Abstract
The ongoing requirement in adult heart for transcription factors with key roles in cardiac development is not well understood. We recently demonstrated that TBX20, a transcriptional regulator required for cardiac development, has key roles in the maintenance of functional and structural phenotypes in adult mouse heart. Conditional ablation of Tbx20 in adult cardiomyocytes leads to a rapid onset and progression of heart failure, with prominent conduction and contractility phenotypes that lead to death. Here we describe a more comprehensive molecular characterization of the functions of TBX20 in adult mouse heart. Coupling genome-wide chromatin immunoprecipitation and transcriptome analyses (RNA-Seq), we identified a subset of genes that change expression in Tbx20 adult cardiomyocyte-specific knockout hearts which are direct downstream targets of TBX20. This analysis revealed a dual role for TBX20 as both a transcriptional activator and a repressor, and that each of these functions regulates genes with very specialized and distinct molecular roles. We also show how TBX20 binds to its targets genome-wide in a context-dependent manner, using various cohorts of co-factors to either promote or repress distinct genetic programs within adult heart. Our integrative approach has uncovered several novel aspects of TBX20 and T-box protein function within adult heart.
Sequencing data accession number (http://www.ncbi.nlm.nih.gov/geo): GSE30943.
INTRODUCTION
Tissue development and function are driven by complex genetic programs that rely on activation and repression of genes by transcription factors. TBX20 is a transcription factor that performs critical roles during early heart development, coordinating cardiac proliferation and regional specification (1), formation of the cardiac chamber (2–4) and valves (4). Tbx20 null mouse embryos display hypoplastic hearts and die at mid-gestation (1–4).
In keeping with the importance of this transcription factor in cardiac development, TBX20 has been associated with congenital heart diseases in humans, including defects in septation, chamber growth and valvulogenesis (5,6). A gain-of-function TBX20 mutation is associated with congenital atrial septal defects, patent foramen ovale and cardiac valve defects (7).
There has been growing interest in the functions of ‘developmental’ transcription factors like TBX20 in adult tissues, since mutations in these genes might explain diseases in mature heart which are not caused by developmental morphological abnormalities (8,9). Genome-wide association studies have identified a number of genomic loci associated with adult-onset diseases which harbor genes encoding transcription factors with critical roles in embryonic development, but as yet unexplored ongoing functional requirements in adulthood (10–12). In the case of Tbx20, genetic variation within this gene has been associated with QRS duration and ventricular conduction phenotypes (10).
The importance of TBX20 in adult heart has been demonstrated in Drosophila, where knockdowns of a Tbx20 homologue display adult heart disease phenotypes (13). Additionally, adult mice which are heterozygous germline null for Tbx20 mice display mild morphological and conduction abnormalities (3). We previously reported that tamoxifen-mediated conditional ablation of Tbx20 in adult cardiomyocytes resulted in an uncommonly severe cardiac phenotype, leading to death within 15 days following tamoxifen induction (14). Functional abnormalities were detected as early as 5 days following Tbx20 ablation, accompanied by rapid progression to heart failure. We showed that expression levels of selected genes that included several potassium and calcium channels (e.g. Kcnd2, Kcnip2, Cacna1c, Cacna1g), gap junction proteins (e.g. Gja1, Gjc1), and other ion transporters including the ATPase Serca2 (Atp2a2), as well as its regulator Pln, were down-regulated in Tbx20 knockout hearts, providing a molecular basis for the severely impaired contractile function of mutant hearts. We also generated a comprehensive catalog of TBX20-binding regions in the genome of adult hearts and found that DNA sites bound by TBX20 were associated with genes involved in ion transport, heart contraction and heart development.
Although these data were critical to providing a mechanistic basis for the observed adult heart phenotype, we were interested in gaining a genome-wide view of the impact of absence of TBX20 on the adult heart transcriptome, to precisely define genes and functional pathways under the direct control of TBX20 and to better understand how this transcription factor regulates expression of genes in different functional pathways. Notably, TBX20 can act as transcriptional activator and repressor, probably depending on context (15). Analysis of TBX20 genome-wide binding data alone does not allow discrimination of functional binding, or to determine which targets are activated or repressed by TBX20. Therefore, we obtained global transcriptome data in adult hearts of conditional Tbx20 knockout and wild-type mice to gain further insights into transcriptional networks directly and indirectly regulated by TBX20 in adult heart.
Here we report that TBX20 is a major regulator of adult cardiac biology, identifying thousands of differentially expressed genes and their biological functions, determining primary and secondary effects of the loss of TBX20 and identifying distinct molecular pathways regulated by TBX20 as either a transcriptional activator or a repressor. We also gained a better understanding of molecular mechanisms by which TBX20 regulates gene expression in different contexts by identifying transcription factors that may act in concert with TBX20 on distinct subsets of genetic pathways.
RESULTS
Transcriptome analysis reveals that TBX20 regulates adult cardiomyocyte structure and function
We have previously characterized genome-wide binding sites for TBX20 in adult mouse heart. We showed that TBX20 binds at sites near genes associated with heart contraction and cardiovascular development functions and, to a lesser extent, energy metabolism (14). We have also confirmed that a number of candidate genes involved in cardiac contractility had decreased expression levels in adult hearts with conditional ablation of Tbx20 in adult cardiomyocytes. This candidate gene approach, however, fails to recognize which of the more than 4000 TBX20 DNA-binding regions reflect a functional role for this protein in the transcriptional regulation of the cognate gene, or simply reflects a binding event with no impact on gene regulation.
To gain a genome-wide view of the functional impact of the abrogation of TBX20 and to perform an unbiased assessment of genes and functions directly or indirectly regulated by this transcription factor, we evaluated, using RNA-seq, the effect of the adult cardiomyocyte-specific ablation of Tbx20 on the transcriptome.
Cardiac-specific ablation was obtained by conditionally excising exon 2 of Tbx20 (mm9 coordinate chr9:24,575,123-24,575,374), by injection of tamoxifen in α-MHC-mER-Cre-mER/Tbx20f/f/R26R-lacZ/lacZ mice, as described in (14). RNA was obtained 4 days following tamoxifen injection, ∼24 h after a 60–80% decrease in TBX20 protein in cardiomyocytes was observed (14). Consistent with the gene ablation strategy used, we detected a significant decrease in sequence reads specifically for exon 2 of Tbx20 knockout hearts compared with wild-type, with the remaining upstream and downstream Tbx20 exons showing no difference between groups. Of the 21 213 RefSeq genes, we identified 10 095 genes expressed in adult mouse heart (47.6%), with an additional 11 118 genes showing no expression or barely measurable expression levels, similar to background levels (see Materials and Methods for details in the classification). A complete annotation of expression levels and ChIP peak-to-gene assignment is given in Supplementary Material, File S1.
