Abstract
Complement receptor 1 (CR1) is an Alzheimer's disease (AD) susceptibility locus that also influences AD-related traits such as episodic memory decline and neuritic amyloid plaque deposition. We implemented a functional fine-mapping approach, leveraging intermediate phenotypes to identify functional variant(s) within the CR1 locus. Using 1709 subjects (697 deceased) from the Religious Orders Study and the Rush Memory and Aging Project, we tested 41 single-nucleotide polymorphisms (SNPs) within the linkage disequilibrium block containing the published CR1 AD SNP (rs6656401) for associations with episodic memory decline, and then examined the functional consequences of the top result. We report that a coding variant in the LHR-D (long homologous repeat D) region of the CR1 gene, rs4844609 (Ser1610Thr, minor allele frequency = 0.02), is associated with episodic memory decline and accounts for the known effect of the index SNP rs6656401 (D′ = 1, r2= 0.084) on this trait. Further, we demonstrate that the coding variant's effect is largely dependent on an interaction with APOE-ɛ4 and mediated by an increased burden of AD-related neuropathology. Finally, in our data, this coding variant is also associated with AD susceptibility (joint odds ratio = 1.4). Taken together, our analyses identify a CR1 coding variant that influences episodic memory decline; it is a variant known to alter the conformation of CR1 and points to LHR-D as the functional domain within the CR1 protein that mediates the effect on memory decline. We thus implicate C1q and MBL, which bind to LHR-D, as likely targets of the variant's effect and suggest that CR1 may be an important intermediate in the clearance of Aβ42 particles by C1q.
INTRODUCTION
Alzheimer's disease (AD) is among the most common age-related conditions, affecting one out of every eight US adults over age 65 (1) with progressive amnesia, dementia, global cognitive failure, and ultimately death. Recent large genome-wide association studies (GWAS) have both identified and replicated several new AD susceptibility loci outside of Apolipoprotein E (APOE), including complement receptor 1 (CR1), which is the primary focus of this study (2–7).
CR1 is a large, polymorphic and multifunctional glycoprotein composed primarily of independently folding domains, known as complement control protein repeats (CCPs), each containing 59–75 amino acids (8,9). The CR1 gene contains many different polymorphisms; its most common version of CR1 (Fig. 1) contains 30 CCPs that form four long homologous repeats (LHR-A, LHR-B, LHR-C and LHR-D), each composed of 7 CCPs (8,9). CR1 has been postulated to impact AD pathogenesis through its key role in regulating complement activity as a receptor of the complement C3b protein (4). Complement activation leads to a proteolytic cascade, which produces activated C3b and C4b fragments that bind covalently to acceptor molecules and mediate phagocytosis through CR1 (4). Some investigators have suggested that this mechanism may mediate CR1's clearance of Aβ particles, which can aggregate to form amyloid plaques, an important component of AD neuropathology (4). The involvement of CR1 in the clearance of soluble Aβ particles may also be mediated through its role as an adherence receptor on red blood cells for immobilizing complement tagged particles in the blood and delivering these particles for removal by resident phagocytes of the liver and spleen (10,11). Our earlier work associating the reported AD susceptibility allele in CR1 with a greater burden of amyloid pathology (12) is consistent with these reports and suggests that the CR1 locus may influence AD-related molecular processes that are also modulated by the well-established APOE susceptibility locus (13,14).
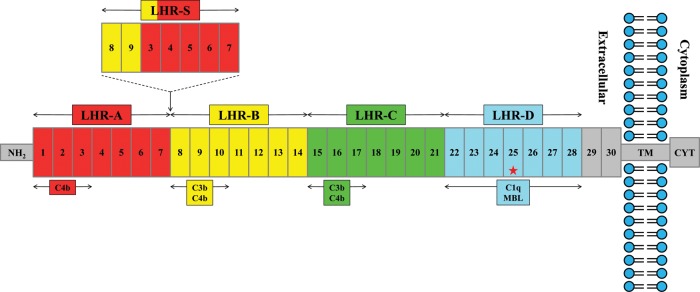
Figure 1.
Schematic representation of CCPs, LHRs and binding sites of the most common CR1 molecule. The most common form of CR1, containing 30 CCPs, is depicted. The first 28 CCPs form four long homologous repeat domains (LHR A−D) are shown above the CR1 molecule. The areas of each LHR containing C4b, C3b, MBL and/or C1q-binding sites are illustrated below the molecule. Also shown above the molecule is the location and structure of the LHR-S polymorphism described by Brouwers et al. (17). NH2 represents the amino terminus; TM, the transmembrane domain; and CYT, the cytoplasmic tail. Finally, the red star denotes the location of the functional variant rs4844609. The schematic was derived from similar figures in Krych-Goldberg and Atkinson (8), Liu and Niu (9) and Brouwers et al. (17).
