric littermate on the right) showing a high amount of contribution from OKM + RepSox line 1 cells constitutively expressing the dTomato red fluorescent protein.
(G) 8 week-old chimeric mouse formed by injection of OK + RepSox line 1 cells (C57BL6 genetic background) into an ICR blastocyst.
(H) Oct4∷GFP+ cells derived from an OKM + RepSox cell line are present in the genital ridge of a male embryo at 13.5 d.p.c.
The Oct4∷GFP+ cells co-expressed alkaline phosphatase (Figure S3C) and the endogenous alleles of the Nanog and Sox2 genes, suggesting pluripotency had been established (Figure 2B). The global transcriptional profile of cells reprogrammed with RepSox was similar to that of an iPS cell line produced with all four transgenes and as similar to those of mES cells (Pearson correlation coefficient = 0.95-0.97) as two distinct mES cell lines profiles were to each other (Pearson correlation coefficient = 0.96) (Figures 2C, S3D, Table S1). The profile differed significantly from that of the somatic MEFs (Figure 2C).
Cells produced with RepSox could readily form both embryoid bodies and teratomas that contained differentiated cell types of the three distinct embryonic germ layers (Figure 2E and S4A). In addition, we observed that these cells could respond to directed differentiation signals in vitro and robustly differentiate into Hb9+/Tuj1+ motor neurons (Figure 2D, Figure S5).
In order to more definitively confirm the pluripotency of cells reprogrammed with RepSox, we tested their ability to contribute to chimeric embryos in vivo. We labeled cells with a lentiviral transgene encoding the red fluorescent Tomato-protein and injected them into blastocysts. Both embryos and adult mice with significant contribution from the iPS cells were obtained (Figures 2F, G). Although adult mice with high contribution from the iPS cells were observed, we found it difficult to assess the contribution of these cells to the germ-line, as the majority of animals developed tumors at or before the time of sexual maturity. However, we did observe that the reprogrammed cells could contribute Oct4∷GFP+ cells to the genital ridges of embryonic chimeras, demonstrating contribution of these pluripotent cells to the germ-line (Figure 2H). Together, these results demonstrate that the RepSox-reprogrammed cells are indeed iPS cells.
RepSox Can Replace Sox2 and c-Myc by Inhibiting Tgf-β Signaling
Previous studies with RepSox suggest that it can act as an inhibitor of the Tgfbr1 kinase (Gellibert et al., 2004). Therefore, we investigated whether the mechanism by which RepSox functions to replace Sox2 is through the inhibition of Tgf-β signaling. If Tgfbr1 is the functional target of RepSox, then a structurally unrelated inhibitor of Tgf-β signaling or depletion of Tgf-β ligands from the culture medium might also replace Sox2. The small molecule SB431542 (Figure 3A) is known to inhibit Tgfbr1 kinase and is structurally distinct from RepSox (Inman et al., 2002). When we treated fibroblasts transduced with Oct4, Klf4, and cMyc with 25 μM SB431542, we observed ∼10 GFP+ colonies per 7500 cells plated (Figure 3B). Likewise, when we transduced fibroblasts in the presence of either an antibody that neutralized a variety of Tgf-β ligands (R&D Systems, AB-100-NA) or an antibody specific to Tgf-β II (R&D Systems, AB-12-NA), Oct4∷GFP+ colonies were generated (Figure 3B). In contrast, we observed no GFP+ colonies in transductions without these Tgf-β inhibitors. These results are consistent with the notion that at least part of the mechanism by which RepSox replaces Sox2 in reprogramming is through the inhibition of Tgf-β signaling.
Figure 3. RepSox Specifically Replaces Sox2 by Inhibiting Tgf-β Signaling.
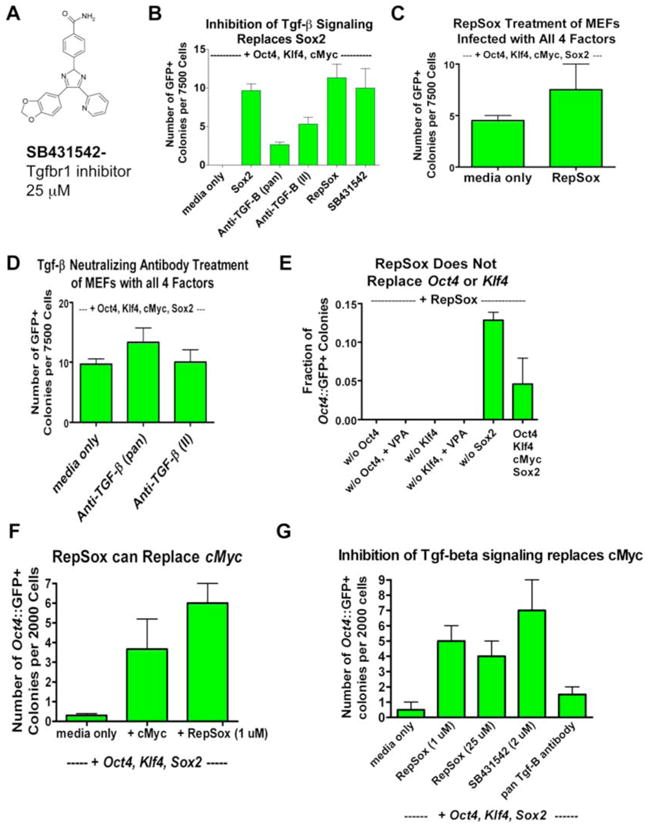
(A) Chemical structure of SB431542, an inhibitor of Tgfbr1 activity.
(B) Inhibition of Tgf-β signaling by treatment of Oct4, cMyc, and Sox2-infected MEFs with SB431542 or TGF-β neutralizing antibodies replaces Sox2.
(C) RepSox does not increase the efficiency of Oct4∷GFP+ colony induction in Oct4, Klf4, cMyc, and Sox2-infected MEFs.
