Abstract
Multiple Hereditary Exostoses (MHE) syndrome is caused by haploinsufficiency in Golgi-associated heparan sulfate polymerases EXT1 or EXT2 and is characterized by formation of exostoses next to growing long bones and other skeletal elements. Recent mouse studies have indicated that formation of stereotypic exostoses requires a complete loss of Ext expression, suggesting that a similar local loss of EXT function may underlie exostosis formation in patients. To further test this possibility and gain greater insights into pathogenic mechanisms, we created heterozygous Ext1+/− and compound Ext1+/−/Ext2+/− mice. Like Ext2+/− mice described previously (Stickens et al. Development 132:5055), Ext1+/− mice displayed rib-associated exostosis-like outgrowths only. However, compound heterozygous mice had nearly twice as many outgrowths and, more importantly, displayed stereotypic growth plate-like exostoses along their long bones. Ext1+/−Ext2+/− exostoses contained very low levels of immuno-detectable heparan sulfate, and Ext1+/−Ext2+/− chondrocytes, endothelial cells and fibroblasts in vitro produced shortened heparan sulfate chains compared to controls and responded less vigorously to exogenous factors such as FGF-18. We also found that rib outgrowths formed in Ext1f/+Col2Cre and Ext1f/+Dermo1Cre mice, suggesting that ectopic skeletal tissue can be induced by conditional Ext ablation in local chondrogenic and/or perichondrial cells. The study indicates that formation of stereotypic exostoses requires a significant, but not complete, loss of Ext expression and that exostosis incidence and phenotype are intimately sensitive to, and inversely related to, Ext expression. The data also indicate that the nature and organization of ectopic tissue may be influenced by site-specific anatomical cues and mechanisms.
Keywords: Multiple Hereditary Exostoses, exostosins, heparan sulfate, mouse mutants, chondrocytes
Introduction
Multiple Hereditary Exostoses (MHE; OMIM 133700, 133701), also known as Multiple Osteochondroma, Osteochondromatosis and Diaphyseal Aclasia, is an autosomal dominant disorder characterized by formation of ectopic, cartilage-capped, growth plate-like exostoses next to long bones and other skeletal elements [1]. The exostoses usually originate proximal to an active growth plate, occur after birth and throughout puberty, and may continue to grow slowly during adulthood [1-3]. Complications of the disease include (i) skeletal deformities, such as shortened or curved limbs and reduced stature, (ii) subluxation of the hand or valgus of the knee due to the asymmetric appearance of exostoses in ulna/radius and tibia/fibula, (iii) morbidity caused by entrapment of blood vessels, nerves, and tendons, and (iv) predilection for conversion to malignant chondrosarcoma (0.5 - 2%). The disease is estimated to occur with an incidence of 1/50,000 in the general population with variable penetrance in affected families [3].
MHE is associated with loss-of-function mutations in EXT1 and EXT2 and most patients have EXT1 or EXT2 haploinsufficiency [4]. Studies of mutant cell lines and enzyme purification showed that Ext1 encodes a dual catalytic enzyme required for polymerization of heparan sulfate, which is a component of several membrane and extracellular matrix proteoglycans [5-8]. Ext1 and Ext2 physically associate and form hetero-oligomers that participate in assembly of Golgi-resident enzymatic complexes [9, 10]. Ext1 and Ext2 are widely expressed in mouse tissues and loss of expression results in diminution of heparan sulfate levels [11-14]. Ext1 and Ext2 belong to a larger family of exostosin-like proteins, some of which may have roles in heparan sulfate formation as well [15-17]. However, to date no cases of MHE have been associated with other exostosin gene family members.
We previously produced heterozygous Ext2 mice to create an animal model of MHE and understand the mechanisms of exostosis formation [14]. The mice, however, failed to develop typical exostoses along their long bones, although they exhibited oval-shaped outgrowths along their ribs near the costochondral junction. To further understand the pathogenesis of MHE, two groups recently used genetic approaches involving two distinct Ext1f/f mouse lines and Col2Cre or Col2CreER mice that resulted in conditional homozygous loss of Ext1 in subpopulations of growth plate chondrocytes [18, 19]. Interestingly, both mutant mice developed numerous exostoses with a stereotypic growth plate-like organization along their long bones that were absent in conditional heterozygous littermates. A major implication of these findings was that full inactivation of Ext expression may be required in subpopulations of chondrocytes for exostosis formation in MHE patients. Such inactivation could involve a variety of mechanisms including loss-of-heterozygosity [20, 21]. To further test these possibilities and gain greater insights into mechanisms, we produced compound Ext1+/−Ext2+/− heterozygous mice. Strikingly, the mice developed outgrowths in their ribs at twice the frequency seen in single Ext heterozygous mice and most importantly, displayed stereotypic growth plate-like exostoses along their long bones. These and other data suggest that a significant, but not complete, decrease in Ext expression is sufficient for formation of stereotypic exostoses and that the nature and organization of ectopic tissue may be influenced by site-specific anatomical cues and mechanisms.
Materials and methods
Mice
Ext1- and Ext2-mutant mice were described previously and were fully backcrossed into C57Bl/6 background [14, 22]. Ext1f/f mice were described previously [13], Dermo1Cre mice were obtained from Dr. D. Ornitz [23] and Col2Cre mice were from Jackson Labs. [24]. Animal were housed in vivaria approved by the Association for Assessment and Accreditation of Laboratory Animal Care located in the School of Medicine, University of California, San Diego, following standards and procedures approved by the local Institutional Animal Care and Use Committee (protocol S99127). Hematological analyses were carried out by the Hematology and Coagulation Core at the University of California, San Diego. Levels of 1,25-dihydroxy vitamin D were measured by Esoterix, Inc. (Austin, TX). In some experiments, compound Ext heterozygous animals were supplemented with 5 mM glucosamine sulfate or 2 mM naphthol-β-D-xyloside in their water supply.
To test if growth plate damage would increase the frequency of exostoses, a 16-gauge needle was inserted twelve times into the tibial growth plate region of anesthetized wild-type and compound heterozygous mutant mice at weaning. Initial experiments included injection of dye to determine the optimal point of insertion of the needle through the skin and into the growth plate. Nicking was repeated every other day for ten days. Mice were euthanized at twelve weeks of age and the hindlimbs examined for outgrowths and exostoses.
