The capacity of human embryonic stem cells (hESCs) to self-renew indefinitely in culture while retaining their ability to differentiate into all cell types suggests that they have enormous potential both in medical applications and as a research tool (Reubinoff et al., 2000; Thomson et al., 1998). Despite their immortal nature, there is a need for derivation of new hESC lines to meet emerging requirements for their use in cell replacement therapies, disease modeling, and basic research. These applications require maximizing the limited resource of donated or experimentally generated human embryos and has motivated our attempts to improve methods for the derivation of new HUES cell lines.
Following the derivation of 17 hESC lines (Cowan et al., 2004), we derived an additional 12 lines using the same method and found that the efficiency of these derivations varied greatly from experiment to experiment (Table S1). To better understand the variables that affect derivation efficiency, we explored methods for ICM isolation, and systematically investigated the relationship between preimplantation biology and the timing of embryonic stem cell derivation. We found that in vitro cultured human preimplantation embryos undergo major changes in morphology as well as expression of OCT4 and CDX2 from days 5 through 9 post-fertilization. We observed a peak of derivation efficiency using day 6 preimplantation embryos, corresponding to restriction of OCT4 to the ICM and CDX2 to the trophectoderm. These comparative studies have led to the derivation of 45 new hESC lines from 140 blastocysts, of which 22 cell lines are derived from sibling embryos. Global gene expression analysis of hESC lines reveal that lines derived on different days do not significantly differ from one another in transcriptional profile, but lines derived from different genetic backgrounds do significantly differ, suggesting that genetic background rather than the timing or method of derivation is a contributing factor in the variability observed among hESC lines.
The most widely used method for hESC derivation involves either chemical or enzymatic removal of the zona pellucida, followed by isolation of the inner cell mass (ICM) of the blastocyst by immunosurgery (Reubinoff et al., 2000; Thomson et al., 1998). In immunosurgery, cells of the trophectoderm (TE) are destroyed by brief exposure to antibodies directed against human cells in tandem with complement activity (Solter and Knowles, 1975). However, only high quality embryos with an intact TE can be subjected to this procedure, as only the structural integrity of the blastocyst prevents the ICM from also being destroyed. We reasoned that isolation of the ICM by laser-mediated ablation of the zona pellucida and TE might reduce exposure of the ICM to potentially cyto-toxic compounds.
The 584 frozen human embryos used in this study were donated for research following informed consent under protocols reviewed and approved by both the Committee on the Use of Human Subjects (IRB) and the Embryonic Stem Cell Research Oversight Committee (ESCRO) at Harvard University. Human zygotes and cleavage stage embryos were thawed and cultured to the blastocyst stage (Figure S1A). ICM isolation was carried out by exposing TE cells to cell-lethal laser pulses from a Xyclone laser (see also Turetsky et al., 2008) and subsequent removal of dead TE cells by using either piezo drill-assisted micromanipulation, or by repeated aspiration into a 50–75 μm glass capillary pipette (Figures S1B, C). The isolated ICM (Figure S1D) was then plated onto γ-irradiated mouse embryonic fibroblasts (MEFs) in hESC-conditioned derivation media.
Isolated ICMs attached to the MEF feeder cell layer within 24 hours, and 4 – 13 days later, gave rise to an ES cell outgrowth (Figures S1E-G) composed of cells with typical hESC morphology (Figure S1H), that could be expanded into cell lines (Figures S1I, J). HESC lines isolated by laser-surgery had a normal karyotype (Figure S1K) and expressed marker antigens found in pluripotent hESCs, including OCT4, SOX2, NANOG, TRA1-81, TRA1-60, and SSEA-4 (Figure S2A-F). Upon differentiation in vitro, via embryoid body formation (Figures S2G-I) and in vivo, via teratoma formation (Figures S2J-O), endoderm, mesoderm and ectoderm lineages were readily observed, demonstrating that these cell lines are bona-fide hESCs (Thomson et al., 1998; The International Stem Cell Initiative, 2007).