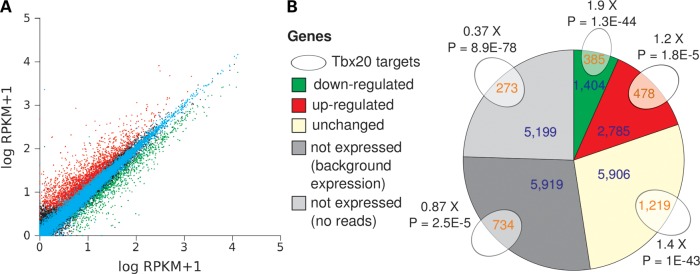
We found that conditional ablation of Tbx20 in adult cardiomyocytes led to global changes in the transcriptome. While Pearson's R2 of the log-normalized expression levels per gene between three RNA-seq replicates of each group (wild-type and knockout) was 0.98 ± 0.01, correlation between wild-type and knockout samples was considerably lower (R2 = 0.88 ± 0.01) (Fig. 1A and Supplementary Material, Fig. S1). Indeed, we identified 4189 genes differentially expressed between wild-type and knockout hearts, using DESeq (16) at an adjusted P-value of <0.001 and >1.5-fold expression difference (Fig. 1B).
Figure 1.
Impact of Tbx20 ablation on the adult mouse cardiac transcriptome. (A) The scatter plot of expression levels per gene in RPKM. To enable calculation of log values of genes with 0 RPKM, 1 was added to all data points. Correlation between two RNA-seq control samples (cyan) was higher than control and Tbx20 conditional knockout samples, reflecting differential expression. Green, down-regulated genes; red, up-regulated. (B) Genes differentially expressed in Tbx20 knockout hearts and TBX20 ChIP-seq target genes at adjusted P-value of <0.001, >1.5-fold expression difference. The pie chart depicts the number of differentially expressed genes, according to RNA-seq data (numbers in blue). White ovals represent genes with a nearby TBX20 ChIP peak (ChIP-seq data). Numbers in orange represent the intersection between each set of differentially expressed genes and genes with a nearby TBX20-binding region, representing putative TBX20 direct targets. All genes with no nearby TBX20 ChIP peak were considered putative indirect targets. The fold-difference between the observed overlap with the ChIP set and number expected (based on the size of each category) is shown near each intersection.
Next we explored the functional categories of genes whose expression levels are affected by ablation of Tbx20, performing an analysis of enrichment of Gene Ontology (GO) terms (17) on the differentially expressed gene sets shown in Fig. 1B. We grouped GO terms in major categories (see Materials and Methods) and compared the occurrence of these categories across different gene sets (Fig. 2A and Supplementary Material, Fig. S2). We did not observe enrichment of cardiac-related functions among genes that were not expressed in the heart, expressed at very low levels, or that did not change expression in Tbx20 conditional knockout hearts.
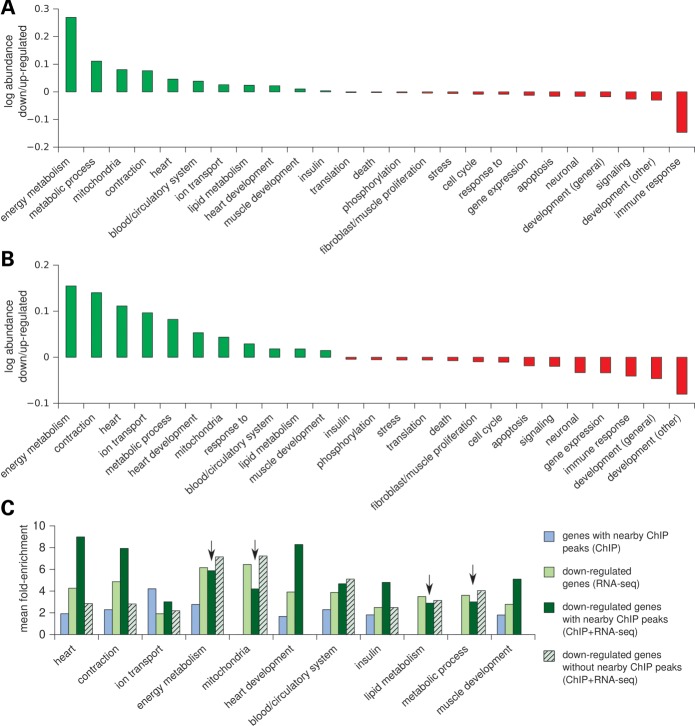
Figure 2.
Distinct molecular functions are associated with TBX20 activation and repression of transcription. (A) The log ratio of the abundance of functional categories (see Materials and Methods) between down- and up-regulated genes (RNA-seq data only). (B) The log ratio of the abundance of functional categories between genes with a nearby TBX20-binding region (putative TBX20 direct targets) that are down- and up-regulated (RNA-seq and ChIP-seq data combined). (C) Comparison of GO term fold-enrichment among (i) genes with a nearby TBX20 ChIP peak, (ii) all down-regulated, (iii) down-regulated with a nearby TBX20 ChIP peak (putative direct targets) and (iv) down-regulated genes with no nearby TBX20 ChIP peak (putative indirect targets). Arrows show instances in which enrichment is lower in direct targets than in indirect targets, suggesting that these categories are enriched for genes that are indirectly regulated in the absence of TBX20.
In contrast, we observed a clear cut difference in GO terms between genes which were up- or down-regulated in Tbx20 adult myocardial mutants (Fig. 2A). While down-regulated genes (1404) were predominantly involved in heart function, contraction, heart development, blood/circulation and energy metabolism, up-regulated genes (2785) showed almost no enrichment for such functions. Rather, up-regulated genes seemed to be primarily involved in neuronal function, immune response, cell death/apoptosis, development and proliferation of various tissues other than heart and muscle/fibroblast proliferation. The full GO annotation is available as Supplementary Material, File S2.
These data suggest that genes involved in excitation/contraction coupling and various aspects of energy metabolism are activated by TBX20, while genes involved in immune response, apoptosis, cell cycle and proliferation and developmental programs of non-cardiovascular tissues and systems are normally repressed by TBX20. This difference in functional categories of genes that are activated or repressed by this dual-role transcription factor has not been previously appreciated. Nevertheless, transcriptome differences alone are not sufficient to distinguish genes that directly rely on TBX20 for their proper expression from those which are adapting to a changing cellular milieu consequent to Tbx20 ablation. To identify genes which are direct targets of TBX20 and change their expression in knockout hearts, we next compared genome-wide TBX20 DNA-binding data (ChIP-Seq) with transcriptome data (RNA-Seq) of wild-type and knockout hearts.
