Abstract
Mutations leading to expansion of a poly-glutamine track in Huntingtin (Htt) cause Huntington's disease (HD). Signs of endoplasmic reticulum (ER) stress have been recently reported in animal models of HD, associated with the activation of the unfolded protein response (UPR). Here we have investigated the functional contribution of ER stress to HD by targeting the expression of two main UPR transcription factors, XBP1 and ATF4 (activating transcription factor 4), in full-length mutant Huntingtin (mHtt) transgenic mice. XBP1-deficient mice were more resistant to developing disease features, associated with improved neuronal survival and motor performance, and a drastic decrease in mHtt levels. The protective effects of XBP1 deficiency were associated with enhanced macroautophagy in both cellular and animal models of HD. In contrast, ATF4 deficiency did not alter mHtt levels. Although, XBP1 mRNA splicing was observed in the striatum of HD transgenic brains, no changes in the levels of classical ER stress markers were detected in symptomatic animals. At the mechanistic level, we observed that XBP1 deficiency led to augmented expression of Forkhead box O1 (FoxO1), a key transcription factor regulating autophagy in neurons. In agreement with this finding, ectopic expression of FoxO1 enhanced autophagy and mHtt clearance in vitro. Our results provide strong evidence supporting an involvement of XBP1 in HD pathogenesis probably due to an ER stress-independent mechanism involving the control of FoxO1 and autophagy levels.
INTRODUCTION
The accumulation of misfolded proteins in the brain is linked to the occurrence of several neurodegenerative disorders, including Alzheimer's, Parkinson's, Huntington's disease (HD), amyotrophic lateral sclerosis (ALS) and prion-related disorders (1). HD is an autosomal dominant neurodegenerative disease characterized by motor abnormalities, and onset of psychiatric symptoms and dementia in early- to mid-adult life. A glutamine expansion of approximately more than 40 repeats within the Huntingtin (Htt) protein confers a dominant gain of toxic function, leading to progressive accumulation of misfolded mutant Htt (mHtt) in the form of intracellular oligomers and inclusions, and to neuronal loss in the striatum (reviewed in 2–4). HD represents one of a growing number of polyglutamine (polyQ) repeat diseases that cause region-specific neuronal degeneration, including spinobulbar muscular atrophy, spinocerebellar ataxias, Machado–Joseph disease and many other diseases (5).
Although the mechanisms through which mHtt expression is deleterious to neuronal function are still highly controversial, recent data in cellular and animal models of HD suggest a direct correlation between disease progression and the occurrence of endoplasmic reticulum (ER) stress (reviewed in 3). ER stress is triggered by a number of conditions that interfere with oxidative protein-folding process in the ER, leading to the accumulation of abnormally folded proteins (6). ER stress engages the unfolded protein response (UPR), an integrated signal transduction pathway that increases the protein-folding capacity and quality control of the ER to reduce the unfolded protein load, or to trigger apoptosis to eliminate irreversibly damaged cells (7,8). The UPR is initiated by the activation of three specialized stress sensors, including IRE1α (inositol-requiring transmembrane kinase/endonuclease), PERK (PKR-like ER kinase) and ATF6 (activating transcription factor 6) (9). IRE1α is a Ser/Thr protein kinase and endoribonuclease that, upon activation, controls the processing of the mRNA encoding the transcriptional factor X-Box-binding protein 1 (XBP1). This event splices out a fragment of 26 nucleotides, changing the reading frame of XBP1 to encode a potent transcription factor (termed XBP1s) (10,11). XBP1s upregulates genes related to protein folding, quality control, ER translocation, ER-mediated degradation (ERAD), among other processes (12,13). Activated PERK inhibits protein translation into the ER through the inactivation of the initiation factor eIF2α, decreasing the overload of misfolded proteins (14). Phosphorylation of eIF2α also allows the expression of ATF4 (activating transcription factor 4), a transcription factor that upregulates UPR genes that function in amino acid and redox metabolism, folding, autophagy and apoptosis (15–17).
Increased expression of downstream UPR effectors including BiP, CHOP and HERP was reported at the mRNA level in human post-mortem HD samples (18). Similarly, some signs of ER stress were observed in two HD mouse models even at early stages of the disease (18,19). Small molecules that target the ER foldase PDI were recently shown to prevent the neurotoxicity of mHtt fragments in vitro (20). In addition, altered ER calcium homeostasis was reported in HD mouse models (21). Attempts to understand the function of wild-type Htt demonstrated that the inhibition of its expression drastically alters the structure of the ER network and trafficking (22), suggesting that its normal biologic function is related to this organelle. Early cellular studies demonstrated that expression of mHtt or expanded polyQ peptides leads to ER stress-mediated apoptosis in cellular models of HD (23–29), whereas a recent report did not detect the engagement of ER stress in cells expressing mHtt (30). At the mechanistic level, the occurrence of ER stress may be related to the impairment of ERAD, leading to the accumulation of misfolded proteins inside the ER (24,30,31). Remarkably, another report suggests that processing of ATF6α is impaired in both animal models and in post-mortem tissue from HD patients (32), which may reduce the ability of neurons to adapt to ER stress. Activation of the PERK/eIF2α UPR branch triggers the degradation of polyQ peptides by macroautophagy (here referred to as autophagy) in vitro (27), a protein degradation pathway suggested relevant for clearance of HD-linked aggregates through lysosome-mediated degradation (33–36). Htt has a membrane association domain capable of partially targeting the protein to the ER and late endosomes as well as autophagic vesicles (37–39). We reported that autophagy activity is partially impaired in mHtt-expressing neurons in part due to a failure of autophagosomes (APG) to recognize their cargos (39), which may lead to general alterations in protein homeostasis. Although disease progression and mHtt aggregation correlate with the engagement of ER stress responses, the actual characterization of UPR signaling in HD in vivo is still incomplete, and the role of the pathway in the disease process has not been addressed directly.
Here we demonstrate that silencing XBP1 expression in the full-length mHtt transgenic mouse strain YAC128 reduces neuronal loss in the striatum and improves motor performance. Cellular studies indicate that these protective effects are related to a strong decrease in mHtt accumulation due to enhanced autophagy. Similar effects on mHtt levels were recapitulated in a knock-in mouse model of HD. Unexpectedly, ATF4 deficiency did not alter mHtt levels, and HD progression was not associated with a global ER stress response. At the mechanistic level, we found an upregulation of the transcription factor Forkhead box O1 (FoxO1) in XBP1-deficient cells, which may contribute to autophagy-mediated clearance of mHtt. Our results reveal an unexpected role of XBP1 in controlling a dynamic crosstalk with the FoxO1 and the autophagy pathway to modulate HD pathogenesis.
RESULTS
XBP1 deficiency protects against HD pathogenesis in the YAC128 mouse model
To establish the contribution of XBP1 to HD in vivo, we generated an XBP1 conditional knockout model where xbp1 was deleted in the nervous system, using the Nestin-Cre system (XBP1Nes−/−) on a C57BL/6 genetic background (40). We cross-bred this strain with the YAC128 HD mouse model on a heterozygous background (XBP1Nes−/−-mHttQ128) to resemble the genetic alterations observed in humans. This transgenic HD model expresses the entire human Htt gene with 128 CAG repeats, spanning the entire genomic region of the human HD gene, including promoter, intronic, upstream and downstream regulatory elements (41). The disease progression of this HD mouse model is associated with a slow Htt aggregation process, accompanied by striatal neuron loss and motor impairment (41).
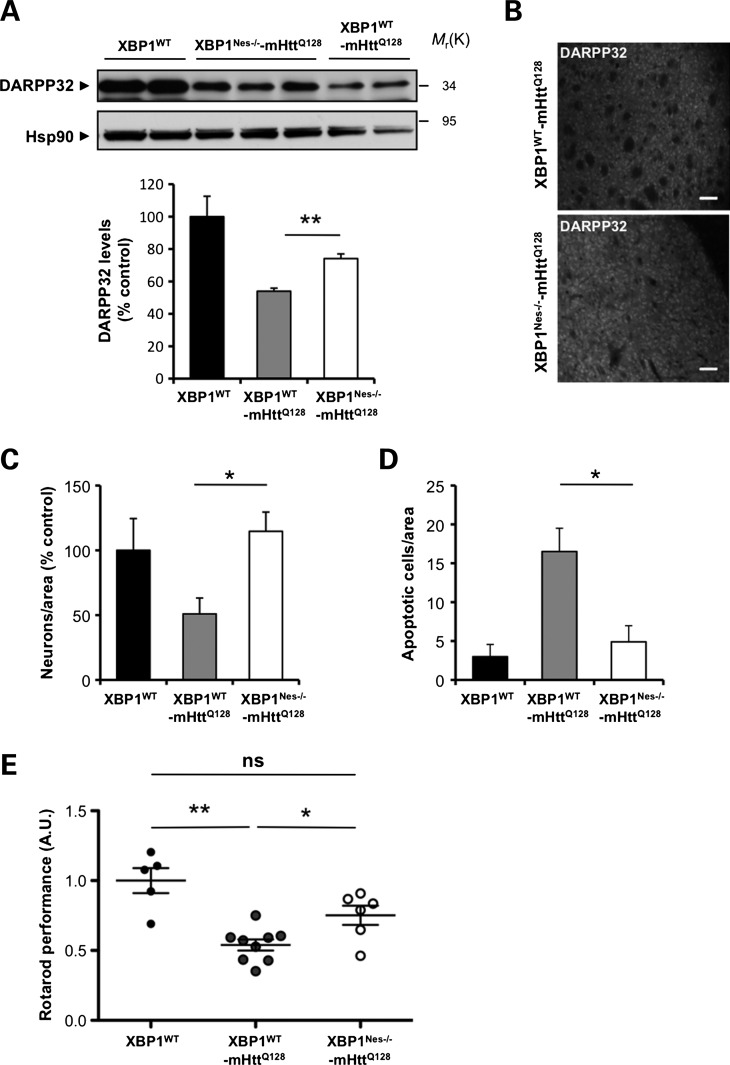
To determine the impact of XBP1 deficiency on neuronal survival, we first monitored the levels of the dopamine-related protein DARPP32 in protein extracts of the dissected striatum. For all biochemical and histologic analysis, littermate controls were employed. As previously reported (42), mHtt transgenic mice presented a decrease of DARPP32 expression, which was partially attenuated in XBP1Nes−/−-mHttQ128 mice at 6 months of age (Fig. 1A and B). We also examined the rate of neuronal survival in YAC128 animals, using electron microscopy (EM) and morphologic analysis in an area of the striatum near the ventricle (Supplementary Material, Fig. S1). This particular region showed significant neuronal loss at 6 months of age when compared with the rest of the striatum that was not significantly affected (Fig. 1C; data not shown). In contrast, XBP1Nes−/−-mHttQ128 animals were markedly protected against neuronal loss, with increased neurons with normal shape when compared with XBP1WT-mHttQ128 animals. In addition, the percentage of neuronal apoptosis was drastically decreased in XBP1Nes−/−-mHttQ128 when compared with control mHttQ128 mice as measured by the presence of classical apoptosis features, including nuclear condensation, fragmentation and cell shrinkage (Fig. 1D, and Supplementary Material, Fig. S1).
Figure 1.
XBP1 deficiency in the nervous system protects against experimental HD. (A) DARPP32 levels were analyzed in the striatum of 6-month-old mice by western blot analysis. Hsp90 was monitored as loading control. Lower panel: The levels of DARPP32 were quantified from XBP1WT (n= 3), XBP1WT-mHttQ128 (n= 5) and XBP1Nes−/−-mHttQ128 (n= 7) mice (left panel). Mean and SEM are presented. (B) Immunohistochemistry analysis of DARPP32 levels in the striatum of 6-month-old mice. Scale bar, 50 μm. Quantification of the number of neurons (C) and (D) apoptotic cells was performed in 10 different regions of the striatum; each region corresponds to 0.08 mm2. Four independent animals were analyzed for each group. (E) Motor performance was monitored with the rotarod assay in XBP1 wild-type or deficient animals bred onto the YAC128 HD mouse model (XBP1WT-mHttQ128 or XBP1Nes−/−-mHttQ128, respectively; 6–9 animals per group) in 4-month-old animals. Rotarod values were normalized to the performance of XBP1WT mice as a value of 1. Mean and SEM are presented. *P< 0.05 and **P< 0.01, calculated with Student's t-test. In all experiments, littermates were employed for comparison.