(D) Inhibition of Tgf-β signaling by TGF-β neutralizing antibodies does not increase the efficiency of Oct4∷GFP+ colony induction in Oct4, Klf4, cMyc, and Sox2-infected MEFs.
(E) RepSox does not replace transgenic Oct4 or transgenic Klf4 in reprogramming. We observed no Oct4∷GFP+ colonies in RepSox-treated Klf4, cMyc, Sox2-infected MEFs or Oct4, cMyc, Sox2-infected MEFs out of 30,000 cells plated both with and without VPA treatment. We routinely observe 30-40 Oct4∷GFP+ colonies when we plate the same number of Oct4, Klf4, cMyc-infected MEFs and treat with RepSox.
(F) RepSox can replace cMyc in reprogramming. Cells were transduced with Oct4, Klf4, and cMyc and treated with RepSox continuously starting at day 5 post-infection.
(G) Inhibition of Tgf-β signaling can replace cMyc in reprogramming. Cells were transduced with Oct4, Klf4, and cMyc and treated with inhibitors of Tgf-β signaling continuously starting at day 5 post-infection.
Our goal was to identify molecules that specifically replace Sox2 instead of generally increasing reprogramming efficiency. If RepSox acts specifically to replace Sox2, then we would not expect it to stimulate reprogramming in the presence of transgenic Sox2. When RepSox- or Tgf-β antibody-treated MEFs were transduced with Oct4, Klf4, cMyc and Sox2, we observed less than a 2-fold increase in the number of GFP+ colonies over the untreated controls (Figures 3C, D). The magnitude by which RepSox stimulated reprogramming in this context was significantly less than the 10-fold increase that we observed following treatment with VPA, a compound thought to increase reprogramming efficiency (Figure 1F).
In order to further investigate the specificity of Sox2 replacement by RepSox, we tested the ability of this molecule to individually replace Oct4, Klf4, and cMyc in reprogramming. We found that RepSox could not induce GFP+ colonies in the absence of either Oct4 or Klf4, even in the presence of VPA (Figure 3E). In contrast, we found that RepSox did increase the number of Oct4∷GFP+ colonies by 20-fold in the absence of cMyc, thereby fully replacing it in reprogramming (Figure 3F). In addition, the structurally distinct Tgf-β inhibitor SB431542 and a Tgf-β-specific neutralizing antibody both increased reprogramming efficiency in the absence of cMyc (Figure 3G). From these experiments, we conclude that RepSox enables the replacement of the reprogramming activities provided by both transgenic Sox2 and cMyc. In both cases, these complementing activities seem to be mediated through the inhibition of Tgf–β signaling.
RepSox Replace Sox2 by Acting on Intermediates Formed During the Reprogramming Process
The development of cocktails of small molecules that can effectively reprogram somatic cells may require a detailed knowledge of the mechanism and kinetics by which each compound acts. Therefore, we determined the optimal duration of time by which inhibition of Tgf-β signaling using RepSox can help induce reprogramming.
Initially, we pretreated MEFs with RepSox, applying the chemical for three days, and then removed it at the time of transduction with Oct4, Klf4, and cMyc. In these experiments, no Oct4∷GFP+ colonies were formed (Figure 4A), suggesting that RepSox does not act on the initial somatic cells to replace Sox2. Consistent with this result, we did not detect a significant increase in the expression of endogenous Sox2 or closely related Sox family members upon RepSox treatment (Figure S6A). In addition, RepSox treatment did not decrease the expression of the mesenchymal gene Snai1 (Figure S6B), which is downregulated 5-40-fold by transduction of the 4 reprogramming factors (Mikkelsen et al., 2008). Thus, RepSox does not destabilize the pre-existing MEF transcriptional program.
Figure 4. A Short Pulse of RepSox is Sufficient for Sox2 Replacement and Most Effective at Later Time Points Post-infection.
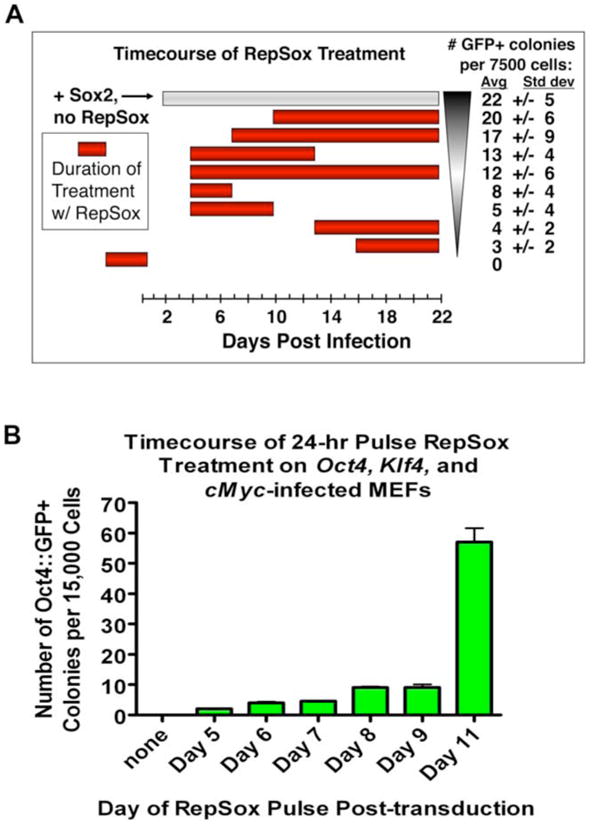
(A) Graph showing the number of Oct4∷GFP+ colonies induced by various timings of RepSox treatment of Oct4, cMyc, and Sox2-infected MEFs in mES medium. Colonies were counted at 24 days post-infection.
(B) Timecourse of RepSox treatment showing the number of Oct4∷GFP+ colonies induced by a 24-hr pulse of RepSox on Oct4, cMyc, and Sox2-infected MEFs in serum-free mES medium with knockout serum replacement (KSR mES). Colonies were counted at 24 days post-infection. Shown are average colony numbers +/− the standard deviation.