Intersection of DNA binding and transcriptome data highlights TBX20 functional targets
To identify genes that are likely primary mediators of TBX20 functions in adult heart, we selected genes that were both differentially expressed in the Tbx20 knockout hearts and harbored a ChIP-seq-identified TBX20 DNA-binding region (TBX20 ChIP peak) in their proximity (Fig. 1B). We refer to these genes as putative direct targets, in contrast to genes with no nearby TBX20 ChIP peak which are referred to as putative indirect targets. We are aware that the presence of a nearby TBX20 ChIP peak does not necessarily imply that a given gene is a direct target. Nonetheless, the results presented below indicate that this set is enriched for real direct targets and supports the validity of this classification.
Genes with a nearby TBX20-binding region were expressed at 4-fold higher levels than all genes expressed in the heart (P < 2.2E-16, Wilcoxon rank-sum test), indicating that the binding regions identified tend to be associated with transcriptionally active loci. Genes down-regulated in Tbx20 conditional knockout hearts and harboring a nearby TBX20-binding region were enriched 1.9-fold (P < 10−43) above random expectation (Fig. 1B). Reassuringly, the number of genes that were not expressed in heart and were TBX20 targets was lower than expected by chance by 2.5-fold (P < 10−77).
TBX20 acts as an activator or repressor to regulate distinct biological processes
Genes that were either up- or down-regulated and had a nearby TBX20-binding region (putative direct targets) displayed enrichment for functional categories similar to those obtained from analysis of transcriptome-only data in Tbx20 mutants and controls (Fig. 2B and Supplementary Material, Fig. S3). Down-regulated genes were preferentially involved in functions related to cardiovascular biology and energy metabolism. In contrast, up-regulated genes showed none of these heart and energy-related terms. Instead, up-regulated genes were involved in distinct biological processes, such as developmental programs in the central nervous system and other non-cardiac tissues, cell cycle and proliferation as well as immune response. These observations reinforce the view that, in adult heart, TBX20 directs a dual-role transcriptional program, activating genes involved in heart contraction and development, while repressing genetic programs of other tissues and systems, and a cellular proliferation program.
Interestingly, the abundance of certain functional categories differed between transcriptome-only data (Fig. 2A) and TBX20 direct targets (Fig. 2B). For example, energy metabolism and immune response were more abundant in the former set, whereas contraction, ion transport and development were more abundant in the latter. Indeed, genes involved in metabolic process, energy metabolism and mitochondrial terms (a proxy for energy metabolism functions) followed a different trend of GO enrichment, compared with contraction and development. The enrichment of the former functional categories in the differential expression/ChIP gene set was lower than that seen in transcriptome-only data or differentially expressed genes without nearby TBX20 ChIP-binding regions (indirect targets), a trend opposite to all other categories (Fig. 2C). These results suggest that whereas TBX20 directly regulates some genes involved in metabolic processes, a number of genes in these functional categories that were differentially expressed in knockout hearts are likely to reflect secondary responses to ensuing cardiac pathology in mutant mice. Functional categories preferentially enriched among up-regulated genes without an assigned TBX20-binding region (indirect targets) included immune response and stress, showing the same pattern described above for energy metabolism (Supplementary Material, Fig. S4).
Comparison of TBX20 ChIP peaks grouped by differential expression patterns
Having established that TBX20 acts as a transcriptional activator and repressor, directly regulating genes with distinct biological functions, we next sought to compare these groups of DNA regions bound by TBX20 to determine features that might explain how this transcription factor differentially regulates gene targets (Fig. 3).
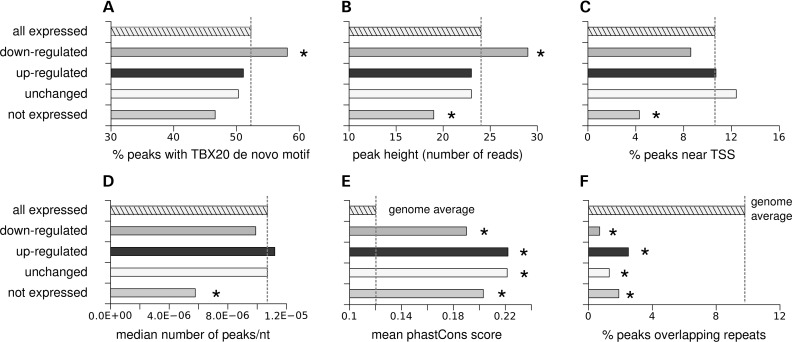
Figure 3.
Comparison of genomic features among peak sets grouped by gene expression pattern. (A) The frequency of TBX20 DNA-binding motif identified de novo from ChIP-seq data (14). (B) The peak height was determined by the peak calling program QuEST (45) and is a proxy for the binding frequency or strength. The higher the peak height, more frequent binding was observed. (C) The frequency of peaks overlapping (±200 bp) with transcription start sites (TSS). (D) TBX20-binding region frequency per gene locus (regions/bp). (E) Conservation as measured by the phastCons algorithm (48). (F) Overlap with repeats. Asterisks indicate statistically significant differences (P < 0.05), compared with all expressed genes (A–D) or to sequences randomly selected from the genome (E and F). A description of the data presented in each panel and the statistical tests used are provided in Materials and Methods. Not expressed consists of the ‘no reads’ category. Results for the not expressed set with background levels were similar and are not shown for clarity sake (Supplementary Material, Fig. S6).
TBX20-binding regions near down-regulated genes contained the TBX20 ChIP-seq DNA-binding motif that we uncovered from our ChIP data (14) more frequently than binding regions elsewhere in the genome and yielded more sequence reads, indicative of stronger or more frequent binding (Fig. 3A and B). However, we found no difference between the percentage of TBX20-binding regions in promoters and in their frequency per loci (regions/bp) across expressed genes, whereas unexpressed genes showed a different trend as discussed below (Fig. 3C and D). Interestingly, all sets of TBX20-binding regions, regardless of expression pattern, displayed evolutionary conservation at clearly higher levels than the rest of the genome, and the proportion of ChIP peaks overlapping low complexity repeats was considerably lower than the rest of the genome in all sets (Fig. 3E and F), suggesting that they are biologically functional.
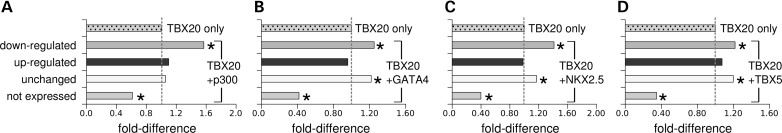
Gene sets differed in their association with TBX20 co-factors. GATA4 and NKX2–5 are known to positively co-regulate gene expression with TBX20 in the heart (13,15) and p300 is a general transcription activator (18). Interestingly, genes whose nearby TBX20 peaks overlapped a peak for GATA4, NKX2–5, TBX5 (19) or p300 (20) were more likely to be down-regulated or remain unchanged, but not more likely to be up-regulated in conditional knockout hearts (Fig. 4). These results suggest that TBX20 may employ distinct cohorts of co-factors to control specific genetic programs within the heart.
