Abstract
Cofilin-2, a small actin-binding protein and member of the AC protein family that includes cofilin-1 and destrin, is predominantly expressed at sarcomeres in skeletal and cardiac muscles. The role of cofilin-2 in muscle development and function is unclear. In humans, recessive cofilin-2 mutations have been associated with nemaline myopathy with minicores. To investigate the functional role of cofilin-2 in vivo, we generated constitutive and muscle-specific cofilin-2-deficient mice using a cre–loxP strategy. Cofilin-2-deficient mice were similar to their wild-type (WT) littermates at birth, but died by day 8. They were significantly smaller, severely weak and had very little milk in their stomachs. The sarcomeric structure was intact at birth, but by Day 7, skeletal muscles showed severe sarcomeric disruptions starting at the Z-line, along with filamentous actin accumulations consistent with a lack of actin depolymerization activity. Cofilin-2-deficient muscles contained elevated numbers of slow fibers and exhibited upregulation of slow fiber-specific genes. Increased amounts of other sarcomeric proteins including α-actinin-2, α-sarcomeric actin and tropomyosin were also present. While destrin was not expressed in either WT or cofilin-2-deficient muscles, cofilin-1 was similarly expressed in developing myofibers of both genotypes. There was no evidence for compensatory changes in expression of either family member in cofilin-2-deficient tissues. The onset of pathology and weakness in cofilin-2-deficient muscles correlated with normal developmental loss of cofilin-1 expression within myofibers, suggesting that cofilin-1 serves as an early developmental sarcomeric isoform. Overall, cofilin-2, although not critical for muscle development, is essential for muscle maintenance.
INTRODUCTION
Cofilin-2 is a small actin-binding protein (mol. wt. 18.7 kDa), encoded by the CFL2 gene on human chromosome 14q12 (MIM 601443). It is a member of the AC group of proteins that include cofilin-1 (encoded by CFL1), cofilin-2 and destrin (encoded by DSTN), formerly called actin depolymerization factor. AC proteins are critical in the regulation of actin filament dynamics in eukaryotes where they function by modulating actin depolymerization and severing (1). They are ubiquitously present throughout eukaryotes, with one AC protein identified in yeast, two in nematodes and three in mammals. The exact function of cofilin-2 is unknown, but a critical role for cofilin-2 in skeletal muscle function was demonstrated by identification of recessive CFL2 mutations in two families with congenital myopathies characterized by the presence of nemaline rods and minicores (2).
In humans, cofilin-2 is mainly expressed in skeletal and cardiac muscles, although it is also present at lower levels in several tissues including the testis, lung, brain, lung, kidney, pancreas, brain, liver and placenta (3). Human CFL2 has four exons with four transcripts resulting from the variable use of the 5′ and 3′ untranslated regions (UTRs). Murine Cfl2 in comparison has four exons, but only two transcripts of 1.8 and 3 kb due to a variable 3′UTR. The 1.8 kb transcript is present in the embryonic day (E) 13, peaks in both the cardiac and skeletal muscles at birth and gradually reduces thereafter, still present at 2 months of age. In comparison, the 3 kb transcript is present only in the skeletal muscles, and its levels go up as the 1.8 kb transcript levels come down (4). At the amino acid level, human cofilin-2 is 99% identical to murine cofilin-2 and 81% identical to human cofilin-1 (3). Functionally, among AC proteins, destrin is most efficient in turning over actin filaments, while cofilin-2 is least effective. In addition, cofilin-2 has a 5–10-fold higher affinity for G-actin, promoting actin filament assembly and suggesting adaptation in muscle cells where actin filaments are more stable (5).
Mutations of AC protein family members have been studied in Saccharomyces, Caenorhabditis elegans, Drosophila and Mus musculus. In Saccharomyces, cofilin-null mutants are lethal and can be rescued by vertebrate destrin/cofilin-1 (6) and Dictyostelium cofilin-1, but not cofilin-2 (7). Point mutations of UNC-60B (cofilin-2 isoform) in C. elegans cause defects in actin organization specifically within the body wall muscle (8). In comparison, although viable, homozygous UNC-60B-null mutants have a more severe motility defect than missense mutants (9). The expression pattern of UNC-60A (cofilin-1 isoform) in this null mutant is similar to that in wild-type (WT) worms, suggestive of no compensatory mechanisms. In Drosophila, homozygous null mutation of the twinstar gene that encodes a homolog of destrin/cofilin leads to death of first-instar larvae while hypomorphs had defects of varying severity including male sterility, abnormal centrosome migration and cytokinesis, semi-lethality and late larval/pupal lethality (10–12). In Mus musculus, non-muscle cofilin (Cfl1) has been targeted to create a null mutant. The homozygous null mutation was embryonic lethal while heterozygous mice were viable. Interestingly, there appeared to be a 3–4-fold upregulation of destrin in the homozygous mutant but that was not sufficient to rescue the phenotype. The predominant defects involved neural tube morphogenesis, neural crest migration and misaligned somites (13). A similar phenotype has been previously observed in several mouse mutants lacking molecules of the Notch signaling pathway, such as Notch, Delta 1 and RBP-JK (14). Mice with autosomal recessive Dstn null and point mutations developed corneal disease with roughened and opaque corneal surfaces, corneal stromal neovascularization and increased content of filamentous actin (F-actin) in the corneal epithelial cells (15).
The function of cofilin-2 in vertebrates is not well known. Based on cofilin-2 expression in mice, it has been postulated that cofilin-2 may be involved in regulation of actin assembly during myofibrillogenesis and actin dynamics in the mature muscle. Postulations about cofilin-2 function vary from it being critical for myofibrillogenesis, to the presumption that the effects of cofilin-2 deficiency may appear only in the adult skeletal muscle (4). To understand cofilin-2 function in the muscle, here, we report creating constitutive and muscle-specific excision of the Cfl2 gene in mice, leading to a markedly shortened life span, severe weakness and the presence of sarcomeric disruptions, nemaline-like bodies and actin accumulations within muscle fibers. Expression of the cofilin-2 paralogs, cofilin-1 and destrin is essentially unchanged in the constitutive Cfl2-knockout (KO) mice (Cfl2−/−). Our data indicate that cofilin-2 plays a critical role in skeletal muscle maintenance but not myogenesis in vivo.
RESULTS
Targeted disruption of cofilin-2 (Cfl2)
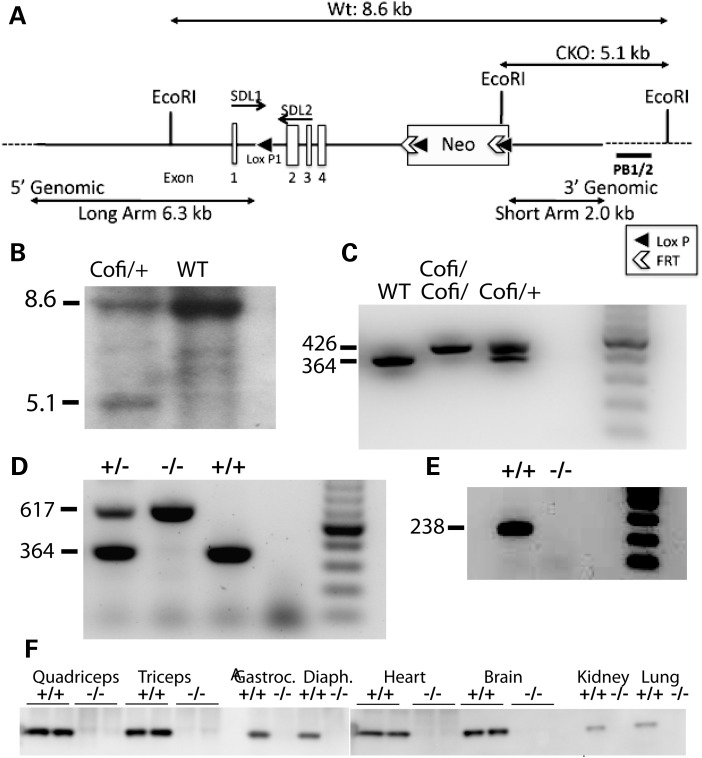
The murine cofilin-2 locus (Cfl2) was targeted by homologous recombination using a vector subcloned from a BAC clone. A loxP site was inserted upstream of Cfl2 exon 2 and a loxP-flanked neomycin (Neo) cassette was inserted downstream of exon 4 to generate a conditional mutant allele, allowing for excision of cofilin-2 in a tissue-specific and constitutive manner (Fig. 1A). The targeting vector was electroporated into C57BL/6 embryonic stem (ES) cells under G418 selection and clones that underwent proper homologous recombination were identified and confirmed by genomic polymerase chain reaction (PCR) and Southern blotting (Fig. 1B). The targeted ES cells were microinjected into Balb/c blastocysts and chimeras that transmitted the loxP/neo-cofilin-2 allele (Cofi) through the germline were identified. Homozygous Cofi mice were obtained (Fig. 1C) and crossed with a strain that expresses cre controlled by regulatory sequences from the mouse zona pellucida 3 (Zp3) gene that directs expression exclusively in the growing oocyte prior to the completion of first meiotic division (16). Females heterozygous for both Cofi and Zp3 alleles were bred with WT males to obtain heterozygous mice with exons 2–4 of the Cfl2 excised. The Cfl2 excision was confirmed by agarose gel analysis (Fig. 1D) and sequencing of PCR products (data not shown). Reverse transcriptase (RT)–PCR (Fig. 1E) and western blot analysis (Fig. 1F) confirmed that cofilin-2 was absent in several tissues. Tissue-specific Cfl2 excision was performed using skeletal muscle-specific (Mef2c-cre) (17), embryonic striated (skeletal and cardiac) muscle-specific (Acta1-cre) (18) and post-natal skeletal and cardiac muscle-specific (Ckmm-cre) cre-expressing mice (19).
Figure 1.