Exostosis detection and analysis
Two methods were used to detect outgrowths and exostoses. In Method 1, rib cages were removed and illuminated to reveal the contours of the growth plate areas, and masses that appeared as shadows were noted. Rib cages and limbs were removed and excess soft tissue was excised in order to detect ectopic tissue masses under a dissecting microscope. In Method 2, rib cages were removed and soaked in 5% potassium hydroxide for 2 hrs at room temperature to remove and clarify soft tissues, and then examined under a dissecting microscope. To prepare whole mouse skeletons, carcasses were mixed with Dermestid (carrion) beetles (Gary Haas, Big Sky Beetleworks, Montana).
Ribs were removed from their surrounding tissue, placed in CalExII (Fisher) overnight for fixation and decalcification and embedded in paraffin. Sections (6 mm) were stained with hematoxylin and eosin or treated overnight with 90 units hyaluronidase (Sigma) at 37°C for immunohistochemical analyses. Some sections were treated with heparin lyases to generate a neo-epitope recognized by monoclonal antibody 3G10 (Seikagaku) as described [25]. In situ hybridization was performed using 35S-labeled riboprobes as described [26].
Cell culture
Costal cartilage was isolated from individual ribs, and digested overnight at 37°C in Dulbecco's Modified Eagles Medium (DMEM, Cellgro) with 0.2% collagenase B (Roche) and 15% fetal bovine serum (HyClone). The digest was poured over a 70 mm filter (Fisher) and the filtrate was centrifuged at 4°C for 10 min at 700 × g. The cell pellet was resuspended in medium with 15% FBS and penicillin/streptomycin (Gibco), added to tissue culture plates at high density, and maintained in a 37°C incubator under an atmosphere of 5% CO2 in 95% air and 100% relative humidity. Pulmonary endothelial cells and fibroblasts were isolated as described [27]. FGF18 (Abcam) stimulation of Erk phosphorylation in chondrocytes was measured by Western blotting with antipErk mAb (Cell Signaling Technology, Inc.).
Analyses of heparan sulfate
Cultured cells were labeled for 24 hrs in the presence of 100 μCi/ml 35SO4 or 3H-glucosamine (Perkin Elmer Life Sciences) in F12 medium (Gibco) supplemented with 15% dialyzed fetal bovine serum (HyClone). [3H]- or [35S]heparan sulfate chains were isolated and analyzed by gel filtration HPLC (Varian) using a TSK4000 column (TosoHaas) with in-line scintillation counting (Radiomatic Flow One) [28].
Results
Heterozygous Ext1+/− mice develop rib exostosis-like outgrowths
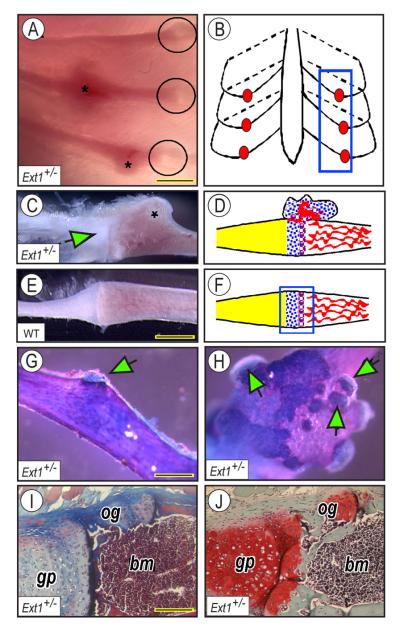
We previously showed that about one-third of Ext2+/− mice display one or more ectopic outgrowths on their ribs [14]. To determine whether heterozygous ablation of Ext1 causes a similar phenotype, we examined the ribcages of Ext1+/− mice at 1 to 2 months of age. Indeed, we detected outgrowths in their ribcages using chest cavity trans-illumination (Fig. 1A and C, asterisks). The outgrowths usually occurred near the growth plate at the costochondral junction along the rib ventral side (schematically shown Fig. 1B, blue rectangle, and 1D), but were absent in ribs from wild-type mice (Fig. 1E and F). The outgrowths were largely cartilaginous as indicated by Alcian Blue/Alizarin Red staining (Fig. 1G and H) and occurred as a single (Fig. 1G, arrow) or multiple ectopic masses (Fig. 1H, arrows). Masson's trichrome (Fig. 1I) and Safranin O (Fig. 1J) staining showed that the apical cartilaginous and basal bony portions of each outgrowth were quite evident [14].
Fig. 1.
Heterozygous Ext1+/− mice develop rib exostosis-like outgrowths. (A-B) Illumination reveals that ectopic outgrowths (asterisks) are usually near the rib costochondral junctions in mutants (circles in A and blue box in B). (C-D) Outgrowth present in mutant rib (asterisk in C) is near the chondro-costal junction (green arrow in C and schematic in D). (E-F) Wild-type rib lacks outgrowths. (G-H) Alcian Blue/Alizarin Red staining reveals presence of single or multiple cartilage capped-outgrowths (green arrows) perched on ectopic bone (blue, cartilage; pink, bone). (I-J) Masson's trichrome (I) and Safranin O (J) staining reveals the juxtaposition of exostosis-like outgrowth (og) to the rib growth plate (gp) and bone and marrow (bm). Bar in A, 1.2 mm; bar in E for C and E, 1 mm; bar in G for G-H, 0.8 mm; bar in I for I-J, 0.15 mm.
The overall frequency of outgrowths in Ext1+/− mice determined by trans-illumination was similar to that seen in Ext2+/− mice previously (Table 1, Method 1, 13.8%-16.6%, respectively). To obtain a more accurate assessment, soft tissues were removed and rib cages were viewed under a dissecting microscope. This method showed that rib outgrowths frequency was higher, but both strains again exhibited similar penetrance (44.4% vs. 36.5% respectively). Examination of long bones and other skeletal elements by necropsy did not reveal outgrowths or exostoses at locations such as long bones where they often occur in MHE patients [2, 3].
Table 1.
Frequency of exostosis-like outgrowths in Ext deficient mice.
| Genotype | Number of Animals (n) | Animals with Rib Outgrowths |
Penetrance (%) | |||
|---|---|---|---|---|---|---|
| Method 1a | Method 2b | Method 1a | Method 2b | Method 1a | Method 2b | |
| Wild-type | 101 | 50 | 0 | 0 | 0 | 0 |
| Ext1 +/− | 101 | 9 | 14 | 4 | 13.8 | 44.4 |
| Ext2 +/− | 120 | 52 | 20 | 19 | 16.6 | 36.5 |
| Ext1 f/+ Col2Cre + | 60 | nd | 6 | nd | 10 | nd |
| Ext1 f/+ | 53 | nd | 0 | nd | 0 | nd |
| Ext1 f/+ Dermo1Cre + | 27 | nd | 4 | nd | 14.8 | nd |
| Ext1 f/+ | 35 | nd | 0 | nd | 0 | nd |
| Ext1 +/− Ext2 +/− | 165 | 14 | 60 | 10 | 36.3 | 71.4 |
Method 1: Backlighting and visualization of growths.