In contrast, we found that RepSox did increase by 5-fold the expression of L-Myc, a close homolog of cMyc that can functionally replace it in reprogramming (Nakagawa et al., 2008)(Figure S6C). Together these data suggest that although RepSox likely functions at the level of the initial somatic cell population to replace cMyc, it does not act on the starting MEF population to replace Sox2.
Because RepSox did not seem to act directly on the fibroblasts to replace Sox2, we investigated whether or not it functioned on intermediates that arose during reprogramming. To address this question, we varied both the duration and timing of RepSox treatment in order to determine when it was most effective. First, we transduced 7500 MEFs with Oct4, Klf4, and cMyc, waited for 4 days, and then treated cultures with RepSox for either 3, 6, 9, or 18 additional days. Although a short 3-day treatment from days 4-7 induced a small number of Oct4∷GFP+ colonies, the 9-day treatment from days 4-13 yielded the most Oct4∷GFP+ colonies (Figure 4A).
Next, we varied the timing at which we initiated RepSox treatment, administering the compound beginning at day 4, 7, 10, 13, or 16 after transduction. We found that delaying the start of RepSox treatment increased its reprogramming potency, with optimal treatment beginning at 10 days post-transduction (Figure 4A). Together these results suggest that RepSox treatment is most effective between days 7-12 post-transduction.
To more precisely define the optimal treatment window, we determined the minimal duration of treatment required to induce reprogramming. We found that a treatment as short as only one day was sufficient to induce detectable reprogramming (Figure 4B). Delaying this short treatment yielded more reprogrammed colonies, with a sharp increase at day 11 (Figure 4B). These results indicate that RepSox is most effective at replacing Sox2 during days 10-11 after transduction and that therefore cultures of Oct4, Klf4, and cMyc-transduced MEFs give rise to intermediates capable of responding to RepSox treatment. These intermediates appear at day 4 post-transduction and peak at days 10-11.
Interestingly, when we tracked the timing of the initial appearance of reprogrammed colonies as a function of the timing of RepSox administration, we found that regardless of whether we began treatment at day 7 or day 10 post-transduction, Oct4∷GFP+ colonies first appeared at day 14 (Figure S7). This suggests that RepSox may not always be the rate-limiting step in this reprogramming process and that other, RepSox-independent events take place during the formation of the RepSox-responsive intermediates.
RepSox-responsive Cell Lines
Our finding that a 24-hr pulse of RepSox can replace Sox2 (Figure 4B) differs strikingly from the 5-10 day period of transgene expression normally required (Sridharan et al., 2009; Wernig et al., 2007) and suggests that RepSox could trigger a switch activating reprogramming. If RepSox acts to flip a switch in semi-stable intermediate cell types that accumulate in the absence of retroviral Sox2 expression, we reasoned that it might also be possible to culture these responsive intermediates for prolonged periods of time. On the other hand, if RepSox acts during a critical window on very transient intermediates, this might not be possible. To distinguish between these models, we transduced Oct4∷GFP MEFs with Oct4, Klf4, and cMyc, waited 10-14 days, and then clonally expanded 10 iPS-like, GFP-negative colonies (Figure 5A). These cell lines continued to proliferate for at least 4 passages and often maintained an iPS-like morphology (Figure 5A) but never further activated expression of Oct4∷GFP. However, when we treated these cell lines with a 48-hour pulse of RepSox, 5-10% of the colonies in 2 of the 10 lines became Oct4∷GFP+ (Figure 5A, B). These results demonstrate that partially reprogrammed cells can accumulate in the absence of Sox2 and that some, but not all, of these cells can be clonally expanded and cultured for prolonged periods while maintaining responsiveness to RepSox.
Figure 5. Stable Intermediates Can Be Reprogrammed by RepSox.
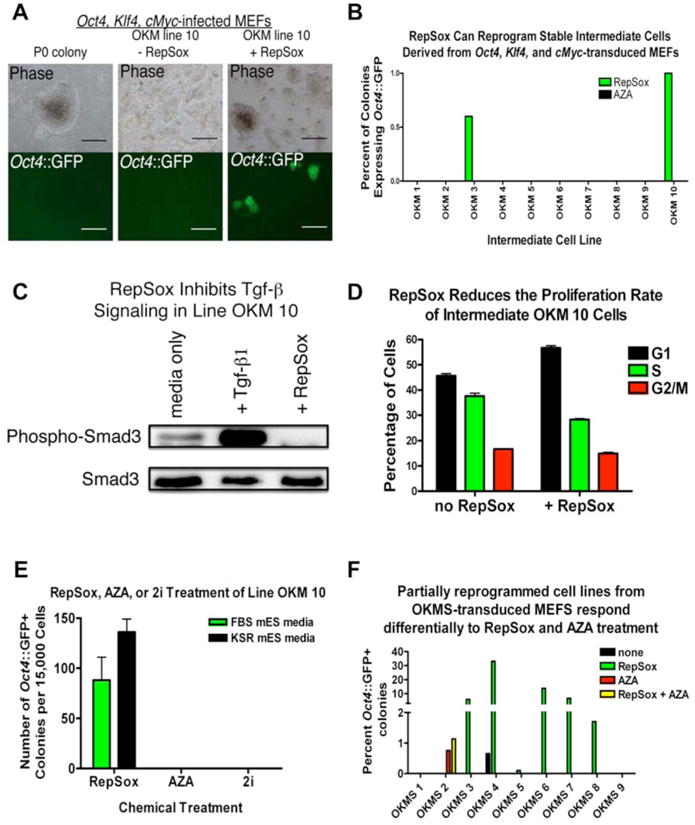
(A) Stable Oct4∷GFP-negative cell lines derived from Oct4∷GFP-negative colonies in Oct4, Klf4, and cMyc-infected MEF cultures can be reprogrammed by RepSox. Scale bars in “OKM line 10 + RepSox” panels = 500 μm, all other scale bars = 200 μm.