Targeting of the Cfl2 gene. (A) Targeting strategy to create Cofi (conditionally targeted Cfl2) allele. An ∼10.4 kb region used to construct the targeting vector was subcloned from a positively identified C57BL/6 BAC clone (RP23-422H22) using a homologous recombination-based technique. The BAC was subcloned into an ∼2.4 kb pSP72 (Promega) backbone vector containing an ampicillin selection cassette. A pGK-gb2 loxP/FRT-flanked Neo cassette was inserted into the gene. The region was designed such that the SA extended 2.0 kb to the 3′ end of loxP/FRT-flanked Neo cassette. The LA extended 6.3 kb to the 5′ end of the single loxP site. The single loxP site was inserted upstream of exon 2, and the loxP/FRT-flanked Neo cassette is inserted downstream of exon 4. The target region was ∼2.1 kb containing exons 2–4. (B) Detection of WT and targeted alleles by Southern blot analysis. DNA isolated from electroporated ES cells was digested with EcoRI and analyzed by Southern blot analysis with a probe against 3′ external region (PB1/2) as shown in (A). The 8.6 and 5.1 kb bands represent WT and targeted alleles, respectively. (C) PCR analysis of DNA isolated from tails of P1 mice showing WT, heterozygous (Cofi/+) and homozygous genotypes (Cofi/Cofi) for the conditionally targeted allele. Primers were designed to include loxP1 present in the Cofi allele to differentiate the two based on sizes, with WT being 364 bp and Cofi 426 bp. (D) Three-primer PCR strategy to confirm excision of two to four exons of Cfl2 along with the Neo cassette. The 364 and 617 bp bands represent the WT (+/+) and Cfl2−/− (−/−) alleles, respectively. (E) RT–PCR using primers for exon 2 (238 bp) confirmed that cofilin-2 was successfully removed in various tissues including the quadriceps. (E) Detection of cofilin-2 protein by western blot analysis. Protein extracted from various tissues of WT (+/+) and Cfl2−/− (−/−) on P7 showed absent cofilin-2 in the quadriceps, triceps, gastrocnemius, diaphragm, heart, brain, kidney and lungs of the Cfl2−/− mice.
Early post-natal lethality of cofilin-2-deficient mice
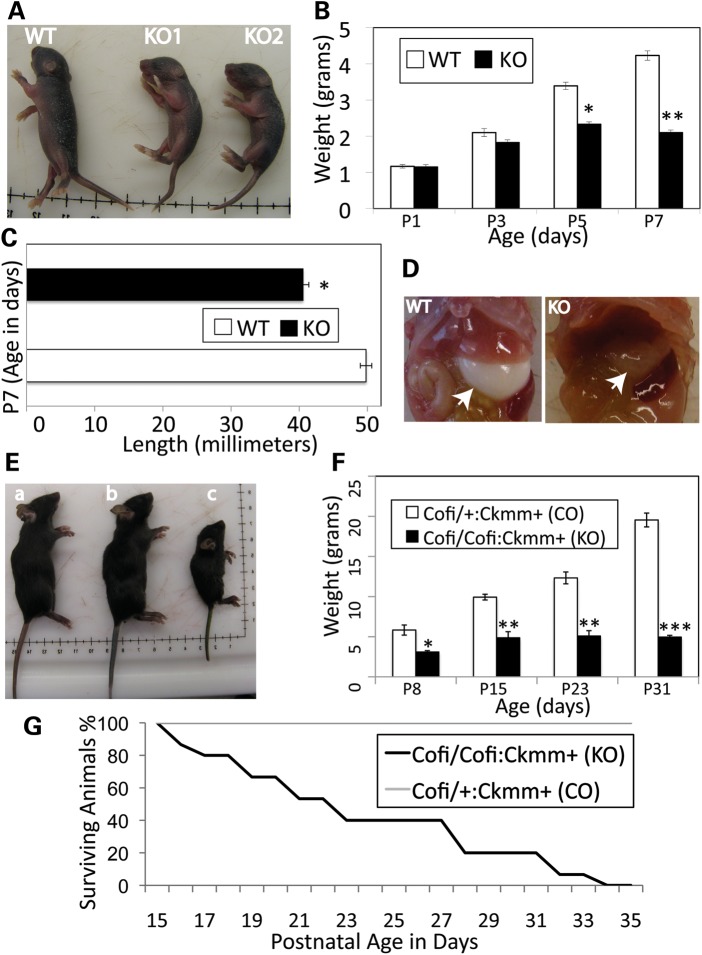
The cofilin-2-null (Cfl2−/−) mice were indistinguishable from their WT littermates at birth and were born in expected Mendelian ratios, but by post-natal day (P) 7, they were significantly smaller (Fig. 2A) and none survived past P8. The mean weights in grams at birth were 1.17 ± 0.12 (mean ± standard deviation) for the WT and 1.15 ± 0.13 for the Cfl2−/− mice (P = 0.84). At P3, the mean weight of Cfl2−/− mice was trending lower, although not statistically significant (1.8 ± 0.15 for Cfl2−/− and 2.1 ± 0.22 for WT, P = 0.11). By P7, the Cfl2−/− mice were significantly smaller and weighed less than half that of the WT mice (Fig. 2B). From P3 to P7, the Cfl2−/− mice gained only 0.3g, while the WT gained 2.1g. In addition, Cfl2−/− mice were significantly smaller in length compared with WT 40.6 ± 0.81 versus 49.8 ± 0.84 mm (P < 0.001) (Fig. 2C). They were unable to turn themselves when placed laterally and had markedly reduced spontaneous movements. Milk intake was evaluated to determine whether it was affected due to the muscle weakness. A similar amount of milk was present in the stomachs of WT and Cfl2−/− mice on P1 and P3, but by P7, Cfl2−/− mice had little or no detectable milk present suggestive of their inability to suckle (Fig. 2D). Cofi/Cofi:Mef2c+ (skeletal muscle-specific cofilin-2-KO) mice and Cofi/Cofi:Acta1+ (embryonic somites and heart-specific cofilin-2-KO) were very similar in presentation to the Cfl2−/− mice and died by P7–12 (data not shown).
Figure 2.
Phenotypic characterization of cofilin-2-deficient mice. (A–D) Cfl2−/− (constitutive cofilin-2-KO) mice. (A) The Cfl2−/− mice (KO1 and KO2) were significantly smaller than a WT littermate. (B) The mean weights for KO (Cfl2−/−) and WT mice were similar at birth (P1) and started to trend lower on P3, and by P5, KO mice were significantly lighter than the WT controls (P < 0.005). On P7, the KO mice weighed less than half that of the WT (P < 0.0001). (C) The length of KO mice was significantly less than that of WT littermates on P7 (P < 0.0001). (D) The stomach bubble (arrows) in KO mice was empty while WT stomachs were distended with milk. (E–G) Cofi/Cofi:Ckmm+ mice (muscle-specific cofilin-2 KO with post-natal excision of Cfl2). (E) The Cofi/Cofi:Ckmm+ mice (c) were significantly smaller compared with the age-matched WT (a) and littermate Cofi/+:Ckmm+ (b) mice as controls. (F) The mean weights of KO mice were significantly lower (Cofi/Cofi:Ckmm+) on P8 (P < 0.05), P15 (P < 0.005), P23 (P < 0.0005) and P31 (P< 0.0001) compared with their Cofi/+:Ckmm+ littermates as controls (CO). (G) The Kaplan–Meier curve showing survival of the Cofi/Cofi:Ckmm+ (KO) mice (median 21 days, range 16–33 days, n = 15).
To perform cofilin-2 excision in muscles (skeletal and cardiac) later in development, Ckmm-cre mice were used, where cre expression starts at E17, is ∼40% of maximal levels at birth and reaches highest levels at P10 and stays high after that (in rats) (19). Cofi/Cofi:Ckmm+ mice were significantly smaller (Fig. 2E), 2–4-fold lighter than Cofi/+:Ckmm+ controls (P8–31) (Fig. 2F) and lived for 16–33 days (Fig. 2G).
