Abstract
Brain machine interfaces (BMIs) transform activity of neurons recorded in motor areas of the brain into movements of external actuators. Representation of movements by neuronal populations varies over time, during both voluntary limb movements and movements controlled through BMIs, due to motor learning, neuronal plasticity, and instability in recordings. To assure accurate BMI performance over long time spans, BMI decoders must adapt to these changes. We propose the Bayesian regression self-training method for updating the parameters of an unscented Kalman filter decoder. This novel paradigm uses the decoder’s output to periodically update the decoder’s neuronal tuning model in a Bayesian linear regression. We use two previously-known statistical formulations of Bayesian linear regression: (1) a joint formulation which allows fast and exact inference, and (2) a factorized formulation which allows the addition and temporary omission of neurons from updates, but requires approximate variational inference. To evaluate these methods, we performed off-line reconstructions and closed-loop experiments with Rhesus monkeys implanted cortically with micro-wire electrodes. Off-line reconstructions used data recorded in areas M1, S1, PMd, SMA, and PP of 3 monkeys while they controlled a cursor using a hand-held joystick. The Bayesian regression self-training updates significantly improved the accuracy of offline reconstructions compared to the same decoder without updates. We performed 11 sessions of real-time, closed-loop experiments with a monkey implanted in areas M1 and S1. These sessions spanned 29 days. The monkey controlled the cursor using the decoder with and without updates. The updates maintained control accuracy and did not require information about monkey hand movements, assumptions about desired movements, or knowledge of the intended movement goals as training signals. These results indicate that Bayesian regression self-training can maintain BMI control accuracy over long time periods, making clinical neuroprosthetics more viable.
Keywords: brain machine interface, decoding, adaptive filtering
1 Introduction
Brain-machine interfaces (BMI) are systems which directly connect brains to external devices, for example prosthetic limbs for persons with paralysis (Nicolelis, 2003). While BMI systems have to perform accurately over long time periods, changes in the properties of recorded neuronal population may cause significant alterations in BMI operation. Past studies have shown that neuronal tuning to arm movement may change over time spans as short as one hour (C. S. Li et al., 2001; Padoa-Schioppa et al., 2004; Lebedev et al., 2005; Kim et al., 2006; Rokni et al., 2007; Chestek et al., 2007). Changes in neuronal tuning over time have been also reported for BMI control (Taylor et al., 2002; Truccolo et al., 2008). Additionally, neuronal and recording instability can cause changes to the shape of recorded spike waveforms. These changes may cause the spike-sorting system to err and lead to changes in the effective tuning of recorded units. Because of these reasons, BMI systems need adaptive decoding to maintain control accuracy over long time periods (Kim et al., 2006; Wu & Hatsopoulos, 2008). In addition to the ability to track neuronal variability, adaptive decoding has a benefit of exploiting plastic changes in neuronal tuning that occur during learning and thus facilitating coadaptive (Taylor et al., 2002) synergies between the user’s brain and the BMI.
Early computational methods for translating recorded neuronal signals into movement commands assumed static neuronal tuning (Wessberg et al., 2000; Serruya et al., 2002; Carmena et al., 2003; Hochberg et al., 2006; Wu et al., 2006; Ganguly & Carmena, 2009). These methods fixed the parameters of the neuronal decoder after an initial period of algorithmic training or parameter fitting. Such decoders required users to adapt to the BMI control system by changing their neuronal modulations, and these changes were not tracked by the decoder.
Some studies have implemented adaptive neuronal decoders (Sykacek & Roberts, 2002; Eden et al., 2004; Rickert et al., 2007; Srinivasan et al., 2007; Wu & Hatsopoulos, 2008; Shpigelman et al., 2009; Gilja et al., 2009; Liao et al., 2009). However, most of these neuronal decoders have assumed that the desired movements – for example, the trajectory from the initial position of the actuator to a target – were available to the update method. The update method then adjusted the decoder parameters such that the BMI output more closely approximated the desired movement. One approach to ensure the availability of desired movements is to instruct the prosthetic user to attempt prescribed movements during periodic retraining sessions.
A more versatile approach to decoder update is to shift the paradigm from assuming a priori knowledge of desired movements to using a computational method that updates the decoder’s model automatically, with little or no external intervention. The goal of this study is to design and validate an update method which can achieve this objective.
We propose the Bayesian regression self-training method for adaptive decoding. This method periodically updates (every 2 minutes in our study) the parameters of the model of neuronal tuning while decoding user intentions. During each update, the procedure combines information from previous model parameters, recent neuronal activity, and outputs of the decoder to compute new model parameters. Instead of using a priori knowledge of the desired movements, the updates use the decoder outputs as substitutes for the desired movements (similar to Gilja et al., 2009; Liao et al., 2009). Bayesian linear regression (Bishop, 2006; Rencher & Schaalje, 2008) between decoder outputs and neuronal activity, with the previous parameters acting as priors, calculates the new parameters. We chose Bayesian linear regression because it combines previous parameters and tuning changes detected from recent neuronal activity in a probabilistic manner, so that the new parameters are the posteriors of a Bayesian update. We also formulate a transition model for the tuning parameters. Together, the Bayesian regression update and transition model form a batch-mode Bayesian filter for the tuning parameters. For the decoding algorithm, we use the n-th order unscented Kalman filter (Z. Li et al., 2009). Our decoder and parameter update system simultaneously predicts the BMI user’s desired movements and tracks tuning changes in the user’s neuronal modulation.
Unlike previous, non-probabilistic, update approaches (Wu & Hatsopoulos, 2008; Shpigelman et al., 2009; Gilja et al., 2009), our approach automatically takes into account the uncertainties of the tuning model parameters and the amount of useful information and noise in the new data to compute Bayesian updates of tuning parameters. Unlike previous studies with parameter updates at every decoding time step (Eden et al., 2004; Rickert et al., 2007; Srinivasan et al., 2007; Wu & Hatsopoulos, 2008), our batch-mode approach allows the use of the Kalman smoother (Rauch et al., 1965) to improve decoder predictions before their use in updates. The Kalman smoother uses the model of movement change over time to refine predictions of desired movements at a time t using predictions of desired movements at times after t. The non-causal nature of this pre-processing step means it is only available in batch-mode operations.
We tested our method with Rhesus monkeys that performed motor tasks manually or through a BMI. We started with offline reconstructions of hand trajectories from neuronal data previously recorded in 3 monkeys. We then conducted experiments in which 1 monkey used our adaptive method in a real-time, closed-loop BMI. Adapting within each session in the offline reconstructions, our decoder was more accurate than the equivalent unscented Kalman filter without updates. Adapting within and across sessions in the closed-loop experiments, our adaptive decoder maintained BMI control accuracy over a period of 29 days, while the unscented Kalman filter without updates degraded in accuracy. These results indicate that Bayesian regression self-training can cope with changes in neuronal tuning or neuronal recording without the need for example movements. Bayesian regression self-training improves the robustness of a BMI to changes in neuronal tuning, making long-term BMI operation more reliable.
2 Neurophysiological Methods
2.1 Neuronal Recordings
We conducted experiments in 3 macaque monkeys (Macaca mulatta) implanted with micro-wire arrays in multiple arm representation areas of the cortex. All surgical and experimental procedures conformed to the National Research Council’s Guide for the Care and Use of Laboratory Animals (1996) and were approved by the Duke University Animal Care and Use Committee. We trained the monkeys to move a cursor on a computer screen either using a hand-held joystick or through a BMI that generated cursor position from neuronal activity.
Figure 1A shows a diagram of electrode geometries and array placement for monkeys C, G, and M. All implants were custom-made by engineers from our lab (Lehew & Nicolelis, 2008). Monkeys C and G had similar implants. Monkey C was implanted with four 32-micro-wire arrays in the primary motor cortex (M1), dorsal premotor cortex (PMd), posterior parietal cortex (PP), and supplementary motor area (SMA) in the right hemisphere. Monkey G was implanted with six 32-micro-wire electrode arrays in M1, primary somatosensory cortex (S1), and PMd of both hemispheres. Electrodes in each array were grouped into pairs and placed on a uniform grid, with a 1 mm separation between pairs. The electrodes in each pair were placed tightly together, and one electrode was 300 microns longer than the other. The recording site on each electrode was at the tip. The material and diameter of the electrodes varied, and the longer electrode had equal or greater diameter. Monkey C was implanted with stainless steel and tungsten electrodes with diameters 46 and 51 microns, respectively, in areas SMA and M1 and tungsten electrodes with a diameter of 51 microns in areas PMd and PP. Monkey G was implanted with stainless steel electrodes measuring 40 and 63 microns in diameter.
Figure 1.
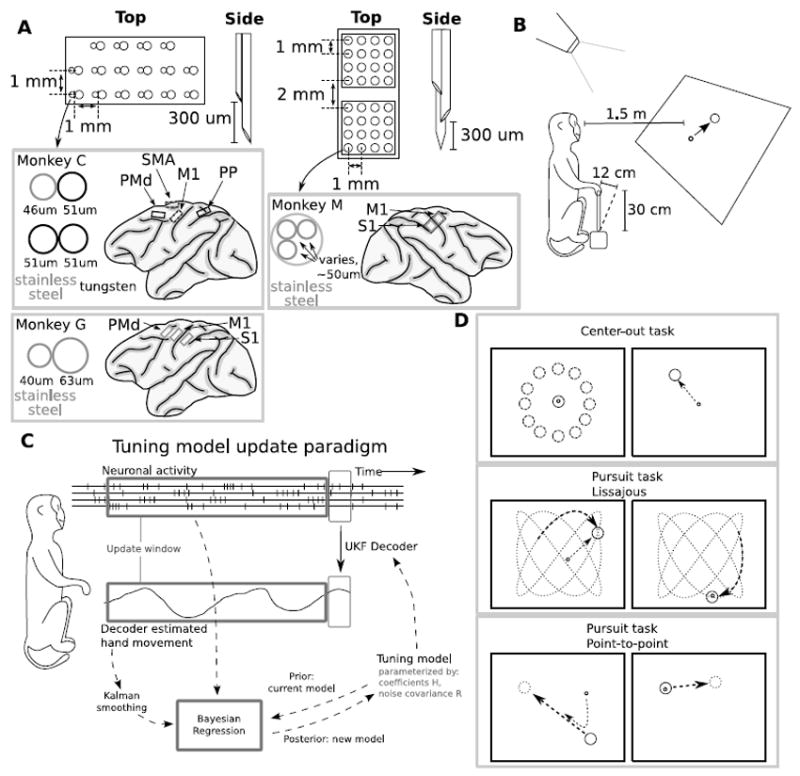
Implants and experimental methods. A: Implant placement and geometries. Micro-wire diameters are specified in microns. To reduce clutter, we only show arrays from which data were used. B: Primate experiment setup. C: Tuning model update paradigm. Updates occur periodically, using the neuronal data and decoder outputs generated since the previous update. D: Three behavioral tasks. Each task required placing the cursor over a target. In center-out, targets were stationary. In the pursuit tasks, targets moved continuously according to a Lissajous curve or a set of invisible, random waypoints.
Monkey M was implanted with four 96-micro-wire arrays in M1 and S1 bilaterally in arm and leg regions so that one array corresponded to the cortical regions associated with each limb. In this study, we only used recordings from the right hemisphere arm array, since monkey M manipulated the joystick with its left hand. Within each array for monkey M, micro-wires were grouped in two 4-by-4 uniformly-spaced grids of 16 triplets of electrodes. One grid was placed over M1 and one grid was placed over S1. The grids were separated by 2 mm which bridged the gap of the central sulcus. The separation between triplets of electrodes within each grid was 1 mm. The electrodes of each triplet had three different lengths, staggered at 300-micron intervals, and recorded from the tips. The penetration depth of each triplet could be adjusted as the triplets could be moved individually in the depth axis, but the depth separation of the triplets was fixed. Electrodes were inserted into the brain 1 to 2 weeks after surgery (during which the array base was implanted on the surface of the brain). Data used in this study were collected while the longest electrodes in all triplets were set to 2 mm in length. The electrodes were constructed of stainless steel. The electrode diameters in the array were heterogeneous, with each micro-wire having a diameter of 50 to 63 microns.