To assess the impact of XBP1 on neuronal function, we then monitored the motor performance of YAC128 mice in control or XBP1Nes−/− mice, using a previously described protocol validated in the YAC128 model (43). XBP1 deficiency led to a significant improvement in motor performance of mHtt transgenic mice (Fig. 1E) that was sustained over time (Supplementary Material, Fig. S2A). XBP1 deficiency was confirmed by quantitative PCR (Supplementary Material, Fig. S2B), in addition to monitor Cre expression in neurons in the striatum, using the ROSA26 system (44) (data not shown).
Reduced mHtt levels in the striatum of XBP1Nes−/− HD transgenic mice, but not ATF4-deficient animals
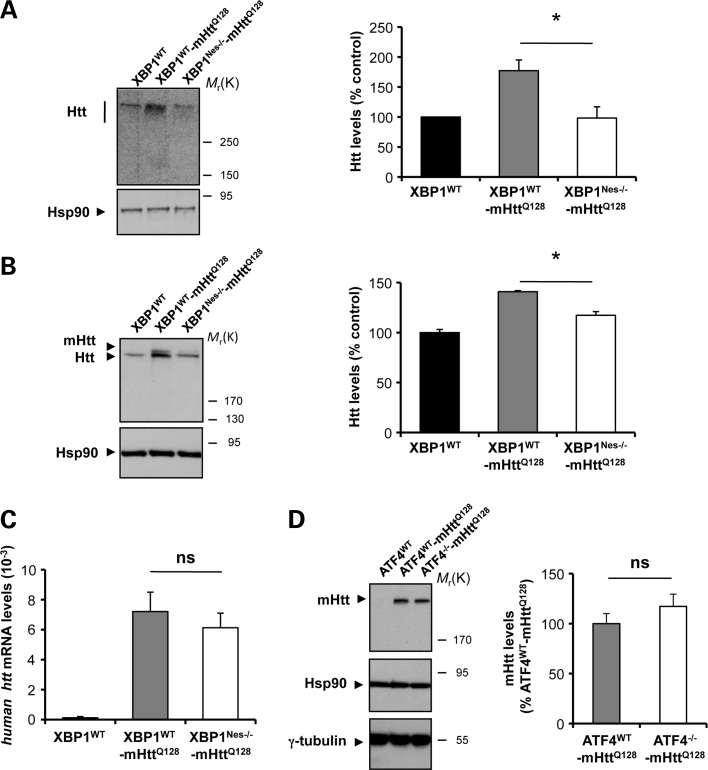
Analysis of early symptomatic mice (6-month-old animals) revealed that XBP1 deficiency resulted in a marked decrease in total full-length Htt levels in the striatum as monitored by western blot analysis (Fig. 2A). Remarkably, heterozygous YAC128 mice displayed a 1.6-fold increase in Htt levels that was reversed in XBP1Nes−/−-mHttQ128 mice (Fig. 2A). This phenotype was observed in both male and female mice (Supplementary Material, Fig. S3). To test whether or not XBP1 deficiency specifically altered the levels of mHtt, we used an antibody that recognizes a different region of human Htt that resolves both mutant and wild-type Htt on western blots (clone mAb2166) (45) (Fig. 2B). In agreement with our previous observations, a significant decrease in mHtt levels was observed in XBP1Nes−/−-mHttQ128 mice when compared with control transgenic mice (Fig. 2B). We confirmed this result using an antibody that only recognizes mHtt (anti-polyQ clone 3B5H10; data not shown). As a complementary readout, we monitored the mRNA levels of the human mHtt transgene, using real-time PCR. Surprisingly, no significant differences were observed in the mRNA levels between XBP1Nes−/−-mHttQ128 and XBP1WT-mHttQ128 mice in striatum brain samples (Fig. 2C). As additional control, we sequenced the poly-glutamine region of the mHtt transgene in XBP1Nes−/−-mHttQ128 and control animals, and did not observe any alteration of the CAG length in either genetic background (Supplementary Material, Table S1A). Taken together, these results suggest that XBP1 deficiency leads to a reduction in mHtt expression at the post-translational level and does not affect the stability of the expanded CAG region.
Figure 2.
XBP1 deficiency in the nervous system decreases mHtt levels in the YAC128 HD model. (A) Levels of Huntingtin (Htt) were measured in the striatum of 6-month-old mice by western blot analysis using the anti-Htt clone mEM48 antibody. XBP1WT animals were analyzed as control. Hsp90 was monitored as loading control. Htt levels were quantified in striatum extracts from XBP1WT (n= 3), XBP1WT-mHttQ128 (n= 8) and XBP1Nes−/−-mHttQ128 (n= 13) mice (left panel). Mean and SEM are presented. (B) In parallel, Htt relative levels were determined using anti-Htt clone 1HU-4C8 antibody. Htt levels were quantified in the brain striatum of XBP1WT (n= 3), XBP1WT-mHttQ128 (n= 3) and XBP1Nes−/−-mHttQ128 (n= 3) mice (left panel). Mean and SEM are presented. *P< 0.05, calculated with Student's t-test. (C) The mRNA level of the human huntingtin gene was analyzed by real-time PCR in total cDNA obtained from the brain striatum of XBP1Nes−/−-mHttQ128 or littermate control mice. All samples were normalized to β-actin levels. Average and SEM of the analysis of three animals per group are shown. (D) mHtt levels were analyzed using anti-polyQ clone 3B5H10 antibody in the striatum of 12-month-old mice (upper panel). Right panel: mHtt levels were quantified in ATF4WT (n= 3), ATF4WT-mHttQ128 (n= 4) and ATF4−/−-mHttQ128 (n= 4) (bottom panel). In addition to Hsp90, γ-tubulin levels were analyzed as loading control. In all experiments, littermates were employed for comparison.
To determine the possible impact of other UPR branches to HD severity, we also generated mHtt transgenic mice deficient for the transcription factor ATF4. Analysis of 1-year-old mice revealed no effects on mHtt levels in ATF4−/−-mHttQ128 mice when compared with control ATF4WT-mHttQ128 animals, using the 3B5H10 antibody (Fig. 2D). Similar results were observed when two additional anti-Htt antibodies (clone mEM48 and clone mAb2166) were employed (data not shown), indicating that the effects of XBP1 on mHtt levels are specific for the IRE1α branch of the UPR. Thus, despite our initial prediction that ablation of xbp1 and atf4 expression would accelerate neuronal dysfunction and disease severity, we observed significant protection against HD pathogenesis only in XBP1Nes−/− animals associated with a drastic reduction in mHtt levels, and improved motor performance and neuronal viability.
Ablation of XBP1 in the nervous system decreases mHtt levels in a knock-in model of HD
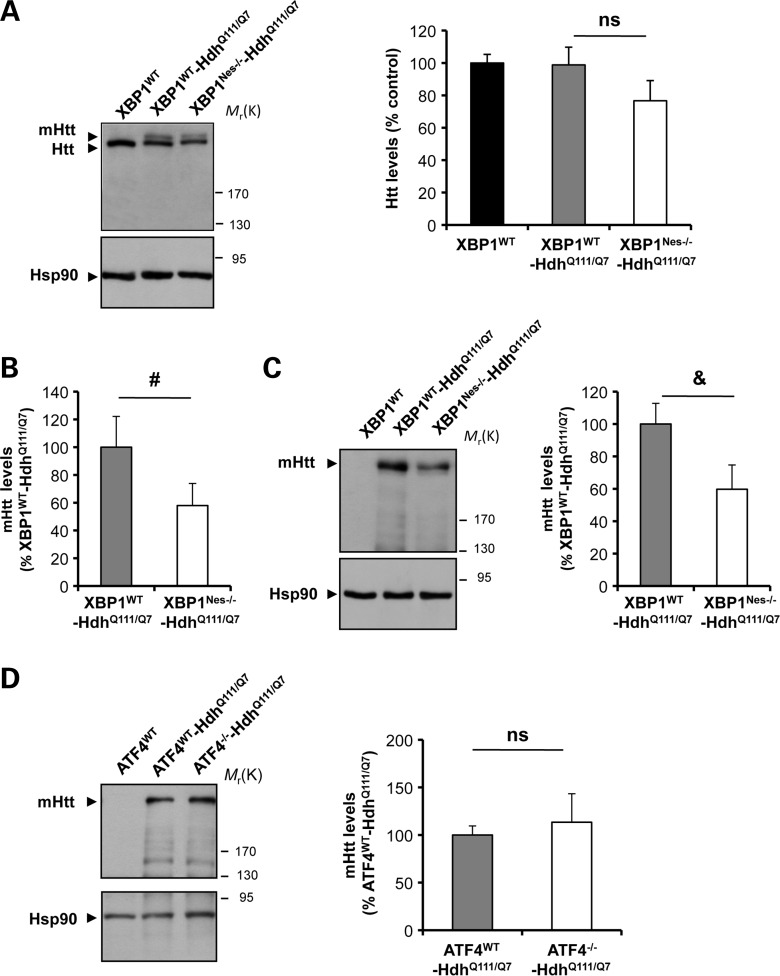
Our previous results suggest that targeting XBP1 decreases the levels of mHtt overexpression in the YAC128 mice, correlating with enhanced neuronal survival and improved motor performance. To test the possible impact of XBP1 deficiency on the levels of mHtt under conditions that do not involve overexpression, we bred XBP1Nes−/− mice with a heterozygous knock-in mouse model of HD expressing mHtt with 111 glutamines (HdhQ111/Q7) (46). This model develops slow progressive histopathology with the accumulation of mHtt inclusions and insoluble aggregates only at 10–15 months of age, without significant changes in motor function or neuronal viability (46). Analysis of Htt expression levels using the mAb2166 antibody in 6-month-old mice revealed a decrease of 20% in Htt total levels (Fig. 3A). Quantification of mHtt levels indicated an average decrease of ∼40% in XBP1Nes−/−-HdhQ111/Q7 when compared with control HdhQ111/Q7 animals (Fig. 3B). Virtually identical results were obtained when mHtt expression was quantified using an anti-polyQ antibody (Fig. 3C). As control, we also monitored the stability of the CAG repeat sequence and did not observe significant differences between control HdhQ111/Q7 and XBP1Nes−/−-HdhQ111/Q7 animals (Supplementary Material, Table S1B). Finally, to further test the specificity of the XBP1 branch on controlling the expression of mHtt, we also generated ATF4−/−-HdhQ111/Q7 animals. Our results indicate no changes in mHtt levels in ATF4-deficient animals (Fig. 3D), confirming our results in the YAC128 model. Thus, using two independent HD animal models, we observed decreased expression of mHtt upon the deletion of xbp1 in the nervous system.