Method 2: Rib cages were removed and soaked in 5% KOH for 2 hrs, then examined under a dissecting microscope.
Penetrance was defined as the percentage of animals that contained one or more outgrowths. Typically, 1-3 rib outgrowths were present in affected animals.
nd, not determined
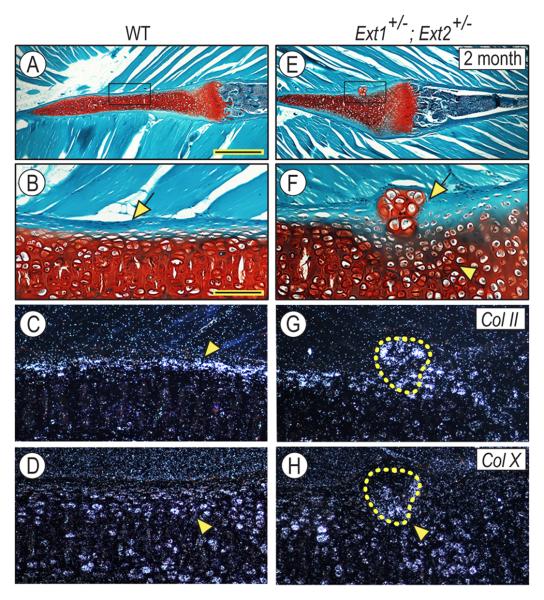
To characterize the phenotype of rib-associated outgrowths, we carried out additional morphological and gene expression analyses. In wild-type mice, the cartilaginous ventral portion of ribs was intensely and uniformly stained with Safranin O (Fig. 2A) and the boundary between cartilage and adjacent perichondrial and connective tissues was well defined and clear as expected (Fig. 2B, arrow). In compound Ext1+/−Ext2+/− mutant littermates, the exostosis-like outgrowths interrupted the continuity of perichondrium (Figs. 2 E and F, arrow); there was also local disorganization of underlying chondrocytes (Fig. 2F, arrowhead). Collagen II, a marker of resting and proliferating chondrocytes, was broadly expressed (Fig. 2G, circled area) and collagen X, a marker of hypertrophic chondrocytes, was expressed over the basal portion adjacent to the rib cartilage (Fig. 2H, arrowhead). In rib cartilage itself, collagen II was expressed by peripheral chondrocytes (Fig. 2C, arrowhead) and collagen X characterized the core chondrocytes (Fig. 2D, arrowhead), a unique pattern of expression typical of the ventral rib region [14] This finding indicated that the ectopic collagen X-positive chondrocytes in the exostosis were not rearranged rib chondrocytes.
Fig. 2.
Rib outgrowths interrupt perichondrium integrity. Longitudinal sections of ribs from 2 month-old wild-type (A-D) and Ext1+/−/Ext2+/− mice (E-H) were stained with Safranin O/fast green (A-B, E-F) or processed for in situ hybridization (C-D, G-H). In wild-type mice, rib cartilage and costochondral growth plate are separated from surrounding muscle by a clear perichondrium (B, arrow). Rib peripheral chondrocytes express collagen II (C, arrowhead), while core chondrocytes express collagen X (D, arrowhead). Boxed area in A is shown at higher magnification in B. In mutants, the outgrowth interrupts the continuity of perichondrium (E-F, arrow), is largely composed of collagen II-expressing chondrocytes (G, circled area) and contains a few collagen X-expressing chondrocytes at its base (H, arrowhead). Boxed area in E is shown at higher magnification in F. Bar in A for A and E, 0.8 mm; and bar in B for B-D and F-H, 0.12 mm.
To determine whether rib outgrowths were induced in mice bearing conditional and tissue-restricted Ext ablation, we crossbred Ext1f/+ animals to Col2Cre mice that express Cre recombinase in cartilage and some perichondrial cells [24] or Dermo1Cre mice that express Cre recombinase in skeletal mesenchymal condensations and perichondrial and periosteal cells [23]. Indeed, rib outgrowths were detected in both Ext1f/+Col2Cre and Ext1f/+Dermo1Cre adult mice (Table 1). Penetrance was similar to that observed in Ext1+/− or Ext2+/− mice (Table 1), providing genetic evidence that cells forming the rib outgrowths are likely to be local and to be members of the perichondrial and/or chondrocytic cell lineages as recently reported [18, 19].
Compound Ext1/Ext2 heterozygote mice develop exostoses on long bones
Ext1 and Ext2 proteins form a hetero-oligomeric complex responsible for polymerization of the carbohydrate backbone of heparan sulfate [9]. Thus, we hypothesized that combining heterozygous mutations might increase the penetrance and severity of the exostosis phenotype because of further reduction of overall Ext expression and function. Mice lacking single copies of both Ext1 and Ext2 (Ext1+/−Ext2+/−) were bred by standard mating schedules. They showed no gross observable phenotypes throughout adulthood, standard hematological parameters (blood counts and chemistry) and urinalysis were unaffected (Supplemental Table 1) and plasma levels of 1,25-dihydroxy-vitamin D were within normal range (18-32 pg/ml). Analysis of rib cages of 165 compound heterozygote adult mice by trans-illumination (Method 1) revealed that 60 mice had developed more numerous rib-associated outgrowths (36.3%) compared to Ext1+/− and Ext2+/− single heterozygous mice (13.8% or 16.6%, respectively). A similar trend was observed using Method 2 (71.4% penetrance compared to 36.5% to 44.4% in single heterozygotes; Table 1). Rib outgrowths were also observed in compound heterozygous mice when the drinking water was supplemented with 5 mM glucosamine sulfate, a building block of glycosaminoglycans (6/12 in its presence versus 4/13 in its absence) [29], or with 2 mM naphthalene β-D-xyloside, an agent that generates free glycosaminoglycan chains and decreases overall glycosaminoglycan addition to proteoglycans (3/7 supplemented mice had exostoses versus 3/8 unsupplemented animals) [30, 31], suggesting that outgrowth formation was not simply the result of generalized alteration of glycosaminoglycan assembly.