To extract motor parameters from neuronal activity, we sampled data from cortical sites contra-lateral to the arm which held the joystick. Data from monkey C (total 5 sessions) were recorded from M1 in 5 daily experimental sessions, PMd in 5 sessions, SMA in 5 sessions, and PP in 1 session. Data from monkey G (total 8 sessions) were recorded from left M1 in 8 sessions, left S1 in 7 sessions, left PMd in 8 sessions, and right PMd in 5 sessions. Data from monkey M (total 16 sessions) were recorded from right M1 and right S1 in all 16 sessions. We included neurons from S1 for predictions because prior work has found that they can be used for decoding arm movements (Carmena et al., 2003). For off-line reconstructions of hand movements using neuronal data, we used 18 experimental sessions (monkey C: 5 sessions, G: 8, M: 5), each with 94 to 240 (average 143) recorded neurons. We conducted 11 closed-loop BMI control sessions (each recording 139 neurons) with monkey M. Extracellular neuronal signals were amplified, digitized, and band-pass filtered using Multichannel Acquisition Processors (Plexon, Inc.). Neuronal action potentials were triggered using thresholds and sorted on-line by the Multichannel Acquisition Processors which ran a template matching algorithm for neuronal waveform templates set by the experimenter.
2.2 Behavioral Tasks
In experimental sessions, monkeys sat in a primate chair in front of a large screen, mounted 1.5 m in front of them (Figure 1B). The recording system was connected to the implants using light, flexible wires, as the monkeys’ heads were not restrained. An analog joystick with two degrees-of-freedom (left-right and forward-backwards) was mounted vertically so that the monkeys grasped the joystick tip with their hands. The joystick was 30 cm in length and allowed a maximum linear deflection of 12 cm. Monkey C and monkey M controlled the joystick with their left hands, and monkey G used its right hand. An electrical resistance touch sensor or an optical touch sensor on the joystick tip determined whether the monkeys were holding the joystick. An LCD projector, controlled by a computer with custom-built BMI experimental software, projected white visual stimuli on a black background on to the screen.
During hand control, monkeys moved a cursor (ring 1.6 cm in diameter) using the joystick. The joystick to cursor gain varied between 3.2x and 6.4x, depending on session. Targets appeared as rings 16 to 20.8 cm in diameter. The median speeds at which monkeys C and G moved the joystick were approximately 3.5 to 5.5 cm/s, depending on the session. The median speed at which monkey M moved the joystick was approximately 0.5 to 1.1 cm/s, depending on the session. The slower movements of monkey M were due to lower target speeds in the behavioral task. We set the target speeds lower because we found that slower movements were decoded more accurately, which is important for model updates when decoded trajectories are used for the training signal. All animals were overtrained (at least one year of experience) in performing the tasks with a joystick. Monkey M had 5 months of experience with BMI control before the closed-loop sessions.
During BMI control, we decoded the monkey’s neuronal modulations using an unscented Kalman filter (Z. Li et al., 2009) to control the cursor. We fitted the unscented Kalman filter’s parameters using data recorded during hand control and updated the filter’s parameters using the Bayesian regression update method (experimental condition) or left them fixed (control condition). The monkey had to grasp the joystick to signal participation (otherwise the task paused and stimuli were not shown), but joystick movements were not used for cursor control.
The behavioral tasks required the monkeys to place the cursor over the target, using either the joystick or BMI. We analyzed data from three behavioral tasks: (1) center-out, (2) pursuit with Lissajous trajectories, and (3) pursuit with point-to-point trajectories (Figure 1D).
In the center-out task (Figure 1D, top), the monkeys had to hold the cursor in a stationary center target and then move to a peripheral target randomly placed on a fixed-radius circle around the center target. After holding at the peripheral target and receiving a juice reward, a new trial began with the disappearance of the peripheral target and the appearance of the center target. The inter-trial interval that followed a successful trial was 500 ms, and the inter-trial interval after a failed trial was 700 to 1000 ms. Hold times varied per session from 350 to 1050 ms. We treated data collected from this and all other tasks as a continuous stream and did not segment by trial or movement onset.
In the pursuit task with Lissajous trajectories (Figure 1D, middle), a continuously moving target followed a Lissajous curve:
| (1a) |
| (1b) |
The variables x and y are the x- and y-axis coordinates and t is time in seconds. We used parameters a = 3, b = 4, δ = 0.5π, υ ∈ {0.1, 0.15, 0.2} Hz, and A = B = 22.4 cm (in screen scale). The x- and y-axis coordinates were uncorrelated because the temporal frequency was different for each. To receive periodic juice rewards, the monkeys had to keep the cursor over the moving target. Rewards were larger when the cursor was closer to the center of the target to motivate the monkeys to pursue the target accurately. This was done by scaling the amount of juice per reward (given every 750 ms of holding inside the target) by the cube of the difference of target radius and distance between cursor and target center.
In the pursuit task with point-to-point trajectories (Figure 1D, bottom), the target moved smoothly from one randomly generated waypoint to another. The waypoint positions were drawn from a uniform distribution on the 76.8 cm wide by 57.6 cm tall (in screen scale) workspace and were not visible to the monkeys. We limited the target’s acceleration (≤ 3.2 cm/sec, screen scale) and velocity (≤ 9:6 cm/sec, screen scale) to ensure smooth trajectories. Rewards were larger when the cursor was closer to the center of the target to motivate the monkey to pursue the target accurately.
2.3 Data Pre-processing
To estimate the instantaneous firing rate, we counted spikes in 100 ms wide, non-overlapping (i.e., spaced at 100 ms) time bins. The joystick position was sampled at 1 KHz and down-sampled to 10 Hz. Velocity was calculated from position by two-point digital differentiation after the down-sampling. We divided position, velocity, and binned spike counts by their standard deviations. We subtracted the means from position, velocity, and binned spike counts during closed-loop BMI experiments; in off-line reconstructions, instead of centering kinematics and neuronal data, we added a bias term to the neuronal tuning (observation) model. During experiments, the monkeys’ heads were not restrained and they were allowed to take breaks from task participation. To exclude data recorded while the monkeys did not participate, we used two methods. First, in both closed-loop and off-line experiments, we excluded data recorded while the monkey did not hold and move the joystick. If the joystick did not move for more than 2 or 3 seconds, depending on the session, the monkey was considered not to be participating. Note that during sessions of BMI control, the disconnected joystick was still held and moved by the monkey (brain control with hand movements). Second, during closed-loop experiments, we found that the monkey would sometimes hold the joystick but look away from the screen and rest. We thus excluded data recorded while the monkey looked away from the screen from update calculations. The experimenter watched the monkey and toggled a button in the experiment software when the monkey looked away.
We conducted off-line analyses using MATLAB software (The MathWorks, Inc). For closed-loop experiments, we implemented the proposed adaptive decoding method in C++ as part of a custom-built BMI software suite running on a desktop computer with an Intel Core i7 processor.
2.4 Evaluation Procedures
We quantified accuracy of reconstructions and control using the signal-to-noise ratio (SNR). SNR is commonly used in engineering and was used in previous studies on BMIs (Sanchez et al., 2002; Kim et al., 2003). SNR is the inverse of the normalized mean squared error, with larger numbers indicating more accurate control. SNR is typically reported in decibels (dB), which transforms the ratio with a logarithm to allow arithmetic averaging. The formula for the SNR is
| (2) |
where VAR is the variance of the desired values and MSE is the mean squared-error of the predicted values from the desired values. For off-line reconstructions, the desired values are the hand trajectories recorded by the joystick, and the predicted values are the outputs of the decoder. For closed-loop control, desired values are the target trajectories of the pursuit task, and the predicted values are the output of the decoder. In this study we analyzed the SNR of the position values of the trajectories. We analyzed x- and y-coordinates separately, and combined them with arithmetic averaging. When we calculated SNR in 1-minute windows of a session, we used the same signal variance (numerator), the value from the entire session, for every window, to allow comparison among windows. When comparing accuracy among conditions, we computed the SNR in decibels for each condition and then subtracted the values.
To ease comparison to previous work, the following equation provides the approximate correlation coefficient (Pearson’s r) corresponding to an SNR value in dB, under two assumptions: (1) the mean of the desired and predicted signals are both zero, and (2) the variances of the desired and predicted signals are equal. Without these assumptions, correlation coefficient and SNR are generally incomparable. The equation is
| (3) |
For the derivation and details on the approximations taken, see the Appendix.
3 Computational Methods
3.1 Overview
Bayesian regression self-training updates the model of neuronal tuning to movements used by the decoder by periodically performing Bayesian regression on the decoder output and neuronal activity (Figure 1C). This paradigm uses the prediction algorithm’s output to update its parameters, hence we call it self-training. Self-training is feasible for updating neuronal tuning models because of the redundancy in motor representation in large populations of neurons (Narayanan et al., 2005). For example, when a neuron changes its tuning, the decoder can still use the tuning of the rest of the population to predict desired movements. The movement information conveyed by the changed neuron’s modulation is drowned out by the movement information conveyed by the entire population. A small error due to the outdated tuning model for the changed neuron will contaminate predictions, since the Kalman filter combines information from all the neurons in a weighted average. However, if the change in tuning is small or the number of neurons which change simultaneously is small compared to the size of the population, the error in predictions using the entire population will be small, allowing accurate enough predictions with which Bayesian linear regression can update the tuning model parameters of the changed neurons.
We use a 5th order unscented Kalman filter (Julier et al., 1995) with a quadratic model of neuronal tuning for the decoder (see Z. Li et al., 2009, for implementational details). This decoder is an extension of the well-known Kalman filter (Kalman, 1960) Bayesian estimation algorithm, which has been used for BMI decoding (Wu et al., 2003, 2006; Gilja et al., 2009, 2010). It differs from previous Kalman filter decoders by incorporating a quadratic model of neuronal tuning (Moran & Schwartz, 1999; Z. Li et al., 2009) and the unscented transform (Julier et al., 1995) approximation mechanism needed to compute posteriors with the non-linear tuning model. It also allows the tuning model to capture tuning across multiple time offsets by keeping multiple time taps of kinematic variables in its state (5 time taps in this study).
Between tuning model updates, we store the output of the unscented Kalman filter (UKF) decoder and the neuronal activity. Before each update, we pre-process the decoder output by performing Kalman smoothing (Rauch et al., 1965). This process improves the accuracy of the estimates by propagating information from estimates of later states to estimates of earlier states using the movement model. For example, if the user is trying to move a BMI-controlled cursor to a particular location, the target location (decoded later in time) can improve the estimate of the movement trajectory. This implements a probabilistic mechanism for improving the data used for updates. This approach is similar in spirit to the heuristic methods found to be successful in prior work: using a point in-between the BMI cursor position and target as the training signal (Shpigelman et al., 2009) and rotating the decoded velocity vector towards the target in the training signal (Gilja et al., 2010).
The smoothed trajectories (independent variable) and the concurrent neuronal activity (dependent variable) are the inputs to the Bayesian linear regression. The Bayesian linear regression uses the previous tuning model parameters as priors and computes the posteriors on the parameters given the stored data. The updates occur in the background (e.g. in a low-priority thread) while the UKF continues to decode in real time using the existing tuning model. Once the update completes, the UKF uses the new tuning model and the storing of outputs for the next update begins. To compute the update, we use two formulations of Bayesian linear regression. One variant uses a joint distribution for the tuning model coefficients and allows analytical calculation of the solution. The second variant is more flexible, allowing temporary omission of some neurons from updates and addition of newly-discovered neurons, but requires a factorized distribution for the tuning model coefficients and an approximate solution using variational Bayes.
