Abstract
Gemcitabine hydrochloride (HCl) is approved for the treatment of a wide spectrum of solid tumors. However, the rapid development of resistance often makes gemcitabine less efficacious. In the present study, we synthesized several novel lipophilic monophosphorylated gemcitabine derivatives, incorporated them into solid lipid nanoparticles, and then evaluated their ability to overcome major known gemcitabine resistance mechanisms by evaluating their in vitro cytotoxicities in cancer cells that are deficient in deoxycytidine kinase (dCK), deficient in human equilibrative nucleoside transporter (hENT1), over-expressing ribonucleotide reductase M1 subunit (RRM1), or over-expressing RRM2. In dCK deficient cells, the monophosphorylated gemcitabine derivatives and their nanoparticles were up to 86-fold more cytotoxic than gemcitabine HCl. The majority of the gemcitabine derivatives and their nanoparticles were more cytotoxic than gemcitabine HCl in cells that over-expressing RRM1 or RRM2, and the gemcitabine derivatives in nanoparticles were also resistant to deamination by deoxycytidine deaminase. The gemcitabine derivatives (in nanoparticles) hold a great potential in overcoming gemcitabine resistance.
Keywords: Gemcitabine resistance, nanoparticles, in vitro cytotoxicity, cancer cells
1. Introduction
Gemcitabine (2′,2′-difluorodeoxycytidine, dFdC) HCl is approved for the treatment of a wide spectrum of solid tumors including pancreatic, non-small-cell lung cancer, breast, and bladder cancers (Candelaria, M. et al., 2010; Barton-Burke 1999; Kalykaki et al., 2008; Sandler et al., 2000; Zucali et al., 2008; Cetina et al., 2004; Candelaria, Myrna et al., 2007). However, tumor cells often acquire resistance during or after gemcitabine treatment (Bergman, Andries M. et al., 2002; Andersson et al., 2009; Sezgin et al., 2005). Gemcitabine is a polar deoxycytidine analogue. It requires nucleoside transporters to translocate across the cellular membrane (Heinemann V et al., 1995). Clinical data showed that patients with tumors with a decreased expression of hENT1, a nucleoside transporter, have a significantly lower survival rate after gemcitabine treatment than those with tumors that express a higher level of hENT1 (Giovannetti et al., 2006; Mey et al., 2006; Spratlin et al., 2004). More than 90% gemcitabine that are internalized into cells are deaminated by deoxycytidine deaminase (dCDA) to form inactive 2′-deoxy-2′,2′-difluorouridine (dFdU) (Immordino et al., 2004). Therefore, deamination affects the efficacy of gemcitabine adversely (Immordino et al., 2004). Gemcitabine is a prodrug, which needs to be phosphorylated to gemcitabine monophosphate (dFdCMP) by dCK (Kroep et al., 2002; Bouffard et al., 1993). Subsequently, dFdCMP is phosphorylated by nucleotide kinases to di- and tri-phosphorylated gemcitabine (dFdCDP and dFdCTP, respectively) that are active metabolites of gemcitabine (Bergman, Andries M. et al., 2002; Mini et al., 2006; Ueno et al., 2007). Therefore, tumor cells deficient in dCK are resistant to gemcitabine. Clinical studies in patients with resected pancreatic adenocarcinoma showed a strong correlation between low level of dCK expression and poor clinical outcomes after gemcitabine-based adjuvant therapy (Maréchal et al., 2010). Disease-free survival was significantly longer in patients having high levels of dCK expression (38.6–77.5 months) than in patients having low levels of dCK expression (2.9–9.6 months) (Maréchal et al., 2010). The anti-proliferative activity of gemcitabine is known to be exerted mainly through the inhibition of DNA synthesis by masked chain termination and inhibition of DNA polymerase by dFdCTP (Huang and Plunkett 1995). RR is required to convert ribonucleotides to deoxyribonucleotides. dFdCDP inhibits RR, leading to the depletion of deoxynucleotide triphosphate (dNTP) pool and the enhancement of the activity of gemcitabine (Heinemann et al., 1992). Active RR composes of two homodimers of non-identical subunits, the large RRM1 subunit and the small RRM2 subunit (Candelaria, M. et al., 2010). Both pre-clinical and clinical data have shown that tumor cells that over-express of RRM1 or RRM2 are resistant to gemcitabine treatment (Jordheim et al., 2005; Boukovinas et al., 2008).
The most commonly reported acquired resistance to gemcitabine is dCK deficiency (Bergman, Andries M. et al., 2002; Gregoire et al., 2002). Inefficient intracellular monophosphorylation of gemcitabine may reduce the efficacy of gemcitabine drastically. However, only a few gemcitabine analogues with a phosphate moiety have been reported to address the problem. The first synthesized compound to address dCK deficiency was a gemcitabine derivative linked with a C-1 thioether, C-2 oxyether phospholipid (Alexander et al., 2003; Alexander et al., 2005; Alexander and Kucera 2005). Wu et al. then introduced the gemcitabine phosphoramidate prodrug, which was shown to be 4 times more cytotoxic than gemcitabine in dCK deficient cell lines (Wu et al., 2007). In the present study, several new nucleoside analogues were designed and synthesized to overcome major gemcitabine resistance mechanisms. All the compounds were linked with a monophosphate group at the 5′C of the deoxyribofuranose ring, and thus, have the potential to bypass the rate limiting step of dCK-dependent gemcitabine activation. Moreover, by attaching a long chain stearoyl group on gemcitabine, it is expected that nucleoside transporters may no longer be required for the lipophilized gemcitabine to enter cells. Furthermore, it is expected that the lipophilic monophosphorylated gemcitabine derivatives may also become resistant to deamination by dCDA (Song et al., 2005).
Finally, we previously reported that the incorporation of another lipophilic gemcitabine derivative, 4-N-stearoyl gemcitabine (GemC18), into solid lipid nanoparticles engineered from lecithin/glycerol monostearate-in-water emulsions rendered the gemcitabine less sensitive to resistance caused by the over-expression of RRM1 (Chung et al., 2012; Sloat et al., 2011). Therefore, the solid lipid nanoparticle formulation was extended to the novel lipophilic monophosphorylated gemcitabine derivatives as well. In order to evaluate the extent to which the novel gemcitabine derivatives, alone or in nanoparticles, can overcome various gemcitabine resistance mechanisms, their in vitro cytotoxicities were determined in cancer cells that are dCK deficient, hENT1 deficient, over-expressing RRM1, or over-expressing RRM2. In addition, the ability of selected gemcitabine derivative-containing nanoparticles to competitively inhibit the deamination activity of dCDA was evaluated and compared to that of gemcitabine HCl to understand the extent to which the gemcitabine derivatives in nanoparticles may become less sensitive to deamination.
2. Materials and methods
2.1. General materials and methods
Proton NMR spectra were recorded on a 300 MHz Varian UNITY Plus or a 500 MHz Varian INOVA. Chemical shifts (δ) of 1H NMR spectra were recorded in parts per million (ppm) relative to tetramethylsilane (TMS), which was the reference (δ = 0 ppm). 1H NMR data are reported according to the following order: chemical shift, integration (i.e., number of hydrogen atoms), multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad, brs = broad singlet), and coupling constant in Hertz (Hz). High resolution mass spectra were acquired in electronspray positive and negative ionization modes by direct injection onto an IonSpec 9.4T QFT-FTMS system in the mass spectrometry facility of the Department of Chemistry and Biochemistry at the University of Texas at Austin. The concentrations of deoxycytidine (dCyd) and deoxyuridine (dUrd) in the dCDA assay were determined using an Agilent 1260 Infinity high performance liquid chromatography (HPLC) with an Agilent ZORBAX Eclipse Plus C18 column (4.6 × 150 mm, 5 μm) attached to a ZORBAX Eclipse Plus guard column (Agilent Technologies, Inc., Santa Clara, CA).
All commercially available chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO) or Thermo Fisher Scientific Inc. (Pittsburgh, PA) and were used as received unless noted. Gemcitabine HCl was from U.S. Pharmacopeia (Rockville, MD). Soy lecithin was from Alfa Aesar (Ward Hill, MA). The 1-hydroxy-7-azabenzotriazole (HOAt) was from CreoSalus, Inc. (Louisville, KY). Air or moisture-sensitive reactions were performed under an atmosphere of argon. Thin-layer chromatography (TLC) on Whatman silica gel plates (UV254) from Fisher Scientific was used to monitor the reaction progress. Silica gel – grade 60 (230–400 mesh) from Fisher Scientific was used for column chromatography to purify reaction products. The chemical structures of final compounds were confirmed using NMR and high resolution mass spectrometry.
2.2. Chemical syntheses (Scheme 1)
Scheme 1.
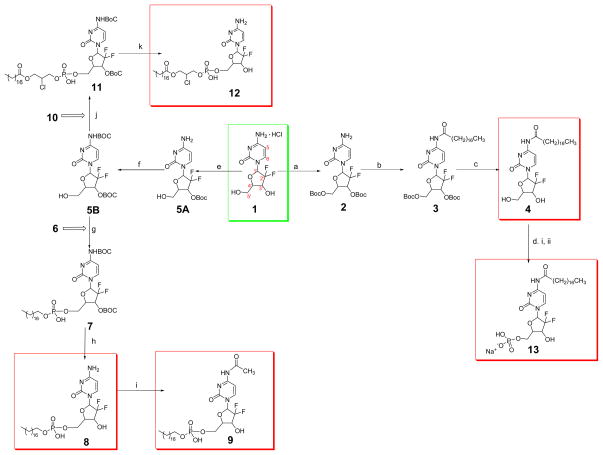
Reagents and conditions: (a) Boc2O, KOH, 1,4 dioxane, 22 °C; (b) CH3(CH2)16COOH, EDCI, HOAt, DCM, rt; (c) TFA, DCM, rt; (d) i. POCl3, triethylamine, DCM, reflux 2 h; ii. NaHCO3, rt, 15 h; (e) Boc2O, Na2CO3, dioxane, H2O; (f) Boc2O, dioxane, 37 °C, 250 rpm, 72 h; (g) 6, TPS, pyridine, 38–40 °C, 24 h; (h) TFA, DCM; (i) Acetic anhydride, CH3OH, reflux; (j) 10, TPS, pyridine, 38–40 °C, 24 h; (k) TFA, DCM.