(B) 2 of 10 stable, non-pluripotent intermediate cell lines derived from MEFs transduced with Oct4, Klf4, and cMyc can be reprogrammed with RepSox treatment but none can be reprogrammed with AZA treatment.
(C) Western blot for phospho-Smad3 showing that RepSox inhibits Tgf-β signaling in line OKM 10 (OKM 10) cells.
(D) RepSox does not increase the proliferation of OKM 10 cells.
(E) Line OKM 10 can be reprogrammed with RepSox treatment but not with AZA or 2i, indicating it is distinct from cell lines that can be reprogrammed by AZA or 2i.
(F) Stable Oct4∷GFP-negative cell lines derived from Oct4∷GFP-negative colonies in Oct4, Klf4, cMyc and Sox2-infected MEF cultures can be reprogrammed by RepSox or by AZA, but lines responsive to RepSox are not responsive to AZA alone and lines responsive to AZA are not responsive to RepSox alone, indicating the presence of two different types of stable intermediates in the reprogramming cultures.
As we had shown that this particular reprogramming molecule seems to replace Sox2 through the inhibition of Tgf-β signaling, we sought to determine whether RepSox treatment affected Tgf-β signal transduction pathways in these responsive cell lines. To this end, we determined the levels of phosphorylated Smad3 by western blot in cell line OKM 10 both with and without RepSox treatment. Without RepSox treatment, we detected relatively high levels of phosphorylated Smad3, suggesting that Tgf-β signaling was active (Figure 5C). In contrast, treatment with 25 μM RepSox almost completely eliminated Smad3 phosphorylation (Figure 5C), indicating that RepSox strongly inhibited Tgf-β signaling in these cells.
Because an increase in cell proliferation can also increase reprogramming efficiency (Hong et al., 2009) and possibly contribute to the replacement of transgenic Sox2, we measured the proliferation rate of partially reprogrammed OKM 10 cells both with and without RepSox. Treatment with RepSox decreased the proportion of cells in G2/M phase of the cell cycle (Figure 5D), indicating it does not increase the proliferation rate of these partially reprogrammed cells.
Cells That Respond To RepSox Treatment Are Distinct From Previously Described Intermediates
It has been shown that certain non-pluripotent, partially reprogrammed cell lines derived from MEFs transduced with Oct4, Klf4, cMyc, and Sox2 can be fully reprogrammed with AZA or a combination of chemical inhibitors of Glycogen Synthase Kinase 3β (GSK-3β) and the Mek signaling pathway (2i conditions) (Mikkelsen et al., 2008; Silva et al., 2008). If the RepSox-responsive cell lines generated by overexpression of Oct4, Klf4, and cMyc were similar to these 4-factor cell lines, then they should also be reprogrammed by AZA or 2i. However, when we treated the 10 stable intermediate lines with either AZA or 2i for 48 hours, we found that none became reprogrammed (Figure 5B), indicating that the RepSox-responsive stable intermediates are distinct from partially reprogrammed cell lines described previously (Mikkelsen et al., 2008; Silva et al., 2009). Consistent with these results, in vitro assays of kinase activity revealed that RepSox does not inhibit the targets of the 2i cocktail (Table S2).
It occurred to us that some non-pluripotent cells derived from MEFs transduced with Oct4, Klf4, cMyc, and Sox2 could potentially be held in a non-pluripotent state due to inappropriate levels of transgene expression and therefore might also be responsive to RepSox treatment. To test this hypothesis, we transduced Oct4∷GFP MEFs with Oct4, Klf4, cMyc, and Sox2, then picked and clonally expanded 9 GFP-negative colonies at day 14 after transduction (Figure S8). After treatment with RepSox, 5 of the 9 cell lines yielded reprogrammed colonies, with 2-33% of the colonies in each line becoming Oct4∷GFP+ (Figures 5F, S8). These results indicate that like the stable intermediate cells generated with only Oct4, Klf4, and cMyc, certain incompletely reprogrammed cells generated by Oct4, Klf4, cMyc, and Sox2 transduction can also be reprogrammed by RepSox.
Next, in order to determine if these RepSox-responsive intermediate cell lines derived after Oct4, Klf4, cMyc, and Sox2 transduction were similar to or distinct from previously described partially reprogrammed cell lines (Mikkelsen et al., 2008), we applied AZA to all 9 lines. After 48 hours of AZA treatment and 12 subsequent days in culture, none of the RepSox-responsive cell lines expressed Oct4∷GFP (Figure 5F). However, one of the lines that had been refractory to RepSox treatment did express Oct4∷GFP after AZA treatment, indicating that it had undergone complete reprogramming (Figure 5F). Together, these results show that there are a variety of intermediates that can form following retroviral transduction and that they vary in their responsiveness to reprogramming molecules.
RepSox Replaces Sox2 by Inducing Nanog Expression
The causal molecular events that drive reprogramming are difficult to detect because of the low efficiency at which somatic cells are successfully reprogrammed (Amabile and Meissner, 2009). However, when we administered RepSox to cell lines that had been partially reprogrammed by retroviral transduction, Oct4∷GFP expression was induced in up to 33% of the resulting colonies (Figure 5F). We used this more efficient reprogramming system to identify the changes in gene expression induced by RepSox that enable it to bypass the requirement for transgenic Sox2 expression.
We treated an Oct4∷GFP-negative, partially reprogrammed cell line (OKMS 6) with RepSox and performed global gene expression analysis at 10, 24, and 48 hours following the initiation of treatment. To confirm that RepSox was inhibiting Tgf-β signaling in this intermediate cell line, we investigated expression changes in known Tgf-β-responsive genes after RepSox treatment. The Inhibition of Differentiation genes Id1, Id2, and Id3 are repressed by Tgf-β signaling in mES cells (Ying et al., 2003). After treating the RepSox-responsive intermediate line OKM 10 with RepSox for 24 hours, we observed increased expression of Id1, Id2, and Id3 (Figure S9A).