3.2 Tuning Model
We used a 5th order UKF with two future taps and three past taps, which means each spike may relate to desired movement occurring within a time difference of 300 ms (Z. Li et al., 2009). This setting is consistent with neurophysiological data and provides accurate decoding without requiring too many parameters. The 5th order UKF used only one tap of kinematics in its movement model, to minimize the decoding contribution from exploiting patterns in the movement. The tuning model described the normalized firing rate of a neuron as a linear combination of the position, distance from center of workspace, velocity, and magnitude of velocity (speed). The magnitude terms have been found to model neuronal activity well (Moran & Schwartz, 1999) and improve decoder accuracy (Z. Li et al., 2009). The tuning model is:
| (4) |
Here, zi, t is the predicted binned spike count of neuron i at time t, xpost and ypost are the x and y coordinates of the cursor at time t, and xvelt and yvelt are the x and y velocities of the cursor at time t. The distance and speed terms make the tuning model quadratic in position and velocity and require the use of an unscented Kalman filter for prediction. hi,1,1, … hi,6,5 are 30 scalar parameters for neuron i. We use the N by 30 matrix H to hold the parameters for all neurons in the population, where N is the number of neurons. The k variable iterates over the time taps in the state variables (see Z. Li et al., 2009, for details of the n-th order model).
The model for the noise in the quadratic tuning model is
| (5) |
where yi, t is the actual binned spike count for neuron i at time t, and υi, t are elements of a noise vector vt which is drawn independently from a multivariate normal distribution with zero mean and covariance R. R is an N by N matrix. The matrices H and R constitute the tuning model or observation model used by the UKF. To facilitate Bayesian updates of these two matrices, we store them as probability distributions and operate on the distribution parameters.
3.3 Bayesian Regression
The model for the multivariate linear regression between the vector of population neuronal activity y and the vector of kinematic features x is
| (6) |
To perform a Bayesian update of the parameters, we need a formulation for the prior and posterior distributions of H and R. It is often desirable for the prior and posterior distributions to be from the same distribution family and only differ in their hyper-parameters (conjugacy). Conjugacy is advantageous because it may allow analytical computation of the hyper-parameters of the posterior (Bernardo & Smith, 2000). After an update, we use the mean of the posterior distribution as the tuning model for subsequent decoding.
We examined two formulations for the distribution on H and R which allow conjugacy. The first formulation represents the prior and posterior joint probability of H and R in a normal-inverse-Wishart distribution. This formulation allows exact, analytical calculation of the Bayesian regression posteriors, and the formulas for computing the posteriors are well-known (Bishop, 2006; Rencher & Schaalje, 2008). However, it does not allow update on subsets of neurons or the addition of newly-discovered neurons. Thus, we explored an alternative formulation. The alternative formulation represents the tuning coefficients of each neuron as a separate multivariate normal distribution. Analytical computation of the regression on this formulation is not possible, and we use a variational Bayesian solution. This method is a multiple-predicted-variable extension of the single-predicted-variable Bayesian linear regression model presented by Bishop (2006).
3.4 Joint Formulation
The probability model for Bayesian regression with a joint distribution for H and R is shown in Figure 2A and specified below:
| (7a) |
| (7b) |
xt is a column vector of length D of regression features at time t, where D is the number of kinematic features (30 for the experiments in this study). Features are calculated from the mean of the smoothed outputs. yt is a column vector of length N of binned spike counts at time t. The vec() function converts a matrix into a column vector by vertically concatenating the columns, with the left-most column at the top. The variables μ (N by D), Λ (D by D), Ψ (N by N), and m (scalar) are hyper-parameters which describe the normal-inverse-Wishart distribution for the tuning model parameters:
| (8) |
Here, ⊗ is the Kronecker product. Due to conjugacy, the posterior parameters are the prior parameters updated by statistics of the neuronal activity and kinematic features. By performing algebraic manipulations on the likelihood and the prior, the following update equations can be found:
| (9a) |
| (9b) |
| (9c) |
| (9d) |
Tildes indicate prior values for the hyper-parameters. X is the D by T matrix of features over the T time points in the update batch. Y is the N by T matrix of binned spike counts. The prior hyper-parameters μ̃, Λ̃, Ψ̃, and m̃ are the posterior hyper-parameters from the previous update or the initial parameter fit (see Section 3.8).
Figure 2.
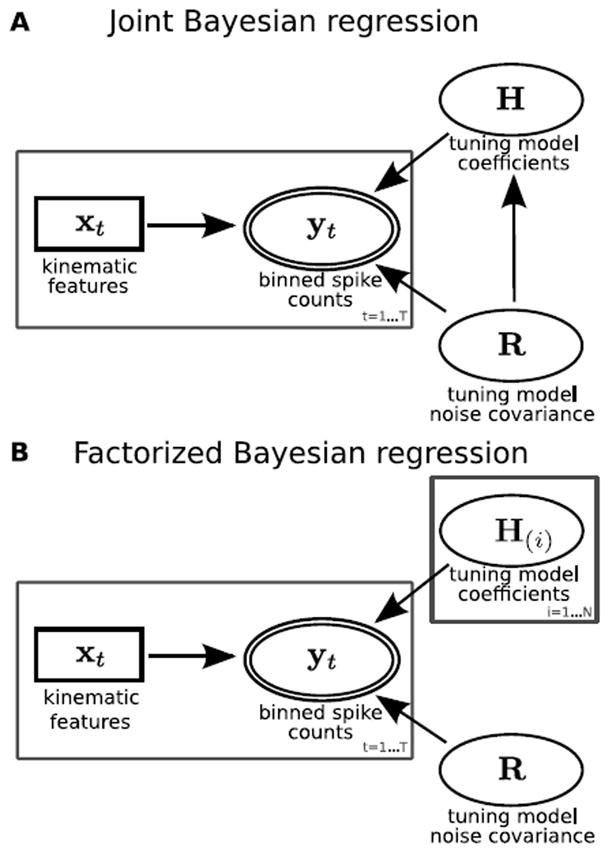
Graphical models for A. joint distribution Bayesian regression, B. factorized distribution variational Bayesian regression. Ellipses indicate random variables, double ellipses indicate observed random variables, rectangles indicate observed point values, and rectangles indicate repeated nodes.
Despite the advantages of offering a fast and exact analytical solution, this formulation has the disadvantage of linking the confidence measure on coefficients from different neurons through the Λ hyper-parameter. This means that we cannot update the coefficients of neurons separately. The ability to update subsets of neurons is important because this allows us to halt updating for a portion of neurons, which is useful if some neurons are affected by transient recording noise or are stable in tuning.
Furthermore, adding newly-discovered neurons to the decoder in a principled manner means using low initial confidence for their coefficients. Adjusting the Λ hyper-parameter for new neurons is not possible since the entire population shares this hyper-parameter. While changing the new neurons’ entries in Ψ affects their initial confidence, this approach does not increase the magnitude of future updates. That is, during subsequent updates, we expect the magnitude of updates for new neurons to be larger, if all other factors are equal, since the confidence about the new neurons’ coefficients should be lower than the confidence about the coefficients of neurons already in the model. Changing Ψ means that in subsequent updates, all neurons are treated as if the confidence in their coefficients is the same (see Equation 9a and b). Alternatively, one may use separate Λ terms for each neuron. Unfortunately, using separate Λ terms for each neuron breaks conjugacy and precludes analytical computation. In the next section, we present an approximate solution for the case where each neuron has a separate Λ hyper-parameter.
3.5 Factorized Formulation
To allow updates on subsets of neurons, we factorized the distribution for the tuning coefficients so that the coefficients for each neuron are represented by a multivariate normal distribution. Since this formulation is no longer conjugate to the likelihood of linear regression, we applied variational Bayes (Jordan et al., 1999), an approximate inference technique for hierarchical probability models. Other studies have applied variational Bayes to decoding neuronal activity (Ting et al., 2005, 2008; Chen et al., 2010).
The variational Bayesian approach assumes a factorization of the joint posterior of the hidden random variables (tuning parameters) given the visible random variables (neuronal activity and smoothed decoder outputs). The factorization facilitates conjugacy in the links of the resulting probability model and allows the use of an iterative, fixed-point procedure to find the approximate posterior on the hidden variables. The fixed-point procedure can be thought of as a Bayesian generalization of the expectation-maximization (Dempster et al., 1977) algorithm. The procedure minimizes the Kullback-Leibler divergence between the true joint posterior on the hidden variables and the factorized posterior. Each step in the procedure will not increase the divergence, which guarantees convergence, but potentially to a local optima. We refer the reader to works by Jordan et al. (1999), Winn & Bishop (2005), and Bishop (2006) for more details on variational Bayes.
We will call the variational Bayesian solution to the factorized Bayesian regression problem variational Bayesian regression (VBR). To facilitate conjugacy, we separated the distributions for the tuning coefficients H and the tuning noise covariance matrix R. The probability model for VBR is shown in Figure 2B and specified below:
| (10a) |
| (10b) |
| (10c) |
Here, i is the index on neurons. hi is row i of H. μi are the mean hyper-parameters for the tuning model coefficients of each neuron (column vectors of length D). Λi are the precision hyper-parameters of the tuning coefficients for each neuron (each D by D). Ψ (N by N) and m (scalar) are hyper-parameters for the tuning noise covariance R. Despite the factorization among neurons of the tuning coefficient distributions, activity among neurons may still be correlated due to other factors. Thus, we use the inverse-Wishart distribution to represent the distribution on a full noise covariance matrix.
3.6 Variational Approximation
In the following, we denote the inferred posterior distributions for hi and R with the Q-functions Q(hi) and Q(R), respectively. Furthermore, we denote the product of Q(hi) as Q(H).
Variational Bayes minimizes the Kullback-Leibler divergence between the true posterior and calculated posterior by maximizing a related lower bound L(Q) (Winn & Bishop, 2005):
| (11) |
Here, Q is shorthand for Q(H)Q(R) and P is shorthand for the true posterior P(H, R∣X, Y) and
| (12a) |
| (12b) |
L(Q) is the lower bound on the log marginal probability ln P(X, Y) as the Kullback-Leibler divergence is non-negative. Since ln P(X, Y) is fixed given the filter outputs and neuronal activity, by maximizing L(Q), we minimize the Kullback-Leibler divergence. Winn & Bishop (2005) show that L(Q) is maximized when
| (13a) |
| (13b) |
where the Z terms are the normalization factors needed to make the Q-functions valid probability distributions. The angled brackets 〈〉 indicate expectations, which are taken with respect to the other Q-functions, as indicated by the subscripts.
The parameters of Q(hi) and Q(R) which maximize the lower bound are found using a set of fixed-point equations derived from the above conditions by factorizing the graphical model and performing algebraic manipulations (Winn & Bishop, 2005). These equations update the hyper-parameters of each Q-function. The updates are executed in turn for each Q-function until convergence is detected by lack of change in L(Q).
The prior hyper-parameters μ̃i, Λ̃i, Ψ̃, and m̃ are the values from the previous VBR update or the initial parameter fit. At the beginning of the fixed point iterations, we initialize μi, Λi, Ψ, and m to their prior values.
The update for H is as follows:
| (14a) |
| (14b) |
Tildes indicate prior values of hyper-parameters. yi is the length T row vector of binned spike counts for neuron i. 〈R−1〉(i, i) is the (i, i) entry of the expectation of R−1:
| (15) |
The update for R is as follows:
| (16a) |
| (16b) |
〈H〉 is the matrix formed by the vertical stacking of the row vectors . W (N by N) is a temporary matrix used to account for the covariance of tuning coefficients:
| (17) |
wi, j is the (i, j) entry of W, and tr() denotes the trace operator.
Once the posterior values of μi, Λi, Ψ, and m are found, the posterior expectations for H and R can be computed for use in subsequent decoding. 〈H〉 is described above. 〈R〉 is:
| (18) |
3.7 Lower Bound Computation
We calculate the lower bound L(Q) to detect convergence. It also serves as a useful diagnostic, since it should not decrease after an update on any random variable. The equation for the lower bound can be rewritten as:
| (19) |
The expectations are taken with respect to Q(H) and Q(R). By factorizing the graphical model (Winn & Bishop, 2005), we decompose the lower bound as the sum of three terms LH, LR, and Ly:
| (20a) |
| (20b) |
| (20c) |
After substituting the probability density functions, re-arranging, and taking expectations, we use the following expressions for LH, LR, and Ly to calculate the lower bound:
| (21) |
| (22) |
| (23) |
where
| (24) |
| (25) |
| (26) |
| (27) |
| (28) |
Σt is an N by N temporary matrix. Γ() is the Gamma function, and ψ() is the digamma function.