2.2.1. 4-N-stearoyl gemcitabine (4)
Compound 4 or GemC18 was synthesized as previously reported (Guo and Gallo 1999; Immordino et al., 2004). Gemcitabine HCl salt (1) (200 mg, 0.67 mmol) in 13.3 mL of 1N potassium hydroxide (KOH) was cooled to 4°C. To this solution, di-tert-butyl dicarbonate (Boc2O, 1.483 g, 6.8 mmol) in anhydrous dioxane (13.3 mL) was added over 10 min under argon atmosphere. The reaction mixture was stirred at 22°C for 1 h, followed by an extraction procedure (i.e., The mixture was extracted with ethyl acetate (EtOAc)) and a workup protocol (i.e., The organic layer was washed with brine, dried over anhydrous sodium sulfate (Na2SO4), filtered and solvent was removed under reduced pressure). The residue was added to Boc2O (1.483 g, 6.8 mmol) in anhydrous dioxane (13.3 mL) and 1N KOH (13.3 mL) at 22°C. The reaction progress was monitored by TLC. After 1 h, the extraction and workup protocols were repeated, and the crude product was purified by column chromatography (dichloromethane (DCM) : acetone, 1:1). The desired product fractions were pooled and dried to yield 219 mg of 2 (yield of 71%). 1H NMR (500 MHz, acetone-d6) δ 7.60 (1H, d, J = 7.6 Hz, 6-CH), 6.34 (1H, brs, 1′-CH), 5.97 (1H, d, J = 7.6 Hz, 5-CH), 5.29 (1H, brs, 3′-CH), 4.53–4.39 (3H, m, 4′-CH, 5′A-CH, 5′B-CH), 2.82 (2H, s, NH2) 1.50, 1.47 (18H, two s, (CH3)3C). A solution of 2 (219 mg, 0.47 mmol), stearic acid (149 mg, 0.52 mmol) and HOAt (70 mg, 0.52 mmol) in anhydrous DCM was pre-cooled to 4°C, and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDCI) (109 mg, 0.57 mmol) was added. The mixture was de-gassed by vacuum sonication and then stirred at room temperature under argon for about 40 h. Water (15 mL) was added to the reaction mixture and extracted with the mixture of EtOAc and hexane (2:1). The combined organic phase was washed with saturated ammonium chloride (NH4Cl) followed by the workup protocol as aforementioned, and the residue was purified by column chromatography (EtOAc : Hexane, 3:7). The conjugated amide 3 was isolated as a white powder (319 mg, 92%). 1H NMR (300 MHz, acetone-d6) δ 9.90 (1H, s, NHCO), 8.03 (1H, d, J = 7.8 Hz, 6-CH), 7.45(1H, d, J = 7.5 Hz, 5-CH), 6.38 (1H, t, J = 8.7 Hz, 1′-CH), 5.40–5.30 (1H, m, 3′-CH), 4.56–4.44 (3H, m, 4′-CH and 5′-CH2), 2.57 (2H, t, J = 7.5 Hz, CO–CH2), 1.71–1.65 (2H, m, CO–CH2–CH2), 1.50, 1.47 (18H, two s, (CH3)3C), 1.40–1.20 (28H, m, CH2), 0.90–0.87 (3H, m, terminal CH3). To a stirred solution of the compound 3 (319 mg, 0.44 mmol) in DCM (7 mL), about 1.5 ml of trifluroacetic acid (TFA) was added. After 2 h, excess TFA was removed under reduced pressure. The concentrated sample was co-distilled with DCM for 5 times. The crude sample was chromatographed on silica gel (DCM : ethanol, 94:6) (Immordino et al., 2004). The final product 4 was a white powder (162 mg, 70%). 1H NMR (500 MHz, pyridine-d5) δ 11.97 (1H, s, NHCO), 8.75 (1H, d, J = 7.6 Hz, 6-CH), 7.74 (1H, d, J = 7.6 Hz, 5-CH), 6.99 (1H, t, J = 7.2 Hz, 1′-CH), 5.18–5.11 (1H, m, 3′-CH), 4.47–4.28 (3H, overlapping m, 4′-CH and 5′-CH2), 2.67 (2H, t, J = 7.4 Hz, CO–CH2), 1.83–1.76 (2H, m, CO–CH2–CH2), 1.34–1.20 (28H, m, CH2), 0.87 (3H, t, J = 6.9 Hz, terminal CH3). ESI-HRMS [M+H] + m/z calculated for C27H46F2N3O5: 530.3406, found: 530.3401.
2.2.2. 2′-2′-difluoro-cytosine-5′-octadecylphosphate (8) (Bligh and Dyer 1959; Guo and Gallo 1999; Perie et al., 1990)
The mixture of gemcitabine HCl (200 mg, 0.67 mmol) and Na2CO3 (354 mg, 3.3 mmol) in water (3.3 mL) and dioxane (13.3 mL) was added to Boc2O (147 mg, 0.67 mmol). After 48 h of stirring at room temperature, 15 mL of water was added, followed by the extraction and workup procedure mentioned above. The crude sample was chromatographed on silica gel (DCM : acetone, 1:2) to obtain 212 mg of 3′-O-(tert-butoxycarbonyl)-2′-2′-difluoro-cytidine (5A) (87%). 1H NMR (300 MHz, acetone-d6) δ 7.75 (1H, d, J = 7.5 Hz, 6-CH), 6.34 (1H, t, J = 9.1 Hz, 1′-CH), 6.04 (1H, d, J = 7.8 Hz, 5-CH), 5.40–5.31 (1H, m, 3′-CH), 4.23–4.19 (1H, m, 4′-CH), 4.00–3.82 (2H, overlapping m, 5′A-CH, 5′B-CH), 1.51(9H, s, (CH3)3C). Boc2O (1.26 g, 5.8 mmol) was added to a stirred solution of 5A (212 mg, 0.58 mmol) in 10 mL of dioxane. The resultant mixture was maintained at 37°C in a rotary shaker at 250 rpm for 3 days. Water (25 mL) was added to the sample and followed by the extraction and workup protocol mentioned above. The concentrated sample was chromatographed on silica gel (EtOAc : hexanes, 1:1) to obtain the desired product of 4-N-3′-O-Bis(tert-butoxycarbonyl)-2′-2′-difluoro-cytidine (5B) (196 mg). 1H NMR (300 MHz, acetone-d6) δ 8.25 (1H, d, J = 7.5 Hz, 6-CH), 7.30 (1H, d, J = 7.5 Hz, 5-CH), 6.36 (1H, t, J = 8.6 Hz, 1′-CH), 5.37–5.28 (1H, m, 3′-CH), 4.34–4.28 (1H, m, 4′-CH), 4.08–3.98 (1H, m, 5′A-CH), 3.87 (1H, m, 5′B-CH), 1.52, 1.50 (18H, two s, (CH3)3C). Octadecanol (5 g, 18.48 mmol) and triethylamine (4.8 g, 47.52 mmol) were partially dissolved in 50 mL of DCM under argon. Phosphorus oxychloride (POCl3) (2.8 g, 18.48 mmol) was added drop-wise and heated to reflux for 2 h. The reaction mixture was filtered to remove triethylamine hydrochloride, and the filtrate was added to 0.2 N sodium bicarbonate (NaHCO3) (370 mL). After 15 h stirring at room temperature, 370 mL of acetone was added, and the white precipitate was recovered by filtration. The precipitate was washed with acetone and re-dissolved in 400 mL of water. Another 260 mL of acetone was added, and the precipitate was recovered, washed with acetone, dissolved in a homogeneous mixture of 200 mL of chloroform (CHCl3), 400 mL of methanol (CH3OH), and 200 mL of 0.1 N HCl, and stirred for 1 h at room temperature. A mixture of 200 mL of CHCl3 and 200 mL of water was added, and the organic layer was isolated (Bligh and Dyer 1959; Perie et al., 1990). The aqueous phase was extracted with CHCl3 (2 × 100 mL). The combined organic layer was evaporated to dryness and lyophilized to obtain desired product of octadecylphosphate (6) (2.2 g, 34%). 1H NMR (300 MHz, CDCl3 : CD3OD, 4:1) δ 3.98 (2H, q, J = 6.8 Hz, CH2OP), 1.70–1.62 (2H, m, CH2CH2OP), 1.39–1.19 (30H, m, CH2 from C18 chain), 0.88 (3H, t, J = 6.8 Hz, terminal CH3). ESI-HRMS [M-H]− m/z calculated for C18H38O4P−: 349.2513, found: 349.2515. The powders of compound 5 (100 mg, 0.22 mmol) and octadecylphosphate (220 mg, 0.62 mmol) were mixed and lyophilized for 15 h. To the lyophilized powder, 2,4,6-triisopropylbenzenesulfonyl chloride (TPS) (154 mg, 0.5 mmol) and 2 mL of pyridine were added under argon environment, and the reaction was stirred at 38–40°C for 24 h. A few drops of water were added, and solvent was removed under reduced pressure. The crude oil was chromatographed on silica gel, eluting first with CHCl3 : CH3OH (24:1) and then with CHCl3 : CH3OH (9:1) (Alexander et al., 2003). The fractions of the desired product (Rf = 0.2 in CHCl3 : CH3OH, 9:1) were pooled, and 4-N-3′-O-Bis(tert-butoxycarbonyl)-2′-2′-difluoro-cytidine-5′-octadecylphosphate (7) was isolated as a white powder (156 mg, 89%). 1H NMR (300 MHz, CDCl3 : CD3OD, 4:1) δ 7.94 (1H, d, J = 7.2 Hz, 6-CH), 7.30 (1H, d, J = 7.5 Hz, 5-CH), 6.35 (1H, t, J = 8.6 Hz, 1′-CH), 5.29–5.18 (1H, m, 3′-CH), 4.29–4.10 (3H, overlapping m, 4′-CH, 5′A-CH, 5′B-CH), 3.89 – 3.78 (2H, m, CH2OP), 1.60 (2H, t, J = 6.6 Hz, CH2CH2OP), 1.52, 1.50 (18H, two s, (CH3)3C), 1.39–1.19 (30H, m, CH2 from C18 chain), 0.88 (3H, t, J = 6.6 Hz, terminal CH3). ESI-HRMS [M-H]− m/z calculated for C37H63F2N3O11P−: 794.4174, found: 794.4162. To a stirred solution of 7 (95 mg, 0.12 mmol) in 6 mL of DCM, about 0.9 ml of TFA was added. This solution was stirred at room temperature for 2 h. Excess TFA was removed under reduced pressure, and the concentrated sample was co-distilled with DCM for 5 times. The crude sample was column-purified on silica gel by eluting with 20%, 40%, and 50% CH3OH in CHCl3, sequentially. The desired fractions with the Rf value of 0.25 (CHCl3 : CH3OH, 3:2) were pooled and evaporated to dryness to yield 41 mg of 8 (58%). 1H NMR (300 MHz, CDCl3:CD3OD, 4:1) δ 7.88 (1H, d, J = 8.1 Hz, 6-CH), 6.17 (1H, t, J = 6.9 Hz, 1′-CH), 6.06 (1H, d, J = 7.2 Hz, 5-CH), 4.42–4.00 (4H, overlapping m, 3′-CH, 4′-CH,5′A-CH, 5′B-CH), 3.94–3.71 (2H, m, CH2OP), 1.62 (2H, t, J = 6.6 Hz, CH2CH2OP), 1.39–1.19 (30H, m, CH2 from C18 chain), 0.88 (3H, t, J = 6.6 Hz, terminal CH3). ESI-HRMS [M-H]− m/z calculated for C27H47F2N3O7P−: 594.3125, found: 594.3125.
2.2.3. 4-N-acetyl-2′-2′-difluoro-cytosine-5′-octadecylphosphate (9) (Watanabe and Fo 1966)
Acetic anhydride (150 μL) and 8 (16.5 mg, 0.03 mmol) in 2 mL of CH3OH was refluxed for 15 h, and the resultant mixture was co-distilled with CH3OH five times. The resultant sample was vacuum dried overnight to obtain 14 mg of 9 (73%). 1H NMR (300 MHz, CDCl3 : CD3OD, 4:1) δ 7.90–8.10 (1H, m, 6-CH), 6.30–6.05 (2H, two m, 1′-CH, 5-CH), 4.42–3.80 (6H, overlapping m, 3′-CH, 4′-CH,5′A-CH, 5′B-CH, CH2OP), 2.04 (3H, s, NHCOCH3) 1.65–1.50 (2H, m, CH2CH2OP), 1.39–1.19 (30H, m, CH2 from C18 chain), 0.88 (3H, t, J = 6.3 Hz, terminal CH3). ESI-HRMS [M-H]− m/z calculated for C29H49F2N3O8P− : 636.3231, found: 636.3220.