Skeletal muscles from cofilin-2-deficient mice reveal marked sarcomeric disruption
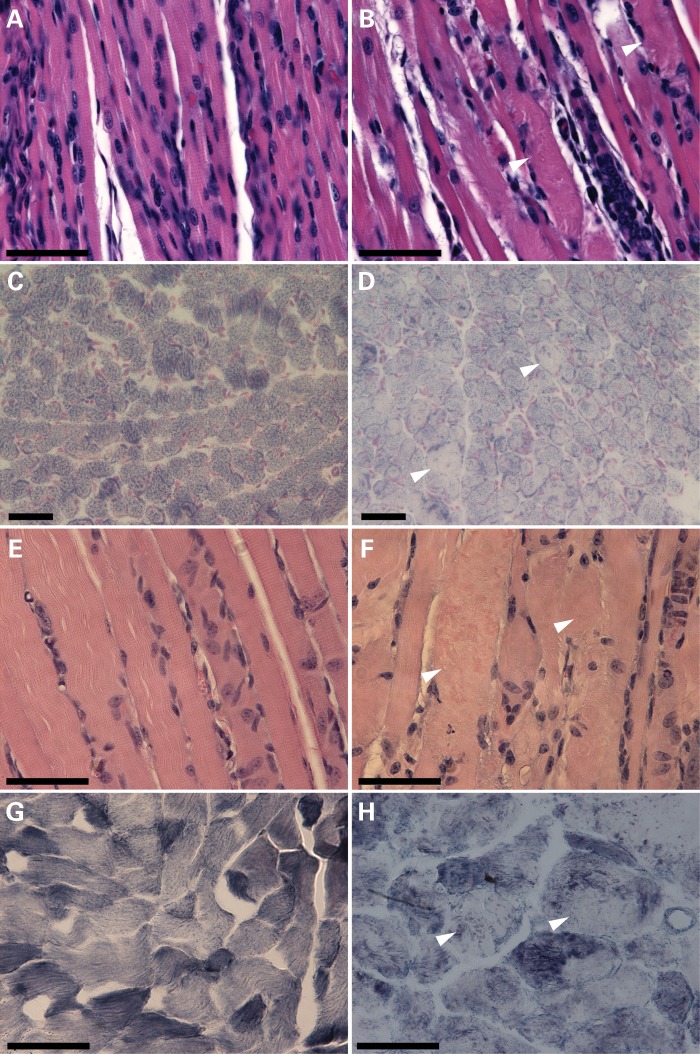
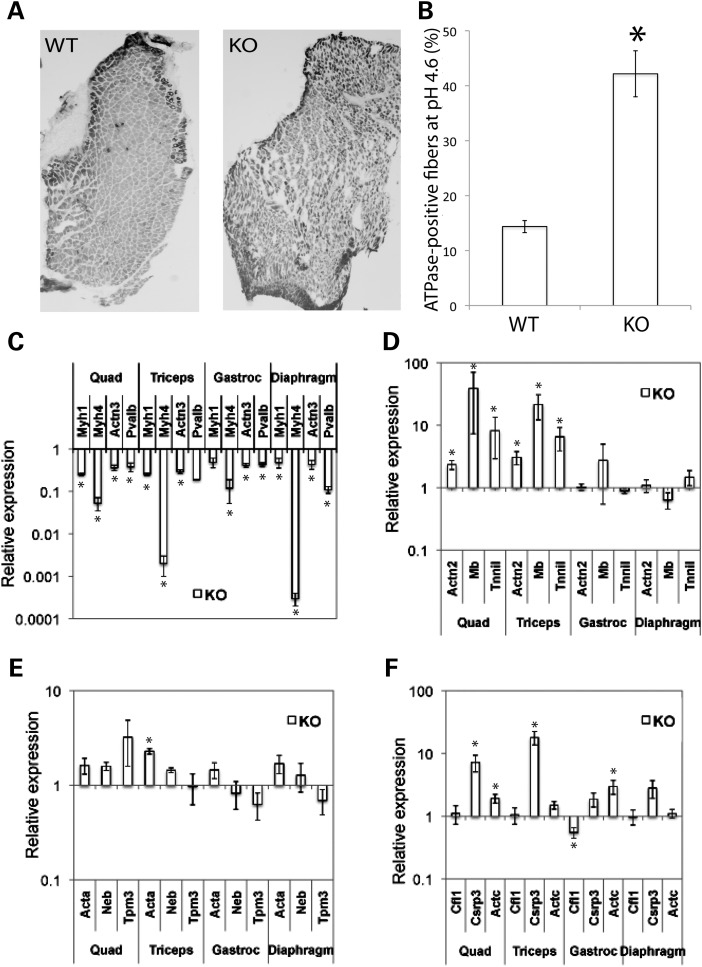
A pathological survey was performed on the various cofilin-2-KO mice at several ages up to and including the end stage. All the major organs including the heart, brain, lungs, intestines, liver, spleen and kidneys (excepting skeletal muscles) were evaluated and no gross abnormalities were identified. Histological examination of these tissues was also normal with the exception of enlarged alveolar spaces in the lungs from Cfl2−/− mice. A gross examination of major limb and trunk skeletal muscles revealed generalized and proportional reductions in size. Histological analysis of various skeletal muscles including the quadriceps, triceps, gastrocnemius and diaphragm from 7-day-old Cfl2−/−, Cofi/Cofi:Mef2c+, Cofi/Cofi:Acta1+ and 3-week-old Cofi/Cofi:Ckmm+ mice and littermate controls was performed. Control muscles were obtained from WT and various cre-expressing Cofi/+ mice (for muscle-specific excision models). Hematoxylin and eosin (H&E) staining of muscles from all four Cfl2-KO mice strains revealed ballooning degeneration of multiple myofibers that were interspersed with relatively normal-looking ones (Fig. 3B and F). Further analysis of frozen quadriceps sections from Cfl2−/− and Cofi/Cofi:Ckmm+ mice and their control littermates using nicotinamide adenine dinucleotide (NADH) staining showed pale core-like areas without staining within the myofibers (Fig. 3D and H) consistent with myofibrillar disruption and absent mitochondria. To evaluate whether myofibrillar disruption was fiber-type-specific, myosin ATPase staining was performed on triceps from WT and Cfl2−/− mice. No obvious differences were noted between fast and slow fibers in relation to the extent of disruption or percentages of disrupted fibers.
Figure 3.
Skeletal muscle histopathology of cofilin-2-deficient mice (Cfl2−/− and Cofi/Cofi:Ckmm+). H&E staining (A, B, E and F) of quadriceps cross-sections from WT (A) and Cfl2−/− (B) at P7, and Cofi/+:Ckmm+ (E) and Cofi/Cofi:Ckmm+ (F) mice at P21 was performed. Sections from Cfl2−/− and Cofi/Cofi:Ckmm+ muscles revealed ballooning sarcomeric degeneration (arrowheads). NADH staining (C, D, G and H) was performed on quadriceps cross-sections from WT (C), Cfl2−/− (D), Cofi/+:Ckmm+ (G) and Cofi/Cofi:Ckmm+ (H) mice. Sections from Cfl2−/− and Cofi/Cofi:Ckmm+ mice revealed pale areas within the myofibers as shown by arrowheads (scale bar = 50 μm).
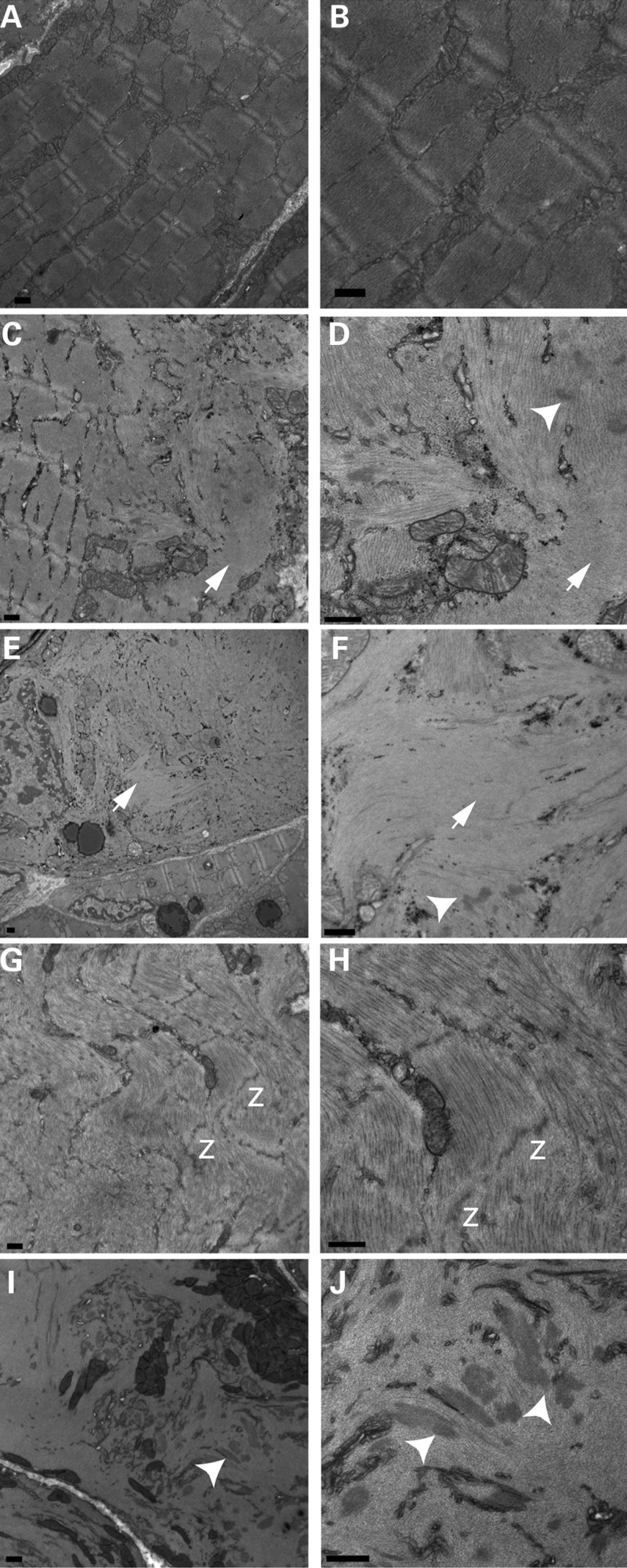
To determine the structure of cofilin-2-deficient skeletal muscle in more detail, electron microscopy (EM) was performed on various muscle groups (quadriceps, triceps, gastrocnemius and diaphragm) from the four KO strains and their littermate controls (at P7 for Cfl2−/−, Cofi/Cofi:Mef2c+, Cofi/Cofi:Acta1+ and P21 for Cofi/Cofi:Ckmm+ mice). These mice exhibited severe disruption of myofibrillar structure including lack of well-defined A- and I-bands alongside areas of relatively normal-looking sarcomeres (Fig. 4C) and normal-looking fibers (Fig. 4E). In areas of myofibrillar disruption, thick, irregular, electron dense structures resembling nemaline bodies were present (Fig. 4D, F, I and J) (20). In some areas, Z-lines were noted to be dissolving (Fig. 4G and H). In addition, F-actin accumulations were noted within the myofibers (Fig. 4C–F). Since myofibrillar disruption and actin accumulations were seen together, we conclude that the actin accumulations were not fiber-type-specific. Abnormal mitochondrial accumulations were also noted (Fig. 4I).
Figure 4.
Transmission EM of cofilin-2-deficient skeletal muscles. Samples were obtained from WT (quad, A and B), Cfl2−/− (triceps, C and D), Cofi/Cofi:Acta1+ (quad, E and F), Cofi/Cofi:Mef2c+ (quad, G and H) and Cofi/Cofi:Ckmm+ (triceps, I and J) mice. Sarcomeric disruptions were present in fibers from cofilin-2-deficient mice (C–J). WT fibers showing intact sarcomeres for comparison (A and B). Sarcomeric disruptions included intact sarcomeres alongside disrupted ones within a fiber (C) or an intact fiber alongside a disrupted fiber (E). Actin accumulations shown by arrows (C, D, G and H) and nemaline bodies shown by arrowheads (D, F, I and J) were present within disrupted fibers. Disintegrating Z-lines were noted, shown by ‘z’ (G and H) (scale bar = 500 nm).
α-Bungarotoxin and luxol blue staining of the quadriceps muscles were performed to evaluate the neuromuscular junctions and myelination of the nerve fibers, respectively. There were no obvious differences in staining patterns between WT and Cfl2−/− muscles (data not shown), suggesting that muscle weakness in these mice is not secondary to neuromuscular junction or nerve myelination defects.