Our analyses focus on the region surrounding the published single-nucleotide polymorphism (SNP) rs6654601 in the CR1 locus (4), for which we have previously described an association with decline in episodic memory and also the burden of neuritic amyloid plaques (12). Since the CR1 locus is implicated in both AD susceptibility and intermediate traits related to AD, we explored the implicated chromosomal segment, containing most of the CR1 gene, for additional SNPs with evidence of being associated with these intermediate traits. Identifying genetic variation that influences the function of a locus will inform our understanding of the targeted gene's biologic function, and some of these functional variants may also be involved in disease susceptibility. Based on our previous study (12) and the fact that we have a large sample size for clinical intermediate phenotypes, we selected decline in episodic memory, a key feature of AD (15), as our primary outcome measure with which to identify additional variants of interest within the CR1 locus. We then validated our results and extended our analyses to include other pertinent traits, including measures of AD pathology.
RESULTS
Demographics
Descriptive characteristics for the Religious Orders Study (ROS) and the Rush Memory and Aging Project (MAP) subjects are presented in Supplementary Material, Table S1; at the time of enrollment, all subjects are non-demented. Genetic information was available for 817 subjects in ROS and 892 subjects from MAP. Of these 1709 subjects, an estimate of episodic memory decline was available for the 1593 (93.2%) with at least two measures of episodic memory. A total of 697 (40.7%) subjects were deceased, and 651 (93.4%) subjects had available neuritic amyloid plaque and neurofibrillary tangle pathology data at the time of our analyses. The average age at enrollment for ROS was about 5 years younger than for MAP (76 versus 81 years), and the average age at death was nearly 2 years younger for ROS (87 versus 89 years). Twenty percent (n = 340) of participants were clinically diagnosed with AD during the course of study.
Descriptive characteristics for subjects from the Alzheimer's Disease Neuroimaging Initiative (ADNI) and the Chicago Health and Aging Project (CHAP) are listed in Supplementary Material, Table S2. Of the 746 individuals for whom data were available in ADNI, 207 (27.8%) were cognitively normal, 365 (48.9%) had mild cognitive impairment and 174 (23.3%) had probable AD at baseline. A total of 311 (41.7%) had clinically diagnosed AD at their last recorded follow-up. Episodic memory decline was available in 716 (96%) of the subjects. The mean [standard deviation (SD)] age was 75.4 (6.9), the mean (SD) years of education was 15.6 (3.0) and 306 (41.0%) were female. Our CHAP sample consisted of 624 European American subjects, 569 (91.2%) of whom had both genetic information and at least 2 measures of episodic memory. At the time of our sample, 50 (11.6%) had a diagnosis of AD, the mean (SD) age was 71.9 (5.2), the mean (SD) years of education was 14.9 (3.3) and 393 (63.0%) were female. Finally, our case–control sample from the Translational Genetics Research Institute (TGEN) consisted of 1004 pathologically confirmed AD cases and 575 controls with both genetic and clinical information. All subjects were included in a recent GWAS for AD susceptibility (2). The mean (SD) age at death was 82.1 (7.7) for cases and 80.9 (8.7) for controls, and 64% of cases and 48% of controls were female.
Decline in episodic memory
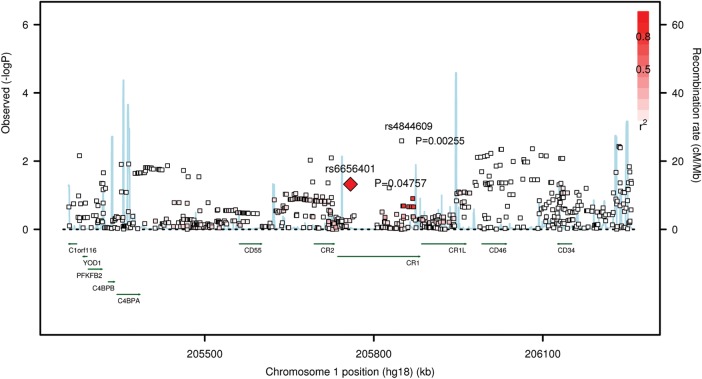
Our primary analysis examined the associations between episodic memory decline and 41 SNPs within the block of linkage disequilibrium (LD) containing the index CR1 SNP, rs6656401, that is associated with AD susceptibility (2,4). These SNPs were the ones that met quality criteria [info score > 0.3 and minor allele frequency (MAF) > 0.01] at the end of our imputation pipeline and were therefore available in our data set of imputed allele doses generated using the HapMap 2 reference (16). Figure 2 and Supplementary Material, Figure S1, give a more comprehensive view of the chromosomal segment containing CR1, illustrating the associations for all SNPs within a 500 kb radius of rs6656401. As previously reported (12), we note a significant association with the index CR1 SNP rs6656401 (P = 0.048), such that, for each additional rs6656401A allele, there is a decrease of 0.009 standard unit of episodic memory per year (Table 1). The most associated SNP in the CR1 LD block is the non-synonymous coding SNP (nscSNP) rs4844609 (P = 0.003), which results in the