Figure 3.
mHtt levels in an HD knock-in model on an XBP1-deficient background. (A) XBP1Nes−/− mice were bred with a knock-in heterozygous HD mouse model (HdhQ111/Q7). Levels of Huntingtin (Htt) and mHtt were measured in the striatum of 6-month-old mice by western blot analysis using the anti-Htt clone 1HU-4C8 antibody (left panel). XBP1WT animals were analyzed as control. Hsp90 was monitored as loading control. Total Htt levels were quantified in striatum extracts from XBP1WT (n= 6), XBP1WT-HdhQ111/Q7 (n= 7) and XBP1Nes−/−-HdhQ111/Q7 (n= 6) mice (right panel). (B) The band corresponding to mHtt levels was quantified from experiments presented in (A) to calculate the relative levels normalized with the expression of HdhQ111/Q7 control mice. (C) In parallel, mHtt relative levels were determined using anti-polyQ clone 3B5H10 antibody, and relative mHtt levels were quantified (left panel). Mean and SEM are presented. *P< 0.05, calculated with Student's t-test. (D) ATF4−/− mice were bred with a knock-in heterozygous HD mouse model (HdhQ111/Q7). Levels of Htt and mHtt were measured in the striatum of 6-month-old mice by western blot analysis using the anti-polyQ clone 3B5H10 antibody. Mean and SEM are presented of the analysis of ATF4WT (n= 3), ATF4WT-HdhQ111/Q7 (n= 6) and ATF4−/−-HdhQ111/Q7(n= 5). In all panels, statistical analysis was performed with Student's t-test. #P= 0.07, &P= 0.06.
Silencing XBP1 decreases the accumulation of mHtt aggregates
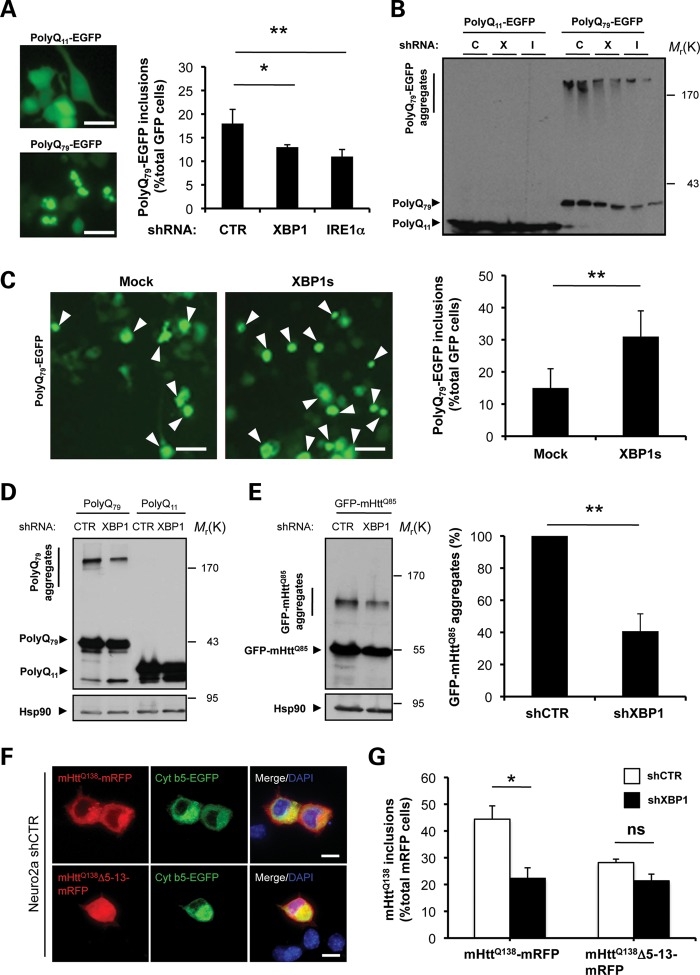
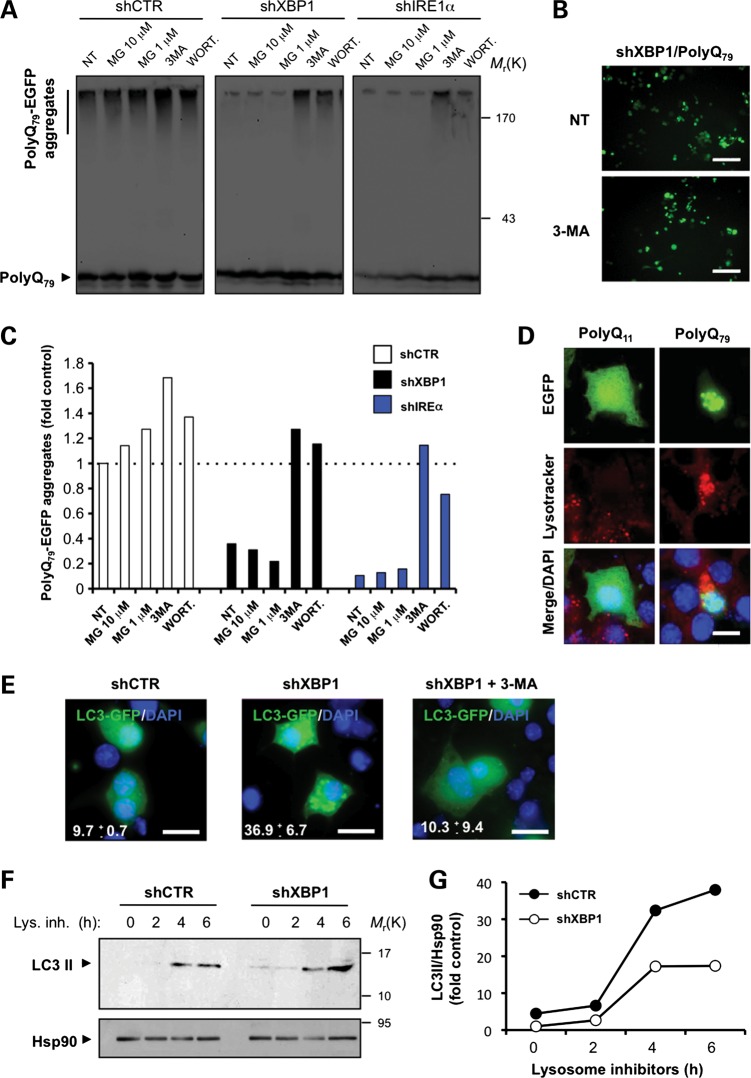
To define the mechanism underlying the decrease in mHtt protein levels by XBP1 deficiency in vivo, we performed cellular studies on several models of HD. Using lentiviruses, we introduced small hairpin RNA (shRNA) sequences to reduce the expression of IRE1α (shIRE1α) and XBP1 (shXBP1) in two neuronal cell lines, NSC34 and Neuro2a (Supplementary Material, Fig. S4A and B; data not shown). We monitored the aggregation of a peptide of 79 polyQs and a control sequence of 11 glutamines as EGFP-fusion proteins, here termed polyQ79-EGFP and polyQ11-EGFP, respectively. We transiently expressed these constructs and examined the accumulation of intracellular polyQ79-EGFP inclusions by fluorescent microscopy. Surprisingly, reduced generation of intracellular polyQ79-EGFP inclusions was observed in XBP1 and IRE1α knockdown NSC34 cells (Fig. 4A). To complement these observations, we monitored the accumulation of monomeric, high molecular weight and detergent-insoluble polyQ79-EGFP species in these cells by western blot analysis in 4–12% gradient gels. A drastic reduction in the aggregation of pathologic polyQ79-EGFP peptides was observed in shXBP1 and shIRE1α cells (Fig. 4B). In addition, a slight reduction of the monomeric polyQ79-EGFP was observed. Conversely, we also investigated the effects of XBP1s overexpression, the active form of the transcription factor, in polyQ79-EGFP aggregation. After co-transfection of an XBP1s expression vector with polyQ79-EGFP constructs, we detected increased generation of intracellular inclusions in NSC34 cells (Fig. 4C).
Figure 4.
XBP1/IRE1α deficiency in neuronal cells reduces abnormal mHtt aggregation. (A) NSC34 motoneuron-like cells were stably transduced with lentiviral vectors expressing shRNA against XBP1, IRE1α or control luciferase mRNA (shXBP1, shIRE1α and shCTR, respectively), followed by transient transfection with expression vectors for polyQ79-EGFP and polyQ11-EGFP as control. After 72 h, polyQ79-EGFP intracellular inclusions were quantified by fluorescent microscopy (right panel). Left panel: The number of cells displaying intracellular inclusions was quantified in a total of at least 300 cells per experiment. Results are representative of four independent experiments. Average and standard deviation are presented. Scale bar, 20 μm. (B) In parallel, detergent-insoluble polyQ79-EGFP aggregates were measured in cell extracts prepared in Triton-X100 buffer followed by western blot analysis in 4–12% polyacrylamide gradient gels. shRNA cells: Control (C), XBP1 (X) or IRE1α (I). Results are representative of at least four independent experiments. Two independent experiments per condition are presented. (C) NSC34 cells were co-transfected with an expression vector for XBP1s or empty vector (mock) in the presence of a polyQ79-EGFP expression vector. After 72 h, polyQ79-EGFP intracellular inclusions (right panel) were quantified by fluorescent microscopy (arrowheads). Left panel: The number of cells displaying intracellular inclusions was quantified in at least 300 cells per experiment. Results are representative of four independent experiments. Mean and SEM are presented. Scale bar, 20 μm. (D) Detergent-insoluble polyQ79-EGFP aggregates were measured in cell extracts prepared in Triton X-100 buffer from Neuro2A shXBP1 or shCTR cells. As control, polyQ11-EGFP was expressed in both cell lines. Levels of Hsp90 served as loading control. (E) Neuro2A shXBP1 or shCTR cells were transiently transfected with expression vectors for human mHtt (exon 1) GFP-mHttQ85. After 72 h, GFP-mHttQ85 aggregates were measured in cell extracts prepared in Triton X-100 and analyzed by western blot. Levels of Hsp90 served as loading control. Left panel: mHtt aggregates were quantified and the results are representative of four independent experiments. Mean and SEM are presented. (F) shCTR Neuro2a cells were transiently transfected with mHttQ138-mRFP or mHttQ138Δ5-13-mRFP constructs in the presence of cytochrome b5-EGFP (Cyt b5-EGFP) to label the ER. After 24 h, cells were visualized by confocal microscopy. Nucleus was stained with DAPI. Scale bar, 20 μm. (G) Neuro2A shXBP1 (black bar) or shCTR (white bar) cells were transiently transfected with mHttQ138-mRFP or mHttQ138Δ5-13-mRFP. After 48 h, mHtt intracellular inclusions were quantified in at least 200 cells per experiment. Mean and SEM are presented. *P< 0.05 and **P< 0.01, calculated with Student's t-test.
We also corroborated our results in Neuro2a cells after knocking down XBP1. Similar to the results obtained in NSC34 cells, silencing XBP1 in Neuro2a cells also decreased the accumulation of polyQ79-EGFP intracellular inclusions (Supplementary Material, Fig. S4C) and levels of protein aggregation (Fig. 4D). Finally, to increase the relevance of these findings to HD, we monitored the aggregation levels of a 171 amino acid N-terminal Htt fragment containing an 85 glutamine expansion fused with GFP (GFP-mHttQ85) construct in shXBP1 cells, using western blot analysis. A 60% reduction in the accumulation of high molecular weight GFP-mHttQ85 species was observed in shXBP1 cells (Fig. 4E), reinforcing the observations obtained by polyQ79-EGFP expression. A slight decrease in the monomeric form of GFP-mHttQ85 was also observed in the analysis. Taken together, these results demonstrate an unexpected role of the IRE1α/XBP1 branch of the UPR in controlling mHtt levels.
Control of mHtt aggregation by XBP1 expression requires the Htt membrane localization signal
The association of Htt to the ER membrane is mediated by the first 18 amino acids, and this association is affected by ER stress (37,38). Mutations in this region lead to the reduction of mHtt aggregation (38). In order to test the potential role of N-terminal membrane-targeting sequence of Htt on the functional effects of XBP1 silencing, we used a previously described Htt construct that includes the region 1–588 amino acids, containing an expansion of 138 glutamines, fused with monomeric RFP (mHttQ138-mRFP) and compared it with a deletion mutant of amino acids 5–13 (mHttQ138Δ5–13-mRFP). As previously described, expression of mHttQ138-mRFP leads to cytosolic distribution, partially co-localizing with the ER marker cytochrome b5-GFP (Fig. 4F). mHttQ138-mRFP aggregation was reduced by knocking down XBP1 (Fig. 4G). As predicted, the generation of mHtt intracellular inclusions was partially reduced by mutations in its N-terminal region when expressed in control cells (Fig. 4F). Interestingly, the basal level of mHttQ138Δ5-13-mRFP aggregation was similar in both shCTR and shXBP1 cells, suggesting that the mHtt membrane localization signal is involved in the protective effects of XBP1 deficiency.
