Abstract
Mitochondrial quality control is increasingly recognized as an essential element in maintaining optimally functioning tissues. Mitochondrial quality control depends upon a balance between biogenesis and autophagic destruction. Mitochondrial dynamics (fusion and fission) allows for the redistribution of mitochondrial components. We speculate that this permits sorting of highly functional components into one end of a mitochondrion, while damaged components are segregated at the other end, to be jettisoned by asymmetric fission followed by selective mitophagy. Ischemic preconditioning requires autophagy/mitophagy, resulting in selective elimination of damaged mitochondria, leaving behind a population of robust mitochondria with a higher threshold for opening of the mitochondrial permeability transition pore. In this review we will consider the factors that regulate mitochondrial biogenesis and destruction, the machinery involved in both processes, and the biomedical consequences associated with altered mitochondrial turnover.
Keywords: mitochondria, mitophagy, autophagy, mitochondrial turnover, cardioprotection
Overview
Mitochondrial quality control is increasingly recognized as an essential element in maintaining optimally functioning tissues. Mitochondrial quality control depends upon a balance between biogenesis and autophagic destruction. Our understanding of mitochondrial turnover is based on 50-year-old studies of radiolabeled mitochondrial proteins [1] which provided information on the bulk turnover of mitochondrial proteins but no insight into the fate of assembled electron transfer complexes or respirasomes. More recent insights into mitochondrial biogenesis, fission, fusion, and autophagy have given support to the idea that these interconnected processes are essential for mitochondrial quality control. What are the predictions from the mitophagy-cardioprotection hypothesis? 1. Any preconditioning intervention will result in a decrease in mitochondrial number. 2. Mitochondrial number, mass, and function will increase rapidly upon refeeding, adaptation, or reoxygenation. 3. The newly regenerated mitochondria will have improved function, will be enriched for respirasomes, and will display other distinctive characteristics. 4. A ‘bottleneck’ will allow selection for the best mitochondrial DNA genome and will permit culling to take place. 5. While mitochondrial turnover may average 17 days (protein half-life) under steady state conditions [2], episodic fasting or repetitive preconditioning will shorten the half-life. Some evidence exists to support these predictions. Caloric restriction induces mitochondrial biogenesis and bioenergetic efficiency [3]. Hypoxia accelerates mitochondrial protein turnover, reflecting increased destruction and biogenesis [4]. Like hypoxia, ischemic preconditioning is expected to accelerate mitochondrial turnover, as it was shown that many mitochondrial inner membrane proteins and heme groups turn over with the same kinetics, reflecting destruction of the entire organelle [5], which has subsequently been shown to be mediated by autophagy.
In this review we will consider the factors that regulate mitochondrial biogenesis and destruction, the machinery involved in both processes, and the biomedical consequences associated with altered mitochondrial turnover.
Mitochondrial biogenesis
The peroxisome proliferator-activated receptor gamma (PPARγ) in tandem with the retinoic acid receptor (RXR) regulates pathways of fatty acid oxidation for both mitochondria and peroxisomes. Free fatty acids and thiazolidinediones are ligands for PPARγ. Mitochondrial biogenesis is controlled by the PPARγ coactivator (PGC) family of transcriptional coactivators, most importantly PGC-1α, PGC-1β, and the PGC-related coactivator PRC. PGC-1α works in tandem with nuclear respiratory factor 2 (NRF-2) to co-activate NRF-1. The NRFs direct the transcription of nuclear encoded mitochondrial proteins, the mitochondrial protein import machinery, and co-factors required for assembly of the respiratory chain complexes, as well as the regulatory factors required for mitochondrial DNA transcription and translation, most importantly mitochondrial transcription factor A (Tfam). Tfam is important not only for mitochondrial gene transcription but also for maintenance of mtDNA copy number [6]. Overexpression of PGC1α is sufficient to drive mitochondrial biogenesis [7]. Acetylation of PGC1α suppresses its transcriptional coactivator activity and can therefore limit mitochondrial biogenesis despite high levels of protein. Therefore an important regulator of mitochondrial biogenesis is the histone deacetylase sirtuin 1 (Sirt1) which can serve to activate PGC1α and mitochondrial biogenesis [8]. Since sirtuins also regulate autophagy through FoxO1 and FoxO3, it appears that mitochondrial removal and resynthesis are linked, at least in the contexts of nutrient deprivation and oxidative stress, two stimuli that activate Sirt1 in the heart [9].
Mitochondrial Dynamics and Mitophagy
Mitochondria are highly dynamic organelles and are constantly undergoing fission and fusion. Mitochondrial dynamics are regulated by at least four conserved dynamin-related GTPases. Drp1 and hFis1 are involved in mitochondrial fission [10, 11], and studies in yeast suggest that Drp1 assembles into rings surrounding the mitochondrial outer membrane with the help of hFis1 [12]. Mitochondrial fusion requires Mitofusins 1 and 2 (Mfn1, Mfn2) in the outer and Opa1 in the inner membrane to control mitochondrial membrane fusion [13, 14]. Fission will produce smaller organelles whereas fusion produces tubular or net-like structures [15]. Interestingly, mitochondrial dynamics not only determines mitochondrial size, but also plays an essential role in mitochondrial turnover. Mitochondrial fission is essential for mitophagy and inhibition of fission results in disruption of mitophagy and accumulation of dysfunctional mitochondria [16, 17]. A recent study by Twig et al. showed that regulated fission and fusion events help segregate dysfunctional mitochondria prior to mitophagy. This study found that fission of mitochondria produced metabolically different fragments with different Δψm. The fragment with high Δψm had a high probability of undergoing fusion, whereas the fragment with low Δψm was more likely to be targeted by autophagy [16].