substitution of a threonine for a serine residue at amino acid position 1610 (Fig. 2). This result is significant since the 41 SNPs tested in the CR1 LD block actually reflect only 10 independent groups of SNPs in strong LD (r2> 0.5); a P-value <0.005 is therefore our threshold of significance after the Bonferroni correction. This nscSNP (rs4844609) lies in exon 37, which encodes a segment of the LHR-D region of the CR1 gene, and has only been observed in European (MAF of 2%) and East Asian populations (MAF of 1–5%) (http://hapmap.ncbi.nlm.nih.gov). The Rsq quality metric (the squared correlation between imputed and true genotypes) for rs4844609 was 0.7212 in ROS and MAP, compared with an Rsq of 0.9997 for rs6656401. Persons with the rs4844609A susceptibility allele had a 4-fold larger decrease in standardized episodic memory per year when compared with subjects with the original susceptibility allele rs6656401A (Table 1). A meta-analysis within two additional sample collections (ADNI and CHAP) confirms the association between rs4844609A and episodic memory decline (P = 0.004) (Table 2). The Rsq quality metric for rs4844609 and rs6656401, respectively, in these samples was 0.6868 and 0.9091 in CHAP and 0.1426 and 0.9270 in ADNI. We do not see any significant association for the index CR1 SNP, rs6656401, in the replication samples. We therefore consider rs4844609 a possible functional variant that influences episodic memory decline within the CR1 locus.
Figure 2.
Association plot for decline in episodic memory in the CR1 region. Each square represents the evidence of association for a particular SNP, the physical position of which is presented on the x-axis. The intensity of red color, presented graphically in the top right corner, represents the LD between each SNP and rs6656401. The blue line reports the recombination rate. The plot highlighting rs4844609 is presented in Supplementary Material, Figure S1.
Table 1.
Associations with episodic memory decline for CR1 SNPs in ROS and MAPa
| Independent models |
Joint model |
|||||
|---|---|---|---|---|---|---|
| β (SE) | P-value | β (SE) | P-value | β (SE) | P-value | |
| rs6656401A | −0.009 (0.005) | 0.048 | — | — | −0.004 (0.005) | 0.397 |
| rs4844609A | — | — | −0.039 (0.013) | 0.003 | −0.035 (0.014) | 0.015 |
SE, standard error.
aCombined ROS and MAP cohort; n = 1593 subjects with non-missing values.
Table 2.
Replication analyses with episodic memory decline for CR1 SNPs
| ADNI (N = 716) |
CHAP (N = 569) |
Meta-Analysis | |||
|---|---|---|---|---|---|
| β (SE) | P-value | β (SE) | P-value | P-valuea | |
| rs6656401A | −0.001 (0.001) | 0.222 | −0.002 (0.003) | 0.618 | 0.162 |
| rs4844609A | −0.007 (0.003) | 0.014 | −0.016 (0.010) | 0.100 | 0.004 |
SE, standard error.
aP-value from fixed effects meta-analysis adjusted for sample size and direction of effect.
To determine whether the two variants have independent effects on our trait of interest, we next fit a joint regression model containing individual terms for both rs6656401 and rs4844609, along with the relevant covariates (Table 1). We see that after adjusting for the effect of rs4844609, the observed association between the index CR1 SNP (rs6656401) and episodic memory decline decreases by more than half and is no longer significant. In contrast, the observed association with our functional variant (rs4844609) was only reduced by 10% and remained significant after adjusting for the rs6656401. These results suggest that the association of episodic memory decline with the CR1 locus is best captured by the rs4844609 variant.
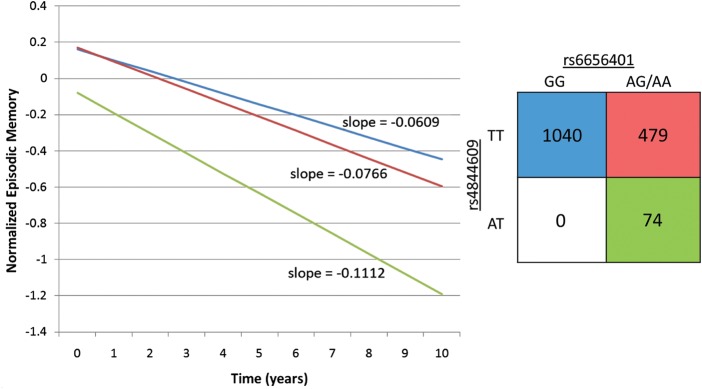
This observation is illustrated in Figure 3, which presents the mean trajectory of different genotypic subject classes based on rs6656401 and rs4844609 genotypes. As previously reported, the effect of the index SNP rs6656401 is best captured using a dominant model (12), which is used in these illustrations. There are no rs4844609AA homozygotes in our data set. We see that all rs4844609A risk alleles are found in subjects with at least one copy of the rs6656401A allele, and that these subjects decline significantly faster than those with neither risk allele (P = 0.013). We also note that the intercept of the mean cognitive trajectory of subjects with an rs4844609A allele is significantly lower than in the other genotype categories (P = 0.010) (Fig. 3). This observation suggests that decline in episodic memory related to rs4844609A has probably been ongoing for many years prior to the start of the study. Illustrations of mean episodic decline for each SNP separately are presented in Supplementary Material, Figure S2.