Figure 4.
Overlap of TBX20-binding regions with p300, GATA4, NKX2–5 and TBX5-binding regions. The set of genes with a nearby TBX20 ChIP-seq peak has different proportions of differentially expressed genes, compared with all genes (Fig. 1). Overlap of TBX20 ChIP peaks with (A) p300 (20), (B) GATA4, (C) NKX2–5 and (D) TBX5 (19) ChIP peaks further biases these proportions, suggesting that these proteins co-regulate gene expression with TBX20. X-axis: fold-difference between (i) the proportion of genes in a given expression category with a nearby TBX20 peak overlapping another ChIP peak and (ii) the proportion of genes in the same category with any nearby TBX20 peak (TBX20 only). Asterisks indicate statistically significant differences (P < 0.05), compared with TBX20 only. A description of the statistical test used is provided in Materials and Methods. Not expressed consists of the ‘no reads’ category. Results for the not expressed set with background levels were similar and are not shown for clarity sake (Supplementary Material, Fig. S6).
Interestingly, TBX20-binding regions in the neighborhood of genes that were not expressed in the heart differed from binding regions assigned to expressed genes, suggesting that they do not play a role in regulation of gene expression. They contained fewer TBX20 motifs, were represented by fewer reads (weaker binding) and occurred at a lower frequency in these loci (binding regions/bp) (Fig. 3A–D). However, as noted above, all sets of TBX20-binding regions, including those near genes not expressed in the heart, were evolutionary conserved and overlapped fewer low-complexity repeats than the rest of the genome (Fig. 3E and F). Under the assumption that these latter features are indicative of function, we cannot completely rule out a biological role for binding events near unexpressed genes. Future studies are needed to establish what functional roles these regions may encompass.
DNA-binding motifs within TBX20-binding regions
To expand our knowledge on possible TBX20 DNA-binding co-factors, we scanned TBX20-binding regions associated with genes that were up- or down-regulated in conditional knockout hearts for over-representation of transcription factor-binding sites (TFBS). Because these gene sets belong to different functional categories and regulatory codes are likely to be specific, we further split down- and up-regulated genes by the functional categories in Figure 2. We then selected peaks associated with genes in each subset and analyzed TFBS over-representation using position weight matrices (PWMs) from Jaspar (21) and UniProbe (22) using Clover (23) against different background sequences.
Among over-represented PWMs, we searched those that were exclusively present in up- or down-regulated gene/binding regions subsets. We employed a highly stringent setup, accepting only PWMs over-represented in three higher order Markov model backgrounds and two mouse genome sequence sets (see Materials and Methods). Although this approach limits the number of TFBS identified, it increases the likelihood that identified factors are biologically relevant. Results of these analyses are summarized in Table 1.
Table 1.
Summary of PWMs over-represented in TBX20 ChIP peaks near differentially expressed genes partitioned into functional categories
| Transcription factor | Over-representation | |
|---|---|---|
| Down-regulated genes | GATA5a, GATA6a, GATA1a | Heart development, muscle development, heart, proliferation, signaling |
| NKX2–5 | Energy and lipid metabolism | |
| NKX2–6 | Lipid metabolism | |
| NKX1–2a, NKX2–4a, NKX2–6a, NKX2–9a, NKX3–1a | Development, immune response, lipid metabolism, translation, metabolism, translation | |
| PPARG:RXRA | Energy metabolism and mitochondria | |
| REST half-site | Metabolism | |
| Up-regulated genes | ESR1 | Muscle development, mitochondria |
| ESR2 | Muscle development, mitochondria, ‘response to’, stress | |
| GATA3a | Muscle development, development (other), energy, heart, metabolism | |
| GATA6a | Heart | |
| REST half-site | Development (general), development (other), metabolism, neuronal, translation |
ESR1: MA0112.2, ESR2:MA0258.1, GATA1: MA0035.2, GATA3: UP00032, GATA5: UP00080, GATA6: UP00100, NKX2–5: UP00249, NKX2–6: UP00147, NKX1–2: UP00139, NKX2–4: UP00107, NKX2–6: UP00147, NKX2–9: UP00119, NKX3–1: UP00017, PPARG: Jaspar MA0065.2, RARA: UP00048, REST full and left half-site: MA0138.2. MA: Jaspar, UP: UniProbe.
aPWM over-representation only verified when considering enrichment in select background model. All the other PWMs were over-represented in all background models.
Motif overrepresentation in genes down-regulated in Tbx20 mutants
As shown in Figure 4, the co-binding of TBX20 with GATA4, TBX5 or NKX2–5 (19) was predominantly associated with genes down-regulated in Tbx20 conditional knockout hearts. Consistently, we also found that GATA and NKX PWM's were predominantly over-represented in subsets of TBX20-binding regions associated with down-regulated genes, reinforcing the idea that these transcription factors act as TBX20 co-factors to activate gene expression in the heart.
We also uncovered an over-representation of a PPARG/RXRA PWM (Jaspar accession number: MA0065.2) only in down-regulated genes within the ‘energy metabolism’ functional category, in line with reports that PPARG is required for fatty acid utilization in the adult heart (24). Interestingly, analysis with GREAT (25) showed that genes in the ‘PPAR signaling pathway’ and ‘Genes up-regulated in differentiating C2C12 cells (myoblasts) upon expression of PPARGC1A [Gene ID = 10891] off an adenoviral vector’ (MSigDB) were exclusively over-represented in the down-regulated/ChIP dataset (Supplementary Material, File S2), reinforcing the observation that PPAR factors may be important TBX20 co-factors for activation of these subsets of genes. To validate PPARG as a TBX20 co-factor in adult heart, we performed ChIP of TBX20-GFP as described in (14) followed by western blot analysis with a PPARG antibody (Supplementary Material, Fig. S7). The result obtained confirmed that PPARG and TBX20 co-occupy DNA fragments and are likely to be functional co-factors in adult heart.
In our previous work, we found that five transcription factors (TBX20, MEF2A, ESRRA/B, CREB1 and TEAD1) seem to be part of a regulatory code for activation of ion transport and contraction genes in adult cardiomyocytes (14). Although the top-down analysis starting with all contraction/ion transport genes did not recover the complete code (only TBX20, MEF2A and TEAD1 were found to be over-represented), the set of genes with nearby TBX20-binding regions containing all the five motifs was 3.1-fold more likely to contain down-regulated genes than expected by chance (P = 2E-7), supporting a functional role for this specific motif combination for gene activation in adult heart.