2.2.4. 2′-2′-difluoro-5′-monophosphate(1-stearate-2-chloro-propyl)cytosine (12) (Alexander et al., 2003; Bligh and Dyer 1959; Perie et al., 1990)
Glycerol monostearate (1 g, 2.79 mmol) and triethylamine (0.726 g, 7.17 mmol) were partially dissolved in 12 mL of DCM under argon. POCl3 (0.428 g, 2.79 mmol) was added drop-wise and heated to reflux for 2 h. The reaction mixture was filtered to remove triethylamine hydrochloride, and the filtrate was added to 0.2 N NaHCO3 (56 mL). After 15 h of stirring at room temperature, 56 mL of acetone was added, and the white precipitate was recovered by filtration. The precipitate was washed with acetone and re-dissolved in 60 mL of water. Another 40 mL of acetone was added, and the precipitate was recovered. It was then washed with acetone, dissolved in a homogeneous mixture of 100 mL of CHCl3, 200 mL of CH3OH, and 100 mL of 0.1 N HCl, and stirred for 1 h at room temperature. A mixture of 100 mL of CHCl3 and 100 mL of water was added, and the organic layer was isolated. The aqueous phase was extracted with CHCl3 (2 × 50 mL). The combined organic layer was evaporated to dryness and lyophilized to obtain 1-stearate-2-hydroxy-3-phosphatidic acid (10) (720 mg, 59%). 1H NMR (300 MHz, CDCl3 : CD3OD, 4:1) δ 4.10–4.03 (2H, m, COOCH2), 4.00–3.90 (3H, overlapping m, CH2OP, CHOH), 2.27 (2H, t, J = 7.4 Hz, COCH2), 1.60–1.48 (2H, m, COCH2CH2), 1.30–1.10 (28H, m, CH2 from C18 chain), 0.80 (3H, t, J = 6.0 Hz, terminal CH3). ESI-HRMS [M-H]− m/z calculated for C21H42O7P−: 437.2674, found: 437.2673. The powders of 5 (50 mg, 0.11 mmol) and 10 (110 mg, 0.25 mmol) were mixed and lyophilized for 15 h. To the lyophilized powder, TPS (74 mg, 0.25 mmol) and 1 mL of pyridine were added under argon environment, and the reaction was stirred at 38–40°C for 24 h. A few drops of water were added, and the solvent was removed under reduced pressure. The crude oil was chromatographed on silica gel, eluting first with CHCl3 : CH3OH (24:1) and then with CHCl3 : CH3OH (9:1) (Alexander et al., 2003). 4-N-3′-O-Bis(tert-butoxycarbonyl)-2′-2′-difluoro-5′-monophosphate(1-stearate-2-chloro-propyl)cytosine (11) was isolated as a white powder (56 mg, 62%). 1H NMR (300 MHz, CDCl3:CD3OD, 4:1) δ 7.95 (1H, d, J = 7.8 Hz, 6-CH), 7.32 (1H, d, J = 6.9 Hz, 5-CH), 6.34 (1H, t, J = 7.6 Hz, 1′-CH), 5.18–5.31 (1H, m, 3′-CH), 4.60–4.10 (7H, overlapping m, 4′-CH, 5′A-CH, 5′B-CH,COCH2, CH2OP), 3.82– 3.71 (1H, m, CHCl), 2.39–2.29 (2H, m, COCH2), 1.68–1.56 (2H, m, CH2CH2CO), 1.53,1.51 (18H, two s, (CH3)3C), 1.38–1.19 (28H, m, CH2 from C18 chain), 0.88 (3H, t, J = 6.6 Hz, terminal CH3). ESI-HRMS [M-H]− m/z calculated for C40H66ClF2N3O13P−: 900.3995, found: 900.3997, [M+H]+ m/z calculated for C40H68ClF2N3O13P+: 902.4141, found: 902.4150. To a stirred solution of 11 (50 mg, 0.055 mmol) in 4 mL of DCM, about 0.5 ml of TFA was added. This solution was stirred at room temperature for 2 h. The excess TFA was removed under reduced pressure, and the concentrated sample was co-distilled with DCM for 5 times. The crude sample was column-purified on silica gel by eluting with 5%, 10%, and 15% CH3OH in CHCl3, sequentially (Alexander et al., 2003). The desired fractions with the Rf value of 0.2 (CH3OH : CHCl3, 2:1) were pooled and evaporated to dryness to yield 18 mg of compound 12 (47%). 1 H NMR (500 MHz, CDCl3 : CD3OD, 4:1) δ 7.78 (1H, d, J = 7.6 Hz, 6-CH), 6.22 (1H, t, J = 6.9 Hz, 1′-CH), 5.95–5.92 (1H, m, 5-CH), 4.24–3.66 (9H, overlapping m, 3′-CH, 4′-CH, 5′A-CH, 5′B-CH,COCH2, CH2OP, CHCl), 2.36–2.29 (2H, m, COCH2), 1.62–1.58 (2H, m, CH2CH2CO), 1.31–1.19 (28H, m, CH2 from C18 chain), 0.88 (3H, t, J = 7.0 Hz, terminal CH3). ESI-HRMS [M-H]− m/z calculated for C20H50ClF2N3O9P−: 700.2947, found: 700.2958, [M+H]+ m/z calculated for C30H52ClF2N3O9P+: 702.3092, found: 702.3089.
2.2.5. 4-N-heptadecylcarbonyl-2′-2′-difluoro-cytosine-5′-sodiummonophosphate (13) (Perie et al., 1990)
Compound 4 (50 mg, 0.094 mmol) and triethylamine (0.072 g, 0.72 mmol) were partially dissolved in 2 mL of DCM under argon. POCl3 (0.0329 g, 0.21 mmol) was added drop-wise, and the same phosphorylation reaction protocol as mentioned in section 2.2.4 was followed. The precipitate was washed with acetone several times and dried under vacuum to obtain 7 mg of pale yellow powder of 13 (12% yields). 1H NMR (300 MHz, CD3OD) δ 8.4–8.3 (1H, m, 6-CH), 7.6–7.5 (1H, m, 5-CH), 6.3–6.2 (1H, m, 1′-CH), 4.2–3.8 (4H, overlapping m, 3′-CH, 4′-CH and 5′-CH2), 2.44 (2H, m, CO–CH2), 1.8–1.7 (2H, m, CO–CH2–CH2), 1.34–1.20 (28H, m, CH2), 0.89 (3H, m, terminal CH3). ESI-HRMS [M-H]− m/z calculated for C27H45F2N3O8P+: 608.2918, found: 608.2933, [M+H]+ m/z calculated for C27H47F2N3O8P+: 610.3063, found: 610.3064.
2.3. Cell lines and cell culture
Human leukemia cell line CCRF-CEM (CCL-119), human pancreatic cancer cell lines PANC-1 (CRL-1469), MIA PaCa-2 (CRL-1420), and BxPC-3 (CRL-1687), human breast adenocarcinoma cell line MCF-7 (HTB-22), and mouse lung cancer cell line TC-1 (CRL-2785) were from the American Type Culture Collection (Rockville, MD). CCRF-CEM-AraC-8C cells (hENT1 deficient) and CCRF-CEM/dCK−/− cells (dCK deficient) were kindly provided by Dr. Buddy Ullmann (Oregon Health & Science University, Portland, OR) and Dr. Margaret Black (Washington State University, Pullman, WA), respectively. L1210 wt and L1210 10K were kindly supplied by Dr. Lars Petter Jordheim (Université Claude Bernard Lyon I, Lyon, France). TC-1-GR cells were previously developed in our lab (Chung et al., 2012). CCRF-CEM, CCRF-CEM-AraC-8C, CCRF-CEM/dCK−/−, L1210 wt, L1210 10K, TC-1, and TC-1-GR cells were cultured in RPMI 1640 medium. MCF-7 and PANC-1 cells were cultured in Dulbecco’s modified Eagle medium (DMEM), and MIA PaCa-2 cells were cultured in DMEM medium with 2.5% horse serum. All media were supplemented with 10% fetal bovine serum (FBS), 100 U/mL of penicillin, and 100 μg/mL of streptomycin (all from Invitrogen, Carlsbad, CA).
2.4. Preparation of nanoparticles
All gemcitabine derivative-containing nanoparticles were prepared as previously described (Sloat et al., 2011; Sloat et al., 2010). Briefly, 3.5 mg of soy lecithin, 0.5 mg of glycerol monostearate, and 1 mg of gemcitabine derivative were added to water (1 mL). The mixture was maintained at 50–62°C while stirring until the formation of homogenous slurry. Tween 20 was added to a final concentration of 1% (v/v). The resultant emulsions were allowed to cool to room temperature while stirring to form nanoparticles. Particle size and zeta potential were determined using a Malvern Zetasizer Nano ZS (Westborough, MA).
2.5. Gel permeation chromatography
Gel permeation chromatography (GPC) was performed to separate un-incorporated 8 from nanoparticles using a 6 mm × 150 mm Sepharose® 4B column, which was equilibrated with phosphate buffered saline (PBS, pH 7.4). Samples (100 μl) were applied into the column and eluted with PBS (Sloat et al., 2011). Elution fractions of 250 μL were collected, and their absorbances at 234 nm were measured using a BioTek Synergy™ HT Multi-Mode Microplate Reader (Winooski, VT). As a control, compound 8 in Tween 20 micelles (100 μg/ml of 8 in 1%, v/v, of Tween 20 in water) were applied to the GPC column.
2.6. In vitro release of 8 from 8-NPs
In vitro release of 8 from 8-NPs was performed as described previously (Sloat et al., 2011). 8-NPs (100 μg/mL) were placed into a 1 mL cellulose ester dialysis tube (MWC 50,000) from Spectrum Chemicals & Laboratory Products (New Brunswick, NJ). The dialysis tube was placed into a conical tube containing 12 mL of PBS with 0.05% (w/v) of sodium dodecyl sulfate (SDS) and incubated in a 37°C shaker incubator. At predetermined time points, 200 μL of the release medium was withdrawn and replaced with 200 μL of fresh release medium. As a control, the diffusion of 8-in-Tween 20 micelles across the dialysis membrane was also measured. The absorbance was measured at 234 nm.