Autophagy-mediated degradation of abnormal polyQ tracks in XBP1-deficient neurons
Diminished mHtt aggregation in XBP1 knockdown cells might be explained by the upregulation of protein degradation pathways involved in polyQ clearance. Both the proteasome and macroautophagy (34,47) have been shown to mediate mHtt degradation in cellular and animal models of HD (34,35). XBP1 down-regulation also enhances autophagy (48,49). To define the contribution of protein degradation pathways to polyQ79-EGFP clearance, we treated shXBP1 and shIRE1 cells with a proteasome inhibitor (MG-132) or phosphatidylinositol-3 (PI3) kinase inhibitors [3-methyladenine (3-MA), and Wortmannin], which block an early step controlling APG formation (47,50), thereby inhibiting autophagy. Blocking PI3-kinases resulted in more polyQ79-EGFP aggregation than proteasome inhibition, with a marked recovery of pathogenic polyQ aggregation in knockdown cells (Fig. 5A and C). Similarly, augmented accumulation of polyQ79-EGFP intracellular inclusions was observed after the treatment of shXBP1 cells with 3-MA (Fig. 5B), whereas proteasome inhibition led only to a slight accumulation of polyQ79-EGFP aggregates in control cells.
Figure 5.
XBP1 deficiency leads to autophagy-mediated degradation of mHtt in cellular models of HD. (A) NSC34 shXBP1, shIRE1α or shCTR cells were transfected with an expression vector for polyQ79-EGFP, and after 48 h cells were treated with MG132 (10 and 1 μm) for 8 h, or with 10 mm 3-MA or 10 μm wortmannin for 16 h and polyQ79-EGFP aggregation analyzed by western blot in 4–12% polyacrylamide gradient gels. (B) Examples of polyQ79-EGFP intracellular inclusions in shXBP1 cells untreated or treated with 3-MA are presented, visualized by fluorescent microscopy. Scale bar, 50 μm. (C) PolyQ79-EGFP aggregates observed in (A) were quantified in each treatment and for comparison normalized to the value obtained in non-treated shCTR cells. (D) NSC34 cells were transiently transfected with expression vectors for polyQ11-EGFP and polyQ79-EGFP. After 24 h, cells were stained with lysotracker and DAPI, and the co-localization with polyQ79-EGFP intracellular inclusions was determined by confocal microscopy. Scale bar, 10 μm. (E) NSC34 shCTR or shXBP1 cells were transiently transfected with expression vectors for LC3-EGFP and stained with DAPI and visualized by confocal microscopy. After 48 h, cells were treated with 10 mm 3-MA for 16 h. Scale bar, 20 μm. (F) To monitor LC3 flux through the autophagy pathway in Neuro2A shCTR or shXBP1, cells were treated or not with a lysosome inhibitor cocktail (lys. inh.) containing 200 nm bafilomycin A1, 10 μg/ml pepstatin and 10 μg/ml E64d for indicated time points and endogenous LC3 levels monitored by western blot. Hsp90 levels served as loading control. (G) Kinetics of the accumulation of LC3-II protein showed in (F) was quantified and normalized to Hsp90 levels and then to non-treated shCTR cells.
APG fuse with lysosomes to form autophagolysosomes (APGL) in which their content is degraded (34,47), and increased lysosomal biogenesis has been recently linked to autophagy induction (51). To study the possible role of lysosomal compartments in the degradation of polyQ79-EGFP, we first analyzed the content of lysosomes in polyQ79-EGFP expressing cells. A clear enhancement of Lysotracker staining was observed in cells expressing intracellular polyQ79-EGFP inclusions (Fig. 5D and Supplementary Material, Fig. S5). In agreement with reduced levels of polyQ79-EGFP aggregates in shXBP1 cells, increased basal levels of autophagy were observed by monitoring the accumulation of LC3-EGFP-positive dots, which was inhibited by treating cells with 3-MA (Fig. 5E). Similarly, enhanced induction of autophagy was observed by monitoring LC3-II flux by western blot after inhibiting lysosomal activity with a cocktail of 200 nm bafilomycin A1, 10 μg/ml pepstatin and 10 μg/ml E64d (Fig. 5F and G).
As control, we also knocked down ATF4 in Neuro2a cells, using an shRNA construct (Supplementary Material, Fig. S4D). Targeting ATF4 did not affect autophagy levels or polyQ79-EGFP aggregation (Supplementary Material, Fig. S4E and F), consistent with our in vivo experiments in the YAC128 mouse model. Taken together, these results indicate that ablating XBP1 expression specifically induces autophagy, enhancing the degradation of pathologic polyQ peptides and mHtt.
Enhanced autophagy-mediated degradation of mHtt in the brain of XBP1Nes−/− mice
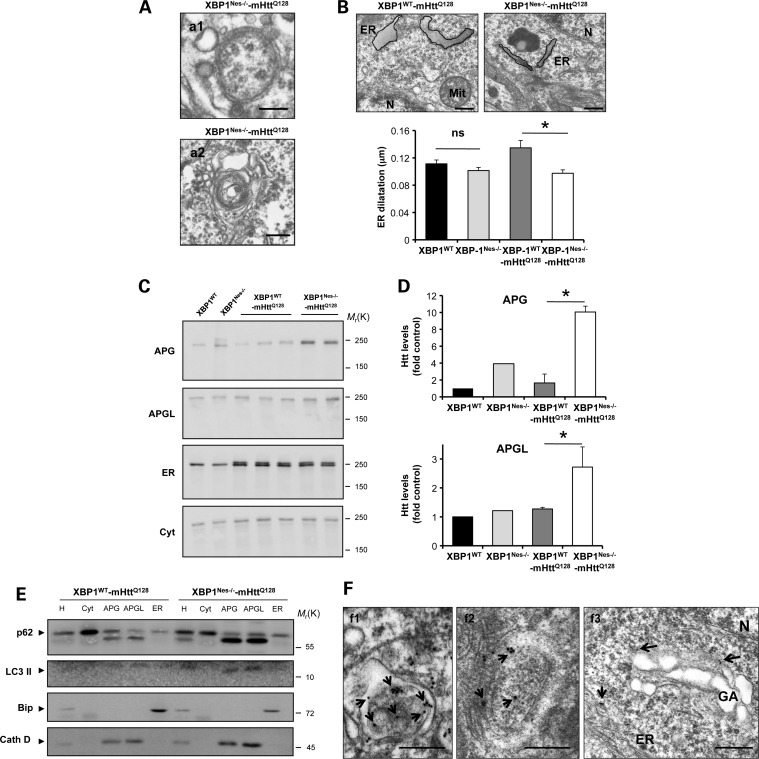
Based on our cellular studies, we monitored the levels of autophagy in XBP1Nes−/−-mHttQ128 animals. This issue is particularly challenging due to the fact that autophagy flux assays are not available for in vivo testing in the nervous system. Through EM ultrastructural analysis of the striatum of 6-month-old mice, we observed an enhanced accumulation of APG, characterized by double-membrane vesicular structures, in addition to the accumulation of multivesicular vacuoles (Fig. 6A). In this ultrastructural analysis, we also noticed the presence of dilated ER in mHttQ128 mice, a phenomena reverted in XBP1Nes−/−-mHttQ128 animals (Fig. 6B). Dilatation of the ER lumen has been described as a pathologic event in other diseases such as ALS (52). To test whether or not mHtt is delivered to APG in vivo in XBP1Nes−/−-mHttQ128 mice, we performed subcellular fractionation experiments. We purified from the brain of 6-month-old mHttQ128 mice fractions enriched in cytosol, ER, APG fraction and APGL fraction. Remarkably, an enhanced accumulation of full-length mHtt was observed in the APG fractions purified from the brain of XBP1Nes−/−-mHttQ128 when compared with control YAC128 transgenic brain samples (Fig. 6C and D). Of note, levels of Htt in APG and APGL fractions of XBP1WT-mHttQ128 and non-transgenic wild-type mice were similar (Fig. 6C and D). In the APGL fraction from XBP1Nes−/−-mHttQ128, Htt levels were reduced compared with APG, suggesting active flux through the pathway leading to mHtt degradation in the lysosome (Fig. 6C and D). We also observed an increase in the levels of LC3-II in APG and APGL fractions from XBP1Nes−/−-mHttQ128 compared with the mHttQ128 control mice fractions (Fig. 6E). The weak signal of LC3-II in these autophagic vesicle-enriched fractions is due to the high tendency to undergo degradation after purification (data not shown).
Figure 6.
Loss of XBP1 targets mHtt to the autophagy pathway. (A) The accumulation of APG-like structures was visualized by EM in neurons of the striatum of XBP1Nes−/−-mHttQ128 mice at 6 months of age. Scale bars, 230 nm (a1) and 600 nm (a2). Images represent the analysis of three independent animals. Neurons were identified by their morphology at low magnification. (B) ER structure was visualized by EM in neurons of the striatum of mice at 6 months of age. Scale bar, 300 nm. Left panel: ER dilatation was quantified from XBP1WT (n= 3), XBP1Nes−/− (n= 3), XBP1WT-mHttQ128 (n= 3) and XBP1Nes−/−-mHttQ128 (n= 3) mice as described in Materials and Methods. Mean and SEM are presented. As an example, the perimeter of the ER was marked with a black line with a gray area. *P<0.05 calculated with Student's t-test. (C) Brain extracts from indicated animals were subjected to subcellular fractionation to purify fractions enriched in different organelles including autophagosomes (APG), autophagolysosomes (APGL), ER and cytosol (cyt). (D) Quantification of Htt levels in the autophagic compartments relative to XBP1WT is shown (XBP1WT-mHttQ128, n= 3; XBP1Nes−/−-mHttQ128, n= 3) using the anti-Htt clone 1HU-4C8 antibody. Mean and SEM are presented. (E) Biochemical characterization of the fractions isolated in (C). Immunoblot for the indicated marker proteins to verify the purity of the fractions was performed for the homogenate (H), cytosol (cyt), autophagosomes (APG), autophagolysosomes (APGL) and endoplasmic reticulum (ER) isolated from cortex and midbrain. (F) Immunogold staining and EM analysis of the brain striatum, using an anti-expanded polyQ antibody (clone 3B5H10) was performed in tissue derived from XBP1Nes−/−-mHttQ128 mice. (f1) Co-localization with ER membrane. Scale bar, 500 nm. (f2) A double-membrane APG-containing positive immunogold staining in its lumen. Scale bar, 300 nm. (f3) An example of co-localization of immunogold staining and Golgi apparatus membrane is shown. Scale bar, 500 nm. Data represent the analysis of three independent animals. Arrows indicate immunogold staining. N, nucleus; GA, Golgi apparatus; ER, endoplasmic reticulum.
To define whether or not mHtt is associated with the internal lumen of the APG or with the external APG membrane, we performed immunogold staining for mHtt and EM studies, using an antibody that recognizes mHtt only but not the wild-type mouse form. Consistent with increased autophagy levels in XBP1Nes−/−-mHttQ128 mice, co-localization of mHtt with the lumen of autophagosomal structures was observed in symptomatic mice (Fig. 6F, panels 1 and 2). In addition, we observed a co-localization of mHtt with or close to other organelle membranes (Fig. 6F, panel 3). Thus, our results indicate that mHtt is present in autophagy-related compartments in vivo in an HD mouse model, a process enhanced by silencing XBP1 expression in the nervous system.