Also supporting the notion that ESRRA/B may be a TBX20 co-factor, our analysis with GREAT (25) showed that the gene sets ‘Genes up-regulated by ESRRA only’ and ‘Genes regulated by ESRRA in MCF-7 cells (breast cancer)’ and the ESRRA motif in ‘MSigDB Predicted Promoter Motifs’ are exclusively over-represented in the down-regulated/ChIP set.
Motif overrepresentation in genes up-regulated in Tbx20 mutants
Among motifs over-represented in genes up-regulated in Tbx20 mutants are ESR1, ESR2, GATA and a REST half-site (Table 1). ESR1 and ESR2 serve as binding sites for estrogen receptors, which have well-defined roles in repressing transcriptional programs in the cardiovascular system, particularly in mitochondria (26), in line with our findings (Table 1). It is intriguing that REST is also among these motifs, given that REST/NRSF is a transcription factor commonly associated with repression of neuronal programs in non-neuronal tissues (27,28). Recent data show that the expression of a dominant-negative REST isoform in the adult mouse heart leads to dilated cardiomyoptahy, arrhythmias and death, illustrating the ongoing requirement of REST for proper adult heart function (29). While our data posit an association between DNA-binding motifs for TBX20 and other transcription factors with well-established roles in repressing transcription programs in the heart, it remains unclear whether the cognate transcription factors in Table 1 and Supplementary Material, File S2 are co-factors of TBX20 in vivo; a subject of future investigation.
Overall, these TFBS signatures are consistent with the idea that TBX20 cooperates with distinct cohorts of transcription factors to act as a transcriptional activator or repressor to regulate distinct genetic pathways.
TBX20 binds, in vitro, to a novel motif with lower affinity than previously characterized T-box motifs
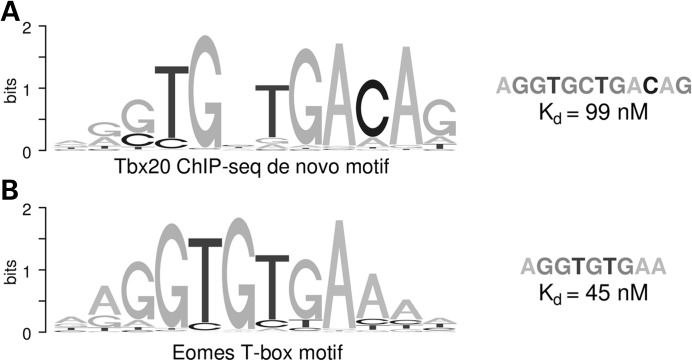
The TBX20 DNA-binding motif that we uncovered from our ChIP data (14) (consensus AGGTGNTGACAG) resembles known T-box DNA-binding motifs identified in vitro [TBX20 and TBX5 motifs identified by SELEX (30) and EOMES motif identified by protein-binding microarray (31), both consensus AGGTGTGA], but contains one position with no nucleotide preference, creating a gap in the motif and ends with CAG instead of AAA (Fig. 5). We hypothesized that the affinity of the in vivo binding motif may be lower than the in vitro motifs, which, by design, select for high-affinity sites. To directly test this, we employed surface plasmon resonance to compare relative dissociation constants (Kd) of TBX20-binding affinity to a representative sequence of the EOMES motif (consensus AGGTGTGAA) and one based on the consensus sequence of the novel TBX20-binding motif (AGGTGCTGACAG) using a non-specific sequence as reference. The binding affinity for the representative EOMES sequence was 45.7 ± 0.1 nm, whereas the sequence for the novel TBX20 motif had lower affinity at 99 ± 13.4 nm (P = 0.011).
Figure 5.
Comparison of T-box DNA-binding motifs and -binding affinities. (A) TBX20 DNA-binding motif obtained de novo from ChIP-seq data (14) and (B) EOMES DNA-binding motif determined by protein-binding microarrays (31). Representative sequences from each motif used to determine their binding affinities and their relative Kd's are shown on the right.
As we noted previously, the higher affinity EOMES motif occurs in considerably lower frequency than the TBX20 motif within TBX20-binding regions (28 vs 51%, respectively) (14). The relative frequency of the EOMES and TBX20 de novo motifs was similar in up- and down-regulated genes. We did not also observe a significant difference in the magnitude of change in expression for genes regulated by TBX20 binding to either motif type. Therefore, differences in binding affinity do not seem to explain the different expression patterns observed.
Nonetheless, the fact that the DNA-binding motif used by TBX20 in vivo has ∼2.5-fold lower affinity than the motif previously described from in vitro experiments is intriguing. It is unclear what, if any, the in vivo functional significance of this relatively modest reduction in affinity is. One possibility is that a reduced binding affinity might allow for a faster dissociation of the bound protein to DNA.
A recent report has demonstrated that Pax6 expression relies on an enhancer with reduced affinity-binding site for PREP1 for its temporal control of expression in the developing eye, although with a 5–7-fold affinity difference between motifs, higher than we observe with the TBX20 motif. Interestingly, it is thought that T-box proteins bind to similar motifs and often compete for the same binding regions in the genome. Toward that end, we observed a significant overlap between TBX20- and TBX5-binding regions in cardiomyocytes (14). It is possible that T-box proteins, notorious for their dosage sensitivity, rely on lower affinity genomic-binding sites as part of their repertoire of strategies to determine the regulatory output of multiple T-box proteins competing for the same binding site, often with widely different outcomes.
Discussion
In the present work, coupling ChIP-seq data with gene expression analysis allowed us to molecularly characterize effects of Tbx20 knockout in adult heart and address regulatory roles of this transcription factor, revealing biological functions affected in the adult cardiomyocyte-specific mutant of Tbx20, and offering mechanistic insights into regulation by TBX20.
Although our criteria for selecting differentially expressed genes in the knockout was conservative (we used both P-value and fold-difference to enrich for biologically significant changes), we found that ∼4200 genes were up- and down-regulated. The strikingly high number of genes highlights the importance of TBX20 in adult heart. Most of the 4200 genes affected in knockout heart do not seem to be directly regulated by TBX20, possibly reflecting indirect propagation of effects caused by the absence of TBX20, as also observed for other transcription factors (32,33). The primary functional categories of secondary-effect differentially expressed genes include energy metabolism, other metabolic processes and immune response, processes with extensive networks of feedback regulation that could explain indirect propagation.