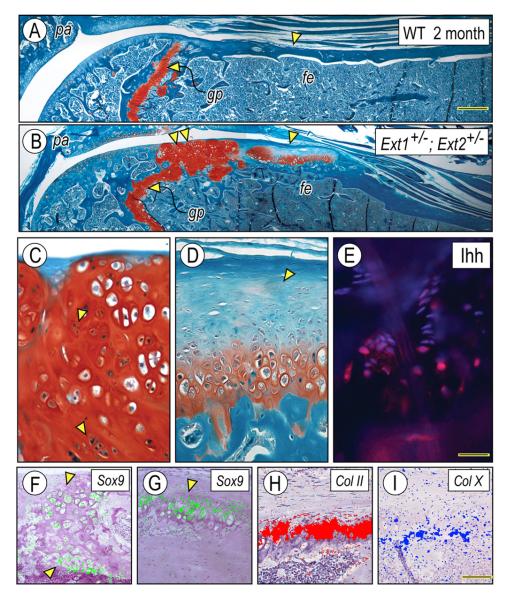
Of significant importance was the finding that in addition to rib outgrowths, compound Ext1+/−Ext2+/− mutants exhibited stereotypic exostoses in long bones (two of three mice available for analysis). One example is shown in Fig. 3A and 3B illustrating the proximal portions of tibias in 2 month-old wild-type and mutant mice, respectively. In the wildtype, the epiphyseal/metaphyseal region displayed a typical growth plate (Fig. 3A, gp) and compact bone flanked by continuous and uninterrupted perichondrium and periosteum all around the outer perimeter (Fig. 3A, arrowhead). In contrast, the periosteal border in the mutant was interrupted by a stereotypic exostosis (Fig. 3B, arrowhead) that consisted of a growth plate-like arrangement of chondrocytes covered with a conspicuous apical cartilage cap (Fig. 3D, arrowhead). Gene expression patterns were typical and sharply defined. Expression of Sox9 and collagen II characterized the cartilaginous cap and top growth plate-like zones (Fig. 3G and H, respectively), expression of collagen X was restricted to hypertrophic chondrocytes (Fig. 3I), and the distribution of Indian hedgehog (Ihh), a marker of pre-hypertrophic chondrocytes, was limited to the thin layer of pre-hypertrophic chondrocytes (Fig. 3E).
Fig. 3.
Ext1+/−/Ext2+/− mutants develop exostoses on long bones. Longitudinal sections of femurs from 2 month-old wild-type (A) and Ext1+/−/Ext2+/− mutants (B) were stained with Safranin O/fast green. Exostoses appear as either hyperplastic (B, double arrowhead) or stereotypic growth plate-like structures (B, arrowhead). The hyperplastic exostosis is locally invasive and contains clusters of proliferating cells (C, arrowheads), and exhibits widespread Sox9 expression (F). The stereotypic exostosis displays a growth plate arrangement and a conspicuous cartilaginous cap (D, arrowhead); Indian hedgehog (Ihh) distribution is limited to the pre-hypertrophic zone (E) and expression of Sox9 (G), collagen II (H) and collagen X (I) is in typical narrow zones. Bar in A for A-B, 0.4 mm; bar in E for C-E, 0.1 mm; and bar in I for F-I, 0.3 mm.
Interestingly, masses of hyperplastic chondrocytes were present in some mutant long bones (such as the one depicted in Fig. 3B, double arrowheads). They lacked a standard apical cartilage cap and displayed a disorganized chondrocyte arrangement (Fig. 3C), exemplified by the scattered expression patterns of Sox9 (Fig. 3F). The hyperplastic masses appeared to be more locally invasive compared to typical exostoses and in fact, proliferating cells visualized as mitotic figures were common (Fig. 3C, arrowheads). It remains unclear whether these masses hint to the origin of, and serve as a platform for, conversion to malignant chondrosarcoma that occur in some MHE patients [32].
To determine if gross skeletal defects were present, carcasses from wild-type and compound Ext1+/−Ext2+/− littermates were cleaned by Dermestid beetles and closely inspected. Amongst skeletons of mutant mice positive for rib outgrowths (n = 20) and wild-type littermates (n = 20), two mutant animals displayed distorted radius and ulna (Fig. S1B) or bony protuberances in scapula (Fig. S1D) that are reminiscent of similar malformations in some MHE patients [2, 3]. Wild-type bones never displayed defects (Fig. S1A and S1C).
The presence of these additional skeletal defects in compound heterozygous mice brought to bear the long-standing issue of why malformations and exostoses appear to occur more frequently at certain sites within the skeleton. Thus, we posed the question as to whether environmental cues such as physical activity or local insults could enhance the frequency of exostoses in compound heterozygous mice. Exercise wheels were provided at weaning and mice were allowed to exert physical activity at will. Necropsy and histological analysis of ribcages and limb bones did not reveal gross abnormalities or exostoses even after an 8-week regimen. Based on previous studies of osteochondroma induction in rats [33], we tested whether microfractures introduced into the tibial growth plate region by a needle might have an effect, but even repeated insults did not lead to exostosis formation (n = 12 for each genotype).