Figure 3.
Episodic memory decline in ROS and MAP subjects, stratified by CR1 genotypes. The effect of the rs6656401A and rs4844609A risk alleles on episodic memory decline is shown by computing the mean trajectory of decline for the different genotype categories. The graph presents the extent of change in episodic memory from baseline (at enrollment). The 2 × 2 table depicted shows both the sample size and the color of the line representing the slope of episodic memory decline for that genotype group. There are no rs4844609AA homozygotes in our data set. The groups are stratified by the presence of both rs6656401 and rs4844609 (P = 0.013 comparing rs6656401GG + rs4844609TT with rs6656401AT/AA + rs4844609AT; P = 0.052 comparing rs6656401GG + rs4844609TT with rs6656401AT/AA + rs4844609TT; P = 0.1253 comparing rs6656401AG/AA + rs4844609TT with rs6656401AT/AA + rs4844609AT).
In secondary analyses, we explored the hypothesis that the effect of CR1 on episodic memory may change over the course of aging; specifically, we assessed whether its effect may be different in non-AD and AD subjects. Given the stratification of subjects, our statistical power is limited in this secondary analysis. We report that the magnitude of the effect of the rs4844609A allele on the decline in episodic memory is similar in both groups of subjects (Supplementary Material, Table S3); however, the analyses are not significant given the reduced sample size. So, although this result suggests that CR1 may affect decline throughout the course of aging, a definitive answer to this question will require larger sample sizes.
Interaction with APOE-ɛ4
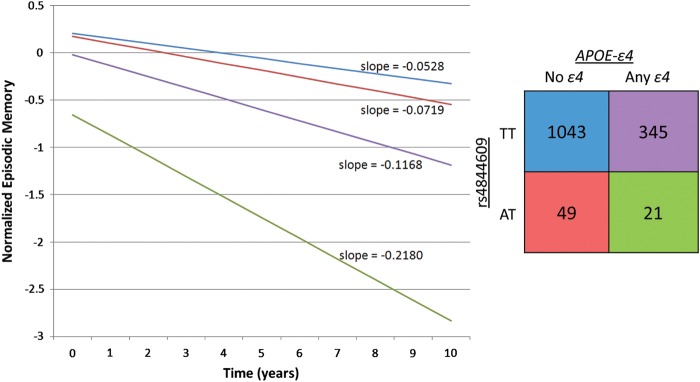
Confirming and extending previous reports for the CR1 locus (4), we find a significant interaction between our candidate functional variant rs4844609 and the presence or absence of APOE-ɛ4 on episodic memory decline (P = 0.027) (Supplementary Material, Table S4). Specifically, among carriers of APOE-ɛ4, subjects with the rs4844609A risk allele have a decrease of 0.084 standard unit of episodic memory per year, more than those with no rs4844609A alleles (P = 0.005). We do not observe a significant association for rs4844609A among non-APOE-ɛ4 carriers, nor do we observe significant interaction between APOE-ɛ4 and the index CR1 SNP rs6656401. As shown in Supplementary Material, Table S4, and as illustrated in Figure 4, we observe a more rapid decline in episodic memory for APOE-ɛ4 carriers with the rs4844609A risk allele, compared with non-APOE-ɛ4 carriers with no rs4844609A risk alleles (P < 0.0001). We note that the slope of decline in the strata with both APOE-ɛ4 and the rs4844609A risk allele is significantly steeper than the other three groups (P≤ 0.005 for all comparisons), although this is based on a small number of subjects with both risk variants.
Figure 4.
Episodic memory decline in ROS and MAP subjects, stratified by APOE-ɛ4 and rs4844609. The slope of episodic memory decline for the four distinct subsets defined by the presence or absence of APOE-ɛ4 and rs4844609A risk alleles is illustrated. The 2 × 2 table depicted shows both the sample size and the color of the line representing the slope of episodic memory decline for each of the four strata. Subjects with both rs4844609AT and APOE-ɛ4 decline significantly faster than all other groups (P < 0.0001 compared with rs4844609TT + no APOE-ɛ4; P < 0.0001 compared with rs4844609AT+ no APOE-ɛ4; P = 0.005 compared with rs4844609TT + APOE-ɛ4).