2.7. In vitro cytotoxicity and apoptosis assay
Cells (5,000/well) were seeded in 96-well plates. After overnight incubation, they were treated with various concentrations of gemcitabine HCl, gemcitabine derivatives, or gemcitabine derivatives in nanoparticles at 37 °C, 5% CO2. TC-1 and TC-1-GR cells were incubated for 48 h. CCRF-CEM, CCRF-CEM-AraC-8C, CCRF-CEM/dCK−/−, L1210 wt, L1210 10K, and MCF-7 cells were incubated for 72 h, and MIA PaCa-2 and PANC-1 for 96 h. Gemcitabine HCl was dissolved in PBS, and gemcitabine derivatives were dissolved in dimethyl sulfoxide (DMSO). The maximum amount of DMSO added per well was 1 μL, which was found non-toxic. Compound 13 was not used in the in vitro cytotoxicity assay because it was not sufficiently solubilized in DMSO. The number of viable cells after the incubation was determined using an MTT assay. Briefly, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (20 μl, 5 mg/mL) was added in each well and incubated for 3 h. Formazan crystals were solubilized with acidified isopropanol (150 μl) (for CCRF-CEM, CCRF-CEM-AraC-8C, CCRF-CEM/dCK−/−, L1210 wt, and L1210 10K cells) or DMSO (150 μl) (for TC-1, TC-1-GR, MCF-7, PANC-1, and MIA PaCa-2). Absorbance was measured using a BioTek Synergy™ HT Multi-Mode Microplate Reader (BioTek Instruments, Winooski, VT) at 570 nm and 630 nm. The fraction of affected (dead) cells (Fa) and the fraction of unaffected (live) cells (Fu) at every dose were calculated, and the Log (Fa/Fu) values were plotted against the Log (concentration of gemcitabine) (Chou and Talalay 1984). IC50 was the dose at Log (Fa/Fu) = 0. The experiment was repeated at least three times. To understand whether the cells underwent apoptosis after treatment with the gemcitabine derivatives in nanoparticles, CCRF-CEM-dCK−/− cells (5 × 104) were incubated at 37°C, 5% CO2 with gemcitabine, 8 in solution, or 8-NPs (10 μM). After 72 h, cells were resuspended in PBS with 2% FBS, stained using a Guava Nexin kit that contains annexin V and 7-amino actinomycin D (7-AAD) according to the manufacturer’s protocol, and analyzed using a Guava Easycyte 8HT Flow Cytometry System (Millipore, Hayward, CA). Flow cytometry data were analyzed using the Guava Analysis Software.
2.8. In vitro cellular uptake
PANC-1 cells (2.5 × 105/well) were seeded in a 6-well plate and incubated overnight at 37°C, 5% CO2. The medium was then replaced with 1 mL of medium containing 8-NPs (40 μM), 8 in solution (40 μM), or gemcitabine HCl (40 μM and 400 μM) and incubated for predetermined time points at 37°C, 5% CO2. Compound 8 in solution was prepared by diluting 8 in DMSO with culture media (DMSO concentration at 0.5%, v/v). The culture medium was removed; cells were washed three times with cold PBS and then lysed with 1% SDS. The cell lysates were lyophilized and re-dissolved in CH3OH for gemcitabine and CHCl3: CH3OH (4:1) for 8-NPs and 8 in solution. The concentrations of gemcitabine and 8 were determined using HPLC. For gemcitabine analysis, the mobile phase was CH3OH in 5 mM sodium acetate (15%, v/v) at the detection wave length of 266 nm, and the flow rate was 1 mL/min. Arabinofuranosyluracil (AraU) was used as an internal standa rd, and the extraction efficiency of AraU was about 100% with the aforementioned extraction procedure. For compound 8, the mobile phase was 1 mM PBS in acetonitrile (45%, v/v) at the detection wave length of 270 nm, and the flow rate was 1 mL/min in reverse phase HPLC.
2.9. Partial purification dCDA and dCDA activity assay
Deoxycytidine deaminase was partially purified from BxPC-3 cells as previously described (Laliberté and Momparler 1994; Bergman, A. M. et al., 2004). The pellet of 1 × 108 cells was suspended in 4 mL of 20 mM Tris buffer (pH 7.5) containing 5 mM potassium chloride (KCl), 1 mM dithiothreitol, 40 μl of streptomycin sulfate (12.74 mg/mL), and 50 μl of protease inhibitor cocktail. The suspended cells were sonicated and centrifuged for 30 min at 20,000 g. Ammonium sulfate was added to reach 40% saturation, stirred for 1 h, and centrifuged at 36,000 g for 20 min at 4°C. Ammonium sulfate was added to the supernatant to reach 55% saturation, mixed for 1 h and centrifuged at 36,000 g for 20 min at 4°C. The pellet was resuspended in 1 mL of 20 mM Tris buffer (pH 7.5) and desalted by overnight dialysis against water. Protein concentration was measured using Bradford reagent from Sigma-Aldrich. The dCDA activity assay was carried out as described previously with slight modifications (Bergman, A. M. et al., 2004; Ruiz van Haperen et al., 1993). Briefly, 55 μl of partially purified dCDA (3.2 mg/mL) and 0.5 mM dCyd (20 μl) in a total volume of 200 μl of 20 mM of Tris buffer (pH 7.5) were incubated at 37°C for 15 min. The reaction was terminated by the addition of 50 μl of trichloroacetic acid (40%, w/v) and chilling on ice for 20 min. Protein was precipitated by centrifugation at 10,000 g for 10 min, and the supernatant was neutralized with 500 μl of trioctylamine and 1,1,2-trichloro-trifluoroethane (1:4). The mixture was centrifuged at 10,000 g for 1 min, and the upper layer was analyzed using HPLC (detection wavelength, 260 nm; mobile phase, 10% CH3OH in water). The relevant peaks were quantified to determine the concentrations of dCyd and dUrd. For the competition assay, 20 μl of gemcitabine HCl or gemcitabine derivative-containing nanoparticles, with molar equivalent concentrations of gemcitabine derivatives, were included in the reaction mixture. Controls include a reaction with substrate (dCyd) but without an inhibitor and a reaction without substrate and inhibitors, but with blank nanoparticles.
2.10. Statistics
Statistical analyses were completed using ANOVA followed by Fisher’s protected less significant procedure. A p value of ≤ 0.05 was considered significant.
3. Results and discussion
3.1. Syntheses of novel lipophilic monophosphorylated gemcitabine derivatives
GemC18 was synthesized as previously reported with slight modification (Scheme 1) (Guo and Gallo 1999; Immordino et al., 2004; Sloat et al., 2011). Briefly, the primary and secondary alcohols of deoxyribofuranose ring of gemcitabine (1) were Boc-protected to prevent potential side reactions. The stearoyl group was conjugated to 4-amino group, and the Boc groups were removed to obtain 4 or GemC18 as a white crystalline powder (Steps a–c; Scheme 1). To facilitate the direct conjugation to the 5′-OH, the primary alcohol and 4-amino groups of gemcitabine were Boc protected (5B) (Guo and Gallo 1999). Octadecanol was phosphorylated to give the desired product of 6 (Bligh and Dyer 1959; Perie et al., 1990). The mixture of lyophilized powders of 5B and 6 were conjugated at the 5′-OH. After removing Boc groups, the crude sample was chromatographed to obtain compound 8. Compound 9 was obtained by the acetylation of 8 on the 4-amino group (Steps e–i; Scheme 1) (Watanabe and Fo 1966). Glycerol monostearate was phosphorylated to obtain compound 10 (Bligh and Dyer 1959; Perie et al., 1990). Compounds 5B and 10 were conjugated (Alexander et al., 2003), and deprotection resulted in compound 12 (Steps j– k; Scheme 1). Finally GemC18 was phosphorylated to obtain compound 13 (Step d; Scheme 1) (Perie et al., 1990). Purities of synthesized compounds 4, 8, 9, 12, and 13 (Scheme 1) were ≥ 95.0% based on NMR data.
3.2. Preparation and characterization of gemcitabine derivatives containing nanoparticles
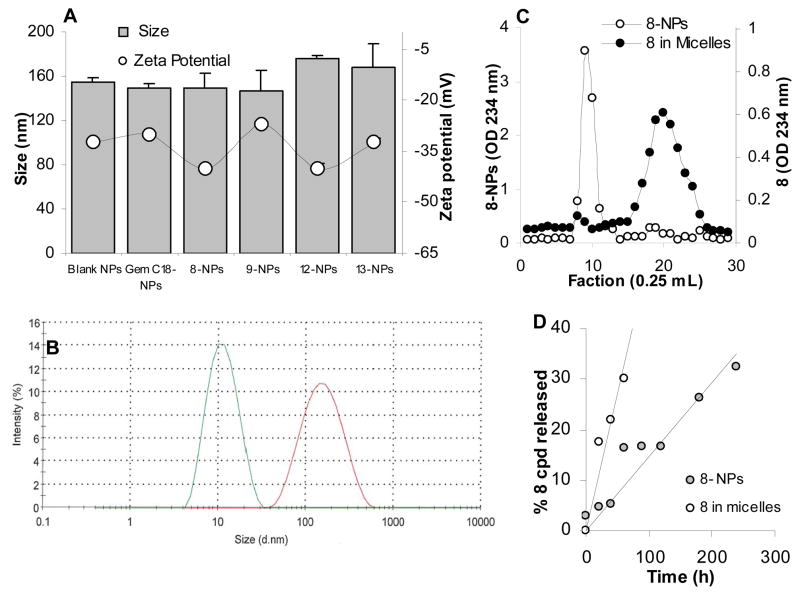
GemC18 and other newly synthesized lipophilic monophosphorylated gemcitabine derivatives, 8, 9, 12, and 13, were incorporated into solid lipid nanoparticles prepared from lecithin/glycerol monostearate-in-water emulsions to prepare GemC18-NPs, 8-NPs, 9-NPs, 12-NPs, and 13-NPs, respectively (Sloat et al., 2010; Sloat et al., 2011). The sizes of the resultant nanoparticles were 150–175 nm (Fig. 1A), with a polydispersity index of 0.2–0.3 and zeta potentials of −27 mV to −40 mV (Fig. 1A). It was likely that all the gemcitabine derivatives were incorporated into the nanoparticles, as supported by the lack of a micelle peak in the dynamic light scattering spectra (Fig. 1B, 8-NPs are shown) and the GPC graph of the nanoparticles (Fig. 1C). In fact, in our previous study, when 5 times more GemC18 (i.e., 5 mg/mL) was used to prepare nanoparticles, the incorporation efficacy remained at close to 100% (Sloat et al., 2011). Shown in Fig. 1D is the in vitro release profile of compound 8 from 8-NPs. Compound 8 was used in Figures 1B–D because data that will be presented in the following sections showed that it had the highest cytotoxicity against most of the gemcitabine resistant tumor cells.
Fig. 1.
Characterization of gemcitabine derivative-containing nanoparticles. (A) The size and zeta potential of the nanoparticles, blank NPs, GemC18-NPs, 8-NPs, 9-NPs, 12-NPs, and 13-NPs. (B) The dynamic light scattering spectra of the 8-in-Tween 20 micelles (green, left peak) and 8-NPs (red, right peak) overlaid. (C) In GPC, 8-NPs (○) eluted in much earlier fractions (fractions 8 to 11) than 8 in Tween 20 micelles (●) (fractions 17 to 24). The concentrations of the 8 in Tween 20 micelles and in 8-NPs were 100 μg/mL and 1 mg/mL, respectively. (D) The release profile of 8 from 8-NPs. As a control, the diffusion of compound 8 (8 in micelles) through the dialysis membrane was also measured. Data shown are mean ± S.D. (n = 3 for A and C, 6 for D). Standard deviations were not shown in C and D for clarity.