Heparan sulfate content and average chain length in growth plates and cell cultures
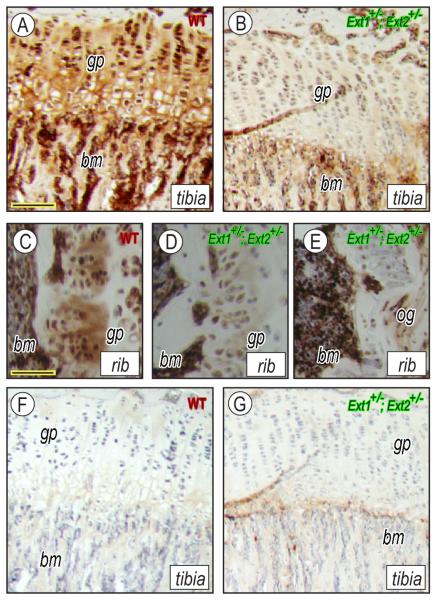
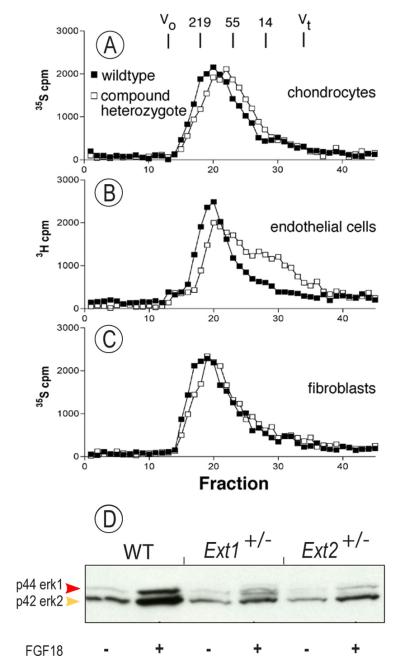
To examine the effects of compound Ext1+/−Ext2+/− ablation on heparan sulfate content, longitudinal sections of wild-type and Ext1+/−Ext2+/− mutant tibias were treated with heparin lyases and immunostained with antibody 3G10 [25]. Wild-type growth plates and surrounding tissues stained heavily for heparan sulfate (Fig. 4A), whereas corresponding tissues in compound Ext1+/−Ext2+/− mutants stained much less (Fig. 4B). The difference in staining intensity was also observed in rib growth plates from wild-type and Ext1+/−Ext2+/− mice (Fig. 4C and 4D, respectively), and rib outgrowths also had little heparan sulfate (Fig. 4E). In the absence of primary antibody, wild-type and Ext1+/−Ext2+/−tibias showed little nonspecific staining (Fig. 4F and 4G, respectively). Analysis of heparan sulfate newly-synthesized by cultured wild-type and compound heterozygote costochondral chondrocytes by gel filtration HPLC showed that chains produced by wild-type cells had an average size of ~115 kDa (Fig. 5A, filled symbols), whereas chains produced by mutant chondrocytes were ~76 kDa (Fig. 5A, open symbols). Similar differences were seen in wild-type and compound mutant endothelial cells and fibroblasts (Fig. 5 B and C).
Fig. 4.
Ext1+/−/Ext2+/− mutant tissues contain very low heparan sulfate. Longitudinal sections of tibia and rib growth plates from wildtypes (A, C, F) and Ext1+/−/Ext2+/− mutants (B, D-E, G) were treated with heparinase and processed for immunostaining in presence or absence of anti-HS antibody. Note that positive staining is much stronger in wild-type (A, C) than mutant tissues (B, D) and was also low the rib outgrowth (E). Omission of primary antibody gave background staining only (F, G). gp, growth plate; bm, bone marrow; and og, outgrowth. Bar in A for A-B and F-G, 0.120 mm; and bar in C-E, 0.1 mm.
Fig. 5.
Ext mutant cells produce shorter heparan sulfate chains and respond less to FGF18. Gel filtration HPLC analysis of heparan sulfate produced by cultured chondrocytes (A), endothelial cells (B) and fibroblasts (C) shows that Ext1+/−/Ext2+/− mutant cells (open squares) produce shorter chains compared to wildtype (closed squares). Immunoblots show that FGF18-treated Ext1+/− or Ext2+/− mutant chondrocytes exhibit lower responsiveness and levels of phosphorylated p44 and p42 Erk proteins compared to wild-type cells.
To determine whether the reduction in heparan sulfate would result in reduced responsiveness to heparan sulfate-dependent growth factors, wild-type and single mutant chondrocytes were treated with 10 ng/ml FGF18, an important growth plate factor that signals via FGFR3 and Erk1/2 phosphorylation [34]. Immunoblot analysis showed that the levels of phosphorylated Erk (pErk) were strongly increased in FGF18-treated wild-type chondrocytes (Fig. 5D, first 2 lanes), but were less so in mutant chondrocytes (Fig. 5D, remaining 4 lanes).
Discussion
Our data demonstrate that stereotypic exostoses closely resembling those in MHE patients, in both anatomical location and organization, form in mouse long bones after compound heterozygous ablation of Ext1 and Ext2. Hyperplastic exostoses possibly representing an intermediate stage toward malignancy were occasionally seen along the long bones, and the mutant mice have additional skeletal deformities. Thus, the compound heterozygous mice mimic major diagnostic and pathologic features of MHE. Smaller and oval-shaped outgrowths are present in the ribs of single Ext1 or Ext2 heterozygous mice and their frequency nearly doubles in compound mutant mice. Together, the data indicate that penetrance and severity of the exostosis phenotype are intimately sensitive to, and inversely related to, Ext gene dosage and that a significant, but not necessarily complete, decrease of Ext expression may be sufficient for formation of stereotypic growth plate-like exostoses. The data also indicate that certain skeletal sites are more suitable for, and prone to, ectopic tissue formation [3] and that the nature and organization of ectopic tissue may be influenced by chondrocyte/mesenchyme-associated cues and mechanisms. The data imply that the varying skeletal phenotypes and defects seen in different MHE patients could reflect differential severity of Ext mutations and/or differential local anatomical consequences of Ext function.
We and others proposed previously that exostosis formation is directly tied to alterations of heparan sulfate biosynthesis. The increased penetrance of the phenotype in compound mouse mutants seen here is consistent with the fact that Ext1 and Ext2 proteins interact to form oligomeric complexes required for heparan sulfate synthesis [9, 10]. Indeed, we find that the tissue levels of heparan sulfate are markedly reduced in compound Ext1+/−/Ext2+/− mice based on immunostaining with mAb 3G10, as observed previously using similar methods in human exostoses [35] and in tissue sections from mice bearing a hypomorphic gene trap allele of Ext1 [36]. This result is somewhat surprising since mAb 3G10 reacts with a neoepitope generated by heparin lyase treatment that remains on the core proteins of heparan sulfate bearing proteoglycans. For unknown reasons, it appears that shortening of the chains, which is evident in chondrocytes and other cells derived from the compound mutant, results in loss of proteoglycans from the tissue. Additional studies are needed to assess the level of heparan sulfate chemically or immunohistochemically in the growth plate and exostoses and to compare these values in heterozygous and compound heterozygous mice. Data in the two recent mouse studies show that multiple exostoses were observed along the long bones following conditional homozygous loss of Ext1, leading to a nearly complete local loss of heparan sulfate [18, 19]. Because MHE patients are heterozygous for EXT1 or EXT2, the dramatic loss of heparan sulfate in affected tissues could result from sporadic loss of heterozygosity as observed in exostosis tissue from some, but not all, patients [21, 37]. Other mechanisms may include epigenetic mechanisms that silence Ext or a second hit in other regulatory genes. Regardless of the mechanism, the more exuberant formation of outgrowths and exostoses appears to correlate with a major drop in heparan sulfate levels [18, 35]; such a drop could represent the key inductive event for formation of stereotypic exostoses.