One way that RepSox could function to replace transgenic Sox2 would be to induce the expression of endogenous Sox2 or a Sox-family member, such as Sox1 or Sox3, that can substitute for it in reprogramming (Nakagawa et al., 2008). However, we again did not observe a significant increase in the expression of Sox1, Sox2, Sox3, or any of the remaining Sox-family transcription factors within the first 48 hours of RepSox treatment (Figure S9B). Additionally, shRNA-mediated depletion of Sox1, the most potent Sox-family member other than Sox2 itself (Nakagawa et al., 2008), did not affect the rate of reprogramming in the presence of RepSox (Figure S9C). These results show that RepSox does not replace Sox2 by directly activating endogenous Sox2 or other closely related genes.
Next, we more broadly investigated changes in transcription factor expression following chemical treatment. We did not observe an increase in endogenous Oct4 or Klf4 expression at early time points following RepSox treatment. However, we found that the expression of the homeodomain factor Nanog was among the most increased following RepSox treatment. Relative to untreated controls, Nanog transcription increased 4-fold within 24 hours and 10-fold after 48 hours of RepSox treatment (Figure 6A). In contrast, we did not observe a rapid increase in Nanog expression in 2 Oct4∷GFP-negative intermediate cell lines that could not be fully reprogrammed using RepSox (Figure S10). Therefore, we hypothesized that RepSox might replace Sox2 by inducing Nanog expression.
Figure 6. RepSox Replaces Sox2 by Inducing Nanog Expression.
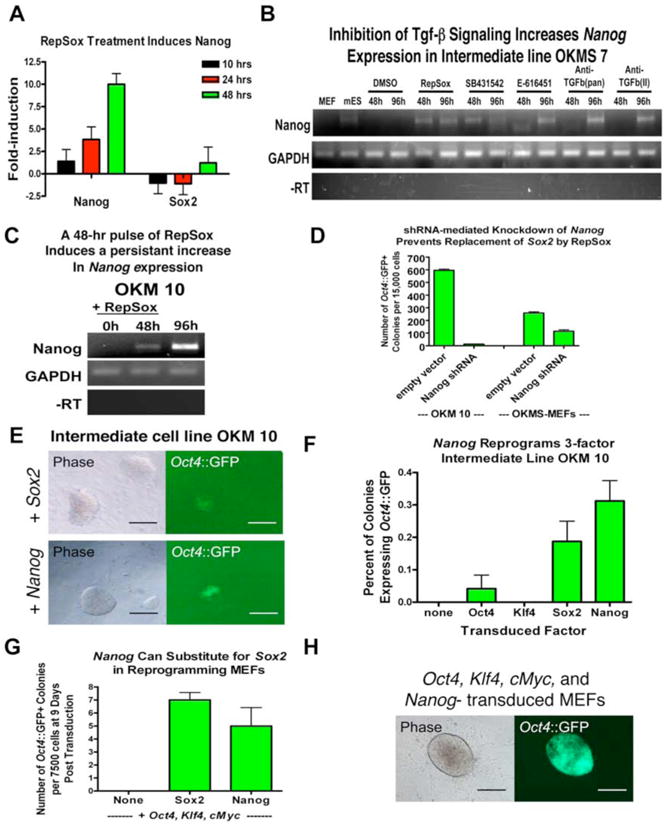
(A)RepSox treatment of RepSox-responsive line OKMS 6 strongly increases Nanog mRNA levels. Data were generated by microarray analysis and are relative to untreated controls. Nanog is induced faster and more significantly than Sox2, indicating it is upregulated before fully reprogrammed cells form.
(B) RT-PCR analysis showing that inhibition of Tgf-β signaling increases Nanog expression in the RepSox-responsive intermediate line OKMS 7.
(C) A pulse of RepSox induces a persistent increase in Nanog expression in the RepSox-responsive intermediate line OKM 10. OKM 10 cells were treated with 25 μM RepSox for 48 hours and RNA samples were taken at 0, 48, and 96 hours (48 hours after removal of RepSox) and analyzed by RT-PCR.
(D) shRNA-mediated knockdown of Nanog in OKM 10 cells inhibits replacement of Sox2 by RepSox.
(E) Pictures of reprogrammed Oct4∷GFP+ colonies induced by Sox2 (A) or Nanog (B) transduction of line OKM 10. Scale bars = 200 μm.
(F) Nanog transduction can reprogram line OKM 10 at a similar efficiency as Sox2 transduction.
(G) Nanog can substitute for Sox2 in defined-factor reprogramming of somatic fibroblasts.
(H) Picture of a reprogrammed Oct4∷GFP+ colony induced by Oct4, Klf4, cMyc and Nanog-transduction of MEFs. Scale bars = 100 μm.
Because we had determined that inhibition of Tgf-β signaling by several different small molecules and antibodies can replace Sox2, we reasoned that if the increase in Nanog expression was critical for Sox2 replacement, the alternative inhibitors of Tgf-β signaling should also upregulate Nanog. To test this hypothesis, we treated the RepSox-responsive cell lines with RepSox, SB431542, or neutralizing antibodies and analyzed Nanog expression after 48 hours. In all cases, Nanog expression was strongly induced within 48-96 hours (Figure 6B).
If RepSox functions by increasing Nanog expression, then a short pulse of RepSox should induce a persistent increase in Nanog expression. To test this, we treated the RepSox-responsive intermediate cell line OKM 10 with RepSox for 48 hours, withdrew RepSox and then analyzed Nanog expression 48 hours later. A control time point taken just before RepSox withdrawal showed a significant increase in Nanog transcription (Figure 6C). 48 hours after RepSox removal (96 hours after the initiation of treatment), Nanog expression continued to increase (Figure 6C).