3.8 Initial Fit
We fit the initial neuronal tuning model using Bayesian regression so that tuning model parameters were described by distributions instead of point estimates. For parameters updated with joint formulation Bayesian regression, we used the joint formulation Bayesian regression for initial fitting; for parameters updated with factorized formulation Bayesian regression, we used the factorized formulation Bayesian regression for initial fitting.
During initial fitting of tuning model parameters, we used priors which have a similar effect as the regularization in ridge regression:
| (29a) |
| (29b) |
| (29c) |
| (29d) |
0 is the zero matrix and I is the D by D identity matrix. For joint formulation Bayesian regression, there is only one μ̃ and one Λ̃. For factorized formulation Bayesian regression, one μ̃i and Λ̃i for each neuron must be initialized. The prior variance λ of the coefficients acts in the same way as the ridge regression regularization parameter. In reconstructions, we chose the value for λ by optimizing for decoder accuracy. For closed-loop experiments, we used the optimal λ from reconstructions. When fitting the movement model for the unscented Kalman filter, we used λ = 10−8 and m̃ = 6.
We fitted the initial tuning model using hand movements and neuronal data collected during joystick control of the task cursor. We did not address the problem of fitting models in the absence of a training signal, as numerous studies (Taylor et al., 2002; Wahnoun et al., 2004, 2006; Hochberg et al., 2006; Donoghue et al., 2007; Velliste et al., 2008) have addressed this problem. Instead, we focused on the problem of adaptive decoding (after the initial fitting) without a training signal, since in a clinical setting, a dedicated training procedure for initial fitting is reasonable, but frequent user re-training for maintaining decoder accuracy is burdensome.
The movement model of the UKF was also fitted using hand movement training data and Bayesian regression using the procedure described above. However, the movement model was not updated. At the start of decoding, we set the UKF’s initial state to the mean state in the training data and the initial covariance to the state covariance in the training data.
3.9 Adding and Omitting Neurons
For the factorized formulation variational Bayesian regression, neurons may be added or omitted by adjusting μi, Λi, and Ψ.
One may add a new neuron i by adding a new μi vector, a new matrix Λi, and increasing each dimension of Ψ by 1. The values of the new entries encode prior knowledge of the new neuron’s tuning, similar to the prior values used for the initial fit described above. An uninformative prior could be encoded with
| (30a) |
| (30b) |
where λ is a small positive number.
For the new entries in Ψ, one may use m × a for the new diagonal (i, i) entry and zeros for the new non-diagonal entries in the i-th row and i-th column. This encodes a strong prior for the value a in the diagonal entry of R for the new neuron. To encode a non-informative prior, one could set the 〈R−1〉(i, i) terms to 1 in Equation 14 in the first VBR iteration (before Ψ is updated), set the i-th row and column of Ψ̃ to 0 in Equation 16, and multiply the i-th row and column of Ψ by m/T after using Equation 16.
One may omit a neuron i from an update (or permanently remove it from decoding) by removing its μi vector, Λi matrix, and the i-th row and i-th column from Ψ. For neurons which are temporarily omitted, one may wish to adjust the i-th row and column of Ψ so that the omitted neuron’s entries in R remain the same, since m changes for the entire population during an update. This can be done by multiplying by mnew/m, where mnew is the new, larger value of m after the update.
Multiple neurons may be added or omitted by repeating the above procedures.
In closed-loop experiments, we omitted neurons from decoding and updating if they were flagged inactive, based on two tests. First, if a neuron produced no spikes during the self-training data window, it was flagged inactive. Second, a likelihood-ratio test was employed during filtering in some sessions. In the likelihood-ratio test, the product over the previous 10 seconds of the likelihood ratio between two models was calculated and compared to two thresholds. In the numerator of the ratio is the likelihood of the active model. In this model, the filter innovations for neuron i follow a normal distribution with zero mean and variance equal to the its diagonal entry in the innovation covariance matrix. In the denominator is the likelihood of the inactive model, in which the observations (not innovations) for neuron i follow a normal distribution with zero mean and the same variance as the active model. If the product of the likelihood ratio over the previous 10 seconds fell below 10−10, the neuron was flagged inactive. If the product of the likelihood ratio rose above 1, the neuron was flagged active. The thresholds were set by hand for a low false-positive detection rate for inactivity.
3.10 Baseline Firing Rates
The quadratic model of tuning described previously operated on binned spike counts with their means, or baseline firing rates, subtracted. Studies have shown that baseline firing rates of neurons change over time (Chestek et al., 2007; Ganguly & Carmena, 2009) and so the means should be updated. We explored two methods for updating the means in off-line reconstructions: (1) using a time-varying bias term in the quadratic model of tuning without the mean subtraction and (2) using the sample means of the binned spike counts in the update window as the new baseline firing rates, updated once per update window. We found little difference in these two approaches. In the results, we report reconstructions made with the bias term approach and closed-loop experiments performed with the sample mean approach.
To check if only updating the baseline firing rates for each neuron was sufficient to retain control accuracy, we updated the baseline firing rates of the static UKF (control condition) in the last two closed-loop sessions. To do this, we computed exponential moving averages on the binned spike counts. The half-lives of the moving averages were between 1 and 3 minutes.
3.11 Transition Model for Tuning Parameters
Since we assume the tuning model parameters to vary over time, we need a model of how they vary in order to track them. This model is a transition model on the tuning model parameters. The update rules of the transition model described below are executed before the Bayesian linear regression of each self-training update.
For the tuning model coefficients H, we used the identity transition model with normally distributed noise. We called the amount of noise the model drift parameter δ. This transition model increases the covariance of the coefficients by the model drift:
| (31) |
I is the D by D identity matrix. For joint formulation Bayesian regression, the single Λ is updated; for factorized formulation Bayesian regression, the Λi for each neuron is updated.
For the tuning model noise covariance matrix R, we increased the variance of the inverse-Wishart distribution without changing the mean by scaling m and Ψ by the same value (less than one). We applied the scaling when m exceeded a degrees of freedom cap m̂:
| (32a) |
| (32b) |
We applied the scaling when m exceeded the cap because this approach allowed the use of more data to estimate R initially and produced slightly more accurate reconstructions. With the correct settings, the two approaches can behave in the same manner asymptotically. However, the scale-at-every-update approach can handle updates at different frequencies by adjusting the scaling factor.
3.12 Kalman smoothing
The Kalman smoother, also known as the Rauch-Ting-Striebel smoother (Rauch et al., 1965), combines the Kalman filter (forward pass) and a backwards smoothing pass to form the forward-backward algorithm for linear-Gaussian systems. The backwards pass of the Kalman smoother starts from the most recent filter output and iterates to the earliest filter output in the update window:
| (33) |
where
| (34) |
| (35) |
x̂t is the smoothed state at time t, Pt is the state covariance at time t, F is the movement model matrix, and Q is the movement model noise covariance matrix. These equations describe one iteration of the backwards pass of the smoother. To initialize the iterations, we set x̂T to xT, the most recent filter output.
Note that, since we use a linear transition (movement) model, the standard Kalman smoother is applicable.
4 Results
4.1 Off-line Reconstructions
We first performed off-line reconstructions using 18 sessions in which monkeys controlled the cursor using a joystick. We discarded the first minute of each session and used the subsequent 2 minutes of hand movement (measured via joystick) and neuronal data to fit the initial tuning and movement models. We used Bayesian regression with priors as described in Section 3.8 to perform the initial fits. Then, the 5th order unscented Kalman filter (UKF) with and without Bayesian regression self-training updates (occurring every 2 minutes) reconstructed the hand movements for the remainder of the session from neuronal data only. We performed reconstructions with both variants of Bayesian regression and repeated for different settings of the model drift parameter (see Section 3.11). While our update method did not require a priori knowledge of the desired movements of the BMI user, we speculated that if we used the recorded hand movements (representing desired movements) for updates, accuracy of the updates would improve, due to reduced noise in the training data. We also wanted to compare our update method to previous work which used hand movements for updates. Thus, we also performed reconstructions using hand movements as input to the Bayesian regression updates.
To summarize, the experimental conditions were: non-updated UKF fitted with joint distribution Bayesian regression (static-BR-fit), non-updated UKF fitted with factorized distribution variational Bayesian regression (static-VBR-fit), UKF updated with joint distribution Bayesian regression self-training (BR), UKF updated with factorized distribution variational Bayesian regression self-training (VBR), UKF updated with joint distribution Bayesian regression on hand movements (BR-hand), and UKF updated with factorized distribution variational Bayesian regression on hand movements (VBR-hand). There are two static conditions (static-BR-fit and static-VBR-fit) because the fits calculated with Bayesian regression and variational Bayesian regression are slightly different.
Figure 3A shows the position reconstruction accuracy of 5 conditions (we omitted static-VBR-fit for clarity) quantified using signal-to-noise ratio (SNR, in decibels, see Section 2.4) and grouped by monkey. Table 1 shows results per session. We tried multiple settings of the model drift parameter; Figure 3A and Table 1 show the accuracy for the empirically best parameter setting found on the test data.
Figure 3.
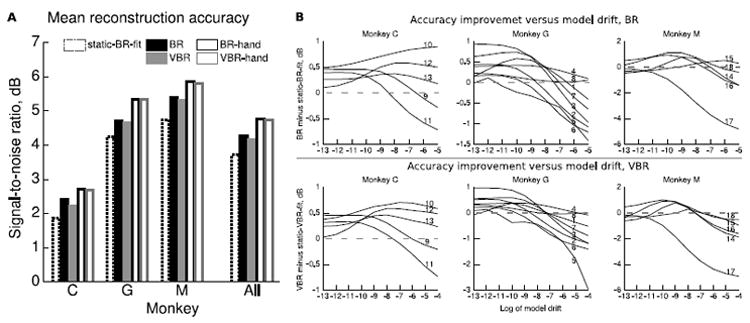
Position reconstruction accuracy of UKF with and without updates. A. Bars summarize mean accuracy for each monkey and condition. The static condition used parameters fit with joint distribution Bayesian regression. BR-hand and VBR-hand conditions used hand movements for updates. Accuracy values are from the best setting of the model drift parameter found using the test data. B. Accuracy improvement versus model drift parameter. Sessions indicated by curve labels.
Table 1.
Reconstruction accuracy of UKF with and without updates. Accuracy values in decibels SNR. Static (non-updated) UKF used parameters fitted with Bayesian regression (Static-BR-fit) or variational Bayesian regression (Static-VBR-fit). BR-hand and VBR-hand conditions used hand movements for updates. Accuracy values are from the best setting of the model drift parameter found using the test data. Task key: C is center-out, L is pursuit task with Lissajous trajectory, P is pursuit task with point-to-point trajectory.