Intriguingly, we found 1219 genes (40% of TBX20 ChIP targets) that had a nearby TBX20-binding region but were not differentially expressed in Tbx20 conditional knockouts. One possibility is that these binding regions are primed by TBX20 for activation in other time points and conditions. High numbers of genes that are bound by a given transcription factor but are unaffected by its absence have been observed (32,34–36), and functional redundancy or compensation by other transcription factors has been proposed as an explanation (34,37). A study on T-bet (Tbx21) also identified genes that were bound by this T-box protein but were unaffected by its over-expression or absence. The authors suggested that this observation could reflect functional redundancy with other T-box proteins and different roles for a given T-box protein at each locus (37).
Distinct activator and repressive functional categories of direct gene targets of TBX20
Genes activated by TBX20
Confirming our analysis of TBX20 chromatin-binding data only (14), we showed that TBX20 is an activator of genes responsible for ion transport/contraction. We also unraveled a previously unappreciated role of TBX20 in energy metabolism, with binding regions near genes involved in glycolysis, beta-oxidation, tricarboxylic acid cycle, electron transport chain and other metabolic processes. Energy requirements during cardiac development change as cardiomyocytes progressively undergo proliferation, differentiation and postnatal growth, shifting from glycolysis as the major energy source to fatty acid beta-oxidation as a preferential source of energy in adult hearts (38). Our data showed that genes belonging to both energy pathways were positively regulated by TBX20 in adult heart, indicating that this factor controls both energy metabolism programs.
TBX20 regulation of energy metabolism genes seems to involve the transcription factor PPARG, a member of a family of proteins known for their roles in regulating lipid metabolism in general and also in adult heart (24,39), as indicated by our bioinformatics and western blot analyses.
Genes repressed by TBX20
In parallel to activation of genes involved in cardiac contraction and energy metabolism, our analysis revealed a repressive role for TBX20, corroborating previous in vitro reports (15,40). We were able to tease out, genome-wide, the identity and functional roles of these genes. We observed that genes repressed by TBX20 included those with roles in development and specification of non-cardiac tissues and systems and repressed in heart.
How TBX20 establishes and maintains a transcriptional repressive program will be the goal of future investigation. It is possible that the repressive function on certain genes might be temporally restricted, such as the repression of genes involved in cell cycle and proliferation, given that TBX20 is a key mediator of cardiac proliferation during embryogenesis but also seems to regulate an active anti-proliferation program in adult heart. It is unclear how the functional outcome of activation or repression is achieved upon binding of TBX20 to DNA. Different isoforms or the presence of different co-factors may play a role. The longest known mouse TBX20 isoform contains both transactivating and transrepressing domains whereas shorter ones carry only the DNA-binding domain and isoforms respond differently to co-factors such as GATA4 and NKX2–5 (15).
CONCLUSIONS
By coupling genome-wide DNA binding and transcriptome data from normal and Tbx20 knockout mouse hearts, we uncovered major biological functions regulated by TBX20. We identified different cohorts of co-factors that, together with TBX20, are likely to differentially regulate distinct sets of genes with specific biological roles. Among these, PPARG may be an important TBX20 co-factor activating energy metabolism genes in the adult heart while REST, a known transcriptional repressor, may act with TBX20 to silence genes controlling genetic programs unrelated to adult heart function or in response to damage to this tissue. Finally, we characterized the binding properties of the main TBX20-binding motifs in vivo, showing that the novel TBX20 motif that we described previously has lower binding affinity than the motif previously identified in vitro. Altogether, comprehensive and integrative approaches and analyses that we employed allowed a detailed molecular characterization of the ongoing functions of TBX20 and its co-factors in maintaining the structure and function of adult heart.
MATERIALS AND METHODS
RNA-seq data generation
Total RNA from adult mouse hearts was extracted using Trizol®. Ten micrograms of total RNA was used to isolate mRNA with Dyna1 oligo dT beads (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions. mRNA was fragmented using Ambion® 10× RNA Fragmentation Reagent and heat to 70°C for 5 min. Cleaved RNA fragments were transcribed into first-strand cDNA using Random Primer Mix (NEB) and Supercript II Reverse Transcriptase (Invitrogen). This was followed by a second-strand synthesis using DNA Polymerase I and RNAseH (Invitrogen). After purification, double-stranded cDNA fragments were repaired by a Klenow reaction, ligated to Illumina adapters and size-selected (200 pb) by gel-excision to generate the library for sequencing. Adapter-modified cDNA fragments were enriched by PCR amplification, purified and sequenced for 36 cycles on the Illumina Genome Analyzer II. Three replicates for each condition (wild-type and Tbx20 conditional knockout) were generated from one mouse heart each.
The sequencing data are available from NCBI GEO (41) (http://www.ncbi.nlm.nih.gov/geo), accession number GSE30943.
Differential gene expression
Genomic coordinates of RefSeq mRNAs aligned to mm9 were downloaded from UCSC Genome Browser (42) on February 2010 and clustered by gene symbol resulting in 21 213 mRNA clusters/genes.
Illlumina sequence reads were aligned to the mm9 genome with Bowtie (43). Gene expression was quantified by counting the number of reads mapping to all unique exons of RefSeq genes and exon junctions and normalizing counts by base pair and total number of reads per library (Reads Per Kilobase per Million, RPKM). DESEq (16) was used to call differentially expressed genes. We accepted only genes whose highest RPKM (wild-type or knockout) was above background levels (adjusted P < 0.01). Background levels were estimated by calculating RPKMs of 20 kb intergenic chunks. The percentage of chunks with RPKM ≥ a given sample RPKM was used as the P-value for being background. P-values were corrected for multiple testing using the ‘fdr’ procedure of the p.adjust function of the R statistical package (44).
When trying different significance cutoffs, we noticed that higher stringency resulted in higher fold-enrichments of heart contraction/development and energy metabolism terms. We chose a cutoff that was statistically stringent, resulted in high GO enrichments and yielded a sizable number of genes for our downstream analyses.
ChIP data
TBX20 ChIP data used in this paper were previously published (NCBI GEO (41) (http://www.ncbi.nlm.nih.gov/geo) accession number GSE29636) (14). Briefly, Illumina reads were aligned to the mouse genome (mm9) using Eland and only reads uniquely mapped to the genome were used. Peaks were called against whole heart input chromatin using QuEST 2.3 (45) with the ‘transcription factor’ setting and ChIP-to-background cutoff of 30. All other parameters were default.
TBX20 ChIP peak assignment
Peaks occurring inside the gene or within 6 kb around the transcription start site were assigned to that gene (multiple assignments allowed) and peaks occurring in intergenic regions were assigned to the nearest gene to avoid ambiguity in assignment to up- and down-regulated genes.
Motif enrichment
Clover (23) was run with default parameters (P-value cutoff = 0.05, 1000 random sets) against five control sequence sets: a mouse promoter set and chromosome 21 sequences provided with Clover and sequences generated from 3rd-, 4th- and 5th-order Markov models derived from each peak set.