XBP1 deficiency triggers enhanced FoxO1 expression
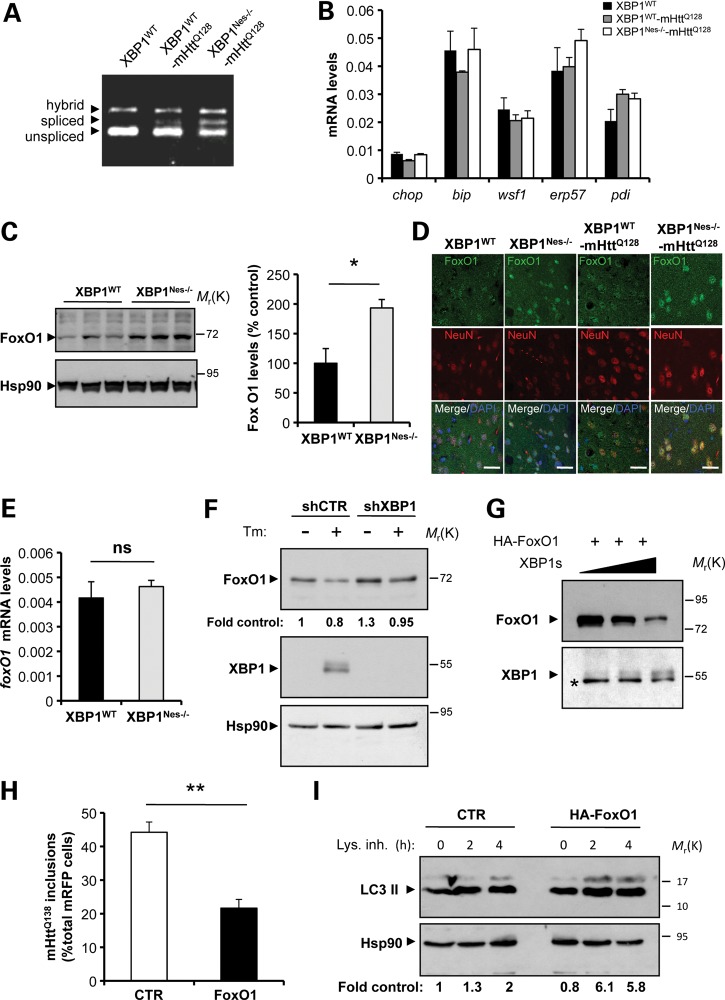
Based on the observation that ATF4 deficiency did not alter mHtt levels, and that targeting XBP1 led to improvement rather than worsening of disease phenotypes as predicted, we examined the levels of UPR target genes in mHtt transgenic mice. Analysis of XBP1 mRNA splicing with two independent assays revealed the activation of the pathway in the striatum of mHtt mice (Fig. 7A and Supplementary Material, Fig. S6A). In addition, XBP1 mRNA splicing was augmented in the mHtt striatum as revealed by processing of the truncated mRNA generated by the delete xbp1 allele (Fig. 7A), which may represent a feedback loop that hyperactivates IRE1α as previously described in other tissues (53,54). Unexpectedly, analysis of classical ER stress markers including bip, chop, wfs1, erp57 and pdi by real-time PCR did not reveal any changes in their mRNA levels in the striatum of mHtt mice, on either an xbp1 wild-type or knockout background (Fig. 7B). Similarly, western blot analysis of ATF4, CHOP, BiP, ERp72, Erp72 and PDI expression did not reveal any changes in their expression in the striatum of mHtt mice (Supplementary Material, Fig. S6B). Thus, these results suggest that although the IRE1α/XBP1 pathway is engaged in the YAC128 HD model, the disease is not associated with a classical ER stress response. This observation is consistent with accumulating evidence suggesting ER stress-independent functions of XBP1 and other UPR components (reviewed in 55).
Figure 7.
XBP1 negatively regulates FoxO1 expression. (A) The levels of XBP1 mRNA splicing were analyzed in the striatum from XBP1Nes−/−-mHttQ128 and littermate control mice at 3 months of age. (B) The mRNA levels of indicated UPR-target genes were measured by real-time PCR in total cDNA obtained from the striatum of four XBP1Nes−/−-mHttQ128 mice or littermate control mice at 6 months of age. All samples were normalized to β-actin levels. Average and SEM of the analysis of three animals per group are shown. (C) FoxO1 levels were analyzed in the striatum of 6-month-old mice by western blot. Hsp90 was used as loading control. Left panel: The relative levels of FoxO1 were quantified from XBP1WT (n= 3) and XBP1Nes−/− (n= 3) mice and normalized with Hsp90 levels. Mean and SEM are presented. (D) The distribution of FoxO1 (green) was analyzed in the striatum of XBP1WT−-mHttQ128, XBP1Nes−/−-mHttQ128, XBP1WT and XBP1Nes−/− animals at 6 months of age. Co-staining with NeuN (red) and DAPI (blue) stain nucleus was performed. Images were visualized with a confocal microscope and represent the analysis of three animals per group. Scale bar, 50 μm. (E) The mRNA level of foxO1 was analyzed by real-time PCR in total cDNA obtained from the brain striatum of XBP1Nes−/− or littermate control mice. All samples were normalized to β-actin levels. Average and SEM of the analysis of three animals per group are shown. (F) Neuro2A cells were stably transduced with lentiviral vectors expressing shRNA against XBP1 or control luciferase mRNA (shXBP1 and shCTR, respectively). FoxO1 and XBP1s levels were monitored after treatment with Tm (5 μg/ml, Tm) for 8 h, using western blot analysis. Levels of Hsp90 were used as loading control. Quantification is presented at the bottom of the gel as fold change. (G) HEK cells were transiently transfected with expression vectors for HA-FoxO1 and different concentrations of XBP1s or empty vector (pCDNA.3). FoxO1 and XBP1s levels were analyzed by western blot. The asterisk indicates unspecific band used as loading control. (H) Neuro2A cells were co-transfect with expression vector for mHttQ138-mRFP with HA-FoxO1 or empty vector, and after 48 h, the number of RFP-positive cells containing mHtt inclusions was quantified. Mean and SEM are presented. **P< 0.01, calculated with Student's t-test. (I) Neuro2A cells were transiently transfected with an HA-FoxO1 expression vector or empty vector, and then treated for the indicated times with a cocktail of lysosomal inhibitors (200 nm bafilomycin A1, 10 μg/ml pepstatin and 10 μg/ml E64d). LC3-II flux was then monitored by western blot analysis and normalized with the loading control and as a fold change to the untreated control cells (quantification at the bottom).
We hence explored ER stress-independent pathways that might explain the neuroprotective effects of XBP1 deficiency by augmented autophagy. A recent report indicates that XBP1s physically interacts with the transcription factor FoxO1, leading to its degradation by the proteasome (56). Since FOXO1 is a central regulator of autophagy in different cell types (57–59), including neurons (60), we hypothesized that the ablation of XBP1 expression in the nervous system may lead to an increase in FoxO1 levels. In agreement with this prediction, analysis of FoxO1 protein levels in the striatum of XBP1Nes−/− mice revealed a significant increase in the protein levels (Fig. 7C). Consistent with this result, enhanced expression and nuclear translocation of FoxO1 was observed in XBP1Nes−/− and XBP1Nes−/−-mHttQ128 mice compared with their respective littermate control animals after co-staining with NeuN by immunofluorescence followed by confocal microscopy analysis (Fig. 7D). In contrast, no changes in the mRNA levels of foxo1 were observed in the same brain samples (Fig. 7E). Remarkably, no changes in FoxO1 levels were observed in ATF4-deficient animals when compared with littermate control mice (Supplementary Material, Fig. S7), indicating that XBP1 specifically regulates FoxO1 expression in the nervous system.
We then investigated the role of FoxO1 on the effects of XBP1 on a cellular model of HD. Knocking down XBP1 in Neuro2a cells resulted in a 30% increase in FoxO1 levels (Fig. 7F). Moreover, induction of ER stress after tunicamycin (Tm) treatment led to the expression of XBP1s, which negatively correlated with FoxO1 levels. Likewise, co-expression of XBP1s with an HA tag version of FoxO1 in 293T cells reduced FoxO1 expression levels (Fig. 7G). To address the possible impact of FoxO1 on mHtt levels, we co-transfected expression vectors for FoxO1 and mHttQ138-mRFP in Neuro2a cells. Decreased accumulation of mHttQ138-mRFP inclusions was observed in FoxO1 overexpressing cells (Fig. 7H). In agreement with this result and previous findings (57–59), expression of FoxO1 in this cellular model enhanced autophagy activity as monitored using an LC3 flux assay by western blot analysis (Fig. 7I). Together, these data suggest that XBP1 deficiency triggers FoxO1 upregulation, which may contribute to the induction of autophagy and mHtt degradation.
XBP1s and LC3-II levels in the striatum of HD patients
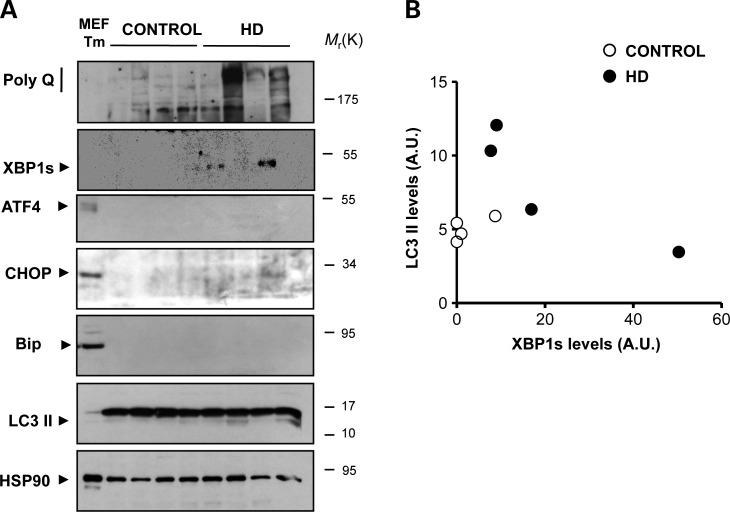
To evaluate the levels of ER stress in the brain of HD patients, we monitored the expression of classical ER stress markers in post-mortem striatum samples from four HD patients and four age-matched healthy control subjects. We confirmed the HD genotype of the patients by PCR (Supplementary Material, Table S2). In two HD cases, there was a detectable expression of XBP1s in the striatum that inversely correlated with LC3-II levels (Fig. 8A and B). These changes were not observed in cortex and cerebellum samples from the same individuals analyzed (data not shown). Similar to the results obtained in the YAC128 model, no clear signs of a general ER stress response were observed in HD-derived tissue as indicated by the absence of alterations in ATF4, CHOP or BiP expression levels (Fig. 8A). As positive control, cells treated with Tm are also shown.
Figure 8.
XBP1s, ER stress and autophagy markers in HD human post-mortem samples. (A) The levels of mHtt (anti-polyQ), XBP1s, ATF4, CHOP, BiP and LC3 were measured in total protein extracts derived from the striatum of post-mortem samples from HD patients at disease stage III and healthy control subjects. These identified samples were obtained from the Harvard Brain Tissue Resource Center. HSP90 levels were analyzed as loading control. As positive control for ER stress, MEFs treated with 1 µg/ml Tm is presented. (B) For comparison, relative levels of XBP1s and LC3-II were quantified and plotted after normalization with HSP90 levels shown as arbitrary units.