F-actin accumulations in cofilin-2-deficient mouse muscles
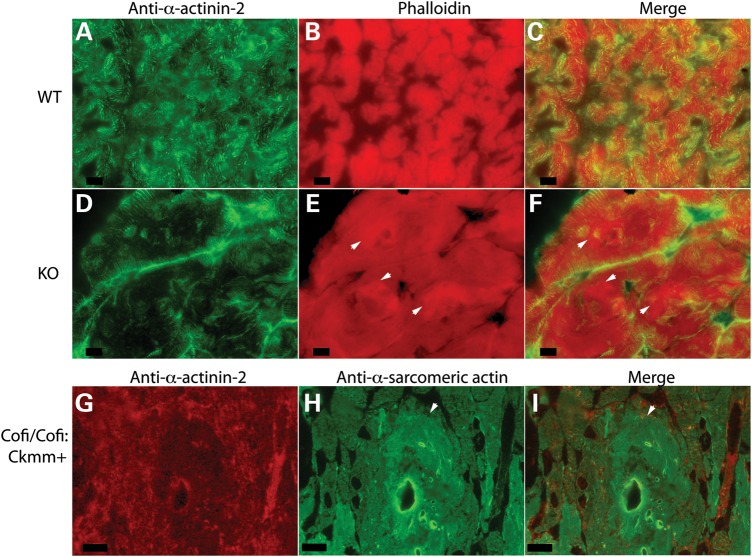
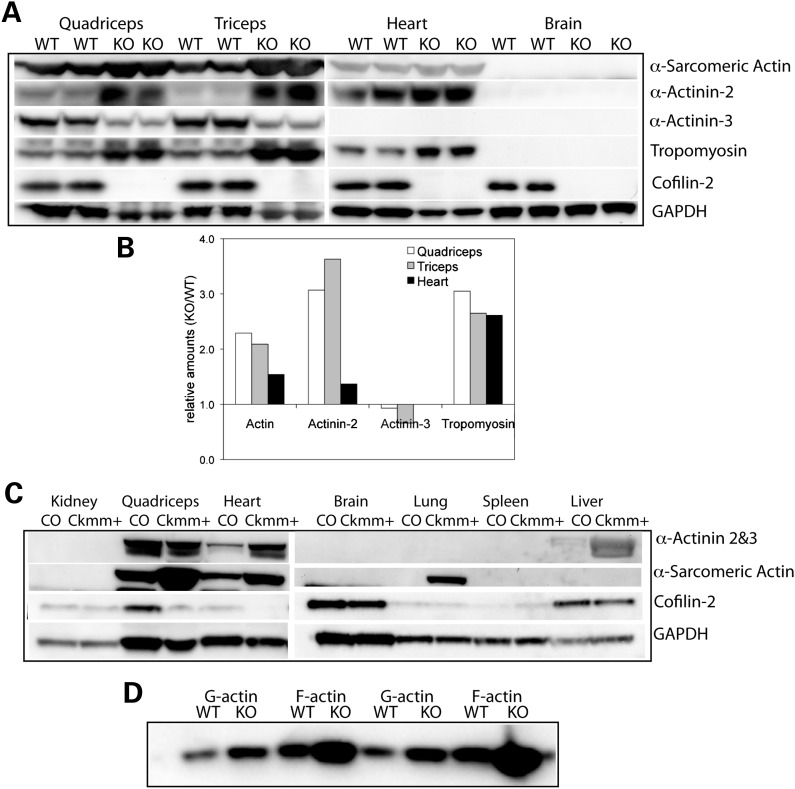
Destrin and the cofilins play essential roles in regulating actin dynamics by severing actin filaments and promoting dissociation of actin monomers from pointed ends (21). Compared with the other two isoforms, cofilin-2 has a relatively weaker actin depolymerization activity and it also promotes filament assembly (5). Since EM of cofilin-2-deficient muscles had revealed the presence of F-actin accumulations in various muscle groups including the quadriceps and triceps, we further evaluated the F-actin accumulations utilizing phalloidin staining of frozen sections to specifically label F-actin as well as an antibody that reacts with both F-actin and monomeric G-actin. Both stains reacted similarly, confirming the presence of F-actin accumulations in the cofilin-2-deficient mice (Cfl2−/− and Cofi:Cofi/Ckmm+) (Fig. 5). To determine whether cofilin-2 deficiency is associated with alterations in Acta1 expression and/or α-sarcomeric actin levels, we examined muscles from constitutive Cfl2−/− and Cofi:Cofi/Ckmm+ mice and their littermate controls. Acta1-mRNA levels were compared in samples taken from the quadriceps, triceps, gastrocnemius and diaphragm muscles of WT and Cfl2−/− mice. There was a small but statistically significant increase in levels of Acta1 transcripts in triceps from Cfl2−/− mice (n = 4) but not in the quadriceps, gastrocnemius and diaphragm, where the increasing trend was not statistically significant (Fig. 6E). In comparison, western blot analysis revealed a large increase in the amounts of actin present in the quadriceps and triceps of Cfl2−/− mice muscles compared with WT (Fig. 7A and B). Similarly, an increased amount of actin was present in the Cofi/Cofi:Ckmm+ mice (Fig. 7C). Further, F-actin and monomeric G-actin amounts were measured in the quadriceps muscles from Cfl2−/− and WT littermates, and G-actin/F-actin ratios were calculated. While there was an increase in both G-actin and F-actin in the Cfl2−/− mice compared with WT, there was a relatively higher increase in F-actin levels compared with G-actin (Fig. 7D). Although cofilin-2 has been shown to promote both polymerization and depolymerization of F-actin, these results are consistent with the primary function of cofilin-2 in vivo being to sever and depolymerize F-actin.
Figure 5.
Fluorescence microscopic analysis revealing actin filament accumulations. Muscle sections from WT (A–C), KO (Cfl2−/−) (D–F) and Cofi/Cofi:Ckmm+ (G–I) mice were immunostained with anti-α-actinin-2 (A, D and G), phalloidin (B and E) and anti-α-sarcomeric actin (H) antibodies. Merged images are shown in (C), (F) and (I). Several actin accumulations are indicated by arrowheads in (E), (F), (H) and (I) (scale bar = 10 µm).
Figure 6.
Fiber-type distribution in triceps of WT and Cfl2−/− mice on P7 and expression analysis of fiber-type-specific genes. Frozen sections of triceps from KO (Cfl2−/−) and WT littermates at P7 assayed for myosin ATPase activity (pH 4.6), demonstrating relative proportions of slow (dark stained) and fast (light stained) fibers. (A) KO triceps had more darkly stained fibers on microscopy. (B) Slow fibers and total number of fibers were counted for sections taken from the triceps (n = 4). The percentage of slow fibers in the WT and KO triceps was calculated, and the mean difference was found to be statistically significant (P < 0.05). Expression of fast fiber-specific genes (C) and slow fiber-specific genes (D) in various muscle groups from WT and KO mice (n = 4) was measured using qRT–PCR and analyzed. Expression of genes encoding thin filament proteins (E) and expression of Cfl1, Csrp3 and Actc (F) were also compared. Relative expression was compared using mean fold change (KO/WT) ± standard error, and P-values using t-test were calculated (*P < 0.05).
Figure 7.
Western blot analysis of sarcomeric protein expression in skeletal muscles, the heart and other tissues from Cfl2−/− and Cofi/Cofi:Ckmm+ mice. (A) Immunoblot analysis of P7 KO (Cfl2−/−) and WT littermate in duplicate. Increased levels of α-sarcomeric actin, α-actinin-2 and sarcomeric tropomyosin were present in the KO quadriceps, triceps and the heart. Decreased levels of α-actinin-3 were present in the KO quadriceps and triceps (absent in the heart). Cofilin-2 was expressed in the brain, but α-sarcomeric actin, α-actinin-2, α-actinin-3 and tropomyosin were absent. (B) Densitometric analysis using Quantity One software confirmed the increase in sarcomeric proteins as described above. (C) Immunoblot analysis of tissues from 3-week-old Cofi/Cofi:Ckmm+ (Ckmm+) and control (CO) littermate. Cofilin-2 levels were markedly reduced in the quadriceps and heart but were similar in other tissues including the kidney, brain, lung, spleen and liver. Increased levels of α-sarcomeric actin in the quadriceps, heart and presence of actin in lungs were noted in the Ckmm+ mice. (D) Assessment of G-actin and F-actin levels in the frozen muscle extracts of WT and KO mice (in duplicate) by separating the two fractions by centrifugation and probing blots using antibody against α-sarcomeric actin. Both G-actin and F-actin were increased in the KO, but an increase in F-actin was relatively higher.
Proportion of slow fibers and fiber-type-specific genes are altered in Cfl2−/− mice
Slow or type I fiber predominance is frequently observed in human congenital myopathies including nemaline myopathy (22–24). Slow fiber predominance has been identified in a mouse model with α-tropomyosin deficiency and NM phenotype (25). To investigate possible alterations in the fiber-type proportions, transverse sections of triceps muscle from four sets of 7-day-old Cfl2−/− and WT littermate mice were assayed for myosin ATPase activity using pH-sensitive histochemical stain (Fig. 6A). The mean percentage of slow fibers in the Cfl2−/− mice was significantly higher than the WT (42.2 ± 4.2 versus 14.4 ± 1.1%, P < 0.001) (Fig. 6B). To evaluate whether the increased proportion of slow fibers correlated with changes in expression of various fiber-type-specific genes, we compared them in the Cfl2−/− and littermate-matched WT mice across various muscle groups (quadriceps, triceps, gastrocnemius and diaphragm). Transcripts for genes expressed predominantly in fast fibers (Myh1, Myh4, Actn3 and Pvalb) and slow fibers (Actn2, Tnni1 and Mb) were compared. The fast fiber-specific genes were significantly downregulated in all the four muscle groups from Cfl2−/− mice (Fig. 6C); in comparison, slow fiber-specific genes were significantly upregulated in the cofilin-2-deficient quadriceps and triceps muscles (Fig. 6D).
We evaluated the transcripts of other genes of interest including those encoding for thin filament proteins (Acta1, Neb and Tpm3), Csrp3 and Actc (Fig. 6E and F). Levels of transcripts for genes encoding thin filament proteins were not significantly altered except upregulation of Acta1 in triceps. Further, Csrp3 was upregulated in the Cfl2−/− muscle compared with the littermate WT. The protein encoded by CSRP3 is known to interact with cofilin-2, thereby regulating F-actin dynamics in the cardiac and skeletal muscle, and it is upregulated in several myopathies (26). In addition, Actc, encoding cardiac actin, was upregulated in the Cfl2−/− muscle, indicative of ongoing regenerative activity (27), a feature that has been noted in muscular dystrophies (28).