| Session/Monkey | Length mins. | Neurons | Task | Static-BR-fit | BR | BR-hand | Static-VBR-fit | VBR | VBR-hand |
|---|---|---|---|---|---|---|---|---|---|
| 1 G | 20 | 139 | L | 3.90 | 4.49 | 5.79 | 3.89 | 4.27 | 5.79 |
| 2 G | 19 | 200 | L | 4.70 | 5.15 | 5.68 | 4.51 | 5.05 | 5.67 |
| 3 G | 15 | 189 | L | 5.53 | 6.47 | 6.68 | 5.44 | 6.42 | 6.66 |
| 4 G | 10 | 157 | L | 6.34 | 6.77 | 6.90 | 6.38 | 6.77 | 6.89 |
| 5 G | 22 | 94 | L | 4.33 | 4.45 | 3.91 | 4.29 | 4.43 | 3.95 |
| 6 G | 13 | 103 | L | 4.27 | 4.45 | 5.34 | 4.26 | 4.49 | 5.33 |
| 7 G | 12 | 103 | L | 3.76 | 4.51 | 5.30 | 3.80 | 4.40 | 5.29 |
| 8 G | 12 | 240 | C | 1.15 | 1.39 | 3.15 | 1.12 | 1.43 | 3.08 |
| 9 C | 10 | 101 | C | 3.59 | 4.06 | 4.03 | 3.38 | 3.84 | 4.03 |
| 10 C | 11 | 101 | C | 1.55 | 2.45 | 2.71 | 1.47 | 2.17 | 2.68 |
| 11 C | 19 | 109 | C | 2.15 | 2.55 | 3.33 | 2.15 | 2.48 | 3.27 |
| 12 C | 18 | 131 | C | 0.63 | 1.21 | 1.55 | 0.34 | 0.93 | 1.54 |
| 13 C | 10 | 157 | C | 1.40 | 1.78 | 2.02 | 1.16 | 1.62 | 1.87 |
| 14 M | 33 | 196 | L | 5.24 | 6.03 | 5.97 | 5.29 | 6.09 | 5.97 |
| 15 M | 17 | 145 | L | 3.93 | 4.47 | 4.80 | 3.87 | 4.18 | 4.79 |
| 16 M | 54 | 134 | P | 4.03 | 5.00 | 5.06 | 3.96 | 4.99 | 4.98 |
| 17 M | 72 | 134 | P | 7.12 | 7.01 | 6.72 | 7.09 | 6.93 | 6.65 |
| 18 M | 59 | 134 | P | 3.35 | 4.49 | 6.77 | 3.43 | 4.36 | 6.67 |
| Mean | 23.7 | 142.6 | 3.72 | 4.26 | 4.76 | 3.66 | 4.16 | 4.73 |
BR was significantly more accurate than static-BR-fit (two-sided, paired sign test, n=18, α = 0.05, p < 0.001) with a mean difference of 0.54 dB. VBR was significantly more accurate than static-VBR-fit (p < 0.001) with a mean difference of 0.50 dB. BR was significantly more accurate than VBR (p < 0.05) with a mean difference of 0.10 dB. Static-BR-fit was significantly more accurate than static-VBR-fit (p < 0.05) with a mean difference of 0.06 dB. BR-hand was significantly more accurate than BR (p < 0.05, 0.50 dB mean difference) and static-BR-fit (p < 0.002, 1.04 dB mean difference). VBR-hand was significantly more accurate than VBR (p < 0.05, 0.57 dB mean difference) and static-VBR-fit (p < 0.002, 1.07 dB mean difference).
Figure 3B shows the accuracy improvement of BR versus static-BR-fit and VBR versus static-VBR-fit for different settings of the model drift parameter. We chose the model drift parameter settings to be equally spaced in logarithmic space. Different sessions had different optimal values of the model drift. We did not optimize the model drift parameter on the training data, as 2 minutes of training data was too brief to have dramatic changes in tuning. Since the best model drift parameter was found using the test data, the best accuracy is an optimistic estimate. However, using the best overall value (based on mean across sessions) of e−10 ≈ 4.5 · 10−5 results in significant improvement versus static (BR: mean 0.36 dB, p < 0.01; VBR: mean 0.33 dB, p < 0.01). Accuracy as a function of model drift appears uni modal.
We examined the time course of accuracy differences between updated and static decoders in each session by comparing the improvement (BR minus static-BR-fit, VBR minus static-VBR-fit), calculated in 1-minute windows, versus the time in the session. The mean (across sessions) slope of the best-fit linear trend line of the improvement versus time was 0.036 dB/min for BR, 0.033 dB/min for VBR, 0.078 dB/min for BR-hand, and 0.082 dB/min for VBR-hand.
We examined the benefit of using the Kalman smoother to smooth decoder outputs before use in self-training. We performed off-line reconstructions without using the Kalman smoother and compared the reconstruction accuracy. For BR, Kalman smoothing improved accuracy by an average of 0.62 dB. For VBR, Kalman smoothing improved accuracy by an average of 0.80 dB. Both improvements were statistically significant (p < 0.002). The BR and VBR adaptive decoders without Kalman smoothing were nominally less accurate than static decoding (by 0.082 dB and 0.30 dB, respectively), but the difference was not statistically significant (p > 0.05).
The Bayesian regression self-training updates were computationally fast. The slowest step during each update was the Kalman smoothing backwards pass. With a MAT-LAB implementation on a Intel Core i7 class desktop computer, each BR update in the reconstructions, including pre-processing, used 1.57 ± 0.002 seconds (mean ± standard deviation). Each VBR update used 1.97 ± 0.13 seconds. While these execution times are longer than the 100 ms binning interval, updates can execute in a low-priority thread in a real-time BMI system, which is the approach taken in our closed-loop experiments.
4.2 Closed-loop BMI
We conducted closed-loop BMI control experiments with monkey M in 11 sessions over the span of 29 days. We used variational Bayesian regression self-training updates to better handle transient recording problems. When a neuron had zero spikes during an update window or if a likelihood ratio test flagged it as temporarily quiet, it was not included in updates and decoding.
Monkey M performed the point-to-point variant of the pursuit task in each session. We set spike-sorting templates by hand using Plexon spike-sorting software at the beginning of the first session and kept the templates fixed for the remaining sessions. We made no attempt to select neurons with stable waveforms; we used all 139 sorted units.
In the first session, we used 2 minutes of hand-controlled behavior to fit the initial tuning and movement model parameters using variational Bayesian regression. In each session, the monkey controlled the cursor using the 5th order UKF and two sets of parameters, in turn. The first set (static condition) consisted of the parameters fit from the hand-controlled behavior. These parameters were not updated and were the same for each session. The second set (VBR condition) was updated by variational Bayesian regression self-training through the course of the experiments. The second set was initially equal to the parameters fit from the hand-controlled behavior, but updated repeatedly with changes saved between sessions. In other words, we stored the VBR condition parameters at the end of each session and used them in the subsequent session, where they were updated further. In each session, monkey M first performed the pursuit task with the static parameters for 6 to 10 minutes. Then the monkey performed the pursuit task with the updated parameters for 10 to 50 minutes. The variance in time was due to the monkey’s participation. An update occurred each time two minutes of self-training data were collected; since we did not use decoder predictions made while the monkey was not looking at the screen and holding the joystick, updates occurred less frequently than every two minutes. The model drift parameter ranged between 10−3 and 10−5. We used larger values at the beginning of each session to compensate for the longer time duration since the previous update.
Chestek et al. (2007) and Ganguly & Carmena (2009) found significant changes in baseline firing rates of neurons. We investigated whether updating only the baseline firing rate parameter for each neuron could maintain control accuracy. We updated baseline firing rates with exponential moving averages of firing rates in the last two sessions for the static condition.
To quantify control accuracy, we measured the signal-to-noise ratio of the decoder output cursor position with respect to the target position. We calculated the SNR across the entire time segment of each condition and the best SNR among 1-minute windows of each condition. We included the latter metric because it is less sensitive to practice and motivational effects.
Figure 4A shows control accuracy for the 11 sessions over 29 days. The dark curves indicate SNR over the entire condition (solid, static all) and best SNR among 1-minute windows (dotted, static best) for static. The light curves indicate SNR over the entire condition (solid, VBR all) and best SNR among 1-minute windows (dotted, VBR best) for VBR. Note that no data was available for the static condition on day 6 because control was so poor that the monkey was not willing to perform the task for a period long enough to gather useful statistics. The UKF with VBR self-training updates maintained control accuracy much better than the static UKF. The slopes of the best-fit linear trend-lines for each curve are: VBR all -0.088 dB/day, VBR best -0.070 dB/day, static all -0.399 dB/day, and static best -0.487 dB/day. The update of the baseline firing rate parameters in the last two sessions for the static condition did not increase control-accuracy to the level provided by the VBR self-training updated UKF, suggesting that updating the baseline firing rate parameters alone is insufficient for maintaining control accuracy.
Figure 4.
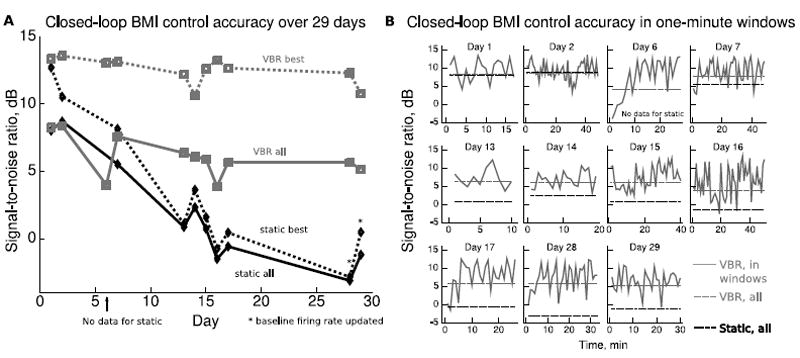
Closed-loop BMI control accuracy with and without updates. A. Solid curves indicate SNR over the entire condition time segment of each session, and dotted curves indicate the best SNR among 1-minute windows in the condition time segment of each session. In the last two sessions, we updated the baseline firing rate parameters of the static UKF using exponential moving averages. B. Closed-loop BMI control accuracy versus time within each session. In four sessions, control accuracy was poor initially, but improved after two to three updates.
The solid curves in Figure 4B show control accuracy of VBR in 1-minute windows for each session. The highest values of these curves are the VBR best values in Figure 4A. The light dotted lines indicate the SNR over the entire VBR condition for each session (VBR all), and the dark dashed lines indicate the SNR over the entire static condition for each session (static all). During days 6, 7, 17, and 28, control accuracy was initially poor but improved after two to three updates, demonstrating the ability of VBR self-training to adapt to changes in tuning or recording which had occurred since the last session. The large variations in accuracy were due to the monkey becoming unmotivated or frustrated and briefly stopping participation. When the monkey restarted participation, it had to re-acquire the pursuit target; this process reduced the control accuracy metric.
Figure 5 shows x-axis position traces of BMI control for the VBR and static conditions from sessions at the beginning, middle, and end of the closed-loop experiments. The traces show minutes 10-12 of control in the VBR condition and minutes 2-4 of control in the static condition. Dark curves indicate the position of the pursuit task target, and light curves indicate the BMI-controlled cursor. VBR self-training maintained control accuracy during the course of the experiments, while control with the non-updated UKF deteriorated. The oscillations seen in later sessions, especially for the static decoder, are about 0.5 to 2 Hz in frequency. We have observed similar oscillations in past experiments. The major departures from target pursuit are due to the monkey becoming frustrated and briefly stopping participation.
Figure 5.
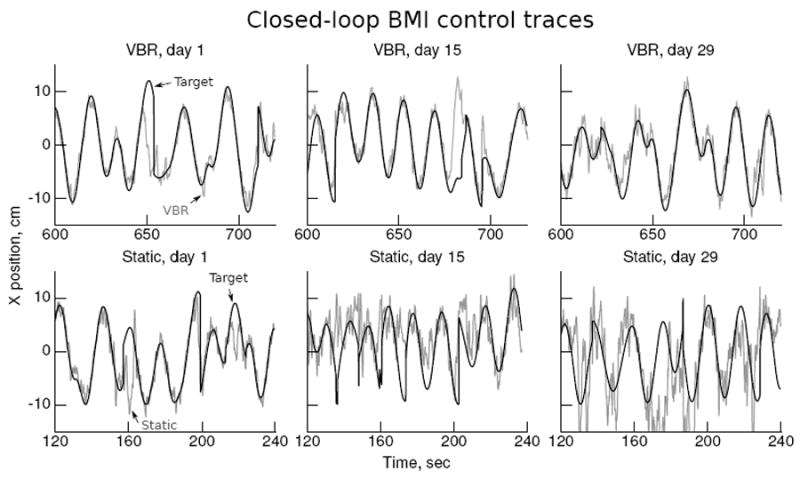
Traces of closed-loop BMI control with the UKF updated with VBR and with a static UKF. Plots show x-axis position, dark curves indicate the target location, and light curves indicate the BMI controlled cursor location.
We investigated the changes in the updated tuning model parameters of the UKF between the initial fit and the parameters on day 29. Figure 6A shows a histogram (normalized) of the absolute changes in tuning model coefficients (entries of H), with all coefficients from all neurons pooled into the sample. As we normalized the binned spike counts and kinematic variables to unit standard deviation before filtering, the units of the x-axis in the histogram are standard deviations of binned spike counts divided by standard deviations of kinematics. Dotted vertical lines indicate median and mean in the histogram. The large peak at zero and long tail indicate that a minority of coefficients changed while the majority did not.