If RepSox replaces Sox2 by increasing Nanog expression, then a forced reduction of Nanog expression should inhibit or even prevent reprogramming by RepSox. To test this hypothesis, we transduced the RepSox-responsive cell line with a lentivirus encoding a short-hairpin RNA specific for Nanog. The Nanog-knockdown cells reprogrammed at a frequency that was 50-fold lower than cells transduced with an empty control vector (Figure 6D). This effect was not due to a general decrease in reprogramming efficiency or differentiation of reprogrammed cells due to Nanog depletion because MEFs transduced with Oct4, Klf4, cMyc, Sox2, and the Nanog shRNA construct only suffered a 50% loss in reprogramming efficiency (Figure 6D). These results demonstrate that increased Nanog expression in this context was only necessary for the replacement of Sox2 by RepSox.
Previous reports have shown that chemical inhibition of Tgf-β signaling by SB431542 increases Bone Morphogenetic Protein (Bmp) signaling in embryonic stem cells (Xu et al., 2008). It has separately been shown that Bmp signaling in the presence of Stat3 induces Nanog expression in mES cells (Suzuki et al., 2006). The cross-talk between the Tgf-β and Bmp signaling pathways may be the result of a common requirement for Smad 4, which mediates transcriptional events in the nucleus (Attisano and Wrana, 2002). Similarly, we observed an increase in the levels of phosphorylated Smad1 protein and Bmp-3 mRNA in incompletely reprogrammed intermediates following RepSox treatment (Figure S11). Furthermore, the stable, partially reprogrammed cells that responded to RepSox expressed the LIF receptor at levels equivalent to those found in mES cells (Figure S12A). Expression of this receptor suggests that its downstream signal transduction pathway could be active in these cells, resulting in the presence of activated Stat3, which is known to induce Nanog expression in conjunction with Bmp signaling.
Since RepSox does not act on the initial population of fibroblasts to replace Sox2, we would not expect Nanog to be upregulated in RepSox-treated MEFs. Indeed, within 7 days of transduction of MEFs with Oct4, Klf4, and cMyc, we did not observe an increase in Nanog expression upon RepSox treatment (Figure S12B). This may be explained in part by the observation that the LIF receptor, and thus activated Stat3, were not highly expressed in these cells (Figure S12A). Because Nanog plays a key role in maintaining ES cells in an undifferentiated state (Chambers et al., 2003) and has been shown to enhance the efficiency of reprogramming (Silva et al., 2006; Silva et al., 2009; Yu et al., 2007), we decided to test whether Nanog could directly replace Sox2 in reprogramming.
If RepSox replaces Sox2 by inducing Nanog expression, then retroviral transduction of RepSox-responsive intermediate cells (line OKM 10, Figures 5A, B) with Nanog should reprogram them. When we transduced line OKM 10 with Sox2 as a control, .2% of the colonies expressed Oct4∷GFP after 10 days, indicating that reprogramming could be induced in this cell line by Sox2 (Figures 6E, F). When we transduced the same stable intermediate cell line with Nanog, it could also be reprogrammed, with .3% of the colonies expressing Oct4∷GFP+ after 10 days (Figures 6E, F). In contrast, transductions with Oct4 or Klf4 resulted in only .04% and 0% reprogramming efficiencies (Figure 6F). These results suggest that Nanog can indeed functionally replace Sox2 and induce reprogramming in these stable intermediates formed from Oct4, Klf4, and cMyc-transduced MEFs.
If Nanog can complement for the omission of Sox2 in defined factor reprogramming, then MEFs transduced with Oct4, Klf4, cMyc, and Nanog might be as efficiently reprogrammed as MEFs transduced with Oct4, Klf4, cMyc, and Sox2. When we transduced MEFs with Oct4, Klf4, cMyc, and Sox2 then scored cultures 9 days later, an average of 7 Oct4∷GFP+ colonies appeared for every 7500 cells plated (Figure 6G). A control transduction with only Oct4, Klf4, and cMyc yielded no Oct4∷GFP+ colonies (Figure 6G). Similar to the positive control transduction, MEFs transduced with Oct4, Klf4, cMyc, and Nanog gave rise to an average of 5 Oct4∷GFP+ colonies for every 7500 cells plated (Figures 6G, H). These colonies could be picked and expanded and remained Oct4∷GFP+ for at least 5 passages (Figure S13A). Immunocytochemistry indicated that these cells strongly activated Sox2 expression from the endogenous allele (Figure S13B). Importantly, QPCR analysis demonstrated that they also transcribed endogenous Oct4, Klf4, Nanog, and Rex1 (Figure S13C), indicating that a pluripotent gene expression program had been established. Furthermore, transgene-specific QPCR analysis showed that these cells had silenced the retroviral Oct4, Klf4, and cMyc transgenes, (Figure S13D). Additionally, Oct4, Klf4, cMyc, and Nanog-reprogrammed cells could readily form embryoid bodies in vitro (Figure S13E). However, we found that leaky expression of transgenic Nanog, which is a potent inhibitor of embryonic stem cell differentiation (Chambers et al., 2003; Chambers et al., 2007), reduced the amount of differentiation in vitro (Figure S13D). We anticipate that efficient differentiation of cells created with Oct4, Klf4, cMyc, and Nanog will eventually require the use of an excisable transgenic Nanog cassette to completely remove ectopic Nanog expression. Although definitive proof of the pluripotency of these cells will be required to conclude that Nanog expression is sufficient to replace Sox2 in defined factor reprogramming, our results suggest this may be the case. Taken together however, our results demonstrate that RepSox inhibition of Tgf-β signaling bypasses the need for Sox2 in defined-factor reprogramming through the induction of Nanog.