Although some of the molecules that control mitochondrial fission and fusion are known, new molecules and pathways that control this process continue to be discovered, suggesting that this process is more complex than was previously thought. Recently, it has been reported that the Bcl-2 family proteins may participate in the regulation of mitochondrial dynamics in healthy cells. For instance, Karbowski et al. reported that in healthy cells, pro-apoptotic Bax or Bak is required for normal fusion of mitochondria by activating assembly of Mfn2 [18]. Bcl-XL was found to perturb mitochondrial dynamics, where Bcl-XL overexpressing cells displayed either fragmented or fused mitochondria networks [19]. Berman et al. recently confirmed the finding that Bcl-XL could increase either fission or fusion [20]. These studies demonstrate that the Bcl-2 family proteins have a function that is completely separate from their role as regulators of outer mitochondrial membrane permeabilization during apoptosis. However, how these proteins regulate mitochondrial morphology is not known and needs to be further investigated.
Removal of Mitochondria Is Accomplished through Autophagy (Mitophagy)
Although individual mitochondrial proteins can be degraded via Lon protease, other AAA proteases, and even proteasomes located in the mitochondrial matrix, the vast majority of mitochondrial degradation is accomplished through the autophagy-lysosome pathway, although some outer membrane proteins are eliminated via the proteasome [21]. While early studies suggested this might be a non-specific bulk removal of cytosolic components, this was soon abandoned as the process was soon shown to be selective: mitochondria may be removed leaving endoplasmic reticulum or contractile assemblies intact. Moreover, mitochondria are not removed randomly: subpopulations of mitochondria are removed through complex mechanisms that are still being delineated. Two important mediators of mitophagy—Nix/Bnip3 and PINK1/Parkin—will be discussed below.
Mitochondrial fission, fusion, and autophagy have been shown to be essential partners in mitochondrial quality control [16]. In our work, only 10% of autophagosomes colocalize with a mitochondrial marker at any given time [Yuan, personal communication], yet autophagy mediates a 70% reduction in mitochondrial number (and surface area) in cells after 3.5 hours of starvation [22]. This suggests that mitophagy is a very rapid process. In mice subjected to overnight fasting, we observed a rapid 50% decrease in the number of mitochondria, accompanied by an inverse increase in the number of autophagosomes, measured by flow cytometry of the postnuclear fraction labeled with mCherryLC3 (autophagosomes) and anti-monoamine oxidase B (mitochondria) (Gottlieb, unpublished data). This surprising result reveals a substantial degree of mitochondrial turnover in response to a modest fast! This magnitude and pace of mitochondrial turnover was previously unsuspected. One consequence of such generalized mitochondrial destruction is replacement of mitochondria with newer, more efficient mitochondria. In rats subjected to 50% caloric restriction for one week followed by one week of re-feeding, the hepatic mitochondria were shown to be more efficient, evidenced by higher state 3 mitochondrial respiratory capacity and increased superoxide dismutase activity [23]. Recently we reported myocardial ischemic preconditioning involved autophagy, and unpublished studies confirm mitochondria to be the target (Gottlieb, unpublished data). One might wonder whether this process of mitochondrial culling might result in reduced ATP production. If depolarized mitochondria are selected for removal, there would be little downside: depolarized mitochondria contribute little to ATP production, and can hydrolyze ATP in a futile effort to restore mitochondrial membrane potential (Fig. 1). Moreover, there appears to be significant metabolic reserve in the normal heart. Inhibition of mitochondrial respiration with amytal reduced phosphocreatine content by 63%, but ATP levels were unchanged. Maximal cardiac output was decreased by 30% [24]. However, it appears the heart would tolerate a significant reduction in mitochondrial content, particularly if the slackers are the first ones to get sacked.
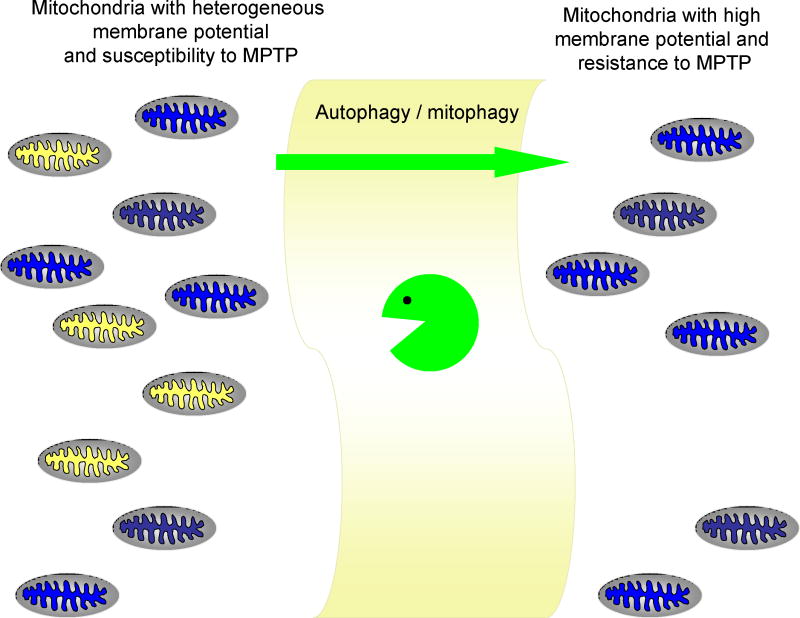
Figure 1.
The mitochondrial population is heterogeneous with respect to membrane potential and susceptibility to MPTP opening. A stress triggers MPTP in the most vulnerable mitochondria, which are removed by selective mitophagy. The remaining mitochondria have high membrane potential, lower ROS production, and greater resistance to MPTP opening.