Figure 6.
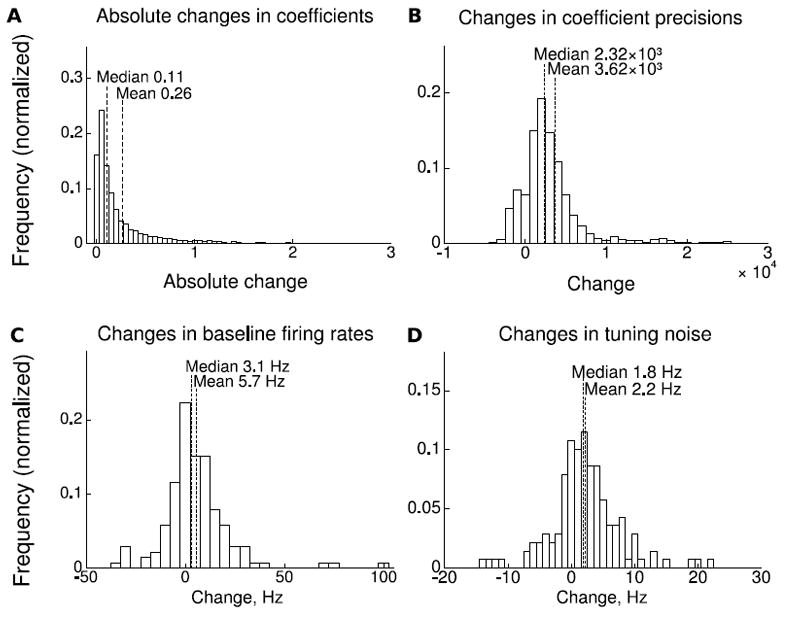
Changes in tuning parameters detected by the adaptive decoder during closed-loop sessions. A. Histogram of absolute changes in tuning model coefficients (H). B. Histogram of changes in coefficient precisions (diagonals of Λi). C. Histogram of changes in baseline firing rates. D. Histogram of changes in noise standard deviation (square root of diagonal of R).
Figure 6B shows a histogram of the changes in tuning model coefficient precisions (diagonal entries of Λi). The precisions quantify the certainty of the coefficient estimate and are equal to the inverse of the variances of the normal distributions for the coefficients. Larger precisions indicate more certainty. Units of the precisions are standard deviations of the kinematics squared divided by standard deviations of binned spike counts squared. Most coefficient precisions increased, indicating that certainty of coefficient estimates increased for most coefficients over the course of the experiments.
Figure 6C shows a histogram of the changes in the baseline firing rate parameters for each neuron. Some neurons did not change baseline firing rate, but there were also increases and decreases in the baseline firing rate. The mean and median of the changes were larger than zero, indicating a general increase in the firing rate.
Figure 6D shows a histogram of the changes in tuning model noise standard deviation entries (square root of diagonal entries of R). The tuning model of the UKF decoder includes both coefficients of tuning and a covariance matrix of the noise in tuning, assuming that the noise follows a multivariate normal distribution. Figure 6D shows the changes in the square root of the diagonal entries of this covariance matrix. Larger standard deviation entries mean more noise in the neuron’s firing, due to modulation unrelated to the kinematic variables of interest, intrinsic randomness, and recording noise. The positive mean and median changes indicate that noise standard deviation entries increased overall. However, the standard deviation of some neurons decreased. At the end of the experiments, entries for three neurons were zero.
In the initial fitting, 2 sorted units (of 139) were flagged inactive because they did not fire during the 2 minute training data. These two units received zeros for all of their tuning coefficients. One of these units was later flagged active and tuning parameters were fitted for it, demonstrating the ability of the adaptive decoder to add neurons. In total, 32 different units were flagged inactive for at least one update. The number of inactive units during each update fluctuated and many inactive-flagged neurons became active again later. This was likely due to recording instability or noise. On days 1 through 7, 1 to 3 units were flagged inactive in each update. On days 13 to 17, 2 to 9 units were flagged inactive in each update. On days 28 and 29, 9 to 16 units were flagged inactive in each update. The inactive neurons may explain some of the decrease in control accuracy for both the adaptive and static decoders.
5 Discussion
Bayesian regression self-training can improve reconstruction accuracy without having access to desired movements or using a priori assumptions about them. When knowledge of desired movements is available for update calculations, our method can produce even more accurate reconstructions (Figure 3A). It is reasonable to suggest that the improvement in reconstruction accuracy seen in sessions with length on the scale of dozens of minutes is partly due to the adaptive decoder improving the initial parameters fitted with the short 2 minutes of training data. This learning ability could exploit newly-discovered neurons or neurons that become more tuned. Our method may also be adapting to changes in motivational or attentional state, as well as recording instabilities.
The closed-loop experiments indicate that our adaptive decoder can maintain control accuracy better than a non-adaptive decoder (Figure 4A) over almost a month. We believe the difference in control accuracy between adaptive and non-adaptive decoders is partly due to tuning changes and partly due to recording instabilities. Though we fixed spike-sorting parameters, we cannot be certain that the neurons recorded in each on-line session are the same. Also, electrode movements may change the shape and amplitude of action potentials, leading to increases in spike-sorting errors. These sorting errors may cause a portion of the spikes from a neuron to be dropped, which change the effective tuning properties of the recorded unit. The results indicate that the adaptive decoder can deal with changes in effective tuning due to recording instability (as compared to static). From this, we believe our adaptive decoder can handle real changes in neuronal tuning, even if they are not already present in our data. In the future, we hope to combine adaptive spike-sorting (Wolf & Burdick, 2008; Gasthaus et al., 2009) with adaptive decoding to address recording instability directly. Alternatively, one may perform spike-sorting in each session, find correspondences between neurons using metrics on neuron identity (Emondi et al., 2004; Wolf & Burdick, 2008), and add or delete neurons using the procedure described in Section 3.9.
While we cannot rule out the contribution of user practice effects to the differences between static and adaptive control accuracy, we note that the differences in accuracy are large even when comparing the best one-minute window during static control to the overall adaptive control accuracy. The best one-minute window metric should mitigate practice effects during the course of the static control condition. Furthermore, since the static and adaptive decoders had different tuning parameters, practice with one is not likely to help with the other. We also note that static decoding was conducted first, when the monkey was most thirsty and motivated, while adaptive decoding went on while the monkey became more and more satiated and less motivated. Near the end of the adaptive decoding condition, the monkey took breaks with increasing frequency and duration, until stopping altogether. These breaks reduce the SNR value, since the BMI cursor leaves the target and the target must be re-acquired upon resumption. Thus, any motivational effects should favor the static decoding condition.
We note that when Kalman smoothing was disabled in the off-line reconstructions, adaptive decoding was not significantly better than static decoding. Since our update paradigm uses decoded trajectories as the training signal, the more accurate the decoded trajectories, the better the update results. We believe that the Kalman smoothing step, in our reconstructions, improved the accuracy of the decoded trajectory above a certain threshold. Above this accuracy threshold, the training signal was good enough so that the updated parameters could provide better subsequent decoding. We conjecture that if the decoder and neuronal recordings gave high enough accuracy before an update, then even without Kalman smoothing, the adaptive decoder would perform better than the static decoder, since the training signal would already be good enough. In other words, we believe that, while Kalman smoothing helps improve adaptive decoding, it is not strictly necessary for adaptive decoding if baseline accuracy was high enough.
The Bayesian regression updates in our decoder are computationally light and, due to their batch-mode operation, can be performed in the background while decoding continues. By using a probabilistic model of how the tuning parameters change and by calculating Bayesian updates, our adaptive decoder essentially performs Bayesian filtering on the tuning parameters. However, the parameter filter operates at a lower frequency than the movement state filter.
5.1 Changes in Neuronal Tuning
Studies have found that neuronal tuning to arm and hand movements may change over time. Taylor et al. (2002) observed changes in neuronal tuning as monkeys learned to operate a BMI based on a coadaptive algorithm. C. S. Li et al. (2001) and Padoa-Schioppa et al. (2004) reported that tuning characteristics of motor cortical neurons change as the result of adaptation to external force-fields. In our previous experiments, we found that neuronal tuning to hand movements and to BMI-controlled cursor movements changed differently during prolonged BMI operation (Lebedev et al., 2005). Kim et al. (2006) used a bootstrap analysis to determine significance in tuning changes. They found significant changes in tuning that accumulated with time. Rokni et al. (2007) found tuning changes due to learning as well as additional slow, random changes. Chestek et al. (2007) found small but significant changes in tuning and baseline firing rates. However, they asserted that these changes were due to measurement noise, learning effects, and subtle behavioral changes rather than intrinsic randomness. Truccolo et al. (2008) examined tuning changes in neuronal data collected from 2 human subjects and found that 42% of neurons exhibited significant changes.
The results from reconstruction and closed-loop experiments indicate a benefit of adaptive versus static decoding. In our closed-loop experiments, the adaptive decoder changed tuning model coefficients, baseline firing rates, and tuning model noise (Figure 6). Though we cannot be certain that changes in tuning found by the method were not the result of recording instability, our results support the existence of changes in neuronal inputs observed by others and attributed to various reasons.
5.2 Static versus Adaptive Decoder
Adaptive decoding, while more complex than static decoding, offers the following advantages. If neuronal tuning changes during motor learning, an adaptive decoder could exploit the changes more than a static decoder, e.g. by assigning more importance to neurons which increase in tuning depth. If neurons change in tuning in a manner unrelated to learning, a static decoder’s accuracy would suffer, while an adaptive decoder could compensate. If the recording system degrades temporarily or permanently, an adaptive decoder can adjust the importance given to affected neurons to mitigate the effects of noise. Coadaptive learning (Taylor et al., 2002) – when neural circuitry and the decoder adapt to each other simultaneously – decreases training time and improves accuracy. An adaptive decoder allows long-term coadaptive learning, which may give larger improvements in control accuracy from practice than if practice was conducted with a static decoder.
Ganguly & Carmena (2009) asserted that a non-changing BMI decoder may be sufficient for long-term control accuracy, as demonstrated by BMI control using a static Wiener filter over 19 days. They found that motor cortical neurons initially changed their preferred direction and tuning depth. As neurons’ tuning stabilized, BMI control performance plateaued. They reported that even small changes to decoder parameters or loss of one or two neurons could harm control accuracy. Unlike the present study, Ganguly and Carmena used only a subset of neurons, picked using criteria for spike waveform stability. This approach limits the communication bandwidth (which we loosely define as the aggregate recording quality i.e. the total number of recorded neurons weighted by how well each is recorded) of the BMI because neurons with useful tuning, but putatively unstable waveforms, may be ignored. Furthermore, if recordings for some of the putatively stable neurons worsen, control accuracy could suffer. Ganguly and Carmena also reported that BMI control accuracy with a static decoder, as measured by percentage correct and time-to-target, increased initially and plateaued after about 10 days of practice. In comparison, the control accuracy in the present study, as measured by signal-to-noise ratio of the cursor trajectory with respect to the target trajectory, decreased slightly over 29 days. The difference in trends may be due to the following factors.
The performance metrics and behavioral tasks may not be comparable between the studies. The center-out task does not prescribe trajectories, unlike the pursuit task. Signal-to-noise ratio of the BMI cursor trajectory should have a higher level of saturation than percent of trials successful and time to reach. That is, improvement to control accuracy which already saturates the percent correct and time to target metrics can still be detected by SNR of the trajectory. Thus, a BMI user may maintain the same (saturated) percent correct and time to target values, while decreasing in SNR of the trajectory.
Our decoder, parameter fitting procedure, and the use of a much larger neuronal population may explain our high initial control accuracy and slight decrease in accuracy over time, as opposed to the very low initial accuracy and improvements over time reported by Ganguly and Carmena. Our regularized initial model fit, combined with a more sophisticated decoder (see Z. Li et al., 2009, for a comparison) and larger population size, may have decreased the possible benefit of extra practice with BMI control. Thus, control accuracy did not increase over time in our study. The simpler decoder, unregularized parameter fitting procedure, and small population used by Ganguly and Carmena could be the reason that initial control was poor and practice over several sessions could improve control.