3.3. Cytotoxicities of gemcitabine derivatives and their nanoparticles in cancer cells
To evaluate the antitumor activity of the gemcitabine derivatives and the extent to which the gemcitabine derivatives and their corresponding nanoparticles can overcome various mechanisms of gemcitabine resistance, the cytotoxicities of them in cancer cells that are dCK deficient, hENT1 deficient, or over-expressing RRM1 or RRM2 were determined. In addition, the ability of selected gemcitabine derivative-containing nanoparticles to competitively inhibit the deamination activity of partially purified dCDA was evaluated and compared to that of gemcitabine HCl as well.
3.3.1. Lipophilic gemcitabine derivatives and their nanoparticles can overcome dCK deficiency
The in vitro cytotoxicities of the gemcitabine derivatives and their nanoparticles in human leukemia cell line, CCRF-CEM, and its derivative line, CCRF-CEM/dCK−/−, were evaluated and compared to that of gemcitabine HCl. The IC50 value of gemcitabine HCl in the parent CCRF-CEM cells was 2.9 ± 1.8 nM (Table 1), which was 6-77-fold smaller than that of the gemcitabine derivatives, GemC18, 8, 9, 12, and the derivatives in nanoparticles, GemC18-NPs, 8-NPs, 9-NPs, 12-NPs, and 13-NPs (Table 1), demonstrating that in CCRF-CEM cells, gemcitabine HCl was more cytotoxic than the gemcitabine derivatives, alone or in nanoparticles. Overall, this finding is in agreement with data from our previous studies, which showed that the GemC18-NPs were less cytotoxic than gemcitabine HCl in various cancer cells including the CCRF-CEM (Chung et al., 2012), likely because the gemcitabine needs to be hydrolyzed from the GemC18 or GemC18-NPs to be effective (Sloat et al., 2011). In fact, doubling the incubation time of the GemC18-NPs with the TC-1 lung cancer cells enabled the GemC18-NPs to kill the same proportion of the cancer cells as gemcitabine HCl (Sloat et al., 2011). Finally, it appears that the IC50 values of GemC18, 8, 9, 12 were not significantly different from that of their corresponding nanoparticles in CCRF-CEM cells (Table 1), indicating that the incorporation of the gemcitabine derivatives into nanoparticles did not improve their cytotoxicities against the CCRF-CEM cells.
Table 1.
The IC50 values of gemcitabine, gemcitabine derivatives, and the derivatives in nanoparticles in CCRF-CEM and CCRF-CEM/dCK−/− cells.
| CCRF-CEM (nM) | CCRF-CEM/dCK−/− (μM) | Ratio of IC50 in CCRF-CEM/dCK−/− to that in CCRF-CEM | |
|---|---|---|---|
| Gemcitabine HCl | 2.9 ± 1.8 | 240.4 ± 29.0 | 82,897 |
| GemC18 | 19.4 ± 13.3 | 47.3 ± 8.7 | 2,438 |
| 8 | 195.9 ± 28.1 | 38.3 ± 1.2 | 196 |
| 9 | 58.1 ± 18.5 | 62.3 ± 5.3 | 1,072 |
| 12 | 49.8 ± 5.9 | 87.4 ± 10.1 | 1,755 |
| GemC18-NPs | 16.3 ± 4.5 | 13.3 ± 2.6 | 816 |
| 8-NPs | 152.1 ± 15.7 | 3.8 ± 0.2 | 25 |
| 9-NPs | 86.5 ± 9.4 | 5.9 ± 1.4 | 68 |
| 12-NPs | 58.8 ± 15.6 | 2.8 ± 0.2 | 48 |
| 13-NPs | 222.2 ± 68.7 | 11.3 ± 3.5 | 51 |
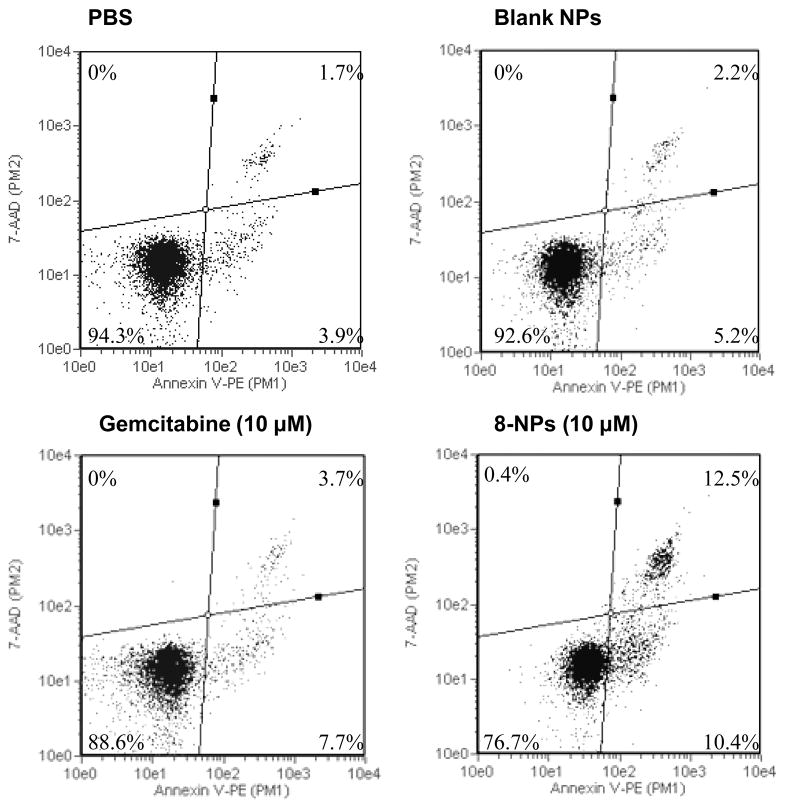
In the CCRF-CEM/dCK−/− cells, the IC50 value of gemcitabine HCl was 240.4 ± 29.0 μM, which was 82,897-fold greater than that in the parent CCRF-CEM cells (Table 1). In contrast, the IC50 values of GemC18, 8, 9, 12, GemC18-NPs, 8-NPs, 9-NPs, 12-NPs, and 13-NPs in the CCRF-CEM/dCK−/− cells were only 25- to 2,438-fold greater than their IC50 values in the parent CCRF-CEM cells (Table 1). In the dCK deficient CCRF-CEM- dCK−/− cells, the IC50 values of the gemcitabine derivatives and their corresponding nanoparticles were 3-86-fold smaller than that of gemcitabine HCl. In other words, the gemcitabine derivatives, alone or in nanoparticles, were more cytotoxic to the CCRF-CEM/dCK−/− cells than gemcitabine HCl. To further confirm this finding, an apoptosis assay was performed in CCRF-CEM/dCK−/− cells by staining them with annexin V and 7-AAD after they were incubated for 72 h with gemcitabine HCl (10 μM), 8-NPs (10 μM), 8-free nanoparticles, or PBS. Similar to gemcitabine HCl, 8-NPs induced tumor cells to undergo apoptosis (Fig. 2), but the 8-NPs were significantly more effective than gemcitabine HCl in inducing apoptosis (Fig. 2). Therefore, the gemcitabine derivatives and their nanoparticles are less dependent on dCK to be active than gemcitabine HCl. The finding with the gemcitabine derivatives in nanoparticles is new, and the finding with the derivatives alone is consistent with previous data generated in dCK over-expressing or dCK deficient cells using other phospholipid gemcitabine derivatives and gemcitabine phosphoramidate (Alexander et al., 2005; Wu et al., 2007).
Fig. 2.
Flow cytometric graphs of CCRF-CEM-dCK−/− cells after 72 h of incubation with gemcitabine HCl (10 M) or 8-NPs followed by staining with Annexin V and 7-AAD. Numbers in the quadrates are % of cells (mean from 3 replicates). Upper right quadrant represents cells in late apoptotic stage; lower right, cells in early apoptotic stage; lower left, viable cells.
Moreover, the ratios of the IC50 value of the same compound in the CCRF-CEM/dCK−/− to that in the parent CCRF-CEM cells seem to show that the monophosphorylated gemcitabine derivatives (i.e., 8, 9, and 12) were less dependent on the dCK to be active than GemC18, which is not monophosphorylated (Table 1). Importantly, it appears that the incorporation of the gemcitabine derivatives into nanoparticles made the monophosphorylated gemcitabine derivatives further less dependent on the dCK to be active (Table 1). For example, for the gemcitabine derivatives alone, the ratio of the IC50 value of CCRF-CEM/dCK−/− to CCRF-CEM was 2,438 for the GemC18, 196–1,755 for the other monophosphorylated derivatives (Table 1). However, for the gemcitabine derivatives in nanoparticles, the ratio was 816 for the GemC18-NPs, but only 25–68 for 8-NPs, 9-NPs, 12-NPs, and 13-NPs (Table 1). Gemcitabine is phosphorylated by dCK, and the monophosphorylation of gemcitabine is the rate limiting step in the activation of gemcitabine (Ueno et al., 2007; Mini et al., 2006). Therefore, it was expected that the monophosphorylated gemcitabine derivatives are less dependent on dCK to be active than the GemC18. Interestingly, it appears that the combination of monophosphorylation of gemcitabine and the incorporation of the lipophilic monophosphorylated gemcitabine derivative into nanoparticles can more effectively bypass the rate limiting step of phosphorylation in gemcitabine activation.
To further validate this finding, the cytotoxicities of gemcitabine HCl and selected gemcitabine derivatives in nanoparticles were evaluated in another dCK deficient cell line, the murine leukemia cells L1210 10K. The IC50 value of gemcitabine HCl in the parent L1210 wt cells was 1.3 ± 0.3 nM, which was 17,046-fold smaller than the IC50 value of gemcitabine HCl in the dCK deficient L1210 10K cells (22.2 ± 3.7 μM) (Table 2). Interestingly, in the L1210 10K cells, GemC18-NPs and 8-NPs were 4- and 8-fold more cytotoxic than gemcitabine HCl, respectively (Table 2). In addition, the IC50 values of GemC18-NPs and 8-NPs in the L1210 10K cells were only 17-431-fold greater than that in the L1210 wt cells (Table 2), further confirming that the incorporation of the gemcitabine derivatives in nanoparticles makes them less dependent on dCK to be active. We did not investigate the mechanism of hydrolysis of the monophosphorylated gemcitabine derivatives, but it is likely that they were hydrolyzed between the lipophilic chain and the phosphate group, similar to the hydrolysis of 1-β-D-arabinofuranosylcytosine (ara-C) or other gemcitabine phospholipid derivatives (Raetz et al., 1977; Alexander and Kucera 2005).
Table 2.
The IC50 values of gemcitabine HCl, gemC18-NPs, and 8-NPs in L1210 wt and L1210 10K cells.
| L1210 wt (nM) | L1210 10K (μM) | Ratio of IC50 values in L1210 10K cells* | Ratio of IC50 in L1210 10K to that in L1210 wt | |
|---|---|---|---|---|
| Gemcitabine HCl | 1.3 ± 0.3 | 22.2 ± 3.7 | 1 | 17,046 |
| GemC18-NPs | 13.1 ± 0.3 | 5.6 ± 0.1 | 4 | 431 |
| 8-NPs | 172.5 ± 55.2 | 2.9 ± 0.3 | 8 | 17 |
Ratio is the IC50 values of gemcitabine HCl divided by that of the nanoparticles.