Roles of Nix and Bnip3 in Mitophagy
Nix/BNIP3L and Bnip3 belong to the BH3-only proteins of the Bcl-2 family. Both Nix and Bnip3 have been implicated in the pathogenesis of cancer [25–27] and heart disease [28–30]. Similar to other BH3-only proteins, they contain a BH3 domain [31, 32], localize to the mitochondria where they can activate Bax/Bak [33–35], and interact with Bcl-2 and Bcl-XL [31, 36]. However, in contrast to other BH3-only proteins, Nix and Bnip3 have an alternative function to apoptosis They have been identified to be important regulators of mitophagy [29, 37, 38]. For instance, erythroid cells go through enucleation and removal of mitochondria during terminal differentiation [39] and the mitochondrial clearance via autophagy was dependent on the presence of Nix. Interestingly, it did not require other BH3-only proteins such as Bim or Puma [37], suggesting that this function is specific for Nix. They found that Nix was not required for the induction of autophagy, but was required for the selective elimination of mitochondria. Similarly, Sandoval et al. found that NIX was required for mitophagy in erythroid cells [40]. Erythrocytes in the peripheral blood of Nix−/− mice exhibited mitochondrial retention and reduced lifespan in vivo. Similarly, Bnip3 has been reported to induce mitochondrial autophagy. For instance, overexpression of Bnip3 induced mitophagy in HL-1 cells [29] and hypoxia-mediated mitophagy required the hypoxia-dependent factor-1-dependent expression of Bnip3 [41]. Exactly how Nix and Bnip3 flag mitochondria for autophagy is unclear, but Sandoval et al. proposed that Nix-dependent loss of Δψm was important in targeting the mitochondria to autophagosomes for clearance during erythroid maturation [40]. They found that mitophagy was restored when Nix−/− cells were treated with compounds that caused mitochondrial depolarization [40]. In addition, Nix was recently reported to interact directly with the autophagy proteins LC3 and GABARAP, and to recruit GABARAP to depolarized mitochondria [42]. This suggest that Nix might be functioning as a receptor for autophagosomes and that binding of Nix to LC3 on the autophagosome tethers the mitochondrion to the autophagosome. It is still unknown how Bnip3 targets mitochondria to the autophagosomes and if Bnip3 can also act as an autophagy receptor. Further studies are needed to answer these questions.
Roles of Parkin, Pink1 and p62/SQSTM1 in Mitophagy
The PTEN-induced kinase 1 (PINK1) and Parkin have been identified as important regulators of mitochondrial function and autophagy in cells. PINK1 is a mitochondrially targeted serine/threonine kinase and Parkin is an E3 ubiquitin ligase. Loss-of-function mutations in the genes coding for PINK1 and Parkin are associated with early-onset familial Parkinson’s disease [43, 44]. Studies in Drosophila melanogaster revealed that PINK1 and Parkin play key roles in regulating mitochondrial function where loss of function causes striking changes in both mitochondrial structure and function. For instance, removal of or mutations in the Drosophila homologue of PINK1 induced mitochondrial impairment which resulted in flight muscle degeneration and male sterility [45, 46]. Similarly, Parkin deficient flies exhibited loss of dopaminergic neurons and swollen and disordered mitochondria appearing before degeneration of their flight muscles [47, 48]. PINK1 or Parkin deficiency in mice also resulted in impaired mitochondrial respiration and oxidative damage of proteins and lipids which were exacerbated with aging [49, 50]. Several studies have shown that PINK1 functions upstream of Parkin [45, 51, 52]. For instance, Park et al. found that PINK1 and Parkin mutants had similar phenotypes and that overexpression of Parkin rescued the mitochondrial phenotype in PINK1 mutant Drosophila but not vice versa [45].
Recently, PINK1 and Parkin have been identified to play important roles in mitochondrial turnover. Narendra et al. reported that Parkin promoted the clearance of impaired mitochondria by autophagy. They found that Parkin accumulated on depolarized mitochondria which promoted their removal by autophagy [53]. Moreover, translocation of Parkin to mitochondria was dependent on PINK1 [54, 55]. Knockdown of PINK1 abolished Parkin recruitment to mitochondria and mitophagy in response to CCCP treatment [54], whereas overexpression of PINK1 promoted translocation of Parkin to mitochondria with normal Δψm [55]. Since Parkin is an E3 ubiquitin ligase, it was surprising that Parkin was found to induce mitophagy by promoting the ubiquitination of proteins on dysfunctional mitochondria. Also, this study found that Parkin-mediated ubiquitination served to recruit ubiquitin-binding deacetylase HDAC6 and p62/SQSTM1, which promotes the assembly of the autophagy machinery to clear impaired mitochondria [56]. p62/SQSTM1 targets substrates for autophagy by binding both ubiquitinated proteins and LC3 on the autophagosome [57]. Geisler et al. found that p62/SQSTM1 acts as an adaptor protein for Parkin-mediated mitophagy where the recruitment of p62/SQSTM1 to poly-ubiquitin-positive clusters of mitochondria was dependent on functional Parkin [54]. Knockdown of p62/SQSTM1 had no influence on mitochondrial translocation of Parkin, but completely blocked mitochondrial clearance of damaged mitochondria [54]. Thus, p62/SQSTM1 is not required for Parkin translocation to damaged mitochondria, but is essential for their final autophagic clearance. Similarly, Lee et al. found that Parkin-mediated ubiquitination served to recruit p62/SQSTM1 and that p62/SQSTM1-deficient cells were defective in Parkin-dependent mitophagy [56]. Thus, this suggests that ubiquitination of mitochondrial outer membrane proteins is a critical basis for the specific and effective clearance of mitochondria by autophagy.
Under normal conditions, mitochondria exist as a tubular interconnected network. Two separate studies noted that PINK1 and Parkin disrupted the mitochondrial network and induced aggregation of mitochondria in the perinuclear region where the fragments were removed by autophagosomes [55, 56]. Mitochondrial transport is dependent on the microtubule dynein motor and treatment of cells with the microtubule destabilizing agent nocodazole inhibited both Parkin-mediated perinuclear mitochondrial aggregation and mitophagy [55, 56]. These studies suggest that transport of mitochondria via the microtubules to the perinuclear region is an essential step in the mitochondrial degradation pathway. However, many components are still unknown in this pathway. For instance, Vives-Bauza found that PINK1 and Parkin interacted but there was no evidence that PINK1 phosphorylated Parkin or that Parkin catalyzed PINK1 ubiquitination [55]. Also, exactly how PINK1 recognizes dysfunctional mitochondria and how Parkin’s E3 ligase activity is activated are still unclear. Thus, there are still many questions regarding the mechanism of PINK1/Parkin-mediated mitophagy.