Increased levels of sarcomeric proteins
Western blotting of striated muscles from Cfl2−/− and Cofi/Cofi:Ckmm+ mice and littermate controls was performed to evaluate amounts of various sarcomeric proteins present in the A-band. Levels of α-sarcomeric actin, α-actinin-2 and sarcomeric tropomyosin were elevated in the quadriceps and triceps muscles from Cfl2−/− mice while the amount of α-actinin-3 was lower (Fig. 7A and B). In addition, the cardiac muscle from Cfl2−/− mice exhibited an increase in α-sarcomeric actin, α-actinin-2 and tropomyosin-3 (Fig. 7A and B). A marked reduction in cofilin-2 in the skeletal muscles and heart was confirmed in the Cofi/Cofi:Ckmm+ mice (Fig. 7C). Antibody cross-reacting with both α-actinin-2 and α-actinin-3 showed similar total protein amounts in the quadriceps from control and Cofi/Cofi:Ckmm+ mice (Fig. 7C). This is consistent with a reduction in α-actinin-3 and an increase in α-actinin-2 seen in Cfl2−/− mice. As α-actinin-2 is mostly present in the slow fibers and α-actinin-3 in the fast fibers (29), an increase in α-actinin-2 and reduction in α-actinin-3 is consistent with slow fiber predominance seen in muscles from Cfl2−/− mice.
Cofilin-2 is required for muscle maintenance
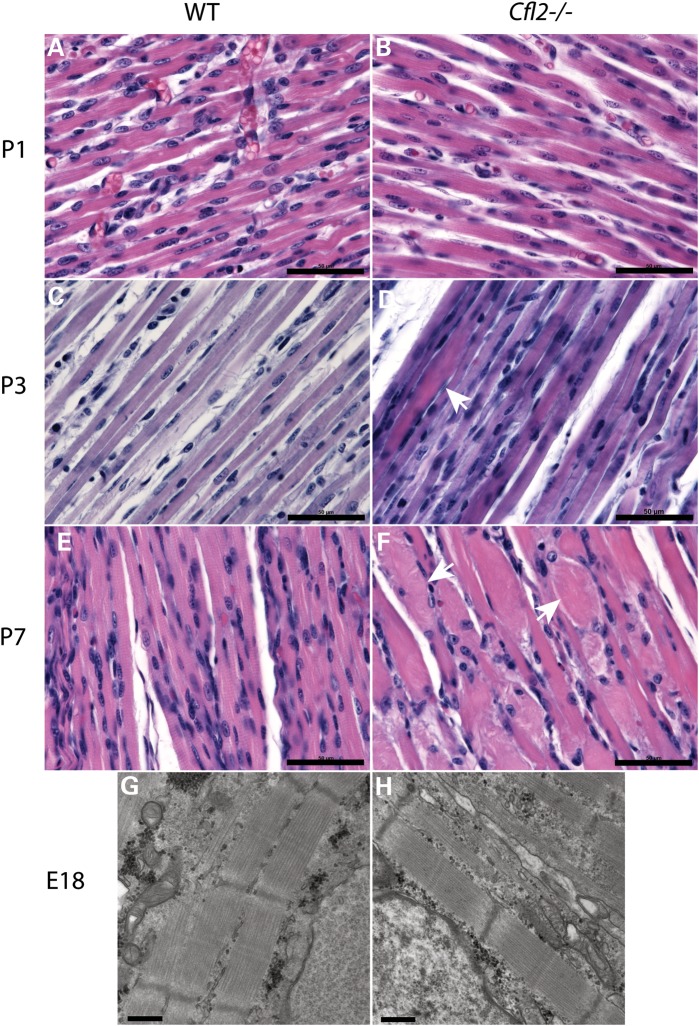
The role of cofilin-2 in myofibrillogenesis is unclear. Its expression in mouse embryos begins at E10.5 and by E13 in the somites and it is present in the myogenic regions of the forelimb buds and myotomes, areas where myogenin, a muscle-specific transcription factor than induces myogenesis, and troponin T, a muscle structural protein, are expressed at the same time (4). To evaluate the possibility that Cfl2 expression is required for myogenesis, sections of quadriceps muscles were prepared from P1, P3 and P7 Cfl2−/− and WT littermate mice. Light microscopy of H&E-stained sections showed no obvious differences between WT and Cfl2−/− muscles at P1 (Fig. 8A and B). On P3, occasional degenerating fibers were noted in the Cfl2−/− mice (Fig. 8D), and by P7, muscles from Cfl2−/− mice showed extensive and severe degenerative changes (Fig. 8F). Thus, while cofilin-2 was not essential for initial myofibrillogenesis, its expression was critical for muscle maintenance soon after birth. To further confirm that myofibrils were indeed well formed in the Cfl2−/− mice, EM was performed on sections from E18 WT and Cfl2−/− embryos. Sarcomeres from E18 Cfl2−/− mice had a normal sarcomeric structure with wide myofibrils, distinct A-bands and narrow, uniformly spaced Z-lines, very similar to the WT (Fig. 8G and H), demonstrating normal myofibrillogenesis.
Figure 8.
Evolution of muscle degeneration in Cfl2−/− mice (B, D, F and H) and comparison with WT littermate (A, C, E and G). On P1, muscles from the WT and Cfl2−/− mice were very similar on H&E staining (A and B). By P3, occasional degenerating fibers were noted (arrow) in Cfl2−/− mice (D). On P7, multiple degenerated fibers (arrows) were present in the Cfl2−/− mice muscles (F) (scale bar = 50 µm). EM on a section from E18 embryos showed normally developed myofibrils in both WT and Cfl2−/− mice (scale bar = 500 nm).
Expression of cofilin-1, destrin and cofilin-2 isoforms is essentially unchanged in Cfl2−/− mice
To determine whether the expression of other AC proteins (cofilin-1 and destrin) is altered in response to cofilin-2 deficiency, their RNA transcripts and protein levels were compared between the WT and Cfl2−/− mice at P7. Quantitative RT (qRT)–PCR was performed on the extracted RNA from various muscle groups (quadriceps, triceps, gastrocnemius and diaphragm) of WT and Cfl2−/− mice (n = 4). Dstn transcripts were not detected in either the WT or Cfl2−/− mouse muscles, whereas Cfl1 mRNA transcript levels were similar in both except gastrocnemius where the Cfl1 transcripts were reduced in the Cfl2−/− mice (Fig. 6E). To evaluate alterations, if any, at the protein level, western blotting of samples from the quadriceps and triceps was performed using specific anti-cofilin-1 antibodies. Levels of cofilin-1 appeared similar in WT and Cfl2−/− muscles on P1, P3 and P7 (Fig. 9O). The levels of phosphorylated cofilin-1 were also noted essentially indistinguishable between WT and Cfl2−/− muscles on P1, P3 and P7 (Fig. 9O). These data suggest that levels of other AC family proteins remain essentially unchanged in mice in response to cofilin-2 deficiency.
Figure 9.
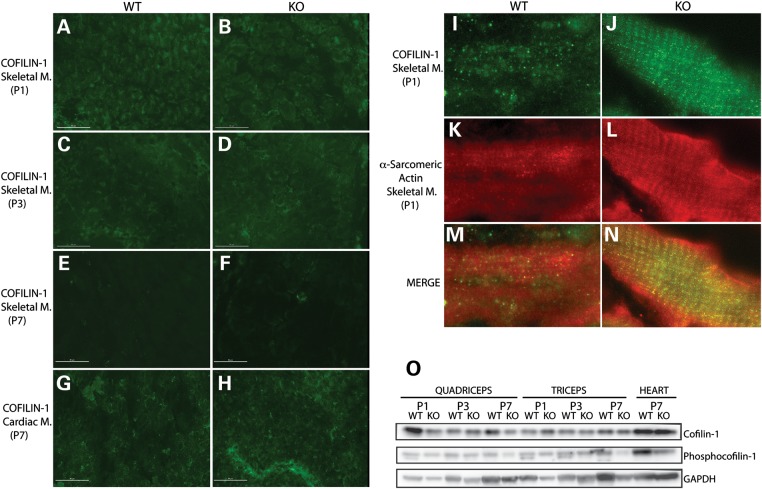
Cofilin-1 expression in WT and KO (Cfl2−/−) mice. Immunofluorescence analysis to evaluate cofilin-1 expression in skeletal muscle samples obtained from WT and KO mice on P1 (A and B), P3 (C and D) and P7 (E and F), and cardiac muscle samples on P7 (G and H) using anti-cofilin-1-specific antibody. In skeletal muscles, progressive reduction in cofilin-1 expression was noted with age (A, C and E for WT and B, D and F for KO), and on P7, cofilin-1 was absent from within myofibers in WT (E) and KO mice (F). In the cardiac muscle, cofilin-1 remained present within the myofibers (G and H) on P7 (scale bar = 50 μm). Evaluation of cofilin-1 localization using anti-cofilin-1 (I and J)- and anti-α-sarcomeric actin (K and L)-specific antibodies, along with merged images (M and N), in skeletal muscles from WT and KO mice on P1. Cofilin-1 was present in a striated pattern, and co-localized with α-sarcomeric actin in both WT (I, K and M) and KO (J, L and N) mice. (O) Western blot analysis of cofilin-1 and phosphocofilin-1 levels in the WT and KO quadriceps, triceps and heart on P1, P3 and P7. The amounts of cofilin-1 and phosphocofilin-1 were similar between the WT and KO quadriceps, triceps and heart on P1, P3 and P7.
Timing of muscle weakness in Cfl2−/− mice coincides with cofilin-1 depletion within myofibers
To further understand cofilin-1 dynamics in the developing muscle, we performed immunofluorescence on muscle samples from Cfl2−/− and littermate WT mice using cofilin-1-specific antibody. On P1, cofilin-1 co-localized with actin (A-band) on longitudinal sections (Fig. 9I–N) and was highly expressed within the myofibers on transverse sections in both WT and Cfl2−/− mice (Fig. 9A and B). On P3, cofilin-1 expression was markedly reduced, and by P7, very little if any cofilin-1 was detectable within the myofibers of WT and Cfl2−/− mice (Fig. 9C–F). The timing of reduction in cofilin-1 coincided with the onset of myofibrillar degeneration and skeletal muscle weakness in Cfl2−/− mice. Further, cofilin-1 expression was evaluated in WT and Cfl2−/− cardiac muscles, as Cfl2−/− mice do not reveal any evidence of degeneration on H&E staining on P7 (data not shown). Interestingly, cofilin-1 was highly expressed within the myofibers of cardiac muscles from WT and Cfl2−/− mice on P7 (Fig. 9G and H). Based on these findings, it is plausible that cofilin-1, if overexpressed, may compensate for cofilin-2 deficiency.