Clinically diagnosed AD
Given the robust effect on decline in episodic memory, we next assessed the effect of the rs4844609A allele on clinically diagnosed AD. In our ROS and MAP discovery cohorts, we observed a 2-fold increase in the odds of clinical AD associated with rs4844609A [odds ratio (OR) = 2.2, P = 0.007; 95% confidence interval (CI): 1.25–3.87] (Table 3). This association remained significant (OR = 2.0, P = 0.028; 95% CI: 1.08–3.81) after adjusting for rs6656401. Interestingly, no significant associations were observed between clinically diagnosed AD and rs6656401 (P = 0.104 in independent model; P = 0.582 after adjusting for rs4844609) in our discovery sample, which is of modest size and is underpowered to replicate the effect of the index SNP on AD susceptibility. The results of our replication analyses for AD susceptibility are presented in Supplementary Material, Table S5. Replication efforts are limited by sample size, study design and quality of the imputation of rs4844609 allele dosage in each stratum of the meta-analysis. Although two of the three strata in the replication meta-analysis demonstrate a direction of effect consistent with the discovery cohorts, the forest plot illustrates the non-significant trend toward replication (Supplementary Material, Fig. S3). A joint analysis of all data remains significant and suggests a strong effect on AD susceptibility (OR 1.40, 95% CI: 1.02–1.94) by rs4844609A; however, more subjects and direct genotyping of the variant will be needed to provide a definitive assessment of the role of rs4844609 in AD.
Table 3.
Associations with secondary phenotypes for CR1 SNPs in ROS and MAP cohorts
| Independent models |
Joint model |
|||||
|---|---|---|---|---|---|---|
| Estimate* | P-value | Estimate* | P-value | Estimate* | P-value | |
| Clinically defined ADa | ||||||
| rs6656401A | 1.20 (0.96, 1.50) | 0.104 | — | — | 1.07 (0.84, 1.38) | 0.582 |
| rs4844609A | — | — | 2.19 (1.25, 3.87) | 0.007 | 2.03 (1.08, 3.81) | 0.028 |
| Neuritic plaquesb | ||||||
| rs6656401A | 0.084 (0.040) | 0.035 | — | — | 0.052 (0.043) | 0.231 |
| rs4844609A | — | — | 0.286 (0.113) | 0.011 | 0.227 (0.123) | 0.065 |
| Neurofibrillary tanglesb | ||||||
| rs6656401A | 0.027 (0.029) | 0.365 | — | — | −0.0002 (0.032) | 0.994 |
| rs4844609A | — | — | 0.192 (0.083) | 0.021 | 0.192 (0.091) | 0.035 |
SE, standard error; OR, odds ratio.
*Estimates presented as OR (95% CI) for clinically defined AD and Beta (SE) for Neuritic plaques and Neurofibrillary tangles.
a340 cases and 1368 controls.
b651 deceased subjects with non-missing measures.
Haplotype analysis
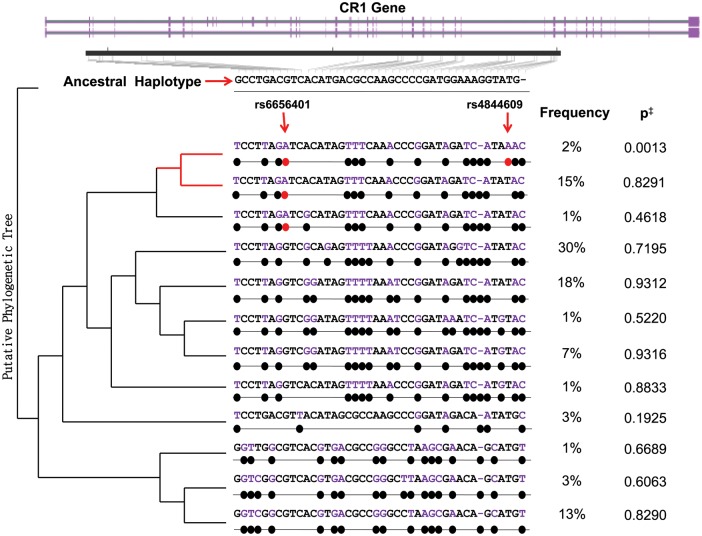
Another approach to assessing the relative effects of the rs4844609 and rs6656401 variants involves implementing a haplotype-based analysis (Fig. 5). Among our ROS and MAP subjects, we observed 12 haplotypes of >1% frequency in this region. The risk allele of the index SNP (rs6656401A) is found in three of these haplotypes, one of which is the only haplotype containing rs4844609A (frequency = 0.02). The haplotype containing both risk alleles is the only one significantly associated with clinical AD in our data (P = 0.001 in univariate analyses, as covariates cannot be introduced in Haploview). In order to conduct a conditional haplotype analysis, we note that our coding variant rs4844609 distinguishes this risk haplotype from the other 11 haplotypes in our sample. Therefore, analyzing rs4844609A as a binary variable (any or none) is equivalent to assessing this risk haplotype against all others. When we perform this analysis, we observe significant associations with clinically diagnosed AD (OR = 2.09, P = 0.006; 95% CI: 1.24–3.53), episodic memory decline [β (SE) = −0.034 (0.012), P = 0.005] and neuritic plaque burden [β (SE) = 0.244 (0.106), P = 0.021], adjusted for age, sex, education, cohort and the first three principal components generated from genome-wide genotype data that capture the effect of sub-structure in European ancestry among our subjects. This analysis suggests that, of the considered polymorphisms, rs4844609A best captures the effect of the CR1 locus on AD susceptibility in our data, and that rs6656401A may be a surrogate marker harbored on the same haplotype (D′ = 1 and r2= 0.084 between rs4844609 and rs6656401). The more frequent (15%) haplotype containing only the risk allele of the index SNP demonstrates no association with AD susceptibility in our data (P = 0.829, Fig. 5).