Mitochondrial Dynamics and Respirasome Maintenance
Synthesis of nuclear and mitochondrial encoded electron transfer subunits must be neatly coordinated so that all necessary components are available for assembly. The electron transfer complexes in turn are incorporated into larger assemblies, called supercomplexes, or respirasomes, which represent the optimal functional units of mitochondrial respiration and ATP production [58]. It is unknown whether supercomplexes undergo reversible assembly and disassembly. However, as subunits acquire oxidative damage, they must be replaced. Several scenarios are possible: individual subunits are removed, degraded (e.g., by Lon or AAA proteases), and replaced asynchronously; supercomplexes undergo dynamic assembly/disassembly, allowing for incorporation of fully functional complexes into respirasomes and exclusion of complexes containing damaged subunits; whole mitochondria are removed and replaced. The existence of Lon protease and other AAA proteases suggests that damaged components can be removed selectively and replaced. One means by which removal of components and targeted replacement could occur would be through dynamic assembly and disassembly of respiratory supercomplexes (respirasomes), which comprise stoichiometrically conserved assemblies of respiratory complexes and F0F1 ATPase assemblies in a paracrystalline array [59]. These assemblies would efficiently exclude misfolded proteins [58]. Indeed, Hoppel’s group has shown that in heart failure, respirasome content diminishes although total content of mitochondrial electron transfer complexes remains unchanged [60]. Mitochondrial fusion events allow for rapid mixing of matrix contents, but exchange of inner membrane components occurs on a slower time scale, based on experiments monitoring the diffusion of photoactivated GFP (PA-GFP) targeted to matrix or inner mitochondrial membrane [61]. However, PA-GFP is not incorporated into ox-phos complexes or respirasomes, and therefore studies using this approach may greatly overestimate the rate of mixing of electron transfer complexes and respirasomes. It is conceivable that some components are not exchanged at all but rather persist for the life of the mitochondrion, while recently synthesized components are assembled de novo into new electron transfer complexes and then into respirasomes. Regions enriched in supercomplexes might have more rigid membrane architecture and conceivably might be less likely to undergo fission, while individual electron transfer complexes not organized into supercomplexes, as well as any misfolded components, would be excluded and would end up in regions of increased membrane fluidity. When fission ensues, the region with supercomplexes would have high membrane potential, while the other segment would contain isolated electron transfer complexes and damaged proteins, and would have lower membrane potential (Fig. 2). Newly synthesized proteins would be more likely to be imported into the mitochondrion with higher membrane potential, since protein import is heavily dependent on membrane potential. This leads to the prediction that some mitochondria would be enriched for newly assembled and highly-functional respirasomes. This proposed sorting process represents the functional equivalent of forming a new mitochondrion, something that might be called mito-neogenesis.
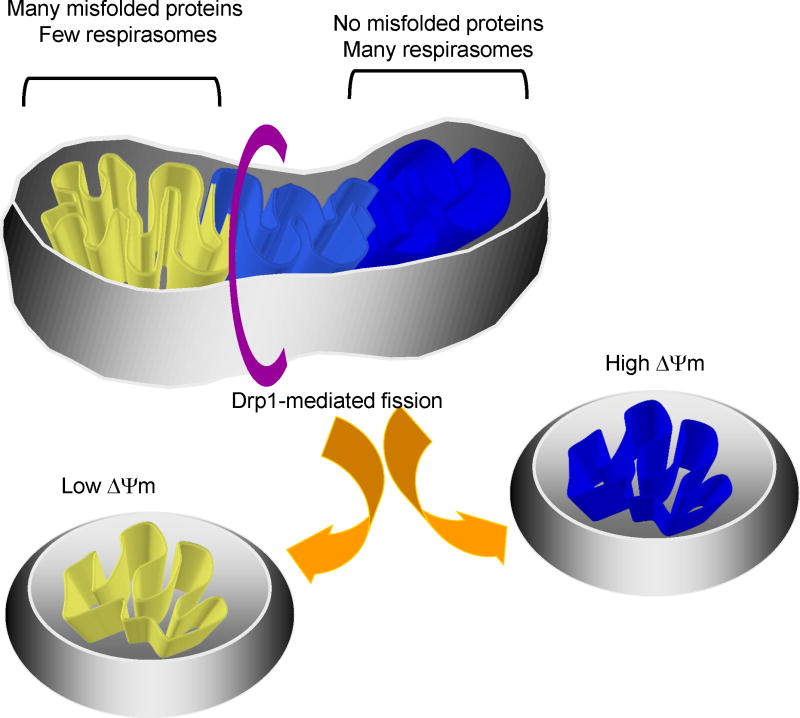
Figure 2.
During fusion, cycles of assembly and disassembly of electron transfer complexes and respirasomes results in sorting, so that misfolded proteins are excluded from re-assembled respirasomes. Regions enriched for respirasomes are shown in darker blue. Fission takes place where the inner membrane is less rigid, i.e., where there are few respirasomes. This results in two daughter mitochondria, one with low membrane potential and defective electron transfer complexes, and another with high membrane potential and many respirasomes.
Consequences of Altered Mitochondrial Fission/Fusion
Although the physiological roles of mitochondrial fusion and fission in cell function and survival are still poorly understood, it is clear that these processes are essential for cellular homeostasis. Inhibition of Drp1 suppresses mitophagy, revealing a close linkage between fission and mitophagy [16]. Fission and fusion are a dynamic dance essential for mitochondrial quality control, while autophagy serves to remove the mitochondria that are no longer competent to participate in fusion.