DISCUSSION
Most neurodegenerative diseases are characterized by the accumulation of abnormally folded proteins in the brain in the form of soluble oligomers and large protein inclusions, associated with impaired neuronal functions. Although the signaling pathways underlying neurodegeneration in protein misfolding disorders are not well understood, correlative evidence indicates that the occurrence of ER stress is a common cellular response in multiple neurodegenerative diseases (reviewed in 1). However, most of the studies linking ER stress to HD are correlative (3). To address the possible contribution of the UPR in mHtt pathogenesis in vivo, we silenced the expression of two main UPR transcription factors in the setting of an HD rodent model that expresses full-length mHtt as a transgene. Despite our initial prediction that XBP1 and ATF4 serve as protective factors in HD by controlling protein quality control and folding, we found the opposite result where targeting XBP1 improved motor performance and neuronal survival in an HD mouse model. These protective effects were associated with enhanced autophagy and diminished levels of mHtt as demonstrated in cellular and animal models of HD. In sharp contrast, ATF4 deficiency did not affect Htt levels in vivo. This is the first time the contribution of these two key UPR transcription factors has been assessed side by side in the same disease under similar experimental paradigms. Surprisingly, although we were able to detect the induction of XBP1 mRNA splicing in the YAC128 mouse model and human brain samples from HD patients, no signs of a general ER stress response were observed on a total striatum extract. However, we cannot exclude the possibility that ER stress occurs transiently and in a non-synchronic manner in HD, making its detection difficult using biochemical approaches. Further more, physiologic levels of UPR activation in vivo are very low and some times could lead, for example, to <5% XBP1 mRNA splicing (8,55). Interestingly, a recent study reported that the expression of full-length or truncated versions of mHtt does not trigger an evident ER stress response, but they actually alter the physiology of the ER (30). This observation may be in line with accumulating evidence suggesting that the IRE1α-XBP1 signaling branch can be activated under many conditions that are not directly related to protein misfolding, consistent with new functions of UPR signaling components/modules in organ physiology (8,55). Indeed, XBP1 has been implicated in the control of innate immunity (61), lipid and cholesterol metabolism (54), insulin resistance (62), cell differentiation (13,63,64) and other processes (65). In agreement with our observations, a recent study suggested that IRE1α signaling might repress autophagy, enhancing mHtt aggregation. Most of the experiments in this study were performed in vitro, using overexpression in cell lines (66). Interestingly, the authors provided correlative evidence indicating that reducing IRE1α levels in a fly model of HD protects against eye degeneration. However, the mechanism of protection in this in vivo model was not investigated (66).
We explored possible mechanisms underlying the protective effects of XBP1 deficiency on HD pathogenesis. A recent report indicates a negative regulation of FoxO1 by XBP1s at the posttranslational level in pancreatic β cells (56). XBP1s is proposed to regulate FoxO1 levels independent of its transcriptional activity, due to targeting of FoxO1 to proteasome-mediated degradation (56). Since FoxO1 is a key regulator of autophagy in neurons due transcriptional regulation of autophagy-related genes (60), we monitored its levels in our experimental system. Ablation of XBP1 expression led to a significant upregulation of FoxO1 in the striatum. In vitro experiments demonstrated that ER stress and XBP1s negatively regulate FoxO1 levels in neuronal cells, consistent with previous findings. Moreover, expression of FoxO1 decreased the levels of mHtt and enhanced autophagy in neuronal cell lines. Based on a large body of literature, we speculate that in addition to controlling autophagy, FoxO1 upregulation may have other broad beneficial effects contributing to neuroprotection in XBP1-deficient animals related to mitochondrial metabolism and oxidative stress (reviewed in 58,67). Importantly, FoxO transcription factors have key roles in promoting longevity (58,68), and a direct epistatic relationship between the FOXO homolog in Caenorhabditis elegans and XBP1 through the insulin/IGF-1 pathway has been recently reported in aging models (69). A very recent report investigated the impact of insulin/IGF-1 signaling in HD, using the R6/2 mouse model (70). Genetic manipulation of the pathway improved motor performance and prolonged life span in this HD model. The protective effects described in this study were explained by the upregulation of FoxO1-dependent autophagy associated with reduced accumulation of mHtt aggregates in the brain (70). Together with our current study, these findings suggest a complex scenario where dynamic cross-talk between UPR and aging/stress pathways acts to maintain protein homeostasis in the nervous system. We have reported that XBP1 deficiency improves motor performance and survival of an ALS mouse model (49). It remains to be determined whether a similar mechanism involving FoxO1 also operates in ALS models. In sharp contrast, XBP1 deficiency enhanced the severity of spinal cord injury on a mouse model (71), whereas it did not alter prion pathogenesis (40), indicating that this pathway contributes only to certain diseases with distinct outputs.
Autophagy is becoming a relevant factor in aging and diseases affecting the nervous system (72). Increasing evidence indicates that several mutant aggregate-prone proteins, but not the corresponding wild-type forms, are efficient targets for autophagy-mediated degradation. These effects have been demonstrated for the amyloid precursor protein, Htt, the prion protein, α-synuclein, ataxin-3, SOD1 and other disease-related proteins (reviewed in 73), which are substrates for autophagy-mediated degradation. Furthermore, several studies in HD animal models suggest that pharmacologic strategies to enhance autophagy decrease mHtt levels, leading to reduced neuronal loss, and improved motor performance (35,74). However, accumulating evidence indicate that defects in autophagy flux or in specific autophagy regulatory processes may actually contribute to neurodegeneration in diverse diseases (75–79). This phenomenon has been reported in HD cellular and animal models, in addition to human HD-derived tissue as we described (39). Impaired cargo recognition by autophagic vacuoles (AV) was observed in HD (39). Our results suggest that XBP1 deficiency may actually bypass this pathologic effect of mHtt expression, restoring the capacity of HD neurons to engage autophagy and induce the clearance of mHtt and damaged organelles. The beneficial effect of XBP1 ablation could be in part because of direct removal of mHtt aggregates but it is more likely that continuous improved removal of the soluble forms of the protein prevents its organization into aggregates in these animals. In fact, most of the effects described here for mHtt clearance in the YAC128 and the knock-in HD models were observed during disease stages where little mHtt aggregation is observed. For example, we did not detect significant mHtt inclusions in 6-month-old heterozygous YAC128 mice (data not shown). Combinatorial strategies to repair the autophagy deficit, reduce ER stress levels and enhance protein degradation pathways may be beneficial in selectively decreasing the expression and accumulation of toxic mHtt species and damaged organelles and proteins in HD.
MATERIALS AND METHODS
Materials and plasmids
Tm, wortmannin, 3-MA and MG-132 were purchased from Calbiochem EMB Bioscience, Inc. Cell media and antibiotics were obtained from Life Technologies (MD, USA). Puromycin, pepstatin and E64D were purchased from Sigma. Fetal calf serum was obtained from Atlanta Biologicals. DAPI and Lysotracker were purchased from Molecular Probes. All transfections were performed using the Effectene reagent (Qiagen). DNA was purified with Qiagen kits. GFPQ82-1-171 Htt is an in-frame N-terminal fusion of GFP to truncated human huntingtin (Htt) comprising human exons 1–3. The fusion protein was generated using recombinant PCR, and the final 1.4 kb product was cloned into pcDNA.3.1 (gift from Paulson Henry, University of Michigan). Htt1-588(Q)138-Δ5-13-mRFP and Htt1-588(Q)138-mRFP vectors were provided by Ray Truant (McMaster University) (LC3-EGFP expression vectors); the rat LC3 cDNA was cloned into the BglII and EcoRI sites of the pEGFP-C1 vector (Clontech Laboratories). The XBP1s expression vector was previously described (12). The XBP1 cDNA was obtained from NIH3T3 cells treated with Tm and cloned into the pCDNA.3 vector between the HindIII and ApaI sites. HA-FoxO1 expression vector was obtained from Addgene.
Knockdown of UPR and autophagy components
We generated stable motoneuron cell lines with reduced levels of XBP1 and IRE1α, using methods previously described (49) by targeting their respective mRNAs with shRNAs, using the lentiviral expression vector pLKO.1 and puromycin selection. As control, empty vector or an shRNA against the luciferase gene was employed. Constructs were generated by The Broad Institute (Boston, MA, USA), based on different criteria for shRNA design (see http://www.broad.mit.edu/genome_bio/trc/rnai.html). We screened a total of five different constructs for each gene and selected the most efficient one for further studies. Targeting sequences identified for the mouse XBP1 and IRE1α, mRNA are 5′-CCATTAATGAACTCATTCGTT-3′, and 5′-GCTCGTGAATTGATAGAGAAA-3′. For ATG5 mRNA knock-down, we employed a combination of five different targeting sequences including 5′-GCCAAGTATCTGTCTATGATA-3′, 5′-CCTTGGAACATCACAGTACAT-3′, 5′-GCAGAACCATACTATTTGCTT-3′, 5′-GCATCTGAGCTACCC AGATAA-3′ and 5′-CCCTGAA ATGGCATTATCCAA-3′.
Assays for mutant protein aggregation and detection of intracellular inclusions
We visualized and quantified the formation of intracellular polyQ79 inclusions in living cells after transient transfection followed by fluorescent confocal microscopy. For automatic quantification of the percentage of cells containing PolyQ79 aggregates, we used the ImageJ software (http://rsbweb.nih.gov/ij/). We applied the threshold tool to determinate the total GFP-positive structure in each image, which were quantified using the ‘Analyze Particles’ tool. Then, to visualize only cells containing high fluorescent particles, we applied a new threshold (higher) to cut off the fluorescent emission of diffusion GFP pattern (determined with the polyQ11 control) and automatically determine the number of cells with PolyQ79 inclusions in the same image using the ‘Analyze Particles’ tool. Finally, we calculated the percentage of positive inclusion cells considering the total GFP-positive structure as 100%. This method was validated by manual counting using a double-blind determination. A similar analysis was performed with the mHttQ138-mRFP construct. PolyQ79 and mHtt oligomers were visualized in total cell extracts prepared in 1% Triton X-100 in PBS containing protease inhibitors, sonicated and then analyzed by western blot using 4–12% custom pre-made gels (Lonza, USA), which leads to the accumulation of a smear of high molecular weight aggregates. For determining mHttQ85, 8 or 10% polyacrylamide gels were employed, which resulted in the presence of small oligomers.
EM studies and immunogold staining
APG were also visualized by transmission EM and morphology examined as described in references (78,80). For immunogold EM staining, the conditions for labeling to detect specific signals were as we previously described using anti-polyQ (clone 3B5H10) antibody (1:200 dilution); post-embedding staining was performed by standard methods as we recently described (81). After incubation with the first antibody, grids were washed and incubated with donkey anti-mouse IgG conjugated to 15 nm of colloidal gold (1:100, Ted Pella).
Isolation of AV from the brain
Fractions enriched in APG and APGL were isolated from mouse brains (200 mg wet tissue), using a modified protocol, originally developed for the liver (82), and markers for each fraction collected were employed to validate purity. Briefly, brain tissue was homogenized and subjected to differential centrifugation to separate a fraction enriched in AV, lysosomes and mitochondria. After centrifugation in a metrizamide discontinuous density gradient, a fraction enriched in APG was recovered in the interphase between the 5 and 10% metrizamide density, and a second fraction enriched in APGL was recovered in the interphase between the 10 and 20% metrizamide density. Samples were extensively washed in 0.25 m sucrose and recovered as pellets by centrifugation for biochemical analysis. Cytosolic fractions were obtained by centrifugation of the supernatant of the fraction enriched in AV, lysosomes and mitochondria at 100 000g for 1 h at 4°C. The pellet of this centrifugation was preferentially enriched in ER components in the form of small vesicles (microsomes).