Next, we investigated the consequences of the presence or absence of TE cells in the derivation culture. We compared the efficiency of deriving hESCs following plating of intact blastocysts without ICM isolation with the efficiency following ICM isolation on days 5–9 of development (with the day of insemination representing day 0). While derivation from plating intact blastocysts has previously been reported (Baharvand et al., 2004; Bongso et al., 1994; Genbacev et al., 2005; Heins et al., 2004), we found that the efficiency was low (10% of blastocysts used), and required close outgrowth monitoring in order to isolate the ICM before it was lost to rapid differentiation (Table 1 and Figure S3).
Table 1.
Derivation Is Most Efficient on Day 6 Post-Fertilization.
| Day of ICM isolation | # blastocysts used for derivation | # attachment sites (% of blastocysts) | # ICM outgrowths | # cell lines | % derivation efficiency* | |||
|---|---|---|---|---|---|---|---|---|
| 5 | 19 | 6 (31) | 1 | 1 | 5 | ‡ | ||
| 6 | 27 | 22 (81) | 15 | 14 | 52 | |||
| 7 | 27 | 22(81) | 9 | 9 | 33 | |||
| 8 | 19 | 17 (90) | 5 | 5 | 26 | † | ||
| 9 | 11 | 10 (91) | 4 | 4 | 36 | |||
| No isolation | 10 | 10 (100) | 1 | 1 | 10 | |||
| Unknown≠ | 27 | ND | ND | 11 | 40 | |||
expressed as % according to the number of cell lines obtained per total number of blastocysts used for derivation. Results summarize derivations using both laser- and immunosurgery. A more detailed presentation of the results is given in Table S2.
P= 0.00002,
P= 0.008 (assuming binomial distribution).
HUES 18–28, (see also Table S1).
To investigate the effects of the timing of ICM isolation on hESC derivation, we systematically tested the derivation efficiencies with ICMs isolated from early blastocysts (day 5) through late blastocysts (day 9). During this prolonged culture period, we observed a number of morphological changes. Between day 5 and day 6, ICM cell number increased while cell size decreased, and the TE of high quality embryos expanded (Figure S4A). Both the ICM and TE of blastocysts continued to grow through day 6 of in vitro culture, but by day 7, the TE frequently collapsed and deteriorated, even in high quality embryos (Figure S4B). In contrast to the TE, cells in the interior of the embryo continued to grow. For embryo culture beyond day 6, a shift from Global medium to hESC-conditioned medium improved development, particularly in poor quality embryos with a small or indiscernible ICM. In extremely compromised embryos, we switched to conditioned media as early as day 5. Surprisingly, poor quality embryos without an ICM on days 5 or 6 often developed a distinct ICM after 1–2 days of additional culture (Figure S4C, D) in hESC-conditioned media. Extended culture of these embryos occasionally resulted in an atypical morphology where the interior cells of the blastocysts expanded to form a solid sphere (Figure S4E, F), but no disadvantage was observed in subsequent hESC derivation (Table 1, Table S2).
We found that hESC llines could be derived from embryos at days 5 through 9 after fertilization, with ICM isolation on day 6 resulting in the most efficient derivation of hESCs (Table 1). HESCs have previously been isolated on various days of development (Hovatta et al., 2003; Mitalipova et al., 2003; Stojkovic et al., 2004; Strom et al., 2007), and even from blastomeres and morula stage embryos (Chung et al., 2008; Klimanskaya et al., 2006; Strelchenko et al., 2004). The low number of cell lines generated, however, did not allow a conclusion to be drawn regarding the efficiency of derivation. Isolation of the ICM on day 6 resulted in a 10-fold increase over the derivation efficiency on day 5 (52%, n=27 versus 5%, n=19, respectively; P= 0.00002). This is also 5-fold higher than derivation without isolation of the ICM (52%, n=27 versus 10%, n=10, respectively; P= 0.008). Derivation efficiency correlated with the total number of ICM attachment sites to the feeder layer after ICM plating. The number of attachment sites and resulting cell lines was low on day 5 but increased on day 6 and remained high on days 7 through 9. Derivation efficiency on days 7 through 9 was slightly lower, but not significantly different from derivation on day 6 (P > 0.01). Poor quality embryos benefited greatly from extended culture in hESC-conditioned media, as only 1–2 days of additional culture promoted ICM growth and allowed hESC derivation (Figures S4C-F, Table S2). This approach allowed for derivation of embryonic stem cells from embryos which would have otherwise been unlikely to give rise to hESC lines (e.g. grade 2CC embryos: HUES 33, 43, 57; grade 3CC embryos: HUES 32, 54, 59, 64). In summary, by combining extended embryo culture in hESC-conditioned media with laser-assisted isolation of the ICM on day 6 of preimplantation development, a derivation efficiency of 50% can be routinely achieved.