Mice lacking Mfn1, Mfn2, or Opa1 die at an early embryonic stage [62, 63]. Moreover, in humans, point mutations in Mfn2 and Opa1 lead to severe neurodegenerative diseases such as Charcot–Marie–Tooth type 2A and dominant optic atrophy, respectively [64–67]. On the other hand, deletion of Drp1 is lethal for Caenorhabditis elegans [68], and a mutation in Drp1 was recently described in an infant with fatal abnormal brain development [69]. Impaired mitochondrial dynamics has also been implicated in neurodegenerative diseases including Parkinson, Alzheimer’s and Huntington diseases [70–72]. These findings demonstrate the importance of mitochondrial dynamics in cell homeostasis.
Alterations in mitochondrial dynamics also occur in response to cellular stress which shifts the balance towards fission [71, 73, 74]. Youle and colleges reported that Drp-1 translocated from the cytosol to defined foci on the mitochondrial membrane where it co-localized with pro-apoptotic Bax at the onset of apoptosis [75]. They found that a dominant negative of Drp-1 inhibited fission and apoptosis, but had no effect on Bax translocation in response to staurosporine treatment, suggesting that Bax can induce apoptosis by activating the mitochondrial fission pathway. Moreover, the pro-apoptotic BH3-only protein EGL-1 induced Drp-1-mediated mitochondrial fission and apoptosis during development in C.elegans [74]. However, another study reported that Bax-induced mitochondrial fission was not necessarily linked to cytochrome c release and apoptosis. Overexpression of Bcl-xL blocked cytochrome c release and apoptosis but did not rescue mitochondrial fragmentation [19]. There is less information on the role of fission/fusion in the heart. Recently, it was reported that increased mitochondrial fission plays a role in myocardial ischemia/reperfusion (I/R) injury [76]. Ong et al. found that enhancing fusion in HL-1 myocytes by overexpressing mitofusin or inhibiting fission with a Drp1 dominant negative protected against simulated I/R-mediated cell death. Moreover, treatment with an inhibitor of Drp-1 (mDivi-1) reduced I/R injury in vivo [76], although they did not examine mitochondrial morphology in the treated hearts. Effects of mDivi-1 can be variable, and can lead to fragmentation or fusion [J Wikstrom, personal communication]. Another study found that failing hearts had reduced OPA1 protein levels and myocytes contained small fragmented mitochondria [77]. Interestingly, overexpression of OPA1 increased mitochondrial fusion but did not protect against simulated I/R-mediated cell death in H9c2 cells [77]. Overexpression of Opa1 can have variable effects, favoring fusion when expressed at low to moderate levels, and driving fission when expressed at higher levels [78]. Experimental variability may also arise from the differences in the experimental cell models (HL-1 cells vs. H9c2 cells.) Whether the proteins governing fission and fusion have additional functions is unclear but must be considered when evaluating the literature. Clearly, an imbalance in fission or fusion due to genetic mutation or in response to stress can be detrimental to the cell. Overexpression of fission proteins leads to extensive fission of mitochondria, can induce excessive mitochondrial autophagy, and in some cases can trigger apoptosis [79]. In general, overexpression of fusion proteins leading to enhanced mitochondrial networking protects cells from certain apoptotic stimuli. It is not clear whether overexpression of fusion proteins prevents removal of dysfunctional mitochondria by autophagy, but it has been noted that megamitochondria are not eliminated even when they appear to be damaged [80].
Metabolic reprogramming through mitophagy and biogenesis
Metabolic reprogramming occurs during cell differentiation, as for example when a stem cell with relatively modest energy needs differentiates into a cardiomyocyte with exceedingly high ATP synthetic requirements. The mitochondrial electron transfer complexes, PDH complex, and fatty acid transport systems (CPT-I and –II) must be altered to meet the different needs of the cardiomyocyte, a process requiring assembly of complexes containing different subunit isoforms in the mitochondria. While it is possible that this is conducted in a piecemeal fashion, considerable evidence supports the notion that most mitochondria are eliminated and a wave of mitochondrial biogenesis follows, during which time the new isoforms are expressed and incorporated into the assembling complexes. In the context of cardioprotective signaling, nitric oxide activates autophagy and mitochondrial biogenesis [81]. Autophagy and mitochondrial biogenesis share common signaling pathways [82]. If this is the case, then it follows that autophagy may be a prerequisite for effective mitochondrial biogenesis during metabolic reprogramming and potentially in the more general context of cellular homeostasis over the Circadian cycle. It is known that many ox-phos genes are expressed differentially in the heart over the 24-hr cycle, reflecting differential needs for ATP production during active and sleep phases [83]. Whether a subset of mitochondria are newly assembled each day, or whether individual subunits are swapped out as needed remains to be established. However, it seems likely that even in terminally differentiated adult cardiomyocytes, a sizeable number of mitochondria must turn over on a daily basis (otherwise the changes in mRNA would be negligible).
Mitophagy and cardioprotection
Our studies and those of others have implicated autophagy in cardioprotection [84, 85], and we have shown that autophagy is essential for cardioprotection [86–89]. How might preconditioning trigger autophagy and mitophagy? Opening of the mitochondrial ATP-sensitive potassium channel (mitoKATP) or transient opening of the mitochondrial permeability transition pore have been proposed as mechanisms of preconditioning. One feature shared by these two phenomena is mild depolarization of the mitochondrial inner membrane. Given that mitochondrial depolarization prevents mitochondrial fusion and will signal for autophagic removal, it seems reasonable to propose that preconditioning may operate through mitophagy. Mitophagy (selective autophagy of mitochondria) has been described by many groups [29, 86, 90–93], and is known to occur in response to ischemic stress. Autophagy is required to eliminate damaged mitochondria; removal of mitochondria that are producing ROS would prevent dissemination of injury to neighboring mitochondria and thereby prevent wholesale mitochondrial permeability transition pore opening. A universal feature of myocardial preconditioning is preservation of ATP stores, calcium homeostasis, and mitochondrial integrity [94, 95]. The mitochondria that remain after autophagic culling would be predicted to exhibit higher functionality and a distinctive proteomic profile. Consistent with this notion, Hausenloy reported that mitochondria isolated from preconditioned hearts have a higher threshold for permeability transition pore opening [95], while distinctive changes in the post-translational modifications of several mitochondrial proteins were observed after pharmacologic preconditioning [96].