Recent studies have suggested that Ext2 can form complexes not only with Ext1 but also with N-acetylglucosamine N-deacetylase-N-sulfotransferase-1 (Ndst1), an enzyme involved in sulfation of heparan sulfate [38]. Thus, there may be pleiotropic effects of altering the level of Ext gene expression on both heparan sulfate polymerization and sulfation patterns. This is of particular relevance since changes in heparan sulfate composition as well as size could affect its roles in regulating the topography, intensity and action of multiple heparan sulfate-binding signaling factors that influence skeletal cells and growth plate chondrocytes [39]. Indeed, heparan sulfate was originally found to act as a limiting barrier for diffusion of morphogens as revealed by Drosophila mutants lacking tout velu, the fly homolog of Ext1 [40]. Relevant signaling factors include members of the fibroblast growth factor, hedgehog, bone morphogenetic protein and Wnt protein families [40-42] all of which need to act very precisely during skeletogenesis and skeletal growth [43-45]. We describe here decreases in Erk phosphorylation levels in single Ext mutant versus wild-type chondrocytes following FGF18 treatment suggesting that cellular responses to cytokines are sensitive to even small changes in HS levels or length. In a similar vein, reduction in chain size either by enzymatic cleavage or by alteration of Ext1 expression elicited profound effects on FGF signaling in cells [46, 47]. Reduced heparan sulfate synthesis and proteoglycan expression were found to be associated with enhanced diffusion of Ihh across the growth plate during embryonic development [36], and we showed that a redistribution of Ihh from growth plate to perichondrium was linked to formation of ectopic cartilage and bone tissue [48]. Clearly then, a significant change in heparan sulfate levels and composition could theoretically alter the function of numerous heparan sulfate-binding growth factors that could contribute to deregulation of skeletal outgrowths and exostoses. However, we cannot exclude the possibility that formation of exostoses and exostosis-like outgrowths may also be influenced by other important functions of Exts not related to heparan sulfate formation [38]. In this regard, it is interesting that there are no reports of MHE patients with mutations in other genes involved in heparan sulfate biosynthesis [49].
In addition to pathogenic mechanisms, the origin of exostosis-forming cells remains unclear. It was previously proposed that exostoses are formed by growth plate chondrocytes that misbehave and malfunction as a result of Ext loss-of-function [14]. A similar conclusion was reached in the recent mouse studies involving homozygous somatic loss of Ext1 [18, 19]. Others had previously proposed that exostoses are produced by perichondrial cells based on placement of exostoses at the periphery of the growth plate [50]. We show here that rib outgrowths form in both Ext1f/+Col2Cre and Ext1f/+Dermo1Cre mice signifying that local Ext deficiency is sufficient to trigger outgrowth formation. However, because Col2Cre and Dermo1Cre target both chondrocytes and perichondrial cells, the data do not allow us to identify conclusively the exact nature of the outgrowth-forming cells. A perichondrial origin of exostosis-forming cells would be particularly attractive because the perichondrium contains cells of mesenchymal origin that can differentiate into chondrocytes and osteoblasts and participate in radial bone growth and fracture healing [51, 52]. Thus, we should certainly entertain the idea that exostoses may in fact arise from heparan sulfate deficiency in a stem cell population. Consistent with this idea is the recent demonstration that heparan sulfate proteoglycans are essential regulators of the germline stem cell niches in Drosophila gonads [53] and regulate stem cell differentiation in mice [54, 55].
Supplementary Material
Acknowledgements
J.D.E. and M.P. designed research; B.M.Z., M.S., C.M. and E.K. performed research; D.E.W. and Y.Y. contributed new tools and to analysis of data; and J.D.E. and M.P. wrote the paper. We thank Dr. Nissi Varki (Histology Core, UCSD Cancer Center) for help in analyzing histological sections, David Ditto (Hematology and Chemistry Core, UCSD), and Dr. Karen Bame (University of Missouri) for TSK4000 gel filtration column calibration [56]. This work was supported by grants R37GM33063 (to J.D.E), 1RC1AR058382 (to M.P. and E.K.), and HL57345 for Core support from the NIH, by an award made through Research Supplements for Underrepresented Minorities initiative by the NIH (to B.Z.), and a gift from Wendy and Richard Lubkin. We thank Dominick Stickens and Zena Werb for reading an early version of this manuscript and providing helpful comments. More information on MHE can be found at the MHE Research Foundation website www.mheresearchfoundation.org.