Using these methods, we succeeded in deriving a total of 22 sibling cell lines from seven donor couples (Table S3). To verify their identical maternal origin and demonstrate their karyotypic individuality, we sequenced the hypervariable regions of the mitochondrial genome, and a combination of nuclear short tandem repeats (STR) and compared them to unrelated hESC lines. While their mitochondrial genome was identical, their nuclear genome was different but highly related, sharing more than 50% of STR alleles (Table S4, Figures S5, 6).
Next, we explored whether differences in efficiency of hESC derivation correlate with a change in localization and expression of the respective ICM and TE markers, OCT4 and CDX2. It has been shown in mouse preimplantation embryos that by the 65–128-cell stage, OCT4 becomes restricted to the ICM, and CDX2, to the TE, at approximately 3.5 days post-fertilization in in vitro cultured blastocysts (Dietrich and Hiiragi, 2007; Ralston and Rossant, 2008). We therefore asked whether OCT4 and CDX2 expression and localization in human preimplantation embryos could explain why derivation after day 6 of in vitro culture is more efficient.
Embryos used in stem cell derivation are most commonly staged in terms of days post-fertilization, with the day of insemination representing day 0. We observed that the majority of human embryos at day 5 (approximately 40–75-cells, blastocyst grade 3) had high levels of OCT4 expression in both the ICM and TE, while some embryos had begun to express CDX2 in some, but not all, of the TE (Figure 1A). By day 6 (approximately 75–145-cells, blastocyst grades 4 and 5), blastocysts exhibited a clear restriction of high levels of OCT4 expression to the ICM and of CDX2 to the TE (Figure 1B). By day 8, OCT4 expression was confined to a small number of cells in the presumptive ICM (Figure 1C). As most of cells of the day 8 embryo did not express OCT4 or CDX2, this observation suggests that a cell type other than ICM or TE proliferates at this late stage in vitro. While the identity of these cells is unclear, their proliferation and survival are minimal upon ICM explant for hESC derivation.
Figure 1. OCT4 and CDX2 Expression During Human Preimplantation Development and Gene Expression Analysis of hESC Lines.