The time span of this study was longer than that of Ganguly and Carmena. Combined with the fact that we did not pick neurons for waveform stability, this means that neuronal recordings may have degraded more in our study. The recorded waveforms of some neurons may have changed, causing an increase in spike-sorting error and a decrease in available communication bandwidth. Though our adaptive decoder could adapt to tuning changes by changing the tuning model and handle increased noise by decreasing the relative contribution of affected neurons, the overall decrease in available communication bandwidth could be the cause of the slight decrease in control accuracy. In contrast, our static decoder did not compensate for recording degradation and thus performed much worse. Reliable detection and attribution of spikes should reduce the large contrast between static and adaptive decoders, by reducing the need to adapt.
5.3 Adaptive Filtering
Adaptive filters deal with estimation of a time series with changing parameters, which describes the adaptive decoding problem. Generally, an adaptive filter treats the unknown, changing parameters as variables to be estimated. Two established approaches are joint filtering and dual filtering. Joint filtering (Kopp & Orford, 1963; Cox, 1964) augments the unknown parameters into the state space and performs estimation on the augmented state. The augmentation generally results in a non-linear system. The dual filtering approach (Nelson & Stear, 1976) uses a separate parameter filter, whose state space holds the unknown parameters of the system. The parameter filter operates in lock-step with the state filter (the filter that estimates the state variables of the system), and each filter uses the output of the other as the truth.
Another approach for identification of parameters is to perform batch-mode computations. Shumway & Stoffer (1982) used the expectation-maximization algorithm to identify parameters of a linear system. Beal (2003) presented a variational Bayesian treatment of the expectation-maximization identification approach; the Bayesian extension incorporated priors on parameters, thus allowing Bayesian parameter updates.
An advantage of the batch-mode update approach is that one can apply Kalman smoothing to the filter output before using it for updating. Kalman smoothing improves the accuracy of the data used for parameter updates, and allows estimates of movements later in time to be used to improve estimates of movements earlier in time. Since the backwards pass of the Kalman smoother iterates backwards in time, it is not possible to use Kalman smoothing with joint or dual filtering. Furthermore, the batch-mode operation allows updates to occur asynchronously, taking as much time as needed and as little computational resource as desired. The iterative procedure in variational Bayesian regression may be stopped early to trade accuracy for speed.
5.4 Adaptive Decoding
Some approaches to learning initial decoder parameters may be applicable to learning changes in parameters. Taylor et al. (2002) used a coadaptive framework for fitting decoder parameters using assumed desired movement trajectories. Musallam et al. (2004) assumed knowledge of correct targets in a target selection task for fitting decoder parameters. Wolpaw & McFarland (2004) used knowledge of the correct target to learn parameters for an EEG decoder. Gage et al. (2005) used the true tone frequency to learn parameters for a decoder in a coadaptive frequency-matching task. Wahnoun et al. (2004, 2006) and Hochberg et al. (2006) used a visual following paradigm to fit decoder parameters. In the visual following paradigm, the BMI user is instructed to imagine moving a cursor which is actually controlled by the computer or a technician, and the cursor trajectory is used to fit decoder parameters. DiGiovanna et al. (2009) used reinforcement learning to fit parameters for a decoder in a two-target robot-arm control task. Darmanjian & Principe (2009) developed a linked-mixtures hidden Markov model for unsupervised clustering of movement states, but used Wiener filters fit from example hand movements to reconstruct trajectories.
Some studies have focused on how to update decoder parameters. Wu & Hatsopoulos (2008) proposed a real-time update technique for linear decoders using recorded hand movements. Shpigelman et al. (2009) demonstrated real-time updating of decoder parameters using the assumption that the BMI user tried to move the cursor closer to a known target. Rotermund et al. (2006) proposed a stochastic search update method which uses a hypothetical performance evaluation signal recorded from the brain.
Unlike the methods mentioned above, our adaptive decoder does not require knowledge of the desired movement. However, our adaptive decoder still offers competitive accuracy. Wu & Hatsopoulos (2008) reported 0.0022 cm2 and 0.0053 cm2 per trial reduction in mean squared-error in two sessions when updating the model parameters of a Kalman filter using recorded hand movements. Using the trial length and static Kalman filter accuracies they reported, the MSE reduction translates to an improvement of 0.016 dB/min and 0.038 dB/min. In comparison, our adaptive decoder produced a 0.036 dB/min mean improvement for the joint formulation and 0.033 dB/min mean improvement for the factorized formulation without using hand movements. When we used hand movements for updates, the improvements were 0.078 dB/min (joint) and 0.082 dB/min (factorized).
Some studies have focused on adapting parameters without knowledge of desired movements. Sykacek & Roberts (2002) and Sykacek et al. (2004) devised a variational Bayesian classifier for EEG signals which updates parameters without knowledge of the desired classification. Additionally, their method inferred the parameter change rate. Eden et al. (2004) and Srinivasan et al. (2007) demonstrated the ability of a point-process filter to adapt to tuning model parameter changes using a joint filtering approach in simulation. Rickert et al. (2007) showed that joint filtering using an unscented Kalman filter can adapt to changing parameters in simulation. Liao et al. (2009) presented a dual filtering solution for adaptive decoding of myoelectric recordings. Gilja et al. (2009) updated the tuning model parameters of a Kalman filter using self-training. Unlike our adaptive decoder, their method replaced old parameters with new parameters. This replacement approach does not take into account the information held in the old parameters.
In our adaptive decoding problem formulation, since we allow all decoder parameters to change, there is no fixed relationship between neuronal recordings and desired movements. There is no guarantee that a decoder would converge back to the correct answer if it gets off-track (in either movement estimates or parameter estimates), without knowing the real desired movements or real tuning parameters. There is no theoretical guarantee that the adaptive decoder finds the real tuning parameters once they change, though it empirically performs better than not adapting at all. If the desired movements are available for use in updates, there are convergence guarantees from the recursive system identification literature (see Ljung & Söderström, 1987). Previous studies have shown successful adaptive decoding using hand movements or assumptions on desired movements (Wu & Hatsopoulos, 2008; Shpigelman et al., 2009). Although there are no convergence guarantees, previous studies have shown successful adaptive decoding in simulations where the real desired movements were not used (Eden et al., 2004; Srinivasan et al., 2007; Rickert et al., 2007). In clinical practice, if the adaptive decoder gets off-track, desired movements (e.g. target trajectory in a pursuit task practice session) can be used for a few updates to “reset” the tuning model parameters.
The accuracy of adaptive decoding improves with the number of tuned neurons in the population. This is because the decoder combines observations from the entire population, including any recently-changed neurons (for which it has incorrect parameters). The larger the ratio of correct signal (from the unchanged neurons) to noise (from the changed neurons), the more accurate the decoder output. More accurate decoder output leads to more accurate parameter updates for the changed neurons. Thus, a larger recorded population will allow adaptation to more changing neurons or to larger changes.
We did not provide specifics for the question of what initial tuning model priors and tuning model transition parameters (e.g. the model drift parameter) to use in a clinical setting. A reasonable and practical approach could be to use the range of values which work well with animal experiments and allow adjustment by the clinician or user. Another approach is to identify the parameter settings using a more sophisticated model, similar to the work of Sykacek & Roberts (2002), Sykacek et al. (2004), and Ting et al. (2005). This challenging problem is left for future work.
During the course of closed-loop experiments, we noticed that the noise variance for some neurons approached zero. This was due to decreases in their firing rate variance. This led to near zero predicted movement variances, which caused numerical problems in the Kalman smoother in a few updates. When this occurred, we simply skipped the Kalman smoothing step. Exploring the use of a lower bound on noise variance or the use of an inversion-free smoother are possible topics for future work.
Acknowledgments
We would like to thank Dragan Dimitrov for conducting the animal surgeries, Gary Lehew and Jim Meloy for device and setup engineering, Timothy Hanson for developing the BMI software suite, and Tamara Phillips, Laura Oliveira, and Susan Halkiotis for technical and editorial support.
This study was supported by grants DARPA N66001-06-C-2019, TATRC W81XWH-08-2-0119, and by Award Number DP1OD006798 from the Office of the Director of NIH to MALN. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Office of the Director or the National Institutes of Health.
7 Appendix
We derive the approximate conversion of signal-to-noise ratio to Pearson’s correlation coefficient below. We denote the desired signal by xi and the predicted signal by yi, where i indexes on the n data points. We use x̄ and ȳ to denote the means, and we use sx and sy to denote the sample standard deviations.
We start with a definition of Pearson’s correlation coefficient r:
| (36) |
Using the assumptions of zero mean and equal variance for x and y:
| (37) |
where υ is the shared sample variance of x and y. Next, we examine the definition of mean squared error:
| (38) |
We expand the squared term in the summation and use the fact that x and y have zero mean to recognize the population variance terms in the results:
| (39a) |
| (39b) |
The approximation is due to the substitution of sample variance for population variance. Next, we note the resemblance of the summation to the equation for r in Equation 37 and substitute:
| (40) |
This is another approximation, due to the substitution of for . Next, we substitute our approximate MSE into the equation for SNR:
| (41a) |
| (41b) |
| (41c) |
Finally, we solve for r in terms of SNR to arrive at Equation 3.