Quantification of autophagy
Different assays and control experiments were employed to monitor autophagy-related processes following the recommendations described in reference (78). Lysosomes or acidic compartments were visualized after staining with 200 nm Lysotracker for 45 min at 37°C and 5% CO2. Cells were washed three times with cold PBS, fixed for 30 min with 4% formaldehyde on ice and then maintained in PBS containing 0.4% formaldehyde for immediate visualization on a confocal microscope. Autophagy was monitored by analyzing LC3-positive dots or the levels of LC3-II by western blot and its flux through the autophagosomal/lysosomal pathway as described (49). APG were visualized after the expression of LC3-EGFP by transient transfection of the lowest amounts of DNA tittered to obtain the best signal-to-noise ratio (one-third amount of recommended concentrations by the transfection kit). In control experiments, cells were treated with 10 mm 3-MA to inhibit autophagy. LC3 is initially synthesized as an unprocessed form, proLC3, which is converted into a proteolytically processed form lacking several amino acids from the C-terminus, LC3-I, and is then modified into the phosphatidylethanolamine-conjugated form. To monitor LC3-II dynamics, protein samples were processed in cold RIPA and immediately analyzed by western blot using 15% SDS–polyacrilamide gels. As internal control, LC3-II levels were compared with Hsp90 levels. To follow the flow of LC3I/II through the autophagy pathway, cells were treated with a mix of 200 nm bafilomycin A1, 10 μg/ml pepstatin and 10 μg/ml E64d.
Animal experimentation
All animal experiments were performed according to procedures approved by the Institutional Review Board's Animal Care and Use Committee of the Harvard School of Public Health (approved animal protocol 04137) and the Faculty of Medicine of the University of Chile (approved protocol CBA 0208 FMUCH). XBP1flox/flox mice were crossed with mice expressing Cre recombinase under the control of the Nestin promoter to achieve deletion of XBP1 in the nervous system (40) (XBP1Nes−/−). XBP1flox/flox mice were backcrossed to B57BL/6 background for eight generations and then crossed with pure background Nestin-Cre transgenic mice (The Jackson Laboratory) for three more generations. We employed the full-length mHtt transgenic mice with 128 CAG repetitions, termed YAC128 (41), as an HD model, obtained from The Jackson Laboratory. To generate experimental animals, XBP1Nes−/− mice were then crossed with the YAC128 model on an FVB background and every generation bred to pure background XBP1Nes−/− mice for four to six generations to obtain experimental animals. For proper comparison, all biochemical, histologic and behavioral analyses were performed on groups of littermates of the same breeding generation. ATF4-deficient mice were previously described on a pure B57BL/6 background (83,84), and the same breeding strategy described for XBP1Nes−/− mice was employed. We also used an HdhQ111/Q7 knock-in mouse model of HD as previously described (46). In this case, breedings of ATF4 or XBP-1Nes−/− and the knock-in HdhQ111/Q7 model were performed since the beginning on pure C57BL/6 genetic background.
Rotarod performance
Disease progression was monitored at least once per week, using the rotarod assay as previously reported (43). In brief, a training period was performed, consisting of three trials per day (with 2 h in between trials) over the course of 3 consecutive days. On the fourth day (test day), the data collected were considered an accurate reflection of the animal's coordination. For each testing, mice acclimated to the room for ∼15 min. The settings for rotator performance were (i) maximum speed, 45 r.p.m.; (ii) time to maximum r.p.m., 120 s; (iii) time to ‘no fall’, 600 s; (iv) starting speed, 5 r.p.m.; (v) trials per day, 3; (vi) inter-trial interval, 2 h.
Tissue analysis
To monitor mHtt pathogenesis in vivo, animals were euthanized and tissue collected for histology at different time points depending on the analysis required. Brain tissue was processed for immunohistochemistry, using standard procedures as described (49). Confocal microscopy was used to acquire images and then analysis was performed using the IP lab v 4.04 software (Beckon and Dickenson).
Biochemistry of brain tissue
The brain striatum was dissected and homogenized in RIPA buffer (20mm Tris, pH 8.0, 150mm NaCl, 0.1% SDS, 0.5% DOC, 0.5% Triton X-100) containing a protease inhibitor cocktail (Roche, Basel, Switzerland), by sonication. Protein concentration was determined by micro-BCA assay (Pierce, Rockford, IL, USA). The equivalent of 30–50 μg of total protein was loaded onto 4–12, 7.5, 12 or 15% SDS–PAGE minigels (Cambrex Biosciences), depending on the analysis as described above. The following antibodies and dilutions were used: anti-DARPP32 1:1000, anti-Htt clone mEM48 1:1000, anti-Htt clone 1HU-4C8 1:1000 (Millipore); anti-polyQ clone 3B5H10 1:1000 (Sigma); anti-XBP-1 1:2000 (Biolegend); anti-GFP 1:1000, anti-ATF4, anti-Hsp90, anti-CHOP 1:2000 (Santa Cruz, CA, USA); anti-LC3 1:1000, anti-Beclin-1 1:2000, anti-phospho-eIF2α 1:2000, anti-eIF2α 1:4000 (Cell Signaling Technology); anti-cathepsin D 1:500 (Santa Cruz); anti-p62 1:2000 (Enzo Life Sciences); anti-BiP 1:10000 (BD Biosciences); anti-FoxO1 1:1000 (Abcam); anti-tubulin 1:3000 (Santa Cruz). Real-time PCR methods and primers were previously described (40).
HD human post-mortem brain samples
Human post-mortem tissue from HD and control subjects was obtained as frozen tissue from the Harvard Brain Tissue Resource Center (Boston, MA, USA, http://www.brainbank.mclean.org/) and then processed for biochemical analysis by homogenization of equivalent amounts of tissue in PBS containing protease and phosphatase inhibitors, with further dilution in RIPA buffer.
Statistical analysis
All data are expressed as mean and SEM. Results were statistically compared using Student's t-test performed for paired groups.
SUPPLEMENTARY MATERIAL
FUNDING
This work was funded by a grant from an anonymous foundation (L.H.G.), the CHDI Foundation, Inc. (C.H.), the V. Harold and Leila Y Mathers Charitable Foundation and NIH grant AI32412 (L.H.G.). In addition, we received support from FONDECYT no. 1100176, FONDAP grant no. 15010006, Millennium Institute No. P09-015-F, Muscular Dystrophy Association, Alzheimer Association, North American Spine Society and the Michael J. Fox Foundation for Parkinson Research (to C.H.); a CONICYT fellowship (M.N.); FONDECYT no. 3100039 (R.L.V.); AG031782 (A.M.C.); FONDECYT no. 1070377 and Millennium Nucleus no. P-07-011-F (F.A.C.) and FONDECYT No. 1100165 (J.S.-A.).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Nir Hacohen and The Broad Institute (Boston, MA, USA) for providing shRNA lentiviral constructs. We thank Ray Truant for providing Htt1-588(Q)138-Δ5-13-mRFP and Htt1-588(Q)138-mRFP vectors. We thank the Harvard Brain Tissue Resource Center for providing HD post-mortem samples.
Conflict of Interest statement. The authors declare no conflict of interest in this study.
REFERENCES
- 1.Matus S., Glimcher L., Hetz C. Protein folding stress in neurodegenerative diseases: a glimpse into the ER. Curr. Opin. Cell Biol. 2011;23:239–252. doi: 10.1016/j.ceb.2011.01.003. doi:10.1016/j.ceb.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Bossy-Wetzel E., Petrilli A., Knott A.B. Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 2008;31:609–616. doi: 10.1016/j.tins.2008.09.004. doi:10.1016/j.tins.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vidal R., Caballero B., Couve A., Hetz C. Converging pathways in the occurrence of endoplasmic reticulum (ER) stress in Huntington's disease. Curr. Mol. Med. 2011;11:1–12. doi: 10.2174/156652411794474419. doi:10.2174/156652411794474419. [DOI] [PubMed] [Google Scholar]
- 4.Rubinsztein D.C. Lessons from animal models of Huntington's disease. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. doi:10.1016/S0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- 5.Williams A.J., Paulson H.L. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. doi:10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. doi:10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 7.Woehlbier U., Hetz C. Modulating stress responses by the UPRosome: a matter of life and death. Trends Biochem. Sci. 2011;36:329–337. doi: 10.1016/j.tibs.2011.03.001. doi:10.1016/j.tibs.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 9.Hetz C., Glimcher L.H. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol. Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. doi:10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. doi:10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. doi:10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 12.Lee A.H., Iwakoshi N.N., Glimcher L.H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. doi:10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N.H., Arias C., Lennon C.J., Kluger Y., Dynlacht B.D. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. doi:10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. doi:10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 15.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. doi:10.1016/S1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 16.Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H., Stevens J.L., Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. doi:10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez D., Rojas-Rivera D., Hetz C. Integrating stress signals at the endoplasmic reticulum: the BCL-2 protein family rheostat. Biochim. Biophys. Acta. 2011;1813:564–574. doi: 10.1016/j.bbamcr.2010.11.012. doi:10.1016/j.bbamcr.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Carnemolla A., Fossale E., Agostoni E., Michelazzi S., Calligaris R., De Maso L., Del Sal G., MacDonald M.E., Persichetti F. Rrs1 is involved in endoplasmic reticulum stress response in Huntington disease. J. Biol. Chem. 2009;284:18167–18173. doi: 10.1074/jbc.M109.018325. doi:10.1074/jbc.M109.018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho K.J., Lee B.I., Cheon S.Y., Kim H.W., Kim H.J., Kim G.W. Inhibition of apoptosis signal-regulating kinase 1 reduces endoplasmic reticulum stress and nuclear huntingtin fragments in a mouse model of Huntington disease. Neuroscience. 2009;163:1128–1134. doi: 10.1016/j.neuroscience.2009.07.048. doi:10.1016/j.neuroscience.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 20.Hoffstrom B.G., Kaplan A., Letso R., Schmid R.S., Turmel G.J., Lo D.C., Stockwell B.R. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat. Chem. Biol. 2010;6:900–906. doi: 10.1038/nchembio.467. doi:10.1038/nchembio.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higo T., Hamada K., Hisatsune C., Nukina N., Hashikawa T., Hattori M., Nakamura T., Mikoshiba K. Mechanism of ER stress-induced brain damage by IP(3) receptor. Neuron. 2010;68:865–878. doi: 10.1016/j.neuron.2010.11.010. doi:10.1016/j.neuron.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Omi K., Hachiya N.S., Tokunaga K., Kaneko K. siRNA-mediated inhibition of endogenous Huntington disease gene expression induces an aberrant configuration of the ER network in vitro. Biochem. Biophys. Res. Commun. 2005;338:1229–1235. doi: 10.1016/j.bbrc.2005.10.061. doi:10.1016/j.bbrc.2005.10.061. [DOI] [PubMed] [Google Scholar]
- 23.Kouroku Y., Fujita E., Jimbo A., Kikuchi T., Yamagata T., Momoi M.Y., Kominami E., Kuida K., Sakamaki K., Yonehara S., et al. Polyglutamine aggregates stimulate ER stress signals and caspase-12 activation. Hum. Mol. Genet. 2002;11:1505–1515. doi: 10.1093/hmg/11.13.1505. doi:10.1093/hmg/11.13.1505. [DOI] [PubMed] [Google Scholar]