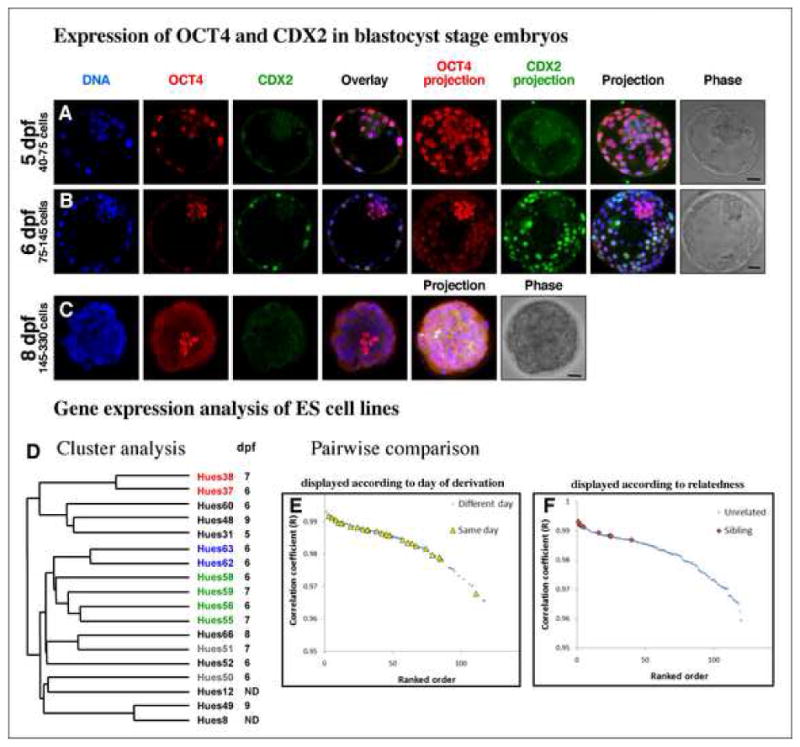
Representative human preimplantation embryos on day 5 (A), day 6 (B) and day 8 (C) stained for the TE marker, CDX2, and ICM marker, OCT4. Total cell numbers for each day indicated. Bar indicates relative size for comparison. (D) Cluster analysis of 16 hESC lines derived on different days of development. Sibling cell lines marked with identical colors. (E, F) Pair-wise comparison of global gene expression profiles between different cell lines. Comparison of 16 cell lines yields 16x15/2=120 data points for all possible pair-wise comparisons. The higher the Pearson correlation coefficient (R), the more similar the gene expression profile between two lines. Pairs were ordered from high to low according to the Pearson correlation coefficient (ranked order). (E) Pairwise comparisons of hESC lines derived on the same versus different days. (F) Pairwise comparisons of sibling hESC lines versus unrelated lines.
Together, these observations suggest that the ICM and TE cells of early day 5 blastocysts may not yet be restricted to either fate, and therefore isolated ICM cells only rarely give rise to embryonic stem cells. The restriction of OCT4 expression to the ICM and of CDX2 to the TE on day 6, together with the increase in ICM cell number, may explain why derivation on day 6 is most efficient. Once segregation of the ICM and TE populations has occurred, derivation efficiency remains high on days 7–9 (Table 1) despite a reduction in the number of OCT4-expressing cells.
The morphological and molecular differences we observed between preimplantation embryos at various developmental time points led us to question whether hESCs derived from embryos on days 5–9 differ in their gene expression programs. It was suggested that pluripotent stem cells isolated from the epiblast of mouse peri-implantation embryos are the mouse equivalent to hESCs. These epiblast stem cells differ in their gene expression profile from mouse ES cells isolated from preimplantation stage embryos and share similarities to hESCs (Brons et al., 2007; Tesar et al., 2007). We therefore examined whether hESC lines derived from days 5–9 generate different types of stem cell lines. We found that hESC lines isolated from different days of development were identical in their growth requirements and expressed the same pluripotency-associated antigens (Table S2). We further analyzed the gene expression profile of 16 hESC lines derived on days 5–9 of development and found that these lines did not group into separate clusters based on their day of derivation (Figure 1D). The distribution of pair-wise correlation coefficients (R) between lines derived from the same day of development was indistinguishable from lines derived from different days (Student’s t-test p=0.12, Figure 1E). In contrast, when all pair-wise correlation coefficients were grouped according to genetically related versus unrelated lines, the similarity between sibling lines was significantly higher than between unrelated lines (Student’s t-test p=1.1×10−8, Figure 1F). These observations suggest that the gene expression differences among hESC lines are due to genetic parentage rather than the day or method of derivation. Such differences likely contribute to the variation in differentiation propensity reported between hESC lines (The International Stem Cell Initiative, 2007; Osafune et al., 2008).