Figure 5.
Haplotypes and associations with clinical AD in the CR1 gene. Using the ‘solid spine of LD’ method of defining LD blocks in the CR1 region, we identified 12 haplotypes of >1% frequency among ROS and MAP subjects, spanning most of the CR1 gene (shown in purple). The ancestral haplotype (derived from the chimp) is shown at the top in black. The 12 haplotypes and frequencies in our sample are shown below the ancestral haplotype, with the derived alleles highlighted in purple and marked with black dots. The presence of rs6656401A and rs4844609A risk alleles is marked with red dots. The index CR1 risk allele appears on three haplotypes, only one of which contains our candidate causal variant. The putative phylogenetic tree is rooted in the ancestral haplotype and illustrates a possible evolutionary relationship of the different haplotypes. ‡P-values for crude associations with clinically diagnosed AD are shown on the right.
Many polymorphisms relating to the function of CR1 have been described, in particular the CR1-S and CR1-F alleles that are defined on the basis of the mobility of CR1 in gel electrophoresis experiments (S = slow; F = fast). This test captures the fact that the CR1-S polymorphism has an extra copy of the long low-copy repeat 1 (LCR1), which results in an additional LHR domain (LHR-S) and, consequently, additional C3b/C4b-binding sites (Fig. 1). Brouwers et al. (17) recently reported that the CR1-S allele, with an MAF of 0.15 in their study, is in strong LD with the index CR1 SNP associated with susceptibility to AD and is therefore found on the haplotype that has been associated with AD susceptibility. Brouwers et al. focus more closely on two variants that are associated with AD risk, rs4844610 and rs1408077, which are in strong LD with both the copy number variant and rs6656401. To provide a more directed assessment of the relative roles of the CR1-S allele and the rs4844609 coding variant, we also performed a haplotype analysis using only these four SNPs (rs6656401, rs4844609, rs4844610 and rs1408077). This analysis returns evidence for six distinct haplotypes in our population (Supplementary Material, Table S6), three of which have a frequency of >1%. Of these haplotypes, only the haplotype containing the risk allele of our coding variant (rs6656401A-rs4844609A-rs4844610A-rs1408077A, frequency = 0.02) is significantly associated with AD susceptibility (P = 0.001). This haplotype contains the CR1-S allele and the risk allele of our coding variant, as well as the risk allele of the index SNP, rs6656401. We do not observe a significant association with AD susceptibility for the most frequent (15%) haplotype which contains the index risk allele, the CR1-S allele but not the coding variant (rs6656401A-rs4844609T-rs4844610A-rs1408077A, P = 0.504). We therefore refine the earlier report on the possible role of CR1-S (17) and suggest that the coding variant rs4844609A may be found within the context of the larger CR1 molecule which contains the additional LHR domain that is encoded in the CR1-S polymorphism.
Exploring the functional consequences of rs4844609A
Given our prior work suggesting that CR1 was associated with amyloid plaque burden in a subset of 553 of the currently available sample of 651 autopsies (12), we next examined the association of rs4844609A with neuritic amyloid plaques and found that rs4844609A was also associated with increased neuritic pathology (P = 0.011, Table 3). The effect size we observe for our coding variant is larger than the one previously observed for the index SNP (12). Joint models including both SNPs once again suggest that rs4844609A best captures the association of this trait with the CR1 locus (Table 3). Further, we also see evidence for an increased burden of neurofibrillary tangles in subjects bearing the rs4844609A allele (P = 0.021), and the association remains significant after adjusting for rs6656401 (Table 3). As previously reported, we do not observe evidence of association between rs6656401 and neurofibrillary tangle burden in our subjects (12).