3.3.2. Lipophilic gemcitabine derivatives and their nanoparticles can overcome gemcitabine resistance related to RRM1 over-expression
RRM1 plays a substantial role in DNA synthesis and gemcitabine resistance (Bergman, Andries M. et al., 2005; Ceppi et al., 2006; Davidson et al., 2004; Goan et al., 1999; Ohtaka et al., 2008; Rosell et al., 2004; Yen 2003). Previously, we developed a tumor cell line that over-expresses RRM1 (TC-1-GR) (Chung et al., 2012). In TC-1-GR cells, GemC18-NPs were significantly more toxic than gemcitabine HCl, although in the parent TC-1 cells, GemC18-NPs were significantly less toxic than gemcitabine HCl (Chung et al., 2012). Importantly, in mice with pre-established TC-1-GR tumors, GemC18-NPs significantly inhibited the tumor growth, but gemcitabine HCl did not show any significant anti-tumor activity (Chung et al., 2012). In the present study, the IC50 values of the new lipophilic monophosphorylated gemcitabine derivatives and their nanoparticles in both TC-1 and TC-1-GR cells were determined to evaluate their ability to overcome gemcitabine resistance caused by RRM1 over-expression. As expected, in TC-1 cells, gemcitabine HCl was more cytotoxic (IC50, 14.7 ± 2.8 nM) than the gemcitabine derivatives and their nanoparticles (Table 3). However, in TC-1-GR cells, the majority of gemcitabine derivatives (except 12) and all the gemcitabine derivatives in nanoparticles were more cytotoxic than gemcitabine HCl (2- to 10-fold) (Table 3). Importantly, in TC-1-GR cells, the IC50 value of gemcitabine HCl was 36.7 ± 5.1 μM, which was 2,497-fold greater than that in TC-1 cells. In contrast, the IC50 values of GemC18, 8, 9, 12 and the nanoparticles, GemC18-NPs, 8-NPs, 9-NPs, 12-NPs, and 13-NPs, in TC-1-GR cells were only 23- to 177-fold greater than that in TC-1 cells (Table 3), demonstrating that the gemcitabine derivatives and their nanoparticles are less sensitive to gemcitabine resistance caused by RRM1-over-expression than gemcitabine HCl. Incorporation of the gemcitabine derivatives into nanoparticles tended to make the derivatives, particularly the GemC18, more cytotoxic to the RRM1-over-expressing TC-1-GR cells. However, unlike what was observed in the dCK deficient cells (Tables 1, 2), it seemed that in RRM1-over-expressing TC-1-GR cells, monophosphorylation of gemcitabine did not add significant additional benefits compare with GemC18.
Table 3.
The IC50 values of gemcitabine HCl, gemcitabine derivatives, and the derivatives in nanoparticles in TC-1 and TC-1-GR cells.
| TC-1 (nM) | TC-1-GR (μM) | Ratio of IC50 values in TC-1-GR cells* | Ratio of IC50 in TC-1-GR to that in TC-1 | |
|---|---|---|---|---|
| Gemcitabine HCl | 14.7 ± 2.8 | 36.7 ± 5.1 | 1 | 2,497 |
| GemC18 | 132.1 ± 17.2 | 7.7 ± 2.4 | 5 | 58 |
| 8 | 245.5 ± 39.9 | 21.1 ± 1.7 | 2 | 86 |
| 9 | 210.7 ± 85.0 | 9.3 ± 2.5 | 4 | 44 |
| 12 | 430.0 ± 52.4 | 76.3 ± 10.5 | 0.5 | 177 |
| GemC18-NPs | 59.8 ± 18.4 | 3.6 ± 0.2 | 10 | 60 |
| 8-NPs | 371.3 ± 27.3 | 8.4 ± 0.5 | 4 | 23 |
| 9-NPs | 258.6 ± 60.9 | 10.1 ± 0.9 | 4 | 39 |
| 12-NPs | 405.7 ± 114.1 | 10.5 ± 2.0 | 3 | 26 |
| 13-NPs | 395.5 ± 40.7 | 9.0 ± 2.6 | 4 | 23 |
Ratio is the IC50 values of gemcitabine HCl divided by that of the derivatives or derivatives in nanoparticles.
3.3.3. Cytotoxicities of gemcitabine derivatives and their nanoparticles in cancer cells over-expressing different levels of RRM2
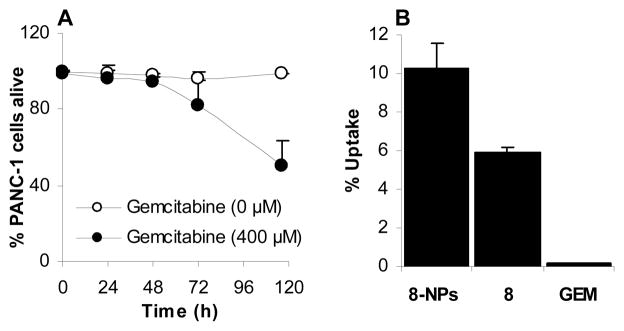
It was reported that MIA PaCa-2 and PANC-1 cells both over-expressed RRM2, but PANC-1 cells express ~70% more RRM2 than MIA PaCa-2 (Duxbury et al., 2003). In MIA PaCa-2 cells, the IC50 value of gemcitabine HCl and GemC18 NPs were 49.7 ± 17.7 nM and 40.6 ± 8.2 nM, respectively (Table 4), all other gemcitabine derivatives and their nanoparticles were less toxic than gemcitabine HCl (Table 4). However, more than 50% of PANC-1 cells were still alive after 96 h of incubation with 400 μM of gemcitabine HCl (Table 1 and Fig. 3A). Data from a trypan blue exclusion assay showed that it took 116 h to kill 50% of PANC-1 cells with 400 μM of gemcitabine HCl (Fig. 3A). The IC50 values of the gemcitabine derivatives and their nanoparticles in PANC-1 cells were 5.8 to 58.7 μM (Table 4). The IC50 values of gemcitabine HCl in PANC-1 cells was more than 8,000-fold greater than that in the MIA PaCa-2 cells, but the IC50 values of gemcitabine derivatives and their nanoparticles in PANC-1 cells were only 24- to 239-fold greater than those in MIA PaCa-2 cells (Table 4). In other words, the gemcitabine derivatives and their nanoparticles were less sensitive than gemcitabine HCl to gemcitabine resistance caused by RRM2 over-expression. Again, monophosphorylation of gemcitabine did not add additional benefits in its cytotoxicity against the RRM2-over-expressing PANC-1 cells, and except for the GemC18-NPs, a conclusion that the incorporation of gemcitabine derivatives into nanoparticles makes them more cytotoxic cannot be drawn. Previously, Duxbury et al. reported that the IC50 values of gemcitabine in MIA PaCa-2 cells and PANC-1 cells were 40 nM and 50 nM, respectively. The IC50 value of gemcitabine HCl in MIA PaCa-2 cells determined in the present study is comparable to what was reported (Duxbury et al., 2003), but the PANC-1 cells were significantly more resistant to gemcitabine HCl in our study. Data in Fig. 3B showed that after 0.5 h of incubation, only about 0.15% of gemcitabine was taken up by PANC-1 cells, in contrast to 10% and 6% of 8 in nanoparticles or in solution. Therefore the low uptake of gemcitabine by PANC-1 cells was probably related to its resistance to gemcitabine. However, it needs to be noted that the low percentage of gemcitabine detected in the PANC-1 cells could also be due to the rapid deamination of gemcitabine. Moreover, it is known that PANC-1 cells overexpress RRM1 and RRM2, which are known determinants of gemcitabine resistance (Jordheim et al., 2005; Boukovinas et al., 2008).
Table 4.
The IC50 values of gemcitabine HCl, gemcitabine derivatives, and the derivatives in nanoparticles in MIA PaCa-2 and PANC-1 cells.
| MIA PaCa-2 (nM) | PANC-1 (μM) | Ratio of IC50 in PANC-1 to that in MIA PaCa-2 | |
|---|---|---|---|
| Gemcitabine HCl | 49.7 ± 17.7 | > 400 | > 8,000 |
| GemC18 | 133.0 ± 60.4 | 6.0 ± 1.1 | 45 |
| 8 | 835.9 ± 163.8 | 50.4 ± 4.3 | 60 |
| 9 | 204.6 ± 39.5 | 38.5 ± 5.7 | 188 |
| 12 | 245.1 ± 38.0 | 58.7 ± 14.2 | 239 |
| GemC18-NPs | 40.6 ± 8.2 | 5.8 ± 0.6 | 143 |
| 8-NPs | 201.7 ± 50.2 | 6.2 ± 1.5 | 31 |
| 9-NPs | 290.0 ± 84.6 | 8.8 ± 1.8 | 30 |
| 12-NPs | 380.0 ± 98.3 | 22.0 ± 3.5 | 58 |
| 13-NPs | 357.6 ± 172.2 | 8.7 ± 2.9 | 24 |
Fig. 3.
(A) Percentage of PANC-1 cells alive after incubation with gemcitabine HCl at 400 μM for up to 116 h. Cell viability was determined using a trypan blue exclusion assay. The experiment was repeated twice. (B) In vitro uptake of 8 in nanoparticles, 8 in solution, or gemcitabine HCl (GEM) by PANC-1 cells. The concentration of compound 8 in the 8-NPs and 8 in solution was 40 μM, and the gemcitabine HCl concentration was 400 μM. Gemcitabine uptake was not detected when cells were incubated with 40 μM of gemcitabine HCl. The incubation time was 0.5 h. Data shown are mean ± S. D. (n = 3).
Finally, similar results were obtained in the MCF-7 human breast adenocarcinoma cell line. Gemcitabine HCl, at a concentration as high as 400 μM, was not sufficient to kill half of the MCF-7 cells after 72 h of incubation (see Supporting Information, Table S1), and data from a trypan blue exclusion assay showed that more than 50% of MCF-7 cells were still alive, even after 116 h of incubation with 400 μM of gemcitabine HCl. However, all the gemcitabine derivatives, except 12, and all gemcitabine derivatives in nanoparticles were significantly cytotoxic to the MCF-7 cells (IC50 values, 2–31 μM) (Table S1). The mechanism of gemcitabine resistance in the MCF-7 cells is unknown, but the data obtained in the MCF-7 cells clearly showed that the gemcitabine derivatives and the derivatives in nanoparticles can overcome that resistance mechanism.
3.3.4. Cytotoxicities of gemcitabine derivatives and their nanoparticles in nucleoside transporter deficient cancer cells
It is known that nucleoside transporters are a prerequisite for the cellular uptake of gemcitabine (Damaraju et al., 2003). Therefore, in the hENT1 deficient CCRF-CEM-AraC-8C cells, the IC50 value of the gemcitabine HCl was 998.8 ± 9.4 nM, 344 times greater than that in the parent CCRF-CEM cells (Table 5). However, the IC50 values of gemcitabine derivatives and their nanoparticles in CCRF-CEM-AraC-8C cells were only 4- to 35-fold greater than that in the parent CCRF-CEM cells (Table 5), indicating that the gemcitabine derivatives, alone or in nanoparticles, are less sensitive to hENT1 deficient than gemcitabine HCl, possibly because the lipophilized gemcitabine derivatives can diffuse into cells without the help of the nucleoside transporters, and the gemcitabine derivatives in nanoparticles can be taken up by cells via endocytosis. However, the cytotoxicities of the monophosphorylated gemcitabine derivatives and their nanoparticles in the CCRF-CEM-AraC-8C cells are not significantly different from that of gemcitabine HCl (Table 5). Only the GemC18 and GemC18-NPs were 2.7- and 12.3-fold more cytotoxic than gemcitabine HCl, respectively, in the hENT1-deficient CCRF-CEM-AraC-8C cells (Table 5), which was consistent with our previous data (Chung et al., 2012). It is likely that the phosphate group on the gemcitabine derivatives made them less effective in entering the hENT1 deficient cells.