Discussion
We have used a phenotypic chemical screen to identify compounds that can replace the reprogramming transcription factor Sox2 and have confirmed the mechanism by which the most potent compound acts: RepSox replaces Sox2 by inhibiting the broadly expressed Tgf-β signaling pathway (Attisano and Wrana, 2002) in cultures containing stable intermediate cells that are trapped in a partially reprogrammed state. This inhibition in turn leads to sustained transcription of Nanog, through which reprogramming is achieved in the absence of Sox2. These results demonstrate the feasibility of replacing the central reprogramming transgenes with small molecules that modulate discrete cellular pathways or processes rather than by globally altering chromatin structure. Furthermore, they show that the mechanisms by which these molecules act in reprogramming can be distinct from those of the factor(s) that they replace.
Importantly, and unlike many other studies (Mikkelsen et al., 2008; Shi et al., 2008a; Shi et al., 2008b; Utikal et al., 2009), the approach that we report here for replacing Sox2 did not rely on procurement of a highly specialized or rare cell type that already expresses Sox2. Furthermore, treatment with RepSox allowed the generation of iPS cells from both adult and embryonic fibroblasts with a frequency comparable to that of transduction with Sox2. Thus, reprogramming efficiency does not need to be compromised by small molecule replacement of transgenic factors.
We observed that instead of working on the initial fibroblast population to replace Sox2, RepSox acts on cellular intermediates formed by overexpression of Oct4, Klf4, and cMyc. Without RepSox treatment, these intermediates are trapped in an unproductive state. Unlike previously described partially reprogrammed cells (Mikkelsen et al., 2008; Silva et al., 2009), the RepSox-responsive intermediates could not be reprogrammed with AZA or 2i treatment, suggesting that they are distinct. In addition, we found that RepSox does not target any of the kinases inhibited by the 2i cocktail, indicating that it works through a different mechanism. Furthermore, 4-factor intermediates that reprogram with RepSox treatment are not responsive to AZA, indicating that they also are distinct.
These findings demonstrate that reprogramming can proceed in a step-wise fashion through different intermediates. Just as in a geographical setting where there are multiple routes to travel from point A to point B, there exist different intermediate states or “way stations” that somatic cells can transit through on the way to complete reprogramming. Interestingly, although our results indicate that defined-factor reprogramming with Oct4, Klf4, cMyc, and Sox2 can occur in the absence of Nanog, its induction is required for chemical reprogramming of both our RepSox-responsive intermediates and the recently described 2i-responsive intermediates made from Oct4, Klf4 and cMyc transduction of cells that express Sox2 endogenously (Silva et al., 2009). This indicates that commonalities can exist in the reprogramming routes used by some sets of distinct intermediates.
Originally, we found it surprising that Nanog was not included in the initial set of defined reprogramming factors (Takahashi and Yamanaka, 2006) given its critical role in maintaining pluripotency in ES cells (Boyer et al., 2005; Chambers et al., 2003) and its ability to stimulate reprogramming by cell-fusion (Silva et al., 2006). However, Takahashi and Yamanaka reported that a combination of 9 factors that included Oct4, Klf4, cMyc, and Nanog, but not Sox2, generated iPS colonies at a detectable rate (Takahashi and Yamanaka, 2006). This combination of factors included other genes that may have inadvertently lowered the rate of reprogramming, causing the combination of Oct4, Klf4, cMyc, and Nanog to be overlooked. Consistent with these data, work by Niwa and co-workers using inducible Sox2-null mES cells demonstrated that Sox2 is dispensable for modulation of the Oct-Sox enhancers that regulate pluripotent-specific gene expression and instead mainly governs pluripotency in ES cells by regulating the expression of Oct4 through other factors (Masui et al., 2007). Therefore, it is possible that Nanog may alleviate the requirement for Sox2 in reprogramming by stimulating or maintaining Oct4 expression. Indeed, Nanog is capable of maintaining Oct4 expression in mES cells (Chambers et al., 2003). Thompson and co-workers also reported that NANOG expression enhanced the reprogramming of human fibroblasts, but that it was not able to replace SOX2 in the presence of only OCT4 and LIN-28 (Yu et al., 2007). This may indicate that Klf4 is required for Nanog to function optimally in reprogramming and suggests that either they or the genes they modulate interact during the reprogramming process.
It is well known that approximately 90% of genes with promoters bound by OCT4 and SOX2 in human ES cells are also bound by NANOG (Boyer et al., 2005). Our result suggests that either Nanog or Sox2 may be sufficient to collaborate with Oct4 to modulate these genes and drive reprogramming. Although Nanog is not required for pluripotency, it safeguards ES cells against neuroectodermal and, to a more limited extent, mesodermal differentiation (Chambers et al., 2007; Vallier et al., 2009). Therefore, it is possible that Nanog functions in reprogramming by repressing differentiation signals, assisting in the transition to an undifferentiated state.
Interestingly, we found that RepSox is also able to functionally replace cMyc in reprogramming. Together, these observations highlight the fact that small molecules may functionally replace reprogramming transcription factors at either early or late stages of the process and that they can act by different mechanisms – by inducing the expression of the gene itself, or a closely related family member, or an unrelated gene that can functionally rescue the omission of the reprogramming transcription factor.
Our observation that a one-day treatment with RepSox can relieve the requirement for transgenic Sox2 indicates that unlike reprogramming using transgenic Oct4, Klf4, and Sox2, where each transgene must be expressed for several days (Sridharan et al., 2009; Stadtfeld et al., 2008), small molecules can act as switches to induce stable changes in gene expression that promote the completion of reprogramming. This could be an important concept for achieving purely chemical reprogramming since our data show that chemicals such as RepSox can affect cellular processes differently depending on the timing of administration.
As we have shown here, there need not always be a discrete, one-to-one mapping between the functions of the reprogramming factors and their chemical replacements. Thus it may be that reiterative screening in the presence of Sox2 replacement molecules will be required to identify compounds that can act in concert to replace Oct4 and Klf4. However, it will be of significant interest to determine whether the novel reprogramming compounds we have identified can collaborate with those previously described (Marson et al., 2008; Shi et al., 2008a; Silva et al., 2008) to replace the remaining reprogramming genes, opening a route to purely chemical reprogramming.