- 24.Duennwald M.L., Lindquist S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008;22:3308–3319. doi: 10.1101/gad.1673408. doi:10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishitoh H., Matsuzawa A., Tobiume K., Saegusa K., Takeda K., Inoue K., Hori S., Kakizuka A., Ichijo H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. doi:10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai Y.C., Fishman P.S., Thakor N.V., Oyler G.A. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J. Biol. Chem. 2003;278:22044–22055. doi: 10.1074/jbc.M212235200. doi:10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- 27.Kouroku Y., Fujita E., Tanida I., Ueno T., Isoai A., Kumagai H., Ogawa S., Kaufman R.J., Kominami E., Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. doi:10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 28.Reijonen S., Putkonen N., Norremolle A., Lindholm D., Korhonen L. Inhibition of endoplasmic reticulum stress counteracts neuronal cell death and protein aggregation caused by N-terminal mutant huntingtin proteins. Exp. Cell Res. 2008;314:950–960. doi: 10.1016/j.yexcr.2007.12.025. doi:10.1016/j.yexcr.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 29.Bertoni A., Giuliano P., Galgani M., Rotoli D., Ulianich L., Adornetto A., Santillo M.R., Porcellini A., Avvedimento V.E. Early and late events induced by polyQ-expanded proteins: identification of a common pathogenic property of polyQ-expanded proteins. J. Biol. Chem. 2010;286:4727–4741. doi: 10.1074/jbc.M110.156521. doi:10.1074/jbc.M110.156521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lajoie P., Snapp E.L. Changes in BiP availability reveal hypersensitivity to acute endoplasmic reticulum stress in cells expressing mutant huntingtin. J. Cell Sci. 2011;124:3332–3343. doi: 10.1242/jcs.087510. doi:10.1242/jcs.087510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H., Liu C., Zhong Y., Luo S., Monteiro M.J., Fang S. Huntingtin interacts with the cue domain of gp78 and inhibits gp78 binding to ubiquitin and p97/VCP. PLoS One. 2010;5:e8905. doi: 10.1371/journal.pone.0008905. doi:10.1371/journal.pone.0008905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez-Fernandez M.R., Ferrer I., Lucas J.J. Impaired ATF6alpha processing, decreased Rheb and neuronal cell cycle re-entry in Huntington's disease. Neurobiol. Dis. 2011;41:23–32. doi: 10.1016/j.nbd.2010.08.014. doi:10.1016/j.nbd.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 33.Sarkar S., Perlstein E.O., Imarisio S., Pineau S., Cordenier A., Maglathlin R.L., Webster J.A., Lewis T.A., O'Kane C.J., Schreiber S.L., et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat. Chem. Biol. 2007;3:331–338. doi: 10.1038/nchembio883. doi:10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubinsztein D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. doi:10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 35.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. doi:10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 36.Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B., Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. doi:10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 37.Atwal R.S., Xia J., Pinchev D., Taylor J., Epand R.M., Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. doi:10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 38.Atwal R.S., Truant R. A stress sensitive ER membrane-association domain in Huntingtin protein defines a potential role for Huntingtin in the regulation of autophagy. Autophagy. 2008;4:91–93. doi: 10.4161/auto.5201. [DOI] [PubMed] [Google Scholar]
- 39.Martinez-Vicente M., Talloczy Z., Wong E., Tang G., Koga H., Kaushik S., de Vries R., Arias E., Harris S., Sulzer D., et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. doi:10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hetz C., Lee A.H., Gonzalez-Romero D., Thielen P., Castilla J., Soto C., Glimcher L.H. Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc. Natl Acad. Sci. USA. 2008;105:757–762. doi: 10.1073/pnas.0711094105. doi:10.1073/pnas.0711094105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slow E.J., van Raamsdonk J., Rogers D., Coleman S.H., Graham R.K., Deng Y., Oh R., Bissada N., Hossain S.M., Yang Y.Z., et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. doi:10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 42.Mazarei G., Neal S.J., Becanovic K., Luthi-Carter R., Simpson E.M., Leavitt B.R. Expression analysis of novel striatal-enriched genes in Huntington disease. Hum. Mol. Genet. 2010;19:609–622. doi: 10.1093/hmg/ddp527. doi:10.1093/hmg/ddp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham R.K., Slow E.J., Deng Y., Bissada N., Lu G., Pearson J., Shehadeh J., Leavitt B.R., Raymond L.A., Hayden M.R. Levels of mutant huntingtin influence the phenotypic severity of Huntington disease in YAC128 mouse models. Neurobiol. Dis. 2006;21:444–455. doi: 10.1016/j.nbd.2005.08.007. doi:10.1016/j.nbd.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 44.Mao X., Fujiwara Y., Chapdelaine A., Yang H., Orkin S.H. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. doi:10.1182/blood.V97.1.324. [DOI] [PubMed] [Google Scholar]
- 45.Metzler M., Gan L., Mazarei G., Graham R.K., Liu L., Bissada N., Lu G., Leavitt B.R., Hayden M.R. Phosphorylation of huntingtin at Ser421 in YAC128 neurons is associated with protection of YAC128 neurons from NMDA-mediated excitotoxicity and is modulated by PP1 and PP2A. J. Neurosci. 2010;30:14318–14329. doi: 10.1523/JNEUROSCI.1589-10.2010. doi:10.1523/JNEUROSCI.1589-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheeler V.C., White J.K., Gutekunst C.A., Vrbanac V., Weaver M., Li X.J., Li S.H., Yi H., Vonsattel J.P., Gusella J.F., et al. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum. Mol. Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. doi:10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- 47.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. doi:10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arsham A.M., Neufeld T.P. A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy-lysosome pathway. PLoS One. 2009;4:e6068. doi: 10.1371/journal.pone.0006068. doi:10.1371/journal.pone.0006068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hetz C., Thielen P., Matus S., Nassif M., Court F., Kiffin R., Martinez G., Cuervo A.M., Brown R.H., Glimcher L.H. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23:2294–2306. doi: 10.1101/gad.1830709. doi:10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. doi:10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. doi:10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasaki S. Endoplasmic reticulum stress in motor neurons of the spinal cord in sporadic amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2010;69:346–355. doi: 10.1097/NEN.0b013e3181d44992. doi:10.1097/NEN.0b013e3181d44992. [DOI] [PubMed] [Google Scholar]
- 53.Kaser A., Lee A.H., Franke A., Glickman J.N., Zeissig S., Tilg H., Nieuwenhuis E.E., Higgins D.E., Schreiber S., Glimcher L.H., et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. doi:10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee A.H., Scapa E.F., Cohen D.E., Glimcher L.H. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. doi:10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rutkowski D.T., Hegde R.S. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J. Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. doi:10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y., Lee J., Reno C.M., Sun C., Park S.W., Chung J., Fisher S.J., White M.F., Biddinger S.B., Ozcan U. Regulation of glucose homeostasis through a XBP-1–FoxO1 interaction. Nat. Med. 2011;17:356–365. doi: 10.1038/nm.2293. doi:10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sengupta A., Molkentin J.D., Yutzey K.E. FoxO transcription factors promote autophagy in cardiomyocytes. J. Biol. Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. doi:10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salih D.A., Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr. Opin. Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. doi:10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y., Yang J., Liao W., Liu X., Zhang H., Wang S., Wang D., Feng J., Yu L., Zhu W.G. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell Biol. 2010;12:665–675. doi: 10.1038/ncb2069. doi:10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 60.Xu P., Das M., Reilly J., Davis R.J. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25:310–322. doi: 10.1101/gad.1984311. doi:10.1101/gad.1984311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martinon F., Chen X., Lee A.H., Glimcher L.H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol. 2011;11:411–418. doi: 10.1038/ni.1857. doi:10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ozcan U., Cao Q., Yilmaz E., Lee A.H., Iwakoshi N.N., Ozdelen E., Tuncman G., Gorgun C., Glimcher L.H., Hotamisligil G.S. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. doi:10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 63.Huh W.J., Esen E., Geahlen J.H., Bredemeyer A.J., Lee A.H., Shi G., Konieczny S.F., Glimcher L.H., Mills J.C. XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology. 2010;139:2038–2049. doi: 10.1053/j.gastro.2010.08.050. doi:10.1053/j.gastro.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reimold A.M., Iwakoshi N.N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E.M., Friend D., Grusby M.J., Alt F., Glimcher L.H. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. doi:10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 65.Hetz C., Martinon F., Rodriguez D., Glimcher L.H. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol. Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. doi:10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- 66.Lee H., Noh J.Y., Oh Y., Kim Y., Chang J.W., Chung C.W., Lee S.T., Kim M., Ryu H., Jung Y.K. IRE1 plays an essential role in ER stress-mediated aggregation of mutant huntingtin via the inhibition of autophagy flux. Hum. Mol. Genet. 2011;21:101–114. doi: 10.1093/hmg/ddr445. doi:10.1093/hmg/ddr445. [DOI] [PubMed] [Google Scholar]
- 67.Accili D., Arden K.C. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. doi:10.1016/S0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 68.Maiese K., Chong Z.Z., Shang Y.C. OutFOXOing disease and disability: the therapeutic potential of targeting FoxO proteins. Trends Mol. Med. 2008;14:219–227. doi: 10.1016/j.molmed.2008.03.002. doi:10.1016/j.molmed.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Henis-Korenblit S., Zhang P., Hansen M., McCormick M., Lee S.J., Cary M., Kenyon C. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc. Natl Acad. Sci. USA. 2010;107:9730–9735. doi: 10.1073/pnas.1002575107. doi:10.1073/pnas.1002575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sadagurski M., Cheng Z., Rozzo A., Palazzolo I., Kelley G.R., Dong X., Krainc D., White M.F. IRS2 increases mitochondrial dysfunction and oxidative stress in a mouse model of Huntington disease. J. Clin. Invest. 2011;121:4070–4081. doi: 10.1172/JCI46305. doi:10.1172/JCI46305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valenzuela V., Collyer E., Armentano D., Parsons G., Court F., Hetz, C. Activation of the Unfolded Protein Response enhances motor recovery after spinal cord injury. Cell Death Dis. 2012 doi: 10.1038/cddis.2012.8. 3, eK. doi:10.1038/cddis.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rubinsztein D.C., Marino G., Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. doi:10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 73.Menzies F.M., Moreau K., Rubinsztein D.C. Protein misfolding disorders and macroautophagy. Curr. Opin. Cell Biol. 2011;23:190–197. doi: 10.1016/j.ceb.2010.10.010. doi:10.1016/j.ceb.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanaka M., Machida Y., Niu S., Ikeda T., Jana N.R., Doi H., Kurosawa M., Nekooki M., Nukina N. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat. Med. 2004;10:148–154. doi: 10.1038/nm985. doi:10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 75.Nassif M., Hetz C. Targeting autophagy in ALS: a complex mission. Autophagy. 2011;7:450–453. doi: 10.4161/auto.7.4.14700. doi:10.4161/auto.7.4.14700. [DOI] [PubMed] [Google Scholar]
- 76.Wong E., Cuervo A.M. Autophagy gone awry in neurodegenerative diseases. Nat. Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. doi:10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sarkar S., Korolchuk V.I., Renna M., Imarisio S., Fleming A., Williams A., Garcia-Arencibia M., Rose C., Luo S., Underwood B.R., et al. Complex inhibitory effects of nitric oxide on autophagy. Mol. Cell. 2011;43:19–32. doi: 10.1016/j.molcel.2011.04.029. doi:10.1016/j.molcel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marino G., Madeo F., Kroemer G. Autophagy for tissue homeostasis and neuroprotection. Curr. Opin. Cell Biol. 2011;23:198–206. doi: 10.1016/j.ceb.2010.10.001. doi:10.1016/j.ceb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 80.Eskelinen E.L. Fine structure of the autophagosome. Methods Mol. Biol. 2008;445:11–28. doi: 10.1007/978-1-59745-157-4_2. doi:10.1007/978-1-59745-157-4_2. [DOI] [PubMed] [Google Scholar]
- 81.Barrientos S.A., Martinez N.W., Yoo S., Jara J.S., Zamorano S., Hetz C., Twiss J.L., Alvarez J., Court F.A. Axonal degeneration is mediated by the mitochondrial permeability transition pore. J. Neurosci. 2011;19:966–978. doi: 10.1523/JNEUROSCI.4065-10.2011. doi:10.1523/JNEUROSCI.4065-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marzella L., Ahlberg J., Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J. Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. doi:10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H.C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T.M., et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. doi:10.1016/S0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 84.Yoshizawa T., Hinoi E., Jung D.Y., Kajimura D., Ferron M., Seo J., Graff J.M., Kim J.K., Karsenty G. The transcription factor ATF4 regulates glucose metabolism in mice through its expression in osteoblasts. J. Clin. Invest. 2009;119:2807–2817. doi: 10.1172/JCI39366. doi:10.1172/JCI39366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.