We next evaluated whether these associations with the burden of neuritic plaques and neurofibrillary tangles mediate the effect of our CR1 coding variant on cognitive decline. Adjusting for the burden of neuritic plaques reduces the observed effect of rs4844609A on episodic memory by 35% [from β = −0.039 (P = 0.003) to β = −0.026 (P = 0.273)], and adjusting for neurofibrillary tangles reduces the effect by 41% [from β = −0.039 (P = 0.003) to β = −0.023 (P = 0.309)]. Thus, we extend our earlier observation that the effect of the CR1 locus on episodic memory decline is mediated, in part, by an effect on amyloid pathology: we now also have evidence implicating Tau pathology in the effect of the CR1 locus. Since both pathologies are closely correlated, additional studies using molecularly specific markers will be needed to distinguish their respective effects. Interestingly, both pathologies are also influenced by APOE-ɛ4 and mediate its effect on cognitive decline (18–20), suggesting a potential molecular correlate to the evidence of statistical interaction that we report between the CR1 and APOE susceptibility alleles.
DISCUSSION
Recent studies have found strong evidence for associations between a CR1 SNP, rs6656401, and both clinically diagnosed AD and aging-related cognitive decline (2,4,12). In this manuscript, we sought to identify variants within the CR1 locus that have functional consequences on traits related to AD. We report evidence supporting the role of a non-synonymous coding variant in the LHR-D region of the CR1 gene (rs4844609) that is strongly associated with episodic memory decline; further, rs4844609 accounts for the previously observed effect of rs6656401 on this intermediate phenotype. In secondary analyses (Supplementary Material, Table S7), we see the same relationships with another intermediate phenotype, global cognitive decline, which incorporates our measures of episodic memory along with assessments of four other cognitive domains. Looking at these other domains separately, we also observe associations with perceptual speed (P = 0.025) and semantic memory (P = 0.015); thus the effect of rs4844609 does not appear to be limited to decline in episodic memory. The rs4844609 variant is an nscSNP found in exon 37 of CR1, which encodes a portion of its LHR-D domain. Taken together, our results suggest that we have found a functional variant that accounts for the effect of the CR1 locus on decline in episodic memory. Our analyses also suggest that this variant may influence AD susceptibility, but this association remains to be further delineated in larger, well-powered studies.
CR1 binds all of the nascent complement opsonins, i.e. C4b, C3b, mannan-binding lectin (MBL) and C1q. C4b, C3b, and MBLs have roles in the clearance of microbes, but C1q has a unique role in the removal of apoptotic/necrotic debris: >90% of subjects with C1q deficiency suffer a severe lupus-like illness with onset in childhood and a high morbidity and mortality (21). CR1 is a critical component of the complement receptor pathway that serves as a scaffold for different complement molecules in its LHR-A, -S, -B and -C domains, but also mediates the removal of opsonized material or cells by binding the opsonins C1q and MBL in its LHR-D domain, which contains the rs4844609A allele associated with cognitive decline (11,22,23). The exact effect of this allele, which results in a threonine for serine substitution at position 1610, on the function of the LHR-D domain in CR1 remains unclear. Nonetheless, it alters the structure of CR1 sufficiently to cause an immune response and anti-CR1 antibody formation following blood transfusion in humans (24), suggesting that the conformational change is substantial.
The identification of the functional unit of CR1 that is involved in decline in episodic memory is an important step forward, implicating several other molecules in the effect of rs4844609A. We speculate that this variant may reduce the binding affinity of the cognate LHR-D ligands, C1q and MBL, reducing the efficacy of the clearance of opsonized material such as amyloid oligomers or amyloid plaques. Although little is known about MBL in AD aside from its decreased level in cerebrospinal fluid of AD subjects (25), C1q is a complement molecule that has been studied extensively in the context of AD. Specifically, in humans, C1q has been reported to be (i) more abundant in AD brains, particularly in brain regions with dense accumulations of amyloid plaques and neurofibrillary tangles (26), (ii) associated with fibrillar Aβ and amyloid plaques (27–30), and (iii) found on certain neurons in AD brains that are associated with microglia (31). Further, in different mouse models of AD, C1q is transcriptionally upregulated during aging and in the context of amyloid deposition (32,33), and it is found to be associated with fibrillar Aβ (34). In addition, C1q null animals, in some transgenic models, have less accumulation of amyloid plaques (28,35). Thus, although the exact role of C1q in the context of AD needs to be further delineated, human tissue and rodent model systems implicate it in the pathophysiology of AD and provide a rich framework from which to design studies to investigate the role of the interaction of CR1's LHR-D domain with C1q in the clearance of amyloid plaques. Although we speculate that clearance of opsonized plaque material may be involved, we note that CR1 ligation also causes, in nucleated cells, phosphorylation of CR1, Ca2+ influx and phospholipase D activation, which may trigger immunomodulatory responses by B cells and possibly other immune cells (10,36,37). Consequently, our coding variant directly connects a gene influencing decline in episodic memory to a rich literature that implicates opsonization in the biology of amyloid plaques in the aging brain.