Experimental Procedures
Retroviral Infection
Retroviral infections were performed as previously described using the pMXs vector (Takahashi et al., 2007a). MEFs were infected with two to three pools of viral supernatant during a 72-hour period. The first day that viral supernatant was added was termed “day 1 post-infection.” For quantification, Oct4∷GFP+ colonies were counted at day 30 post-infection unless otherwise stated.
Small Molecule Screens
On day 4 post-infection, infected MEFs were trypsinized and re-seeded on irradiated feeders in 96-well plates at 2000 cells/well and cultured in mouse ES cell media (Knockout DMEM,15% Hyclone FBS, L-glutamine, penicillin/streptomycin, nonessential amino acids, β-mercaptoethanol, and 1000 U/ml LIF). The next day, compound stock solutions diluted in DMSO and VPA (Sigma) were added at a final concentration of 1 μM and 2 mM, respectively. VPA was removed after 1 week, and compound was re-applied every other day with each media change. Plates were scored for GFP+ colonies after 11 days of compound treatment.
Quantification of Oct4∷GFP+ iPS Cells Generated with Small Molecule Hit Compounds, SB431542, and Tgf-β antibodies
Retroviral infection and compound or antibody treatment was performed as in the original chemical screen. To quantify the numbers of GFP+ colonies produced in different conditions, the number of colonies in each well was counted and at least 2 different wells were counted and averaged. Concentrations of compounds and antibodies were the following: VPA (Sigma)- 2 mM, RepSox (Calbiochem)- 25 μM or 1 μM as noted, E-616451 (Calbiochem)- 3 μM, EI-275 (Biomol)- 3 μM, SB431542 (Sigma)- 25 μM or 2 μM as noted, TgfβII-specific antibody (R&D Systems, AB-12-NA)- 10 μg/ml, pan-Tgfβ antibody (R&D Systems, AB-100-NA)- 10 μg/ml. Unless otherwise noted, all chemical treatments were continuous from initial administration at day 4-5 post-infection until GFP+ colonies were scored at day 30 post-transduction. Fresh chemical was added at each media change.
Chemical Reprogramming of Stable Intermediate Cell Lines
Oct4∷GFP-negative colonies in Oct4, Klf4, and cMyc or Oct4, Klf4, cMyc, and Sox2-infected MEF cultures were picked, plated on irradiated feeders, and single colonies were picked after 1 week. The resulting cell lines were passaged with trypsin and grown in mES media on feeders until passage 4, at which time they were treated with RepSox (25 μM), AZA (500 μM), or both for 48 hours. For 2i treatment, CHIR99021 (Stemgent) was used at 3 μM and PD0325901 (Stemgent) was used at 1 μM. Oct4∷GFP+ colonies were scored 12 days after the beginning of chemical treatment. Treatments were performed in mES media containing FBS unless otherwise noted.
Supplementary Material
Acknowledgments
We thank E. Kiskinis, A. Arvanites, S. Bobrowicz, Weisenthal, R. Maehr, A. Kweudjeu, R. Gali, M. Yamaki, E. Massassa, R. Martinez, K. and Rosowski for technical assistance and S. Sullivan, K. Niakan, K. Rodolfa, S. Mekhoubad, I. Tabansky, C. Sasaki, D. Melton, and S. Chen for helpful discussions. This work was made possible by support provided by the Harvard Stem Cell Institute to L.R. and K.E. and by support from the NIH grant R01 HD046732-01A1 to K.E. E.S. and D.L. acknowledge support from the NIH (GM065400) and from the Howard Hughes Medical Institute. J.K.I. and F.P.D. are New York Stem Cell Foundation postdoctoral fellows. D.H. is a Helen Hay Whitney postdoctoral fellow. K.E. is a fellow of the John D. and Catherine T. MacArthur Foundation.
The authors are filing a patent based on the results reported in this paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amabile G, Meissner A. Induced pluripotent stem cells: current progress and potential for regenerative medicine. Trends Mol Med. 2009;15:59–68. doi: 10.1016/j.molmed.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- Boiani M, Kehler J, Scholer HR. Activity of the germline-specific Oct4-GFP transgene in normal and clone mouse embryos. Methods Mol Biol. 2004;254:1–34. doi: 10.1385/1-59259-741-6:001. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Gellibert F, Woolven J, Fouchet MH, Mathews N, Goodland H, Lovegrove V, Laroze A, Nguyen VL, Sautet S, Wang R, et al. Identification of 1,5-naphthyridine derivatives as a novel series of potent and selective TGF-beta type I receptor inhibitors. J Med Chem. 2004;47:4494–4506. doi: 10.1021/jm0400247. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hanke JH, Gardner JP, Dow RL, Changelian PS, Brissette WH, Weringer EJ, Pollok BA, Connelly PA. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck- and FynT-dependent T cell activation. J Biol Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008a;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008b doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009 doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim C, Moon J, Chung Y, Chang M, Han B, Ko S, Yang E, Cha KY, Lanza R, KIm K. Generation of Human Induced Pluripotent Stem Cells by Direct Delivery of Reprogramming Proteins. Cell Stem Cell. 2009;4 doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nature Cell Biology. 2007;9:11. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of Mouse Induced Pluripotent Stem Cells Without Viral Vectors. Science. 2008 doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008a;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008b;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Raya A, Kawakami Y, Morita M, Matsui T, Nakashima K, Gage FH, Rodriguez-Esteban C, Izpisua Belmonte JC. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:10294–10299. doi: 10.1073/pnas.0506945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007a;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007b;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thrasher AJa, G HB. Severe adverse event in clinical trial of gene therapy for X-SCID. 2007 http://wwwasgtorg/UserFiles/XSCIDstatementpdf.
- Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009;122:3502–3510. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, Rugg-Gunn P, et al. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