Table 5.
The IC50 values of gemcitabine HCl, gemcitabine derivatives, and the derivatives in nanoparticles in CCRF-CEM and CCRF-CEM-AraC-8C cells.
| CCRF-CEM (nM) | CCRF-CEM-AraC-8C (nM) | Ratio of IC50 values in CEM-AraC-8C cells* | Ratio of IC50 in CCRF-CEM-AraC-8C to that in CCRF-CEM | |
|---|---|---|---|---|
| Gemcitabine HCl | 2.9 ± 1.8 | 998.8 ± 9.4 | 1 | 333 |
| GemC18 | 19.4 ± 13.3 | 369.4 ± 235.1 | 2.7 | 19 |
| 8 | 195.9 ± 28.1 | 806.5 ± 111.8 | 1.2 | 4 |
| 9 | 58.1 ± 18.5 | 575.3 ± 97.1 | 1.7 | 10 |
| 12 | 49.8 ± 5.9 | 1766.9 ± 532.8 | 0.6 | 35 |
| GemC18-NPs | 16.3 ± 4.5 | 81.0 ± 17.9 | 12.3 | 5 |
| 8-NPs | 152.1 ± 15.7 | 942.6 ± 163.8 | 1.1 | 4 |
| 9-NPs | 86.5 ± 9.4 | 1325.1 ± 167.2 | 0.8 | 13 |
| 12-NPs | 58.8 ± 15.6 | 712.9 ± 342.9 | 1.4 | 12 |
| 13-NPs | 222.2 ± 68.7 | 1166.6 ± 293.1 | 0.9 | 5 |
Ratio is the IC50 values of gemcitabine HCl divided by that of the derivatives or derivatives in nanoparticles.
3.4. Gemcitabine HCl, but not gemcitabine derivatives in nanoparticles, inhibits dCDA activity
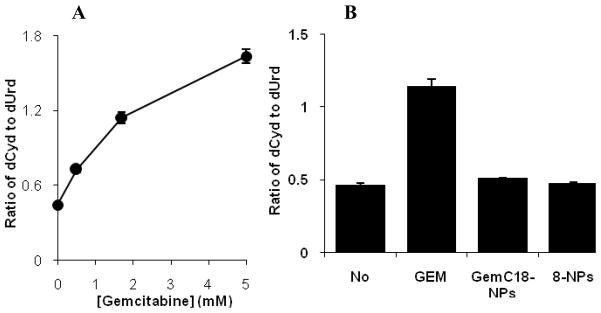
Previously, Bouffard et al. reported that gemcitabine HCl, as a substrate to dCDA, competitively inhibits the deamination of dCyd by dCDA (Bouffard et al., 1993). In order to test whether the gemcitabine derivatives are still good substrates of dCDA and inhibit its activity, dCDA was partially purified from BxPC-3 human pancreatic cancer cells, and its deamination activity against dCyd was determined in the presence or absence of gemcitabine HCl or selected gemcitabine derivatives in nanoparticles (Bergman, A. M. et al., 2004; Laliberté and Momparler 1994; Ruiz van Haperen et al., 1993). The nanoparticles, but not the gemcitabine derivatives alone, were used due to the poor water solubility of the gemcitabine derivatives. GemC18-NPs and 8-NPs were chosen because of their greater cytotoxicity in previous in vitro cytotoxicity assays. dUrd was not detected in the control reaction (with dCyd, but no dCDA) indicating that any observed dUrd would be due to the deamination of dCyd by dCDA. As shown in Fig. 4A, dCyd was converted to dUrd in the presence of the partially purified dCDA. Gemcitabine HCl competitively inhibited the conversion of dCyd to dUrd, and the extent of the inhibition was increased by increasing the concentration of gemcitabine HCl (Fig. 4A). However, GemC18-NPs and 8-NPs did not significantly inhibit the deamination activity of dCDA (Fig. 4B), confirming that gemcitabine HCl, but not GemC18-NPs and 8-NPs, can competitively inhibit the deamination of dCyd by dCDA. Therefore, the gemcitabine derivatives in nanoparticles are no longer good substrates of dCDA, and it is expected that they can potentially overcome gemcitabine resistance caused by deamination. This finding is in agreement with data from previous studies, showing that other gemcitabine derivatives were no longer good substrates of dCDA as well (Bergman, A. M. et al., 2004; Song et al., 2005).
Fig. 4.
dCDA assay. (A) Effect of gemcitabine concentration on the conversion of dCyd to dUrd (ratio, w/w) by dCDA; (B) Effect of GemC18-NPs and 8-NPs on the conversion of dCyd to dUrd by dCDA (No, no inhibitor; GEM, gemcitabine HCl, 1.7 mM). The molar concentration of GemC18 and 8 in the nanoparticles was 1.7 mM. Blank nanoparticles did not inhibit the conversion. Data shown are mean ± S.D. The experiment was repeated at least twice with similar results.
4. Conclusions
In the present study, four novel lipophilic monophosphorylated gemcitabine derivatives were synthesized and incorporated into solid lipid nanoparticles. All the gemcitabine derivatives and their nanoparticles showed a significantly higher cytotoxicity than gemcitabine HCl in cells that are deficient in dCK, and the gemcitabine derivatives in nanoparticles were more cytotoxic than the corresponding gemcitabine derivatives. The majority of the gemcitabine derivatives and all nanoparticles are also more cytotoxic than gemcitabine HCl to cancer cells that over-express RRM1 or RRM2. Finally, the gemcitabine derivatives in nanoparticles were no longer good substrates to dCDA and thus became resistance to deamination. Collectively, the 2′-2′-difluoro-cytosine-5′-octadecylphosphate (8) in nanoparticles showed the highest cytotoxicity to cells that are deficient in dCK, over-expressing RRM1, or over-expressing RRM2, and were resistant to deamination. Future in vivo studies to evaluate its ability to overcome multiple gemcitabine resistant mechanisms are warranted.
Supplementary Material
Acknowledgments
This work was supported in part by a National Cancer Institute grant (CA135274) to Z.C.
Footnotes
Supporting Information: The IC50 values of gemcitabine HCl, gemcitabine derivatives, and the derivatives in nanoparticles in MCF-7 cells are available in Table S1.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander RL, Greene BT, Torti SV, Kucera GL. A novel phospholipid gemcitabine conjugate is able to bypass three drug-resistance mechanisms. Cancer Chemother Pharmacol. 2005;56:15–21. doi: 10.1007/s00280-004-0949-0. [DOI] [PubMed] [Google Scholar]
- Alexander RL, Kucera GL. Lipid Nucleoside Conjugates for the Treatment of Cancer. Curr Pharm Des. 2005;11:1079–1089. doi: 10.2174/1381612053507602. [DOI] [PubMed] [Google Scholar]
- Alexander RL, Morris-Natschke SL, Ishaq KS, Fleming RA, Kucera GL. Synthesis and Cytotoxic Activity of Two Novel 1-Dodecylthio-2-decyloxypropyl-3-phosphatidic Acid Conjugates with Gemcitabine and Cytosine Arabinoside. J Med Chem. 2003;46:4205–4208. doi: 10.1021/jm020571x. [DOI] [PubMed] [Google Scholar]
- Andersson R, Aho U, Nilsson BI, Peters GJ, Pastor-Anglada M, Rasch W, Sandvold ML. Gemcitabine chemoresistance in pancreatic cancer: Molecular mechanisms and potential solutions. Scand J Gastroenterol. 2009;44:782–786. doi: 10.1080/00365520902745039. [DOI] [PubMed] [Google Scholar]
- Barton-Burke M. Gemcitabine: A pharmacologic and clinical overview. Cancer Nurs. 1999;22:176–183. doi: 10.1097/00002820-199904000-00011. [DOI] [PubMed] [Google Scholar]
- Bergman AM, Eijk PP, Ruiz van Haperen VWT, Smid K, Veerman G, Hubeek I, van den IJssel P, Ylstra B, Peters GJ. In vivo Induction of Resistance to Gemcitabine Results in Increased Expression of Ribonucleotide Reductase Subunit M1 as the Major Determinant. Cancer Res. 2005;65:9510–9516. doi: 10.1158/0008-5472.CAN-05-0989. [DOI] [PubMed] [Google Scholar]
- Bergman AM, Kuiper CM, Voorn DA, Comijn EM, Myhren F, Sandvold ML, Hendriks HR, Peters GJ. Antiproliferative activity and mechanism of action of fatty acid derivatives of arabinofuranosylcytosine in leukemia and solid tumor cell lines. Biochem Pharmacol. 2004;67:503–511. doi: 10.1016/j.bcp.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Bergman AM, Pinedo HM, Peters GJ. Determinants of resistance to 2′,2′-difluorodeoxycytidine (gemcitabine) Drug Resist Updat. 2002;5:19–33. doi: 10.1016/s1368-7646(02)00002-x. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. Lipid extraction of c. Elegans Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bouffard DY, Laliberté J, Momparler RL. Kinetic studies on 2′,2′-difluorodeoxycytidine (gemcitabine) with purified human deoxycytidine kinase and cytidine deaminase. Biochem Pharmacol. 1993;45:1857–1861. doi: 10.1016/0006-2952(93)90444-2. [DOI] [PubMed] [Google Scholar]
- Boukovinas I, Papadaki C, Mendez P, Taron M, Mavroudis D, Koutsopoulos A, Sanchez-Ronco M, Sanchez JJ, Trypaki M, Staphopoulos E, Georgoulias V, Rosell R, Souglakos J. Tumor BRCA1, RRM1 and RRM2 mRNA Expression Levels and Clinical Response to First-Line Gemcitabine plus Docetaxel in Non-Small-Cell Lung Cancer Patients. PLoS ONE. 2008;3:e3695. doi: 10.1371/journal.pone.0003695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candelaria M, Cetina L, de la Garza J, Dueñas-Gonzálz A. Clinical implications of gemcitabine in the treatment of cervical cancer. Eur J Cancer Suppl. 2007;5:37–43. [Google Scholar]
- Candelaria M, de la Cruz-Hernández E, Pérez-Cárdenas E, Trejo-Becerril C, Gutiérrez-Hernández O, Dueñas-González A. Pharmacogenetics and pharmacoepigenetics of gemcitabine. Med Oncol. 2010;27:1133–1143. doi: 10.1007/s12032-009-9349-y. [DOI] [PubMed] [Google Scholar]
- Ceppi P, Volante M, Novello S, Rapa I, Danenberg K, Danenberg P, Cambieri A, Selvaggi G, Saviozzi S, Calogero R, Papotti M, Scagliotti G. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17:1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- Cetina L, Rivera L, Candelaria M, de la Garza J, Dueñas-González A. Chemoradiation with gemcitabine for cervical cancer in patients with renal failure. Anti-Cancer Drugs. 2004;15:761–766. doi: 10.1097/00001813-200409000-00004. [DOI] [PubMed] [Google Scholar]
- Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Chung W-G, Sloat BR, Sandoval MA, Lansakara-P DSP, Cui Z. Stearoyl gemcitabine nanoparticles overcome resistance related to the over-expression of ribonucleotide reductase subunit M1. J Control Release. 2012;157:132–140. doi: 10.1016/j.jconrel.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaraju VL, Damaraju S, Young JD, Baldwin SA, Mackey J, Sawyer MB, Cass CE. Nucleoside anticancer drugs: the role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene. 2003;22:7524–7536. doi: 10.1038/sj.onc.1206952. [DOI] [PubMed] [Google Scholar]
- Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA. An Increase in the Expression of Ribonucleotide Reductase Large Subunit 1 Is Associated with Gemcitabine Resistance in Non-Small Cell Lung Cancer Cell Lines. Cancer Res. 2004;64:3761–3766. doi: 10.1158/0008-5472.CAN-03-3363. [DOI] [PubMed] [Google Scholar]
- Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene. 2003;23:1539–1548. doi: 10.1038/sj.onc.1207272. [DOI] [PubMed] [Google Scholar]
- Giovannetti E, Del Tacca M, Mey V, Funel N, Nannizzi S, Ricci S, Orlandini C, Boggi U, Campani D, Del Chiaro M, Iannopollo M, Bevilacqua G, Mosca F, Danesi R. Transcription Analysis of Human Equilibrative Nucleoside Transporter-1 Predicts Survival in Pancreas Cancer Patients Treated with Gemcitabine. Cancer Res. 2006;66:3928–3935. doi: 10.1158/0008-5472.CAN-05-4203. [DOI] [PubMed] [Google Scholar]
- Goan Y-G, Zhou B, Hu E, Mi S, Yen Y. Overexpression of Ribonucleotide Reductase as a Mechanism of Resistance to 2,2-Difluorodeoxycytidine in the Human KB Cancer Cell Line. Cancer Res. 1999;59:4204–4207. [PubMed] [Google Scholar]
- Gregoire V, Rosier JF, De Bast M, Bruniaux M, De Coster B, Octave-Prignot M, Scalliet P. Role of deoxycytidine kinase (dCK) activity in gemcitabine’s radioenhancement in mice and human cell lines in vitro. Radiother Oncol. 2002;63:329–338. doi: 10.1016/s0167-8140(02)00106-8. [DOI] [PubMed] [Google Scholar]
- Guo Z-w, Gallo JM. Selective Protection of 2′,2′-Difluorodeoxycytidine (Gemcitabine) J Org Chem. 1999;64:8319–8322. doi: 10.1021/jo9911140. [DOI] [PubMed] [Google Scholar]
- Heinemann V, Schulz L, Issels RDWP. Gemcitabine: a modulator of intracellular nucleotide and deoxynucleotide metabolism. Semin Oncol. 1995;22:11–18. [PubMed] [Google Scholar]
- Heinemann V, Xu Y-Z, Chubb S, Sen A, Hertel LW, Grindey GB, Plunkett W. Cellular Elimination of 2′,2′-Difluorodeoxycytidine 5′-Triphosphate: A Mechanism of Self-Potentiation. Cancer Res. 1992;52:533–539. [PubMed] [Google Scholar]
- Huang P, Plunkett W. Induction of apoptosis by gemcitabine. Semin Oncol. 1995;4:19–25. [PubMed] [Google Scholar]
- Immordino ML, Brusa P, Rocco F, Arpicco S, Ceruti M, Cattel L. Preparation, characterization, cytotoxicity and pharmacokinetics of liposomes containing lipophilic gemcitabine prodrugs. J Control Release. 2004;100:331–346. doi: 10.1016/j.jconrel.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Jordheim LP, Guittet O, Lepoivre M, Galmarini CM, Dumontet C. Increased expression of the large subunit of ribonucleotide reductase is involved in resistance to gemcitabine in human mammary adenocarcinoma cells. Mol Cancer Ther. 2005;4:1268–1276. doi: 10.1158/1535-7163.MCT-05-0121. [DOI] [PubMed] [Google Scholar]
- Kalykaki A, Papakotoulas P, Tsousis S, Boukovinas I, Kalbakis K, Vamvakas L, Kotsakis A, Vardakis N, Papadopoulou P, Georgoulias V, Mavroudis D. Gemcitabine plus Oxaliplatin (GEMOX) in Pretreated Patients with Advanced Ovarian Cancer: A Multicenter Phase II Study of the Hellenic Oncology Research Group (HORG) Anticancer Research. 2008;28:495–500. [PubMed] [Google Scholar]
- Kroep JR, Loves WJP, van der Wilt CL, Alvarez E, Talianidis I, Boven E, Braakhuis BJM, van Groeningen CJ, Pinedo HM, Peters GJ. Pretreatment Deoxycytidine Kinase Levels Predict in Vivo Gemcitabine Sensitivity 1 Supported by Eli Lilly & Co, International and The Netherlands.1. Mol Cancer Ther. 2002;1:371–376. [PubMed] [Google Scholar]
- Laliberté J, Momparler RL. Human Cytidine Deaminase: Purification of Enzyme, Cloning, and Expression of Its Complementary DNA. Cancer Res. 1994;54:5401–5407. [PubMed] [Google Scholar]
- Maréchal R, Mackey JR, Lai R, Demetter P, Peeters M, Polus M, Cass CE, Salmon I, Devière J, Van Laethem JL. Deoxycitidine kinase is associated with prolonged survival after adjuvant gemcitabine for resected pancreatic adenocarcinoma. Cancer. 2010;116:5200–5206. doi: 10.1002/cncr.25303. [DOI] [PubMed] [Google Scholar]
- Mey V, Giovannetti E, Braud FD, Nannizzi S, Curigliano G, Verweij F, Cobelli OD, Pece S, Tacca MD, Danesi R. In vitro synergistic cytotoxicity of gemcitabine and pemetrexed and pharmacogenetic evaluation of response to gemcitabine in bladder cancer patients. Br J Cancer. 2006;95:289–297. doi: 10.1038/sj.bjc.6603242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17:v7–v12. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- Ohtaka K, Kohya N, Sato K, Kitajima Y, Ide T, Mitsuno M, Miyazaki K. Ribonucleotide reductase subunit M1 is a possible chemoresistance marker to gemcitabine in biliary tract carcinoma. Oncol Rep. 2008;20:279–286. [PubMed] [Google Scholar]
- Perie J, Monsan PF, Willson MJ, Klaebe A, Lauth NJ, Thibault PA. Process for the preparation of lysophosphatidic acids and of salts of the latter FR. 2636331. Patent. 1990
- Raetz CR, Chu MY, Srivastava S, Turcotte JG. A phospholipid derivative of cytosine arabinoside and its conversion to phosphatidylinositol by animal tissue. Science. 1977;196:303–305. doi: 10.1126/science.191910. [DOI] [PubMed] [Google Scholar]
- Rosell R, Danenberg KD, Alberola V, Bepler G, Sanchez JJ, Camps C, Provencio M, Isla D, Taron M, Diz P, Artal A. Ribonucleotide Reductase Messenger RNA Expression and Survival in Gemcitabine/Cisplatin-Treated Advanced Non-Small Cell Lung Cancer Patients. Clin Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- Ruiz van Haperen VW, Veerman G, Braakhuis BJ, BVJ, Boven E, Leyva A, Peters GJ. Deoxycytidine kinase and deoxycytidine deaminase activities in human tumour xenografts. Eur J Cancer. 1993;29A:2132–2137. doi: 10.1016/0959-8049(93)90048-k. [DOI] [PubMed] [Google Scholar]
- Sandler AB, Nemunaitis J, Denham C, von Pawel J, Cormier Y, Gatzemeier U, Mattson K, Manegold C, Palmer MC, Gregor A, Nguyen B, Niyikiza C, Einhorn LH. Phase III Trial of Gemcitabine Plus Cisplatin Versus Cisplatin Alone in Patients With Locally Advanced or Metastatic Non–Small-Cell Lung Cancer. J Clin Oncol. 2000;18:122. doi: 10.1200/JCO.2000.18.1.122. [DOI] [PubMed] [Google Scholar]
- Sezgin C, Karabulut B, Uslu R, Sanli UA, Goksel G, Yuzer Y, Goker E. Gemcitabine treatment in patients with inoperable locally advanced/metastatic pancreatic cancer and prognostic factors. Scand J Gastroenterol. 2005;40:1486–1492. doi: 10.1080/00365520510023819. [DOI] [PubMed] [Google Scholar]
- Sloat BR, Sandoval MA, Hau AM, He Y, Cui Z. Strong antibody responses induced by protein antigens conjugated onto the surface of lecithin-based nanoparticles. J Control Release. 2010;141:93–100. doi: 10.1016/j.jconrel.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloat BR, Sandoval MA, Li D, Chung W-G, Lansakara-P DSP, Proteau PJ, Kiguchi K, DiGiovanni J, Cui Z. In vitro and in vivo anti-tumor activities of a gemcitabine derivative carried by nanoparticles. Int J Pharm. 2011;409:278–288. doi: 10.1016/j.ijpharm.2011.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Lorenzi PL, Landowski CP, Vig BS, Hilfinger JM, Amidon GL. Amino Acid Ester Prodrugs of the Anticancer Agent Gemcitabine:_ Synthesis, Bioconversion, Metabolic Bioevasion, and hPEPT1-Mediated Transport. Mol Pharm. 2005;2:157–167. doi: 10.1021/mp049888e. [DOI] [PubMed] [Google Scholar]
- Spratlin J, Sangha R, Glubrecht D, Dabbagh L, Young JD, Dumontet C, Cass C, Lai R, Mackey JR. The Absence of Human Equilibrative Nucleoside Transporter 1 Is Associated with Reduced Survival in Patients With Gemcitabine-Treated Pancreas Adenocarcinoma. Clin Cancer Res. 2004;10:6956–6961. doi: 10.1158/1078-0432.CCR-04-0224. [DOI] [PubMed] [Google Scholar]
- Ueno H, Kiyosawa K, Kaniwa N. Pharmacogenomics of gemcitabine: can genetic studies lead to tailor-made therapy? Br J Cancer. 2007;97:145–151. doi: 10.1038/sj.bjc.6603860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe KA, Fo JJ. A Simple Method for Selective Acylation of Cytidine of the 4-Amino Group. J Angew Chem, Int Ed. 1966;5:579–580. [Google Scholar]
- Wu W, Sigmond J, Peters GJ, Borch RF. Synthesis and Biological Activity of a Gemcitabine Phosphoramidate Prodrug. J Med Chem. 2007;50:3743–3746. doi: 10.1021/jm070269u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Y. Ribonucleotide Reductase Subunit One as Gene Therapy Target. Clin Cancer Res. 2003;9:4304–4308. [PubMed] [Google Scholar]
- Zucali PA, Ceresoli GL, Garassino I, De Vincenzo F, Cavina R, Campagnoli E, Cappuzzo F, Salamina S, Soto Parra HJ, Santoro A. Gemcitabine and vinorelbine in pemetrexed-pretreated patients with malignant pleural mesothelioma. Cancer. 2008;112:1555–1561. doi: 10.1002/cncr.23337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.