DISCUSSION
Cofilin-2, a member of the AC group of proteins that include cofilin-1 and destrin, regulates actin dynamics by severing actin filaments and causing actin depolymerization (1,30). Cofilin-2 is primarily expressed in skeletal and cardiac muscles and its molecular function in myogenesis and/or muscle maintenance is unknown. We previously identified a homozygous p.A35T-CFL2 mutation in two sisters with congenital myopathy from a consanguineous family who presented with a pathological diagnosis of nemaline myopathy with minicores (2). Since then, a second consanguineous family with congenital myopathy and a novel homozygous missense CFL2 mutation has been identified (C. Ockeloen, personal communication). Our analysis of the various Cfl2-conditional KO mouse strains revealed parallels to the human condition. Mice with constitutive excision of cofilin-2 were born alive and noted to be very similar to WT littermates, but by P3, affected animals exhibited dramatic reductions in size and activity and subsequently worsened rapidly progressing to death by P8. Affected pups did not move much and their stomachs were empty, consistent with their inability to suckle. Similar phenotypes were noted with Cofi/Cofi:Mef2c+ (skeletal muscle-specific cofilin-2-KO) and Cofi/Cofi:Acta1+ (embryonic somites and cardiac-specific cofilin-2-KO). Cofi/Cofi:Ckmm+ mice (skeletal and cardiac muscle-specific) survived a little longer (10–31 days) due to delayed onset of cre expression. Histopathological and EM analysis of the skeletal muscle from cofilin-2-deficient mice revealed ballooning degeneration of several fibers, core-like lesions, extensive sarcomeric disruptions, nemaline bodies and actin accumulations. Skeletal muscle weakness with nemaline bodies, core-like lesions and actin accumulations were also present in the human patient with p.A35T-CFL2 mutation, the mutation postulated to cause cofilin-2 misfolding and consequent reduction in its levels. The degree of weakness in the murine KOs is clearly greater, as the affected girls were ambulant until adolescence, suggesting some partial or residual activity of the mutated cofilin-2 in the human patients. Nevertheless, we conclude that cofilin-2 is critical for muscle function soon after birth, and its deficiency is associated with a unique myopathy in mice similar to that seen in humans with CFL2 mutations.
Cofilin-2 expression in mice starts at E10.5 on the medial side of somites (myotomal myogenic region), and by E13, it is present in the limb buds (along with myogenin, a muscle-specific transcription factor, and troponin T, a structural protein) (4,31). This suggests that cofilin-2 may be critical in muscle development. Interestingly, Cfl2−/− mice had well-formed myofibrils at E18 and normal myofibers on P1, indicating that cofilin-2 was not critical for myofibrillogenesis. Given its complementary pattern of expression and sarcomeric subcellular localization, we feel it is likely that cofilin-1 serves as a fetal skeletal muscle isoform, compensating for cofilin-2 deficiency during initial myofibrillogenesis in the Cfl2−/− mice.
To evaluate expression of other AC proteins in response to cofilin-2 deficiency, we compared expression of Cfl1 and Dstn in muscle samples from WT and Cfl2−/− mice using qRT–PCR and western blot. On qRT–PCR analysis, Cfl1 transcripts were present, while Dstn transcripts were undetectable in both WT and Cfl2−/− mice. Further, Cfl1 expression was very similar in the WT and Cfl2−/− mice across several muscle groups. Western blot analysis to evaluate levels of cofilin-1 in several muscle groups revealed similar levels of cofilin-1 in WT and Cfl2−/− mice on P1, P3 and P7. Thus, we found no evidence that cofilin-1 expression was increased in response to cofilin-2 deficiency. However, it was interesting to note that residual cofilin-1 remained present in muscle samples from Cfl2−/− mice when they died on P7. Previously, it has been postulated that cofilin-2 deficiency effects may appear only in the adult skeletal muscle to coincide with the timing when mice skeletal muscles lack cofilin-1 (by 2 months of age) (4). To evaluate whether there was an alteration in cofilin-1 localization in muscles during the first 7 days after birth, immunofluorescence using cofilin-1-specific antibody was performed on WT and Cfl2−/− muscles, sampled on P1, P3 and P7 after birth. On P1, cofilin-1 was highly expressed, localized to the A-band of the myofibers, but by P7, cofilin-1 expression within the myofibers was absent (reduced on P3), and instead, it localized mainly to the connective tissue and blood vessels (data not shown). No differences in the cofilin-1 expression patterns were noted between the WT and Cfl2−/− mice. Thus, lack of cofilin-1 and cofilin-2 within the skeletal myofibers of Cfl2−/− mice may explain the rapid deterioration in muscle structure and function soon after birth. Interestingly, high expression of cofilin-1 within cardiac myofibers from 7-day-old WT and Cfl2−/− mice heart samples correlated well with the lack of cardiac muscle degeneration in the Cfl2−/− mice. Overall, cofilin-1 may be able to compensate for cofilin-2 deficiency if it is expressed within the myofibers. Breeding cofilin-2-deficient mice with lines overexpressing cofilin-1 in the skeletal muscle may help understand this issue further.
Increased proportions of slow myofibers have been described with various human myopathies and in a mouse model of NM with M9R-Tpm3-knockin mutation and, in many instances, are probably a non-specific response to myopathic conditions (20,32). Similarly, we identified an increased proportion of slow fibers (type I/type IIa) in Cfl2−/− mouse skeletal muscles on ATPase staining. In addition, transcripts for several genes that are primarily expressed in fast fibers (type IIx/IIb) were significantly reduced in abundance, and transcript levels for slow fiber-specific genes were elevated in Cfl2−/− mice compared with WT littermates. Additionally, western blotting revealed increased levels of α-actinin-2 protein (slow fiber-specific) and reduced amounts of α-actinin-3 (fast fiber-specific) in Cfl2−/− mice. Thus, a significant increase in the proportion of slow fibers in Cfl2−/− mice is consistent with other human and murine myopathies. The molecular mechanisms responsible for myopathy-related alterations in the fiber type are yet to be understood.
The presence of F-actin accumulations in cofilin-2-KO mice seen by EM, immunofluorescence and western blot experiments can be explained by reduced or abnormal actin depolymerization activity due to cofilin-2 deficiency. Similar F-actin accumulations in muscles have been noted with certain ACTA1 mutations that alter actin polymerization contributing to insoluble actin filaments (33). F-actin accumulations in corneal epithelial cells have been described in mice deficient in destrin, a member of the AC group of proteins (15).
Z-line disintegration and marked sarcomeric disruption was a critical finding in these cofilin-2-deficient mice. The underlying mechanism is unclear, although we suspect that extensive sarcomeric disruption may be preceded by Z-line disintegration. Z-lines are formed from the interdigitating barbed ends of the antiparallel actin filaments from adjoining sarcomeres cross-linked by α-actinin. Recently, cross-linking of actin filaments by α-actinin was noted to be increased by cofilin-1 in in vitro experiments (34). It was hypothesized that this action was due to cofilin's ability to alter the actin filament twist. It is plausible that cofilin-2 may play a stabilizing role in the muscle Z-line by strengthening the actin–actinin cross-link, and its deficiency may weaken and consequently disrupt sarcomeres starting at the Z-line during muscle contraction. Myofibrillar myopathy (MFM), a muscle disorder associated with similar Z-line disruption, myofibrillar degeneration and accumulation of abnormal proteins, is caused by mutations in genes that encode proteins involved in maintaining the structural integrity of the Z-line including desmin, αB-crystallin, myotilin and LIM domain binding 3 (35). While a majority of MFM cases are autosomal dominant and adult-onset, CFL2 may be a candidate gene for atypical MFM patients with an autosomal recessive inheritance and early-onset disease.
MATERIALS AND METHODS
Animal studies
All studies were performed with approval from the institutional animal care and use committee at Children's Hospital Boston (Boston, MA, USA).
Gene targeting and generation of cofilin-2-conditional KO (Cofi) mice
Cfl2-KO mouse generation was performed by inGenious Targeting Laboratory (Ronkonkoma, NY, USA). An ∼10.4 kb region used to construct the targeting vector was first subcloned from a positively identified C57BL/6 BAC clone (RP23-422H22) using a homologous recombination-based technique. The BAC was subcloned into an ∼2.4 kb pSP72 (Promega) backbone vector containing an ampicillin selection cassette. A pGK-gb2 loxP/FRT-flanked Neo cassette was inserted into the gene. The region was designed such that the short homologous arm (SA) extends 2.0 kb to the 3′ end of loxP/FRT-flanked Neo cassette. The long homologous arm (LA) extends 6.3 kb to the 5′ end of the single loxP1 site (Fig. 1A). This single loxP1 site is inserted upstream of exon 2, and the loxP/FRT-flanked Neo cassette is inserted downstream of exon 4. The target region is ∼2.1 kb containing exons 2, 3 and 4. The targeting vector is confirmed by restriction analysis after each modification step and by sequencing using primers designed to read from the Neo selection cassette into the 3′ end of the LA and the 5′ end of the SA. The loxP1 site was confirmed by sequencing. Ten micrograms of the targeting vector were linearized by ClaI and then transfected by electroporation of IC1 C57BL/6 ES cells. After selection with G418 antibiotic, surviving clones were expanded for PCR analysis to identify recombinant ES clones. Secondary confirmation of positive clones identified by PCR was performed by Southern blotting analysis. In brief, DNA was digested with EcoRI and electrophoretically separated on a 0.8% agarose gel. After transfer to a nylon membrane, the digested DNA was hybridized with a probe targeted against the 3′ external region. DNA from C57Bl/6 (B6) mouse strains was used as WT controls. The expected sizes were 8.6 kb for WT and 5.1 kb for conditional Cfl2-KO allele (Cofi) (Fig. 1B). The PB1/2 probe primers were PB1: AAC GCA GGG AAA TTC TAA CCC and PB2: ACA GGG AAA CAG CTC TAC TAT C.
The targeted IC1 C57BL/6 ES cells were microinjected into Balb/c blastocysts. Resulting chimeras with a high-percentage black coat color were mated to WT C57BL/6 mice to generate heterozygous offspring. Tail DNA was analyzed for mice heterozygous for conditional Cfl2-KO allele (Cofi/+) using the forward primer (SDL1): GGT TGG AGT TTA CAC TTG ATT CTC TT and reverse primer (SDL2): CAA CGT AAG CAG TCC TAT TAG GTC to include loxP1 site present in the conditionally KO allele. With the addition of the loxP site, the amplified conditionally KO allele was 426 bp while the WT allele was 364 bp. The heterozygotes were bred to obtain the homozygous mice (Cofi/Cofi) as shown in Figure 1C. Routine genotyping was achieved on genomic DNA isolated from tail tips of mice as described for each stain below.