Abbreviations
- MHE
Multiple Hereditary Exostoses
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Solomon L. Hereditary Multiple Exostosis. British J Bone Joint Surg. 1963;45:292–304. [Google Scholar]
- 2.Pierz KA, Stieber JR, Kusumi K, Dormans JP. Hereditary multiple exostoses: One center's experience and review of etiology. Clin Orthop. 2002:49–59. doi: 10.1097/00003086-200208000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Schmale GA, Conrad EUr, Raskind WH. The natural history of hereditary multiple exostoses. J Bone Joint Surg Am. 1994;76:986–92. doi: 10.2106/00004623-199407000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Zak BM, Crawford BE, Esko JD. Hereditary multiple exostoses and heparan sulfate polymerization. Biochim Biophys Acta Gen Subj. 2002;1573:346–355. doi: 10.1016/s0304-4165(02)00402-6. [DOI] [PubMed] [Google Scholar]
- 5.Lind T, Lindahl U, Lidholt K. Biosynthesis of heparin/heparan sulfate. Identification of a 70-kDa protein catalyzing both the D-glucuronosyl- and the N-acetyl-D-glucosaminyltransferase reactions. J Biol Chem. 1993;268:20705–20708. [PubMed] [Google Scholar]
- 6.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J Biol Chem. 1998;273:26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 7.McCormick C, Leduc Y, Martindale D, Mattison K, Esford LE, Dyer AP, Tufaro F. The putative tumour suppressor EXT1 alters the expression of cell- surface heparan sulfate. Nat Genet. 1998;19:158–161. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- 8.Wei G, Bai XM, Gabb MMG, Bame KJ, Koshy TI, Spear PG, Esko JD. Location of the glucuronosyltransferase domain in the heparan sulfate copolymerase EXT1 by analysis of Chinese hamster ovary cell mutants. J Biol Chem. 2000;275:27733–27740. doi: 10.1074/jbc.M002990200. [DOI] [PubMed] [Google Scholar]
- 9.McCormick C, Duncan G, Goutsos KT, Tufaro F. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci USA. 2000;97:668–673. doi: 10.1073/pnas.97.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi S, Morimoto K, Shimizu T, Takahashi M, Kurosawa H, Shirasawa T. Association of EXT1 and EXT2, hereditary multiple exostoses in Golgi apparatus. Biochem Biophys Res Commun. 2000;268:860–867. doi: 10.1006/bbrc.2000.2219. [DOI] [PubMed] [Google Scholar]
- 11.Lin X, Gan L, Klein WH, Wells D. Expression and functional analysis of mouse EXT1, a homolog of the human multiple exostoses type 1 gene. Biochem Biophys Res Commun. 1998;248:738–743. doi: 10.1006/bbrc.1998.9050. [DOI] [PubMed] [Google Scholar]
- 12.Stickens D, Brown D, Evans GA. EXT genes are differentially expressed in bone and cartilage during mouse embryogenesis. Dev Dyn. 2000;218:452–464. doi: 10.1002/1097-0177(200007)218:3<452::AID-DVDY1000>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 13.Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- 14.Stickens D, Zak BM, Rougier N, Esko JD, Werb Z. Mice deficient in Ext2 lack heparan sulfate and develop exostoses. Development. 2005;132:5055–5068. doi: 10.1242/dev.02088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim BT, Kitagawa H, Tamura J, Saito T, Kusche-Gullberg M, Lindahl U, Sugahara K. Human tumor suppressor EXT gene family members EXTL1 and EXTL3 encode a1,4-N-acetylglucosaminyltransferases that likely are involved in heparan sulfate/heparin biosynthesis. Proc Natl Acad Sci USA. 2001;98:7176–7181. doi: 10.1073/pnas.131188498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busse M, Feta A, Presto J, Wilen M, Gronning M, Kjellen L, Kusche-Gullberg M. Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J Biol Chem. 2007;282:32802–10. doi: 10.1074/jbc.M703560200. [DOI] [PubMed] [Google Scholar]
- 17.Karibe T, Fukui H, Sekikawa A, Shiratori K, Fujimori T. EXTL3 promoter methylation down-regulates EXTL3 and heparan sulphate expression in mucinous colorectal cancers. J Pathol. 2008;216:32–42. doi: 10.1002/path.2377. [DOI] [PubMed] [Google Scholar]
- 18.Jones KB, Piombo V, Searby C, Kurriger G, Yang B, Grabellus F, Roughley PJ, Morcuende JA, Buckwalter JA, Capecchi MR, Vortkamp A, Sheffield VC. A mouse model of osteochondromagenesis from clonal inactivation of Ext1 in chondrocytes. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.0910875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto K, Irie F, Mackem S, Yamaguchi Y. A mouse model of chondrocyte-specific somatic mutation reveals a role for Ext1 loss of heterozygosity in multiple hereditary exostoses. Proc Natl Acad Sci U S A. 2010;107:10932–7. doi: 10.1073/pnas.0914642107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hecht JT, Hogue D, Strong LC, Hansen MF, Blanton SH, Wagner M. Hereditary multiple exostosis and chondrosarcoma: linkage to chromosome II and loss of heterozygosity for EXT-linked markers on chromosomes II and 8. Am.J.Hum.Genet. 1995;56:1125–1131. [PMC free article] [PubMed] [Google Scholar]
- 21.Hall CR, Cole WG, Haynes R, Hecht JT. Reevaluation of a genetic model for the development of exostosis in hereditary multiple exostosis. Am J Med Genet. 2002;112:1–5. doi: 10.1002/ajmg.10635. [DOI] [PubMed] [Google Scholar]
- 22.Lin X, Wei G, Shi ZZ, Dryer L, Esko JD, Wells DE, Matzuk MM. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev Biol. 2000;224:299–311. doi: 10.1006/dbio.2000.9798. [DOI] [PubMed] [Google Scholar]
- 23.Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–74. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 24.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–6. [PubMed] [Google Scholar]
- 25.David G, Bai XM, Van der Schueren B, Cassiman JJ, Van den Berghe H. Developmental changes in heparan sulfate expression: in situ detection with mAbs. J Cell Biol. 1992;119:961–975. doi: 10.1083/jcb.119.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koyama K, Uobe K, Tanaka A. Highly sensitive detection of HPV-DNA in paraffin sections of human oral carcinomas. J Oral Pathol Med. 2007;36:18–24. doi: 10.1111/j.1600-0714.2006.00490.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Fuster M, Sriramarao P, Esko JD. Endothelial heparan sulfate deficiency impairs Lselectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat Immunol. 2005;6:902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 28.Bame KJ, Esko JD. Undersulfated heparan sulfate in a Chinese hamster ovary cell mutant defective in heparan sulfate N-sulfotransferase. J Biol Chem. 1989;264:8059–8065. [PubMed] [Google Scholar]
- 29.Bassleer C, Rovati L, Franchimont P. Stimulation of proteoglycan production by glucosamine sulfate in chondrocytes isolated from human osteoarthritic articular cartilage in vitro. Osteoarthr Cartilage. 1998;6:427–434. doi: 10.1053/joca.1998.0146. [DOI] [PubMed] [Google Scholar]