It is worth noting that preconditioning is impaired in conditions in which autophagy is also diminished, including diabetes and advanced age, again supporting the idea that the two processes are linked.
PERSPECTIVES
Recent studies reveal that starvation results in a profound reduction in mitochondrial number, which is followed by vigorous mitochondrial biogenesis upon refeeding. This cycle of destruction and resynthesis may allow for culling of the least-fit mitochondria, but also permits metabolic reprogramming of mitochondria [97]. It seems reasonable to propose that cycles of intermittent fasting and resultant mitophagy may represent critical elements of mitochondrial quality control essential to cardiac health.
Mitophagy may be relevant to more than just preconditioning; it could be an important element in lifespan extension, where periodic removal of poor-quality mitochondria would enable their replacement with new, more efficient ones. This could be compared to eliminating 10% of the oil-burning clunkers on the freeway and replacing them with fuel-efficient hybrids. The more often clunkers are removed, the quicker the fleet can be cleaned up, as long as production can match demand. Empiric studies have shown that alternate day fasting results in substantial lifespan extension; thus under normal conditions, production can keep up with alternate-day destruction. Caloric restriction or alternate-day fasting has been shown to confer cardioprotection. Cardioprotection may therefore reflect mitochondrial quality. Autophagy induced by caloric restriction is closely linked to longevity [98, 99], and this may be due to the episodic culling of inefficient mitochondria followed by their replacement with new mitochondria.
It is important to understand the mechanistic relationship between caloric restriction, autophagy, mitochondrial quality, cardiac health and longevity. Developing better tools to monitor mitochondrial turnover will yield new insights into the cellular processes that determine mitochondrial function and oxidative stress, which are key determinants of cardiac function and longevity.
Acknowledgments
RAG is supported by NIH R01 HL060590, R01 AG033283, R01 HL092136, R01 HL034579, and P01 HL085577.
ABG is supported by NIH R01 HL087023, R01 HL101217, and R01 HL092136.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Roberta A. Gottlieb, Email: robbieg@sciences.sdsu.edu.
Åsa B. Gustafsson, Email: abgustafsson@ucsd.edu.
LITERATURE CITED
- 1.Fletcher MJ, Sanadi DR. Turnover of rat-liver mitochondria. Biochim Biophys Acta. 1961;51:356–360. doi: 10.1016/0006-3002(61)90177-9. [DOI] [PubMed] [Google Scholar]
- 2.Menzies RA, Gold PH. The Turnover of Mitochondria in a Variety of Tissues of Young Adult and Aged Rats. Journal of Biological Chemistry. 1971;246:2425–2429. [PubMed] [Google Scholar]
- 3.Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, de Cabo R. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aschenbrenner V, Zak R, Cutilletta A, Rabinowitz M. Effect of hypoxia on degradation of mitochondrial components in rat cardiac muscle. Am J Physiol. 1971;221:1418–1425. doi: 10.1152/ajplegacy.1971.221.5.1418. [DOI] [PubMed] [Google Scholar]
- 5.Beattie DS, Basford RE, Koritz SB. The turnover of the protein components of mitochondria from rat liver, kidney, and brain. J Biol Chem. 1967;242:4584–4586. [PubMed] [Google Scholar]
- 6.Scarpulla RC. Transcriptional Paradigms in Mammalian Mitochondrial Biogenesis and Function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 7.Wende AR, Schaeffer PJ, Parker GJ, Zechner C, Han DH, Chen MM, Hancock CR, Lehman JJ, Huss JM, McClain DA, Holloszy JO, Kelly DP. A Role for the Transcriptional Coactivator PGC-1α in Muscle Refueling. Journal of Biological Chemistry. 2007;282:36642–36651. doi: 10.1074/jbc.M707006200. [DOI] [PubMed] [Google Scholar]
- 8.Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, Yamaguchi M, Namiki S, Nakayama R, Tabata M, Ogata H, Kubota N, Takamoto I, Hayashi YK, Yamauchi N, Waki H, Fukayama M, Nishino I, Tokuyama K, Ueki K, Oike Y, Ishii S, Hirose K, Shimizu T, Touhara K, Kadowaki T. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca, (2+) and AMPK/SIRT1. Nature. 464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- 9.Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 14.Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 16.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 19.Sheridan C, Delivani P, Cullen SP, Martin SJ. Bax- or Bak-induced mitochondrial fission can be uncoupled from cytochrome C release. Mol Cell. 2008;31:570–585. doi: 10.1016/j.molcel.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Berman SB, Chen YB, Qi B, McCaffery JM, Rucker EB, 3rd, Goebbels S, Nave KA, Arnold BA, Jonas EA, Pineda FJ, Hardwick JM. Bcl-x L increases mitochondrial fission, fusion, and biomass in neurons. J Cell Biol. 2009;184:707–719. doi: 10.1083/jcb.200809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeda K, Yoshida T, Kikuchi S, Nagao K, Kokubu A, Pluskal TÅ, Villar-Briones A, Nakamura T, Yanagida M. Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proceedings of the National Academy of Sciences. 107:3540–3545. doi: 10.1073/pnas.0911055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carreira RS, Lee Y, Ghochani M, Gustafsson AB, Gottlieb RA. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 2010;6 doi: 10.4161/auto.6.4.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crescenzo R, Lionetti L, Mollica MP, Ferraro M, D’Andrea E, Mainieri D, Dulloo AG, Liverini G, Iossa S. Altered skeletal muscle subsarcolemmal mitochondrial compartment during catch-up fat after caloric restriction. Diabetes. 2006;55:2286–2293. doi: 10.2337/db06-0312. [DOI] [PubMed] [Google Scholar]