Creating constitutive and tissue-specific mice and PCR screening strategy
Constitutive KO (Cfl2−/−) mice
Cofi/Cofi mice were bred with homozygous C57BL/6-Tg(Zp3-cre)93Knw/J (zp3-cre/zp3-cre) mice to create Cofi/+:zp3-cre/+ mice. Female Cofi/+:zp3-cre/+ were crossed with WT male (Cfl2+/+) to create heterozygous constitutive KO mice (Cfl2+/−) who were then inbred to obtain homozygous constitutive KO (Cfl2−/−) mice. The Zp3-cre-positive mice were screened using the following primers: WT_F: CTA GGC CAC AGA ATT GAA AGA TCT, WT_R: GTA GGT GGA AAT TCT AGC ATC ATC C, Cre_F: GCG GTC TGG CAG TAA AAA CTA TC and Cre_R: GTG AAA CAG CAT TGC TGT CAC TT. The size of WT allele is 324 bp and of Zp3-cre allele is 100 bp. Cofilin-2 excision was confirmed using the following primers: SDL1, SDL2 (as described before) and SDL5_R: TCC GGC TAA TAG CAG AGA GC. The WT allele PCR product size is 364 bp (SDL1 and SDL2) and the KO allele size is 617 bp (SDL1 and SDL5_R) (Fig. 1D). SDL2 is excised in the KO allele and the PCR product size for SDL1–SDL5_R primers in WT allele is 2.6 kb. Sequencing of the PCR products confirmed the excision of exons 2–4 and the Neo cassette.
Cofi/Cofi:Ckmm-cre/+
Cofi/Cofi mice were bred with B6.FVB(129S4)-Tg(Ckmm-cre)5Khn/J hemizygous mice (Ckmm-cre/+) obtained from Jackson labs to create Cofi/+:Ckmm-cre/+ mice which were then bred with Cofi/Cofi mice to create Cofi/Cofi:Ckmm-cre/+ mice. The primers used to genotype Ckmm-cre were: WT_F: CAA ATG TTG CTT GTC TGG TG, WT_R: GTC AGT CGA GTG CAC AGT TT, Cre_F: TAA GTC TGA ACC CGG TCT GC and Cre_R: GTG AAA CAG CAT TGC TGT CAC TT. The WT control size is 200 bp and the Ckmm-cre is 450 bp.
Cofi/Cofi:Mef2c-cre/+
Hemizygous Mef2c-cre mice were resuscitated from cryopreserved material from MMRRC (Item no. 030261-UNC-RESUS). Cofi/Cofi mice were bred with them to create Cofi/+:Mef2c-cre/+ mice that were then bred with Cofi/Cofi mice to create Cofi/Cofi:Mef2c-cre/+ mice. The following genotyping primers were used: forward: TGC CAC GAC CAA GTG ACA GC and reverse: CCA GGT TAC GGA TAT AGT TCA TG. A 700 bp band is present in the Mef2c-cre transgenic mice.
Cofi/Cofi:Acta1-cre/+
Cofi/Cofi mice were bred with B6.Cg-Tg(Acta1-cre)79Jme/J hemizygous mice (Ckmm-cre/+) obtained from Jackson labs to create Cofi/+:Acta1-cre/+ mice which were then bred with Cofi/Cofi mice to create Cofi/Cofi:Acta1-cre/+ mice. The primers used to detect Acta1-cre were the same as used for the Zp3-cre allele.
Tissue collection
Mice were euthanized at various ages (P1, P3, P7 and P28) using inhaled CO2 followed by decapitation immediately prior to tissue collection as per the regulations of the institutional animal care and use committee at Children's Hospital Boston. The quadriceps, gastrocnemius, triceps, soleus and diaphragm muscles were removed in addition to other tissues including the heart, lung, liver, kidneys, brain and spleen. Euthanized mice were also sent for complete autopsy performed at the Rodent Histopathology Core, Harvard Medical School, Boston, MA, USA. In addition, embryos 18-day post-coitus (E18) were dissected and muscle tissue harvested for EM.
Histological analysis
Eight-micrometer cross-sections of isopentane-frozen tissue were obtained and stained with H&E for evaluation using an Eclipse 50i microscope (Nikon Instruments Inc., Melville, NY, USA). Light microscopic images were captured using a SPOT Insight 4 Meg FW Color Mosaic camera and SPOT 4.5.9.1 software (Diagnostic Instruments Inc., Sterling Heights, MI, USA). H&E, NADH and ATPase stainings were performed using standard protocols.
Electron microscopy
Skeletal muscle samples (1–2 mm cubes) were fixed in 2.5% glutaraldehyde, 1.25% paraformaldehyde and 0.03% picric acid in 0.1 M sodium cacodylate buffer (pH 7.4) overnight in room temperature. They were then washed in 0.1 M cacodylate buffer and post-fixed with 1% osmium tetroxide (OsO4)/1.5% potassium ferrocyanide (KFeCN6) for 1h, washed in water 3× and incubated in 1% aqueous uranyl acetate for 1 h followed by two washes in water and subsequent dehydration in grades of alcohol (10 min each; 50, 70, 90%, 2 × 10 min 100%). The samples were then put in propylene oxide for 1h and infiltrated ON in a 1:1 mixture of propylene oxide and TAAB Epon (Marivac Canada Inc., St Laurent, Canada). The following day, the samples were embedded in TAAB Epon and polymerized at 60°C for 48h. Ultrathin sections (∼60 nm) were cut on a Reichert Ultracut-S microtome, picked up on to copper grids stained with lead citrate and examined in a JEOL 1200EX Transmission electron microscope or a TecnaiG² Spirit BioTWIN, and images were recorded with an AMT 2k CCD camera. This was performed at the EM Core of the Harvard Medical School.
RT–PCR analysis
RNA was extracted from various muscle groups using RNeasy Fibrous Tissue Mini Kit (Qiagen, Valencia, CA, USA). Extracted RNA was converted to cDNA using SuperScript III First-Strand Synthesis System (Invitrogen, Grand Island, NY, USA). Specific primers for Cfl2 exons 2–4 (forward: CAT TGG TGA CAC TGT AGA G; reverse: CAT TTA CTT GCC ACT CAT GT) were used. For qRT–PCR analysis, 100 ng of the cDNA was added to 10 μl of the Taqman Universal PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA) and 1 μl of the appropriate Taqman probe (Applied Biosystems) to create the qRT–PCR samples. VIC-labeled Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the control probe and primers for genes of interest had a FAM probe. qRT–PCR was performed and analyzed using the 7300 Real Time PCR System (Applied Biosystems) and 7500 Real-Time PCR System Sequence Detection Software v1.4. The PCR program used was 50°C for 2min, 95°C for 10min and 40 repetitions of 95°C for 0.15min and 60°C for 1min. The relative amount of target gene expression in Cfl2−/− animals was calculated as fold changes relative to WT littermate controls that were normalized to GAPDH.
Immunofluorescence
For immunofluorescence studies, 8 μm frozen transverse sections of the quadriceps muscle were double stained with rabbit α-actinin-2 (36) (3A or 3B at 1:200) and either mouse anti-α-sarcomeric actin (5C5, 1:200 dilution, Sigma, St Louis, MO, USA) or AlexaFluor-conjugated phalloidin 546 (1:40, Molecular Probes, Carlsbad, CA, USA). For phalloidin staining, tissue sections were fixed with 4% formaldehyde solution, and for others, methanol was used. In addition, 1-, 3- and 7-day-old skeletal muscle and 7-day-old heart sections were stained with anti-cofilin-1 antibody (D3F9, 1:40 dilution, Cell Signaling, Beverly, MA, USA). Secondary antibodies included AlexaFluor-conjugated anti-rabbit and anti-mouse IgG (1:100; Molecular Probes). Cover slips were mounted using Vectashield Mounting Medium with or without DAPI (Vector Laboratories, Burlingame, CA, USA). Staining was evaluated using a Nikon Eclipse 90i microscope using NIS-Elements AR software (Nikon Instruments Inc.).
Western blots
Tissues from various muscles and organs were frozen at necropsy and stored at −80°C until analysis. Protein isolation and western blot procedures were performed as described previously (37). Transferred proteins were probed with antibodies against cofilin-2 (C7506-50C, 1:250 dilution; US Biological, Swampscott, MA, USA), cofilin-1 (D3F9, 1:250 dilution, Cell Signaling), phosphorylated cofilin-1 (H-2, 1:250 dilution, Santa Cruz Biotechnology, Santa Cruz, CA, USA), α-sarcomeric actin (5C5, 1:1000 dilution, Sigma), sarcomeric tropomyosin (CH1, Abcam, Cambridge, MA, USA), α-actinin-2 (3A or 3B) (36), α-actinin-3 (4A or 4B) (36), sarcomeric α-actinin (reacts with both α-actinin-2 and α-actinin-3) (Sigma A5044) and GAPDH (6C5, 1:10,000 dilution; Abcam PLC) and visualized using enhanced chemiluminescence. Adequacy of transfer was determined using Ponceau S staining. Quantification of protein levels normalized to GAPDH was performed using the program Quantity One, version 4.2.1 (Bio-Rad Laboratories, Inc., Hercules, CA, USA) on an Image Station 440 (Kodak DS; Eastman Kodak Co., Rochester, NY, USA). G-actin/F-actin assay was performed using kit from Cytoskeleton (Cat. # BK037, Denver, CO, USA).
FUNDING
This work was made possible by generous support from the Lee and Penny Anderson Family Foundation and by grants from the Muscular Dystrophy Association, USA (MDA201302); the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (K08 AR055072 to P.B.A. and R01 AR044345 to A.H.B.); and by a Children's Hospital Boston Career Development Award to P.B.A.
ACKNOWLEDGEMENTS
We wish to thank Brandon Rider and Susie Kim for assistance with mouse dissections and Roderick Bronson of the Rodent Histopathology Core, Harvard Medical School, and Maria Ericsson, Louise Trakimas and Elizabeth Benecchi of the Electron Microscopy Core, Department of Cell Biology, Harvard Medical School, as well as Behzad Moghadaszadeh, Vandana Gupta and Michael Lawlor for their critical advice and assistance in performing histopathology and electron microscopy. Thanks also to Amanda Beggs for critical reading of the manuscript.
Conflict of Interest statement. None declared.