- 30.Fritz TA, Lugemwa FN, Sarkar AK, Esko JD. Biosynthesis of heparan sulfate on beta-Dxylosides depends on aglycone structure. J Biol Chem. 1994;269:300–307. [PubMed] [Google Scholar]
- 31.Miao H-Q, Fritz TA, Esko JD, Zimmermann J, Yayon A, Vlodavsky I. Heparan sulfate primed on β-D-xylosides restores binding of basic fibroblast growth factor. J Cell Biochem. 1995;57:173–184. doi: 10.1002/jcb.240570202. [DOI] [PubMed] [Google Scholar]
- 32.Bovee JV. Multiple osteochondromas. Orphanet J Rare Dis. 2008;3:3. doi: 10.1186/1750-1172-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delgado E, Rodriguez JI, Rodriguez JL, Miralles C, Paniagua R. Osteochondroma induced by reflection of the perichondrial ring in young rat radii. Calcif Tissue Int. 1987;40:85–90. doi: 10.1007/BF02555710. [DOI] [PubMed] [Google Scholar]
- 34.Liu Z, Lavine KJ, Hung IH, Ornitz DM. FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Dev Biol. 2007;302:80–91. doi: 10.1016/j.ydbio.2006.08.071. [DOI] [PubMed] [Google Scholar]
- 35.Hecht JT, Hall CR, Snuggs M, Hayes E, Haynes R, Cole WG. Heparan sulfate abnormalities in exostosis growth plates. Bone. 2002;31:199–204. doi: 10.1016/s8756-3282(02)00796-2. [DOI] [PubMed] [Google Scholar]
- 36.Koziel L, Kunath M, Kelly OG, Vortkamp A. Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Dev Cell. 2004;6:801–813. doi: 10.1016/j.devcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Bernard MA, Hall CE, Hogue DA, Cole WG, Scott A, Snuggs MB, Clines GA, Ludecke HJ, Lovett M, Van Winkle WB, Hecht JT. Diminished Levels of the putative tumor suppressor proteins EXT1 and EXT2 in exostosis chondrocytes. Cell Motil Cytoskel. 2001;48:149–162. doi: 10.1002/1097-0169(200102)48:2<149::AID-CM1005>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Presto J, Thuveson M, Carlsson P, Busse M, Wilen M, Eriksson I, Kusche-Gullberg M, Kjellen L. Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc Natl Acad Sci USA. 2008;105:4751–6. doi: 10.1073/pnas.0705807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada S, Busse M, Ueno M, Kelly OG, Skarnes WC, Sugahara K, Kusche-Gullberg M. Embryonic fibroblasts with a gene trap mutation in ext1 produce short heparan sulfate chains. J Biol Chem. 2004;279:32134–41. doi: 10.1074/jbc.M312624200. [DOI] [PubMed] [Google Scholar]
- 40.The I, Bellaiche Y, Perrimon N. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol Cell. 1999;4:633–639. doi: 10.1016/s1097-2765(00)80214-2. [DOI] [PubMed] [Google Scholar]
- 41.Bellaiche Y, The I, Perrimon N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature. 1998;394:85–88. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- 42.Takei Y, Ozawa Y, Sato M, Watanabe A, Tabata T. Three Drosophila EXT genes shape morphogen gradients through synthesis of heparan sulfate proteoglycans. Development. 2004;131:73–82. doi: 10.1242/dev.00913. [DOI] [PubMed] [Google Scholar]
- 43.Karsenty G. Genetics of skeletogenesis. Dev Genet. 1998;22:301–313. doi: 10.1002/(SICI)1520-6408(1998)22:4<301::AID-DVG1>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 44.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 45.Pacifici M, Shimo T, Gentili C, Kirsch T, Freeman TA, Enomoto-Iwamoto M, Iwamoto M, Koyama E. Syndecan-3: a cell-surface heparan sulfate proteoglycan important for chondrocyte proliferation and function during limb skeletogenesis. J Bone Miner Metab. 2005;23:191–199. doi: 10.1007/s00774-004-0584-1. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Coomans C, David G. Membrane heparan sulfate proteoglycan-supported FGF2-FGFR1 signaling - Evidence in support of the “cooperative end structures” model. J Biol Chem. 2001;276:41921–41929. doi: 10.1074/jbc.M106608200. [DOI] [PubMed] [Google Scholar]
- 47.Osterholm C, Barczyk MM, Busse M, Gronning M, Reed RK, Kusche-Gullberg M. Mutation in the heparan sulfate biosynthesis enzyme EXT1 influences growth factor signaling and fibroblast interactions with the extracellular matrix. J Biol Chem. 2009;284:34935–43. doi: 10.1074/jbc.M109.005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koyama E, Young B, Nagayama M, Shibukawa Y, Enomoto-Iwamoto M, Iwamoto M, Maeda Y, Lanske B, Song B, Serra R, Pacifici M. Conditional Kif3a ablation causes abnormal hedgehog signaling topography, growth plate dysfunction, and excessive bone and cartilage formation during mouse skeletogenesis. Development. 2007;134:2159–69. doi: 10.1242/dev.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bornemann DJ, Duncan JE, Staatz W, Selleck S, Warrior R. Abrogation of heparan sulfate synthesis in Drosophila disrupts the Wingless, Hedgehog and Decapentaplegic signaling pathways. Development. 2004;131:1927–38. doi: 10.1242/dev.01061. [DOI] [PubMed] [Google Scholar]
- 50.Porter DE, Simpson AH. The neoplastic pathogenesis of solitary and multiple osteochondromas. J Pathol. 1999;188:119–25. doi: 10.1002/(SICI)1096-9896(199906)188:2<119::AID-PATH321>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 51.Colnot C, Lu C, Hu D, Helms JA. Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev Biol. 2004;269:55–69. doi: 10.1016/j.ydbio.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Skoog V, Widenfalk B, Ohlsen L, Wasteson A. The effect of growth factors and synovial fluid on chondrogenesis in perichondrium. Scand J Plast Reconstr Surg Hand Surg. 1990;24:89–95. doi: 10.3109/02844319009004526. [DOI] [PubMed] [Google Scholar]
- 53.Hayashi Y, Kobayashi S, Nakato H. Drosophila glypicans regulate the germline stem cell niche. J Cell Biol. 2009;187:473–80. doi: 10.1083/jcb.200904118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baldwin RJ, ten Dam GB, van Kuppevelt TH, Lacaud G, Gallagher JT, Kouskoff V, Merry CL. A developmentally regulated heparan sulfate epitope defines a subpopulation with increased blood potential during mesodermal differentiation. Stem Cells. 2008;26:3108–18. doi: 10.1634/stemcells.2008-0311. [DOI] [PubMed] [Google Scholar]
- 55.Johnson CE, Crawford BE, Stavridis M, Ten Dam G, Wat AL, Rushton G, Ward CM, Wilson V, van Kuppevelt TH, Esko JD, Smith A, Gallagher JT, Merry CL. Essential alterations of heparan sulfate during the differentiation of embryonic stem cells to Sox1-enhanced green fluorescent protein-expressing neural progenitor cells. Stem Cells. 2007;25:1913–23. doi: 10.1634/stemcells.2006-0445. [DOI] [PubMed] [Google Scholar]
- 56.Tumova S, Bame KJ. The interaction between basic fibroblast growth factor and heparan sulfate can prevent the in vitro degradation of the glycosaminoglycan by Chinese hamster ovary cell heparanases. J Biol Chem. 1997;272:9078–9085. doi: 10.1074/jbc.272.14.9078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.