- 24.Kapelko VI, Lakomkin VL, Korchazhkina OV, Pisarenko OI. Cardiac pump function of the isolated rat heart at two modes of energy deprivation and effect of adrenergic stimulation. Mol Cell Biochem. 1996;163–164:131–136. doi: 10.1007/BF00408649. [DOI] [PubMed] [Google Scholar]
- 25.Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–6673. [PubMed] [Google Scholar]
- 26.Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK, Buzzai M, Dicker DT, McKenna WG, Bernhard EJ, El-Deiry WS. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2004;6:597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Okami J, Simeone DM, Logsdon CD. Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res. 2004;64:5338–5346. doi: 10.1158/0008-5472.CAN-04-0089. [DOI] [PubMed] [Google Scholar]
- 28.Yussman MG, Toyokawa T, Odley A, Lynch RA, Wu G, Colbert MC, Aronow BJ, Lorenz JN, Dorn GW., 2nd Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat Med. 2002;8:725–730. doi: 10.1038/nm719. [DOI] [PubMed] [Google Scholar]
- 29.Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, Gottlieb RA, Gustafsson AB. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 30.Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, Li H, Kirshenbaum LA, Hahn HS, Robbins J, Jones WK, Dorn GW. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest. 2007;117:2825–2833. doi: 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imazu T, Shimizu S, Tagami S, Matsushima M, Nakamura Y, Miki T, Okuyama A, Tsujimoto Y. Bcl-2/E1B 19 kDa-interacting protein 3-like protein (Bnip3L) interacts with bcl-2/Bcl-xL and induces apoptosis by altering mitochondrial membrane permeability. Oncogene. 1999;18:4523–4529. doi: 10.1038/sj.onc.1202722. [DOI] [PubMed] [Google Scholar]
- 32.Yasuda M, Theodorakis P, Subramanian T, Chinnadurai G. Adenovirus E1B-19K/BCL-2 interacting protein BNIP3 contains a BH3 domain and a mitochondrial targeting sequence. J Biol Chem. 1998;273:12415–12421. doi: 10.1074/jbc.273.20.12415. [DOI] [PubMed] [Google Scholar]
- 33.Chen G, Cizeau J, Vande VC, Park JH, Bozek G, Bolton J, Shi L, Dubik D, Greenberg A. Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J Biol Chem. 1999;274:7–10. doi: 10.1074/jbc.274.1.7. [DOI] [PubMed] [Google Scholar]
- 34.Kubli DA, Ycaza JE, Gustafsson AB. Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem J. 2007;405:407–415. doi: 10.1042/BJ20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Lewis W, Diwan A, Cheng EH, Matkovich SJ, Dorn GW., 2nd Dual autonomous mitochondrial cell death pathways are activated by Nix/BNip3L and induce cardiomyopathy. Proc Natl Acad Sci U S A. 107:9035–9042. doi: 10.1073/pnas.0914013107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray R, Chen G, Vande VC, Cizeau J, Park JH, Reed JC, Gietz RD, Greenberg AH. BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J Biol Chem. 2000;275:1439–1448. doi: 10.1074/jbc.275.2.1439. [DOI] [PubMed] [Google Scholar]
- 37.Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, Ney PA. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fader CM, Colombo MI. Multivesicular bodies and autophagy in erythrocyte maturation. Autophagy. 2006;2:122–125. doi: 10.4161/auto.2.2.2350. [DOI] [PubMed] [Google Scholar]
- 40.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 44.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 45.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 46.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 47.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitworth AJ, Theodore DA, Greene JC, Benes H, Wes PD, Pallanck LJ. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc Natl Acad Sci U S A. 2005;102:8024–8029. doi: 10.1073/pnas.0501078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 51.Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 55.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrane J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 58.Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 59.Wittig I, Schagger H. Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes. Biochim Biophys Acta. 2009;1787:672–680. doi: 10.1016/j.bbabio.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 60.Rosca MG, Hoppel CL. New aspects of impaired mitochondrial function in heart failure. J Bioenerg Biomembr. 2009;41:107–112. doi: 10.1007/s10863-009-9215-9. [DOI] [PubMed] [Google Scholar]
- 61.Wikstrom JD, Twig G, Shirihai OS. What can mitochondrial heterogeneity tell us about mitochondrial dynamics and autophagy? Int J Biochem Cell Biol. 2009;41:1914–1927. doi: 10.1016/j.biocel.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davies VJ, Hollins AJ, Piechota MJ, Yip W, Davies JR, White KE, Nicols PP, Boulton ME, Votruba M. Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum Mol Genet. 2007;16:1307–1318. doi: 10.1093/hmg/ddm079. [DOI] [PubMed] [Google Scholar]
- 64.Kijima K, Numakura C, Izumino H, Umetsu K, Nezu A, Shiiki T, Ogawa M, Ishizaki Y, Kitamura T, Shozawa Y, Hayasaka K. Mitochondrial GTPase mitofusin 2 mutation in Charcot-Marie-Tooth neuropathy type 2A. Hum Genet. 2005;116:23–27. doi: 10.1007/s00439-004-1199-2. [DOI] [PubMed] [Google Scholar]