References
- Beal MJ. Dissertation. University College London; 2003. Variational algorithms for approximate Bayesian inference. [Google Scholar]
- Bernardo JM, Smith AFM. Bayesian theory. Wiley; 2000. [Google Scholar]
- Bishop CM. Pattern recognition and machine learning. New York: Springer; 2006. [Google Scholar]
- Carmena JM, Lebedev MA, Crist RE, O’Doherty JE, Santucci DM, Dimitrov DF, et al. Learning to control a brain-machine interface for reaching and grasping by primates. PLoS Biology. 2003;1(E42) doi: 10.1371/journal.pbio.0000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Kloosterman F, Wilson MA, Brown EN. Variational Bayesian inference for point process generalized linear models in neural spike trains analysis. Acoustics speech and signal processing (ICASSP), 2010 IEEE international conference on. 2010:2086–2089. [Google Scholar]
- Chestek CA, Batista AP, Santhanam G, Yu BM, Afshar A, Cunningham JP, et al. Single-neuron stability during repeated reaching in macaque premotor cortex. Journal of Neuroscience. 2007;27:10742–10750. doi: 10.1523/JNEUROSCI.0959-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox H. On the estimation of state variables and parameters for noisy dynamic systems. IEEE Transactions on Automatic Control. 1964;9:5–12. [Google Scholar]
- Darmanjian S, Principe J. Spatial-temporal clustering of neural data using linked-mixtures of hidden Markov models. EURASIP Journal on Advances in Signal Processing 2009 [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society. Series B (Methodological) 1977;39(1):1–38. [Google Scholar]
- DiGiovanna J, Mahmoudi B, Fortes J, Principe JC, Sanchez JC. Coadaptive brain-machine interface via reinforcement learning. IEEE Transactions on Biomedical Engineering. 2009;56(1):54–64. doi: 10.1109/TBME.2008.926699. [DOI] [PubMed] [Google Scholar]
- Donoghue JP, Nurmikko A, Black M, Hochberg LR. Assistive technology and robotic control using motor cortex ensemble-based neural interface systems in humans with tetraplegia. Journal of Physiology. 2007;579:603–611. doi: 10.1113/jphysiol.2006.127209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden UT, Frank LM, Barbieri R, Solo V, Brown EN. Dynamic analysis of neural encoding by point process adaptive filtering. Neural Computation. 2004;16:971–998. doi: 10.1162/089976604773135069. [DOI] [PubMed] [Google Scholar]
- Emondi AA, Rebrik SP, Kurgansky AV, Miller KD. Tracking neurons recorded from tetrodes across time. Journal of Neuroscience Methods. 2004;135(1-2):95–105. doi: 10.1016/j.jneumeth.2003.12.022. [DOI] [PubMed] [Google Scholar]
- Gage GJ, Ludwig KA, Otto KJ, Ionides EL, Kipke DR. Naive coadaptive cortical control. Journal of Neural Engineering. 2005;2:52–63. doi: 10.1088/1741-2560/2/2/006. [DOI] [PubMed] [Google Scholar]
- Ganguly K, Carmena JM. Emergence of a stable cortical map for neuroprosthetic control. PLoS Biol. 2009;7:e1000153. doi: 10.1371/journal.pbio.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasthaus J, Wood F, Grr D, Teh YW. Advances in neural information processing systems. Vol. 22. MIT Press; 2009. Dependent Dirichlet process spike sorting. [Google Scholar]
- Gilja V, Nuyujukian P, Chestek CA, Cunningham JP, Fan JM, Yu BM, et al. A high-performance continuous cortically-controlled prosthesis enabled by feedback control design; Presented at the 2010 Society for Neuroscience Annual Meeting; San Diego, CA. 2010. [Google Scholar]
- Gilja V, Nuyujukian P, Chestek CA, Cunningham JP, Yu BM, Ryu SI, et al. Addressing the impact of instability on closed loop neural prosthetic control; Poster presented at the 2009 Society for Neuroscience Annual Meeting; Chicago, IL. 2009. [Google Scholar]
- Hochberg LR, Serruya MD, Friehs GM, Mukand JA, Saleh M, Caplan AH, et al. Neuronal ensemble control of prosthetic devices by a human with tetraplegia. Nature. 2006;442:164–171. doi: 10.1038/nature04970. [DOI] [PubMed] [Google Scholar]
- Jordan MI, Ghahramani Z, Jaakkola TS, Saul LK. An introduction to variational methods for graphical models. Machine Learning. 1999;37(2):183–233. [Google Scholar]
- Julier SJ, Uhlmann JK, Durrant-Whyte HF. A new approach for filtering nonlinear systems. Proceedings of the american control conference. 1995;3:1628–1632. [Google Scholar]
- Kalman RE. A new approach to linear filtering and prediction problems. Transactions of the ASME–Journal of Basic Engineering. 1960;82(Series D):35–45. [Google Scholar]
- Kim SP, Sanchez JC, Erdogmus D, Rao YN, Principe JC, Nicolelis MA. Modeling the relation from motor cortical neuronal firing to hand movements using competitive linear filters and a MLP. International joint conference on neural networks. 2003;1:66–70. [Google Scholar]
- Kim SP, Sanchez JC, Rao YN, Erdogmus D, Carmena JM, Lebedev MA, et al. A comparison of optimal MIMO linear and nonlinear models for brain-machine interfaces. Journal of Neural Engineering. 2006;3:145–161. doi: 10.1088/1741-2560/3/2/009. [DOI] [PubMed] [Google Scholar]
- Kopp RE, Orford RJ. Linear regression applied to system identication for adaptive control systems. American Institute of Aeronautics and Astronautics Journal. 1963;1:2300–06. [Google Scholar]
- Lebedev MA, Carmena JM, O’Doherty JE, Zacksenhouse M, Henriquez CS, Principe JC, et al. Cortical ensemble adaptation to represent velocity of an artificial actuator controlled by a brain-machine interface. Journal of Neuroscience. 2005;25(19):4681–4693. doi: 10.1523/JNEUROSCI.4088-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehew G, Nicolelis MAL. State-of-the-art microwire array design for chronic neural recordings in behaving animals. In: Nicolelis MAL, editor. Methods for neural ensemble recordings. 2. CRC Press; 2008. p. Chapter 1. [PubMed] [Google Scholar]
- Li CS, Padoa-Schioppa C, Bizzi E. Neuronal correlates of motor performance and motor learning in the primary motor cortex of monkeys adapting to an external force field. Neuron. 2001;30(2):593–607. doi: 10.1016/s0896-6273(01)00301-4. [DOI] [PubMed] [Google Scholar]
- Li Z, O’Doherty JE, Hanson TL, Lebedev MA, Henriquez CS, Nicolelis MAL. Unscented Kalman filter for brain-machine interfaces. PLoS ONE. 2009;4(7):e6243. doi: 10.1371/journal.pone.0006243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Fischer J, Milekovic T, Rickert J, Aertsen A, Mehring C. Co-adaptivity during learning to control a myoelectric interface; Poster presented at the 2009 Society for Neuroscience Annual Meeting; Chicago, IL. 2009. [Google Scholar]
- Ljung L, Söderström T. Theory and practice of recursive identification. Cambridge, MA: MIT Press; 1987. [Google Scholar]
- Moran DW, Schwartz AB. Motor cortical representation of speed and direction during reaching. Journal of Neurophysiology. 1999;82:2676–2692. doi: 10.1152/jn.1999.82.5.2676. [DOI] [PubMed] [Google Scholar]
- Musallam S, Corneil BD, Greger B, Scherberger H, Andersen RA. Cognitive control signals for neural prosthetics. Science. 2004;305:258–262. doi: 10.1126/science.1097938. [DOI] [PubMed] [Google Scholar]
- Narayanan NS, Kimchi EY, Laubach M. Redundancy and synergy of neuronal ensembles in motor cortex. Journal of Neuroscience. 2005;25(17):4207–4216. doi: 10.1523/JNEUROSCI.4697-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LW, Stear E. The simultaneous on-line estimation of parameters and states in linear systems. IEEE Transactions on Automatic Control. 1976;21:94–98. [Google Scholar]
- Nicolelis MA. Brain-machine interfaces to restore motor function and probe neural circuits. Nature Reviews Neuroscience. 2003;4(5):417–422. doi: 10.1038/nrn1105. [DOI] [PubMed] [Google Scholar]
- Padoa-Schioppa C, Li CS, Bizzi E. Neuronal activity in the supplementary motor area of monkeys adapting to a new dynamic environment. Journal of Neurophysiology. 2004;91:449–473. doi: 10.1152/jn.00876.2002. [DOI] [PubMed] [Google Scholar]
- Rauch HE, Tung F, Striebel CT. Maximum likelihood estimates of linear dynamic systems. AIAA Journal. 1965;3(8):1445–1450. [Google Scholar]
- Rencher AC, Schaalje GB. Linear models in statistics. 2. Hoboken, NJ: Wiley-Interscience; 2008. [Google Scholar]
- Rickert J, Braun D, Mehring C. Unsupervised adaptive Kalman filter for decoding non-stationary brain signals; Poster presented at the 2007 Society for Neuroscience Annual Meeting; San Diego, CA. 2007. [Google Scholar]
- Rokni U, Richardson AG, Bizzi E, Seung HS. Motor learning with unstable neural representations. Neuron. 2007;54:653–666. doi: 10.1016/j.neuron.2007.04.030. [DOI] [PubMed] [Google Scholar]
- Rotermund D, Ernst UA, Pawelzik KR. Towards on-line adaptation of neuro-prostheses with neuronal evaluation signals. Biological Cybernetics. 2006;95(3):243–257. doi: 10.1007/s00422-006-0083-7. [DOI] [PubMed] [Google Scholar]
- Sanchez JC, Kim SP, Erdogmus D, Rao YN, Principe JC, Wessberg J, et al. Input-output mapping performance of linear and nonlinear models for estimating hand trajectories from cortical neuronal firing patterns. 12th IEEE workshop on neural networks for signal processing. 2002:139–148. [Google Scholar]
- Serruya MD, Hatsopoulos NG, Paninski L, Fellows MR, Donoghue JP. Instant neural control of a movement signal. Nature. 2002;416:141–142. doi: 10.1038/416141a. [DOI] [PubMed] [Google Scholar]
- Shpigelman L, Lalazar H, Vaadia E. Advances in neural information processing systems. Vol. 21. MIT Press; 2009. Kernel-ARMA for hand tracking and brain-machine interfacing during 3D motor control. [Google Scholar]
- Shumway RH, Stoffer DS. An approach to time series smoothing and forecasting using the EM algorithm. Journal of Time Series Analysis. 1982;3(4):253–264. [Google Scholar]
- Srinivasan L, Eden UT, Mitter SK, Brown EN. General-purpose filter design for neural prosthetic devices. Journal of Neurophysiology. 2007;98:2456–2475. doi: 10.1152/jn.01118.2006. [DOI] [PubMed] [Google Scholar]
- Sykacek P, Roberts S. Advances in neural information processing systems. Vol. 15. MIT Press; 2002. Adaptive classification by variational Kalman filtering. [Google Scholar]
- Sykacek P, Roberts SJ, Stokes M. Adaptive BCI based on variational Bayesian Kalman filtering: an empirical evaluation. IEEE Transactions on Biomedical Engineering. 2004;51(5):719–727. doi: 10.1109/TBME.2004.824128. [DOI] [PubMed] [Google Scholar]
- Taylor DM, Tillery SI, Schwartz AB. Direct cortical control of 3D neuroprosthetic devices. Science. 2002;296:1829–1832. doi: 10.1126/science.1070291. [DOI] [PubMed] [Google Scholar]
- Ting JA, D’Souza A, Yamamoto K, Yoshioka T, Hoffman D, Kakei S, et al. Advances in neural information processing systems. Vol. 18. MIT Press; 2005. Predicting EMG data from M1 neurons with variational Bayesian least squares. [Google Scholar]
- Ting JA, D’Souza A, Yamamoto K, Yoshioka T, Hoffman D, Kakei S, et al. Variational Bayesian last squares: An application to brain-machine interface data. Neural Networks. 2008;21(8):1112–1131. doi: 10.1016/j.neunet.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truccolo W, Friehs GM, Donoghue JP, Hochberg LR. Primary motor cortex tuning to intended movement kinematics in humans with tetraplegia. Journal of Neuroscience. 2008;28(5):1163–1178. doi: 10.1523/JNEUROSCI.4415-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velliste M, Perel S, Spalding MC, Whitford AS, Schwartz AB. Cortical control of a prosthetic arm for self-feeding. Nature. 2008;53:1098–1101. doi: 10.1038/nature06996. [DOI] [PubMed] [Google Scholar]
- Wahnoun R, He J, Helms Tillery SI. Selection and parameterization of cortical neurons for neuroprosthetic control. Journal of Neural Engineering. 2006;3(2):162–171. doi: 10.1088/1741-2560/3/2/010. [DOI] [PubMed] [Google Scholar]
- Wahnoun R, Helms Tillery SI, He J. Neuron selection and visual training for population vector based cortical control. Engineering in medicine and biology society, iembs ’04 26th annual international conference of the IEEE. 2004;2:4607–4610. doi: 10.1109/IEMBS.2004.1404277. [DOI] [PubMed] [Google Scholar]
- Wessberg J, Stambaugh CR, Kralik JD, Beck PD, Laubach M, Chapin JK, et al. Real-time prediction of hand trajectory by ensembles of cortical neurons in primates. Nature. 2000;408:361–365. doi: 10.1038/35042582. [DOI] [PubMed] [Google Scholar]
- Winn J, Bishop CM. Variational message passing. Journal of Machine Learning Research. 2005;6:661–694. [Google Scholar]
- Wolf MT, Burdick JW. Bayesian clustering and tracking of neuronal signals for autonomous neural interfaces. Cdc 2008 47th IEEE conference on decision and control. 2008:1992–1999. [Google Scholar]
- Wolpaw JR, McFarland DJ. Control of a two-dimensional movement signal by a noninvasive brain-computer interface in humans. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(51):17849–17854. doi: 10.1073/pnas.0403504101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Black MJ, Gao Y, Bienenstock E, Serruya MD, Shaikhouni A, et al. Advances in neural information processing systems. Vol. 15. MIT Press; 2003. Neural deocoding of cursor motion using a Kalman filter. [Google Scholar]
- Wu W, Gao Y, Bienenstock E, Donoghue JP, Black M. Bayesian population decoding of motor cortical activity using a Kalman filter. Neural Computation. 2006;18:80–118. doi: 10.1162/089976606774841585. [DOI] [PubMed] [Google Scholar]
- Wu W, Hatsopoulos NG. Real-time decoding of nonstationary neural activity in motor cortex. IEEE Transactions on Neural Systems and Rehabilitation Engineering. 2008;16(3):213–222. doi: 10.1109/TNSRE.2008.922679. [DOI] [PMC free article] [PubMed] [Google Scholar]