References
- 1.Bamburg J.R. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal P.B., Greenleaf R.S., Tomczak K.K., Lehtokari V.L., Wallgren-Pettersson C., Wallefeld W., Laing N.G., Darras B.T., Maciver S.K., Dormitzer P.R., et al. Nemaline myopathy with minicores caused by mutation of the CFL2 gene encoding the skeletal muscle actin-binding protein, cofilin-2. Am. J. Hum. Genet. 2007;80:162–167. doi: 10.1086/510402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thirion C., Stucka R., Mendel B., Gruhler A., Jaksch M., Nowak K.J., Binz N., Laing N.G., Lochmuller H. Characterization of human muscle type cofilin (CFL2) in normal and regenerating muscle. Eur. J. Biochem. 2001;268:3473–3482. doi: 10.1046/j.1432-1327.2001.02247.x. [DOI] [PubMed] [Google Scholar]
- 4.Mohri K., Takano-Ohmuro H., Nakashima H., Hayakawa K., Endo T., Hanaoka K., Obinata T. Expression of cofilin isoforms during development of mouse striated muscles. J. Muscle Res. Cell Motil. 2000;21:49–57. doi: 10.1023/a:1005682322132. [DOI] [PubMed] [Google Scholar]
- 5.Vartiainen M.K., Mustonen T., Mattila P.K., Ojala P.J., Thesleff I., Partanen J., Lappalainen P. The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol. Biol. Cell. 2002;13:183–194. doi: 10.1091/mbc.01-07-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iida K., Moriyama K., Matsumoto S., Kawasaki H., Nishida E., Yahara I. Isolation of a yeast essential gene, COF1, that encodes a homologue of mammalian cofilin, a low-M(r) actin-binding and depolymerizing protein. Gene. 1993;124:115–120. doi: 10.1016/0378-1119(93)90770-4. [DOI] [PubMed] [Google Scholar]
- 7.Aizawa H., Kishi Y., Iida K., Sameshima M., Yahara I. Cofilin-2, a novel type of cofilin, is expressed specifically at aggregation stage of Dictyostelium discoideum development. Genes Cells. 2001;6:913–921. doi: 10.1046/j.1365-2443.2001.00470.x. [DOI] [PubMed] [Google Scholar]
- 8.Ono S., Baillie D.L., Benian G.M. UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol. 1999;145:491–502. doi: 10.1083/jcb.145.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ono K., Parast M., Alberico C., Benian G.M., Ono S. Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J. Cell Sci. 2003;116:2073–2085. doi: 10.1242/jcs.00421. [DOI] [PubMed] [Google Scholar]
- 10.Edwards K.A., Montague R.A., Shepard S., Edgar B.A., Erikson R.L., Kiehart D.P. Identification of Drosophila cytoskeletal proteins by induction of abnormal cell shape in fission yeast. Proc. Natl. Acad. Sci. USA. 1994;91:4589–4593. doi: 10.1073/pnas.91.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J., Godt D., Gunsalus K., Kiss I., Goldberg M., Laski F.A. Cofilin/ADF is required for cell motility during Drosophila ovary development and oogenesis. Nat. Cell Biol. 2001;3:204–209. doi: 10.1038/35055120. [DOI] [PubMed] [Google Scholar]
- 12.Gunsalus K.C., Bonaccorsi S., Williams E., Verni F., Gatti M., Goldberg M.L. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J. Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurniak C.B., Perlas E., Witke W. The actin depolymerizing factor n-cofilin is essential for neural tube morphogenesis and neural crest cell migration. Dev. Biol. 2005;278:231–241. doi: 10.1016/j.ydbio.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Oka C., Nakano T., Wakeham A., de la Pompa J.L., Mori C., Sakai T., Okazaki S., Kawaichi M., Shiota K., Mak T.W., et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda S., Cunningham L.A., Boggess D., Hawes N., Hobson C.D., Sundberg J.P., Naggert J.K., Smith R.S., Nishina P.M. Aberrant actin cytoskeleton leads to accelerated proliferation of corneal epithelial cells in mice deficient for destrin (actin depolymerizing factor) Hum. Mol. Genet. 2003;12:1029–1037. doi: 10.1093/hmg/ddg112. [DOI] [PubMed] [Google Scholar]
- 16.de Vries W.N., Binns L.T., Fancher K.S., Dean J., Moore R., Kemler R., Knowles B.B. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- 17.Heidt A.B., Black B.L. Transgenic mice that express Cre recombinase under control of a skeletal muscle-specific promoter from mef2c. Genesis. 2005;42:28–32. doi: 10.1002/gene.20123. [DOI] [PubMed] [Google Scholar]
- 18.Miniou P., Tiziano D., Frugier T., Roblot N., Le Meur M., Melki J. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res. 1999;27:e27. doi: 10.1093/nar/27.19.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruning J.C., Michael M.D., Winnay J.N., Hayashi T., Horsch D., Accili D., Goodyear L.J., Kahn C.R. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 20.Ryan M.M., Ilkovski B., Strickland C.D., Schnell C., Sanoudou D., Midgett C., Houston R., Muirhead D., Dennett X., Shield L.K., et al. Clinical course correlates poorly with muscle pathology in nemaline myopathy. Neurology. 2003;60:665–673. doi: 10.1212/01.wnl.0000046585.81304.bc. [DOI] [PubMed] [Google Scholar]
- 21.Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 2008;87:649–667. doi: 10.1016/j.ejcb.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 22.North K.N., Laing N.G., Wallgren-Pettersson C. Nemaline myopathy: current concepts. The ENMC International Consortium and Nemaline Myopathy. J. Med. Genet. 1997;34:705–713. doi: 10.1136/jmg.34.9.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilmshurst J.M., Lillis S., Zhou H., Pillay K., Henderson H., Kress W., Muller C.R., Ndondo A., Cloke V., Cullup T., et al. RYR1 mutations are a common cause of congenital myopathies with central nuclei. Ann. Neurol. 2010;68:717–726. doi: 10.1002/ana.22119. [DOI] [PubMed] [Google Scholar]
- 24.Lawlor M.W., Dechene E.T., Roumm E., Geggel A.S., Moghadaszadeh B., Beggs A.H. Mutations of tropomyosin 3 (TPM3) are common and associated with type 1 myofiber hypotrophy in congenital fiber type disproportion. Hum. Mutat. 2010;31:176–183. doi: 10.1002/humu.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corbett M.A., Robinson C.S., Dunglison G.F., Yang N., Joya J.E., Stewart A.W., Schnell C., Gunning P.W., North K.N., Hardeman E.C. A mutation in alpha-tropomyosin(slow) affects muscle strength, maturation and hypertrophy in a mouse model for nemaline myopathy. Hum. Mol. Genet. 2001;10:317–328. doi: 10.1093/hmg/10.4.317. [DOI] [PubMed] [Google Scholar]
- 26.Papalouka V., Arvanitis D.A., Vafiadaki E., Mavroidis M., Papadodima S.A., Spiliopoulou C.A., Kremastinos D.T., Kranias E.G., Sanoudou D. Muscle Lim Protein interacts with Cofilin 2 and regulates F-actin dynamics in cardiac and skeletal muscle. Mol. Cell. Biol. 2009;29:6046–6058. doi: 10.1128/MCB.00654-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toyofuku T., Hoffman J.R., Zak R., Carlson B.M. Expression of alpha-cardiac and alpha-skeletal actin mRNAs in relation to innervation in regenerating and non-regenerating rat skeletal muscles. Dev. Dyn. 1992;193:332–339. doi: 10.1002/aja.1001930406. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y.W., Zhao P., Borup R., Hoffman E.P. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J. Cell Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills M., Yang N., Weinberger R., Vander Woude D.L., Beggs A.H., Easteal S., North K. Differential expression of the actin-binding proteins, alpha-actinin-2 and -3, in different species: implications for the evolution of functional redundancy. Hum. Mol. Genet. 2001;10:1335–1346. doi: 10.1093/hmg/10.13.1335. [DOI] [PubMed] [Google Scholar]
- 30.Chen H., Bernstein B.W., Sneider J.M., Boyle J.A., Minamide L.S., Bamburg J.R. In vitro activity differences between proteins of the ADF/cofilin family define two distinct subgroups. Biochemistry. 2004;43:7127–7142. doi: 10.1021/bi049797n. [DOI] [PubMed] [Google Scholar]
- 31.Hasty P., Bradley A., Morris J.H., Edmondson D.G., Venuti J.M., Olson E.N., Klein W.H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 32.Ferreiro A., Quijano-Roy S., Pichereau C., Moghadaszadeh B., Goemans N., Bonnemann C., Jungbluth H., Straub V., Villanova M., Leroy J.P., et al. Mutations of the selenoprotein N gene, which is implicated in rigid spine muscular dystrophy, cause the classical phenotype of multiminicore disease: reassessing the nosology of early-onset myopathies. Am. J. Hum. Genet. 2002;71:739–749. doi: 10.1086/342719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laing N.G., Dye D.E., Wallgren-Pettersson C., Richard G., Monnier N., Lillis S., Winder T.L., Lochmuller H., Graziano C., Mitrani-Rosenbaum S., et al. Mutations and polymorphisms of the skeletal muscle alpha-actin gene (ACTA1) Hum. Mutat. 2009;30:1267–1277. doi: 10.1002/humu.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonet C., Maciver S.K., Mozo-Villarias A. The regulatory action of alpha-actinin on actin filaments is enhanced by cofilin. Eur. Biophys. J. 2010;39:1143–1153. doi: 10.1007/s00249-009-0566-2. [DOI] [PubMed] [Google Scholar]
- 35.Luther P.K. The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. J. Muscle Res. Cell Motil. 2009;30:171–185. doi: 10.1007/s10974-009-9189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan Y., Tong H.Q., Beggs A.H., Kunkel L.M. Human skeletal muscle-specific alpha-actinin-2 and -3 isoforms form homodimers and heterodimers in vitro and in vivo. Biochem. Biophys. Res. Commun. 1998;248:134–139. doi: 10.1006/bbrc.1998.8920. [DOI] [PubMed] [Google Scholar]
- 37.Wattanasirichaigoon D., Swoboda K.J., Takada F., Tong H.Q., Lip V., Iannaccone S.T., Wallgren-Pettersson C., Laing N.G., Beggs A.H. Mutations of the slow muscle alpha-tropomyosin gene, TPM3, are a rare cause of nemaline myopathy. Neurology. 2002;59:613–617. doi: 10.1212/wnl.59.4.613. [DOI] [PubMed] [Google Scholar]