- 65.Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schroder JM, Vance JM. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 66.Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, Bhattacharya SS, Wissinger B. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 67.Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 68.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 69.Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 70.Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakamura T, Lipton SA. Redox regulation of mitochondrial fission, protein misfolding, synaptic damage, and neuronal cell death: potential implications for Alzheimer’s and Parkinson’s diseases. Apoptosis. doi: 10.1007/s10495-010-0476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reddy PH, Mao P, Manczak M. Mitochondrial structural and functional dynamics in Huntington’s disease. Brain Res Rev. 2009;61:33–48. doi: 10.1016/j.brainresrev.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pinton P, Ferrari D, Rapizzi E, Di Virgilio F, Pozzan T, Rizzuto R. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. Embo J. 2001;20:2690–2701. doi: 10.1093/emboj/20.11.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jagasia R, Grote P, Westermann B, Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- 75.Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 2002;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 121:2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- 77.Chen L, Gong Q, Stice JP, Knowlton AA. Mitochondrial OPA1, apoptosis, and heart failure. Cardiovasc Res. 2009;84:91–99. doi: 10.1093/cvr/cvp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Molina AJ, Wikstrom JD, Stiles L, Las G, Mohamed H, Elorza A, Walzer G, Twig G, Katz S, Corkey BE, Shirihai OS. Mitochondrial networking protects beta-cells from nutrient-induced apoptosis. Diabetes. 2009;58:2303–2315. doi: 10.2337/db07-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gomes LC, Scorrano L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim Biophys Acta. 2008;1777:860–866. doi: 10.1016/j.bbabio.2008.05.442. [DOI] [PubMed] [Google Scholar]
- 80.Terman A, Brunk UT. The aging myocardium: roles of mitochondrial damage and lysosomal degradation. Heart Lung Circ. 2005;14:107–114. doi: 10.1016/j.hlc.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 81.Lira VA, Brown DL, Lira AK, Kavazis AN, Soltow QA, Zeanah EH, Criswell DS. Nitric oxide and AMPK cooperatively regulate PGC-1 in skeletal muscle cells. J Physiol. 588:3551–3566. doi: 10.1113/jphysiol.2010.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gottlieb RA, Carreira RS. Autophagy in health and disease. 5. Mitophagy as a way of life. Am J Physiol Cell Physiol. 299:C203–210. doi: 10.1152/ajpcell.00097.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Durgan DJ, Young ME. Linking the cardiomyocyte circadian clock to myocardial metabolism. Cardiovasc Drugs Ther. 2008;22:115–124. doi: 10.1007/s10557-008-6086-y. [DOI] [PubMed] [Google Scholar]
- 84.Gurusamy N, Lekli I, Gherghiceanu M, Popescu LM, Das DK. BAG-1 induces autophagy for cardiac cell survival. Autophagy. 2009;5:120–121. doi: 10.4161/auto.5.1.7303. [DOI] [PubMed] [Google Scholar]
- 85.Yan L, Sadoshima J, Vatner DE, Vatner SF. Autophagy in ischemic preconditioning and hibernating myocardium. Autophagy. 2009;5:709–712. doi: 10.4161/auto.5.5.8510. [DOI] [PubMed] [Google Scholar]
- 86.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 87.Huang C, Liu W, Perry CN, Yitzhaki S, Lee Y, Yuan H, Tsukada YT, Hamacher-Brady A, Mentzer RM, Jr, Gottlieb RA. Autophagy and Protein Kinase C Are Required for Cardioprotection by Sulfaphenazole. Am J Physiol Heart Circ Physiol. 2009 doi: 10.1152/ajpheart.00716.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yitzhaki S, Huang C, Liu W, Lee Y, Gustafsson AB, Mentzer RM, Jr, Gottlieb RA. Autophagy is required for preconditioning by the adenosine A1 receptor-selective agonist CCPA. Basic Res Cardiol. 2009;104:157–167. doi: 10.1007/s00395-009-0006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang C, Yitzhaki S, Perry CN, Liu W, Giricz Z, Mentzer RM, Jr, Gottlieb RA. Autophagy Induced by Ischemic Preconditioning is Essential for Cardioprotection. J Cardiovasc Transl Res. doi: 10.1007/s12265-010-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 92.Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005 doi: 10.1038/sj.cdd.4401697. [DOI] [PubMed] [Google Scholar]
- 93.Hamacher-Brady A, Brady NR, Gottlieb RA, Gustafsson AB. Autophagy as a protective response to Bnip3-mediated apoptotic signaling in the heart. Autophagy. 2006;2:307–309. doi: 10.4161/auto.2947. [DOI] [PubMed] [Google Scholar]
- 94.Murry CE, Richard VJ, Reimer KA, Jennings RB. Ischemic preconditioning slows energy metabolism and delays ultrastructural damage during a sustained ischemic episode. Circ Res. 1990;66:913–931. doi: 10.1161/01.res.66.4.913. [DOI] [PubMed] [Google Scholar]
- 95.Hausenloy DJ, Yellon DM, Mani-Babu S, Duchen MR. Preconditioning protects by inhibiting the mitochondrial permeability transition. Am J Physiol Heart Circ Physiol. 2004;287:H841–849. doi: 10.1152/ajpheart.00678.2003. [DOI] [PubMed] [Google Scholar]
- 96.Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, Pedersen PL, Robinson J, Murata M, Murphy AM, Marban E, Van Eyk JE. Proteomic analysis of pharmacological preconditioning: novel protein targets converge to mitochondrial metabolism pathways. Circ Res. 2006;99:706–714. doi: 10.1161/01.RES.0000243995.74395.f8. [DOI] [PubMed] [Google Scholar]
- 97.Gottlieb RA, Carreira RS. AUTOPHAGY IN HEALTH AND DISEASE: V. Mitophagy as a Way of Life. Am J Physiol Cell Physiol. doi: 10.1152/ajpcell.00097.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bergamini E, Cavallini G, Donati A, Gori Z. The role of autophagy in aging: its essential part in the anti-aging mechanism of caloric restriction. Ann N Y Acad Sci. 2007;1114:69–78. doi: 10.1196/annals.1396.020. [DOI] [PubMed] [Google Scholar]
- 99.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]