The 50% derivation efficiency we achieved using day 6 embryos, laser surgery, and modified embryo culture parameters was higher than previously reported from either our or other laboratories (Cowan et al., 2004; Lerou et al., 2008; Thomson et al., 1998) (Table S5). The increased efficiency and reliability of this method has also allowed us to derive cohorts of stem cell lines that would not have previously been obtainable, including 22 sibling cell lines. These sibling cell lines will be a valuable resource for further investigation of the effects of genetic background on the growth characteristics, pluripotency and differentiation potential of hESCs.
Our findings increase the probability of successful derivation from rare embryos such as those obtained after preimplantation genetic diagnosis or somatic cell nuclear transplantation. A detailed understanding of the naturally occurring variations among hESC lines will also be important for insight into the genetic regulation of human development as well as for evaluating pluripotent stem cells generated by reprogramming.
Supplementary Material
Acknowledgments
We thank E.Trish for technical assistance, K. Osafune for help with ES cell culture, J. Sprague and G. Hardiman for assistance in microarray analysis. A.E.C. is a fellow of the Jane Coffin Childs Memorial Fund for Medical Research and Merck Research Laboratories. H.A. is funded by Health and Labour Sciences Research Grants, Japan. K.E. is a fellow of the John D. and Catherine T. MacArthur Foundation. K.Z. and J.D. are funded by the UCSD new faculty startup fund. J.D. is a CIRM post-doctoral fellow. This work was supported by the Harvard Stem Cell Institute as well as the Stowers Medical Institute and Howard Hughes Medical Institute.
Footnotes
Supplemental data including experimental procedures, 6 supplemental figures, and 5 supplemental tables can be found with this article online at: http://www.cellstemcell.com/cgi/content/full
To request stem cell lines, consult Table S2 for availability for distribution. Note that Harvard University-derived lines are designated as HUESX, with X representing the cell line number. More information can be found at: (http://www.mcb.harvard.edu/melton/hues).
COMPETING INTEREST STATEMENT
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akutsu H, Cowan CA, Melton D. Human embryonic stem cells. Methods Enzymol. 2006;418:78–92. doi: 10.1016/S0076-6879(06)18005-2. [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Baharvand H, Ashtiani SK, Valojerdi MR, Shahverdi A, Taee A, Sabour D. Establishment and in vitro differentiation of a new embryonic stem cell line from human blastocyst. Differentiation. 2004;72:224–229. doi: 10.1111/j.1432-0436.2004.07205005.x. [DOI] [PubMed] [Google Scholar]
- Bongso A, Fong CY, Ng SC, Ratnam S. Isolation and culture of inner cell mass cells from human blastocysts. Hum Reprod. 1994;9:2110–2117. doi: 10.1093/oxfordjournals.humrep.a138401. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Chen AE, Melton DA. Derivation of Human Embryonic Stem Cells by Immunosurgery. Journal of Visualized Experiments. 2007 doi: 10.3791/3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Klimanskaya I, Becker S, Li T, Maserati M, Lu SJ, Zdravkovic T, Ilic D, Genbacev O, Fisher S, et al. Human embryonic stem cell lines generated without embryo destruction. Cell Stem Cell. 2008;2:113–117. doi: 10.1016/j.stem.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, et al. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril. 2000;73:1155–1158. doi: 10.1016/s0015-0282(00)00518-5. [DOI] [PubMed] [Google Scholar]