Gene Ontology analyses
MGI associations of GO terms (17) to genes and the GO database were downloaded in 1 August 2010. MGI GO terms and all upstream terms were associated with RefSeq (46) genes and half of both flanking intergenic regions (locus) and using these associations, a ‘genome fraction’ for each term was calculated [(gene + intergenic regions length/2)/total genome length]. Peaks were assigned with a given GO term via the locus annotated with the term. Significance of GO enrichment was assessed with the binomial test using the ‘genome fraction’ as probability of success to correct for the locus length bias (47). Concomitantly, a hypergeometric test was conducted on genes to which peaks were assigned. Only terms with P < 0.05 in both sets were accepted as significant. The Benjamini–Hochberg method in the R package (44) was used to correct for multiple tests. For analyses involving differentially expressed genes, only the hypergeometric test was used, as RNA-seq data are not as biased for the locus length.
Results for other ontologies were obtained with the online tool GREAT (25), assigning peaks to the nearest gene up to 100 kb against the whole mm9 genome. GO terms obtained with GREAT agreed well with our own method.
To compare GO enrichments in Fig. 2, we pooled all 2300 GO terms enriched in differentially expressed genes and ChIP genes and manually classified 1283 terms in the categories shown in Fig. 2 (Supplementary Material, File S3 and Supplemental Material). The fractions of GO terms for a given gene set are the numbers of terms in a given category enriched in the set divided by the total number of enriched terms in the set. As not only the fraction of terms, but also the fold-enrichment varies across datasets, fractions were multiplied by the mean fold-enrichment to provide a more comprehensive measurement (abundance).
Comparison of genomic features
Peak heights were compared with Wilcoxon rank-sum tests. The percentage of peaks within 200 bp around the TSS and containing a TBX20 ChIP-seq de novo motif were compared with chi-square tests.
Evolutionary conservation 100 bp around peak centers was measured using the 30-way phastCons (48) track from UCSC Genome Browser. For the analysis of repeats, a minimum overlap of 10% of the peak length with low-complexity and simple repeats from the RepeatMasker (49) track from UCSC Genome Browser was required. Since the different ChIP peak sets occurred near the TSS in different frequencies, to avoid nucleotide composition biases associated with the TSS that would affect conservation and frequency of repeats, only peaks located in intergenic regions were considered. To calculate expected phastCons scores and overlap with repeats, 1000 random sets containing the same number of TBX20 ChIP peaks of interest were generated by sampling size-matched coordinates within the same loci of each ChIP peak from the uniquely mappable regions of the genome. These regions were extracted from the mm9 Mapability track of the UCSC Genome Browser (crgMapabilityAlign36 mer.bw).
To estimate how likely a given change in fold-difference associated with overlap with other ChIP peaks could be randomly obtained (Fig. 4), 10 000 sets with the same number of genes of a given test set, randomly sampled from all TBX20 putative target genes, were generated and the number of genes found in a given differential expression gene set was counted. P-values were calculated by counting the number of sets with at least the number of genes from the test set in the same differential expression set.
The BH method in the p.adjust R package function was used to correct for multiple comparisons.
Determination of relative binding affinity
Surface plasmon resonance analysis was performed on a Biacore 3000 instrument. Biotinylated double-stranded oligonucleotides were purified using MinElute spin columns (Qiagen) and were immobilized onto a streptavidin-coated Sensor Chip SA (Biacore); a target response of 200 RU was used during DNA immobilization step. Protein samples were diluted in ice-cold running buffer (10 mm Tris–HCl, pH 7.4; 2 mm β-mercaptoethanol; 0.02% Triton-X-100; 120 mm NaCl, 10% glycerol) and maintained on ice until being applied to the Sensor Chip. Proteins were passed through four flow cells at room temperature at a flow rate of 50 μl/min, using the KINJECT option, 250 μl sample (500 s dissociation phase). Protein samples, at multiple concentrations, were applied to the chip 2–3 times and all response curves of the series for each run were independently fit to kinetic-binding models using Scrubber2 software (BioLogic Software). They are determined by first fixing off-rates (kd), fits of the dissociation phase of the response curves and then using the ka/kd option to fit the on-rates. Nearly identical results were obtained using the km/ka/kd fitting option that accounts for mass transport. Reported Kd= ka/kd values are the mean and standard deviation of these measurements. All SPR DNA probe sequences were 60 bp long. Synthetic SPR non-specific probe is used as a reference for calculating relative Kd values and is designed from 8-mers with PBM E-scores <0.2. Other probes containing Eomes and TBX20 ChIP-seq de novo binding sites, shown in bold, along with their common primer sequence, underlined, are as follows:
Non-specific: ACTAAGCTTGGATCGCGTATATGGAATAAGCCTACGACGAAGGGAGGCTTGCGCAGATAC
Eomes: TAATGGATCGCGTAGGTGTGAAGGAATAA GCCTACGACGAAGGGAGGCTTGCGCAGATAC
TBX20 de novo: TGGATCGCGTAGGTGCTGACAGGGAATAAGCCTACGACGAAGGGAGGCTTGCGCAGATAC
More details on this experiment can be found in Supplementary Material.
SUPPLEMENTARY MATERIAL
FUNDING
This work was partially support by the US National Institute of Health [grant number HG004428, HL088393 and DK078871 to MAN] and [grant number HG003985 to MLB]. N.J.S. and I.A. are recipients of American Heart Association post-doctoral fellowships. L.S. is a recipient of a US National Institute of Health postdoctoral fellowship [number GM090645].
Supplementary Material
ACKNOWLEDGEMENTS
We thank Trevor Siggers for advice, assistance and helpful discussions regarding protein expression, SPR experiments and Scrubber2 analysis.
Conflict of Interest statement. None declared.
References
- 1.Cai C.-L., Zhou W., Yang L., Bu L., Qyang Y., Zhang X., Li X., Rosenfeld M.G., Chen J., Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh M.K., Christoffels V.M., Dias J.M., Trowe M.-O., Petry M., Schuster-Gossler K., Bürger A., Ericson J., Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132:2697–2707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- 3.Stennard F.A., Costa M.W., Lai D., Biben C., Furtado M.B., Solloway M.J., McCulley D.J., Leimena C., Preis J.I., Dunwoodie S.L., et al. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- 4.Takeuchi J.K., Mileikovskaia M., Koshiba-Takeuchi K., Heidt A.B., Mori A.D., Arruda E.P., Gertsenstein M., Georges R., Davidson L., Mo R., et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 5.Liu C., Shen A., Li X., Jiao W., Zhang X., Li Z. T-box transcription factor TBX20 mutations in Chinese patients with congenital heart disease. Eur. J. Med. Genet. 2008;51:580–587. doi: 10.1016/j.ejmg.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Kirk E.P., Sunde M., Costa M.W., Rankin S.A., Wolstein O., Castro M.L., Butler T.L., Hyun C., Guo G., Otway R., et al. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am. J. Hum. Genet. 2007;81:280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Posch M.G., Gramlich M., Sunde M., Schmitt K.R., Lee S.H.Y., Richter S., Kersten A., Perrot A., Panek A.N., Khatib I.H.A., et al. A gain-of-function TBX20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. J. Med. Genet. 2010;47:230–235. doi: 10.1136/jmg.2009.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pashmforoush M., Lu J.T., Chen H., Amand T.S., Kondo R., Pradervand S., Evans S.M., Clark B., Feramisco J.R., Giles W., et al. Nkx2–5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 9.Garg V., Muth A.N., Ransom J.F., Schluterman M.K., Barnes R., King I.N., Grossfeld P.D., Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 10.Holm H., Gudbjartsson D.F., Arnar D.O., Thorleifsson G., Thorgeirsson G., Stefansdottir H., Gudjonsson S.A., Jonasdottir A., Mathiesen E.B., Njolstad I., et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat. Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 11.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sotoodehnia N., Isaacs A., de Bakker P.I., Dorr M., Newton-Cheh C., Nolte I.M., van der Harst P., Muller M., Eijgelsheim M., Alonso A., et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 2010;42:1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian L., Mohapatra B., Akasaka T., Liu J., Ocorr K., Towbin J.A., Bodmer R. Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc. Natl. Acad. Sci. USA. 2008;105:19833–19838. doi: 10.1073/pnas.0808705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen T., Aneas I., Sakabe N., Dirschinger R., Wang G., Smemo S., Westlund J., Cheng H., Dalton N., Gu Y., et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J. Clin. Invest. 2011;121:4640–4654. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stennard F.A., Costa M.W., Elliott D.A., Rankin S., Haast S.J.P., Lai D., McDonald L.P.A., Niederreither K., Dolle P., Bruneau B.G., et al. Cardiac T-box factor Tbx20 directly interacts with Nkx2–5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev. Biol. 2003;262:206–224. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 16.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vo N., Goodman R.H. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 19.He A., Kong S.W., Ma Q., Pu W.T. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. USA. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visel A., Blow M.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryne J.C., Valen E., Tang M.-H.E., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic. Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newburger D.E., Bulyk M.L. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frith M.C., Fu Y., Yu L., Chen J.-F., Hansen U., Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo J., Wu S., Liu J., Li Y., Yang H., Kim T., Zhelyabovska O., Ding G., Zhou Y., Yang Y., et al. Conditional PPARgamma knockout from cardiomyocytes of adult mice impairs myocardial fatty acid utilization and cardiac function. Am. J. Transl. Res. 2010;3:61–72. [PMC free article] [PubMed] [Google Scholar]
- 25.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Lone R., Knorr K., Jaffe I.Z., Schaffer M.E., Martini P.G., Karas R.H., Bienkowska J., Mendelsohn M.E., Hansen U. Estrogen receptors alpha and beta mediate distinct pathways of vascular gene expression, including genes involved in mitochondrial electron transport and generation of reactive oxygen species. Mol. Endocrinol. 2007;21:1281–1296. doi: 10.1210/me.2006-0497. [DOI] [PubMed] [Google Scholar]
- 27.Chong J.A., Tapia-Ramírez J., Kim S., Toledo-Aral J.J., Zheng Y., Boutros M.C., Altshuller Y.M., Frohman M.A., Kraner S.D., Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 28.Schoenherr C.J., Anderson D.J. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 29.Kuwahara K., Saito Y., Takano M., Arai Y., Yasuno S., Nakagawa Y., Takahashi N., Adachi Y., Takemura G., Horie M., et al. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 2003;22:6310–6321. doi: 10.1093/emboj/cdg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macindoe I., Glockner L., Vukasin P., Stennard F.A., Costa M.W., Harvey R.P., Mackay J.P., Sunde M. Conformational stability and DNA binding specificity of the cardiac T-box transcription factor Tbx20. J. Mol. Biol. 2009;389:606–618. doi: 10.1016/j.jmb.2009.04.056. [DOI] [PubMed] [Google Scholar]
- 31.Badis G., Berger M.F., Philippakis A.A., Talukder S., Gehrke A.R., Jaeger S.A., Chan E.T., Metzler G., Vedenko A., Chen X., et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu M., Riva L., Xie H., Schindler Y., Moran T.B., Cheng Y., Yu D., Hardison R., Weiss M.J., Orkin S.H., et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 34.Gitter A., Siegfried Z., Klutstein M., Fornes O., Oliva B., Simon I., Bar-Joseph Z. Backup in gene regulatory networks explains differences between binding and knockout results. Mol. Syst. Biol. 2009;5:276. doi: 10.1038/msb.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon Y.-S., Garcia-Bassets I., Hutt K.R., Cheng C.S., Jin M., Liu D., Benner C., Wang D., Ye Z., Bibikova M., et al. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc. Natl. Acad. Sci. USA. 2007;104:4852–4857. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le P.P., Friedman J.R., Schug J., Brestelli J.E., Parker J.B., Bochkis I.M., Kaestner K.H. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet. 2005;1:e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beima K.M., Miazgowicz M.M., Lewis M.D., Yan P.S., Huang T.H.M., Weinmann A.S. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J. Biol. Chem. 2006;281:11992–12000. doi: 10.1074/jbc.M513613200. [DOI] [PubMed] [Google Scholar]
- 38.Lopaschuk G.D., Jaswal J.S. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 2010;56:130–140. doi: 10.1097/FJC.0b013e3181e74a14. [DOI] [PubMed] [Google Scholar]
- 39.Berger J.P., Akiyama T.E., Meinke P.T. PPARs: therapeutic targets for metabolic disease. Trends Pharmacol. Sci. 2005;26:244–251. doi: 10.1016/j.tips.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Miller S.A., Huang A.C., Miazgowicz M.M., Brassil M.M., Weinmann A.S. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R Development Core Team. 2009 http://www.R-project.org. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 45.Valouev A., Johnson D.S., Sundquist A., Medina C., Anton E., Batzoglou S., Myers R.M., Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruitt K.D., Tatusova T., Klimke W., Maglott D.R. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taher L., Ovcharenko I. Variable locus length in the human genome leads to ascertainment bias in functional inference for non-coding elements. Bioinformatics. 2009;25:578–584. doi: 10.1093/bioinformatics/btp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smit A.F.A., Hubley R., Green P. RepeatMasker Open-3.0. 1996–2010. http://www.repeatmasker.org. (date last accessed on 13 February, 2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.