- Genbacev O, Krtolica A, Zdravkovic T, Brunette E, Powell S, Nath A, Caceres E, McMaster M, McDonagh S, Li Y, et al. Serum-free derivation of human embryonic stem cell lines on human placental fibroblast feeders. Fertil Steril. 2005;83:1517–1529. doi: 10.1016/j.fertnstert.2005.01.086. [DOI] [PubMed] [Google Scholar]
- Heins N, Englund MC, Sjoblom C, Dahl U, Tonning A, Bergh C, Lindahl A, Hanson C, Semb H. Derivation, characterization, and differentiation of human embryonic stem cells. Stem Cells. 2004;22:367–376. doi: 10.1634/stemcells.22-3-367. [DOI] [PubMed] [Google Scholar]
- Hovatta O, Mikkola M, Gertow K, Stromberg AM, Inzunza J, Hreinsson J, Rozell B, Blennow E, Andang M, Ahrlund-Richter L. A culture system using human foreskin fibroblasts as feeder cells allows production of human embryonic stem cells. Hum Reprod. 2003;18:1404–1409. doi: 10.1093/humrep/deg290. [DOI] [PubMed] [Google Scholar]
- Klimanskaya I, Chung Y, Becker S, Lu SJ, Lanza R. Human embryonic stem cell lines derived from single blastomeres. Nature. 2006;444:481–485. doi: 10.1038/nature05142. [DOI] [PubMed] [Google Scholar]
- Lerou PH, Yabuuchi A, Huo H, Takeuchi A, Shea J, Cimini T, Ince TA, Ginsburg E, Racowsky C, Daley GQ. Human embryonic stem cell derivation from poor-quality embryos. Nat Biotechnol. 2008;26:212–214. doi: 10.1038/nbt1378. [DOI] [PubMed] [Google Scholar]
- Mitalipova M, Calhoun J, Shin S, Wininger D, Schulz T, Noggle S, Venable A, Lyons I, Robins A, Stice S. Human embryonic stem cell lines derived from discarded embryos. Stem Cells. 2003;21:521–526. doi: 10.1634/stemcells.21-5-521. [DOI] [PubMed] [Google Scholar]
- Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- Solter D, Knowles BB. Immunosurgery of mouse blastocyst. Proc Natl Acad Sci U S A. 1975;72:5099–5102. doi: 10.1073/pnas.72.12.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojkovic M, Lako M, Stojkovic P, Stewart R, Przyborski S, Armstrong L, Evans J, Herbert M, Hyslop L, Ahmad S, et al. Derivation of human embryonic stem cells from day-8 blastocysts recovered after three-step in vitro culture. Stem Cells. 2004;22:790–797. doi: 10.1634/stemcells.22-5-790. [DOI] [PubMed] [Google Scholar]
- Strelchenko N, Verlinsky O, Kukharenko V, Verlinsky Y. Morula-derived human embryonic stem cells. Reprod Biomed Online. 2004;9:623–629. doi: 10.1016/s1472-6483(10)61772-5. [DOI] [PubMed] [Google Scholar]
- Strom S, Inzunza J, Grinnemo KH, Holmberg K, Matilainen E, Stomberg AM, Blennow E, Hovatta O. Mechanical isolation of the inner cell mass is effective in derivation of new human embryonic stem cell lines. Human Reprod. 2007;22:3051–3058. doi: 10.1093/humrep/dem335. [DOI] [PubMed] [Google Scholar]
- Sullivan S, Cowan CA, Eggan K. Human Embryonic Stem Cells - A Practical Handbook. Wiley; 2007. [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, et al. The International Stem Cell Initiative. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Turetsky T, Aizenman E, Gil Y, Weinberg N, Shufaro Y, Revel A, Laufer N, Simon A, Abeliovich D, Reubinoff BE. Laser-assisted derivation of human embryonic stem cell lines from IVF embryos after preimplantation genetic diagnosis. Hum Reprod. 2008;23:46–53. doi: 10.1093/humrep/dem351. [DOI] [PubMed] [Google Scholar]
- Wang Q, Fang ZF, Jin F, Lu Y, Gai H, Sheng HZ. Derivation and growing human embryonic stem cells on feeders derived from themselves. Stem Cells. 2005;23:1221–1227. doi: 10.1634/stemcells.2004-0347. [DOI] [PubMed] [Google Scholar]
- Zhang X, Stojkovic P, Przyborski S, Cooke M, Armstrong L, Lako M, Stojkovic M. Derivation of human embryonic stem cells from developing and arrested embryos. Stem Cells. 2006;24:2669–2676. doi: 10.1634/stemcells.2006-0377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