Extending the prior observation of interaction between these two loci in terms of AD susceptibility (4), our data suggest that the effect of our coding CR1 variant on cognitive decline may be dependent on the presence of the APOE-ɛ4 allele. The confluence of these results with the observation that APOE-ɛ4 also affects both amyloid and tau pathologies in our cohorts (18–20) suggests that these two molecules may be affecting a single molecular process that has a large effect on susceptibility to AD.
One important limitation of our current analyses is that data for the coding variant are imputed: we use allele dosage files in our analyses in order to mitigate the uncertainty that is inherent to imputation techniques. Although the quality of this imputation varies among our discovery and replication data sets, we note that poor imputation quality does not overestimate our associations but actually decreases our power and biases our results toward the null. Moreover, the authors recognize that it is important to interpret associations with an imputed genotype of low frequency cautiously. To address this issue, we performed direct genotyping of the rs4844609 and rs6656401 variants in a subset of subjects with available DNA, obtaining genotyping information on 1594 (93% of the sample with genome-wide imputed data) after quality control (QC). Within this sample, we found a strong correlation between the imputed dosages and actual genotypes (r2= 0.814, P < 0.0001). Of these subjects, information on episodic memory was available on 1485 (93%), and 318 (20%) were clinically diagnosed with AD. The mean (SD) age at baseline in this sample was 78.5 (7.5), and 495 (31%) were male. We observed associations between these primary phenotypes and the genotyped SNPs that are consistent with our results based on the imputed dosages (Supplementary Material, Table S8). Finally, since the original submission of the manuscript, imputed dosage data using a different reference map, the 1000 Genomes reference panel (dated 23 November 2010 and downloaded in July 2011) (38), became available in our subjects, and 131 additional SNPs can now be interrogated within the CR1 LD block. However, none of the newly imputed SNPs provide more extreme evidence of association to our outcome measures than we have observed with rs4844609 (Supplementary Material, Fig. S4). Ultimately, resequencing of the CR1 locus in a large number of subjects will be required to directly catalog all genetic variation in this genomic segment and to definitively identify the causal variant(s) affecting CR1 function.
Another limitation of these analyses is that our population consists of highly educated individuals of European ancestry, and thus we should be careful about generalizing our results to other populations. Nonetheless, many aspects of our study design can be viewed as strengths. Subjects in both ROS and MAP have high rates of both follow-up and autopsy, limiting the opportunity for bias. Our analyses relied on structured clinical and pathologic procedures and quantitative measures of AD pathology, which have excellent metric properties and increase both our ability and power to discover functional pathways associated with AD.
Overall, our results suggest that we have found a coding variant in the CR1 locus that influences decline in episodic memory, and that this allele's effect on decline in episodic memory may be mediated in large part by enhancing the accumulation of amyloid and tau pathologies. The path forward is clear: future studies are needed to explore the role of this variant in AD susceptibility and to examine the functional consequences of rs4844609A in order to explore the hypothesis that this allele, in the context of APOE-ɛ4, alters responses to AD pathology. With the identification of the rs4844609A allele, manipulation of exon 37 in CR1 or, more broadly, of the function of the LHR-D domain is clearly suggested as an approach to understand the role of CR1 in amyloid and tau-mediated pathologies that are found in AD.
MATERIALS AND METHODS
Discovery sample
The primary analyses used data from participants from two cohort studies, ROS and MAP, described in detail in Supplementary Material, Methods, and published literature (39,40). Briefly, ROS was established in 1994 and consists of Catholic clergy from the USA aged 55 or older and free of known dementia at enrollment. Since January 1994, 1100 participants (1001 self-reported non-Hispanic white) have completed their baseline evaluation, and follow-up and autopsy rate exceed 90%. MAP was established in 1997 and consists of older men and women from continuous care retirement centers in the Chicagoland area, aged >55 and free of known dementia. Since October 1997, 1400 participants (1118 self-reported non-Hispanic white) have completed their baseline questionnaire; follow-up rate exceeds 90% and autopsy rate exceeds 80%. As in prior studies (12,13,39), we analyzed ROS and MAP subjects jointly, adjusting for possible confounders, since (i) they were designed to be combined, (ii) the outcome measures and data collection procedures required for these analyses are identical, and (iii) they are managed by a single investigative team. Both studies were approved by the Institutional Review Board of Rush University Medical Center.
Replication samples
Our replication samples consisted of two prospectively followed samples, ADNI and CHAP, and one large case–control sample from TGEN. Detailed descriptions of these cohorts are presented in Supplementary Material, Methods.
Phenotypic data
Our primary measure of cognition was episodic memory, a composite score of seven instruments in ROS and MAP (word list memory, recognition and recall from CERAD, immediate and delayed recall of Story A from the logical memory subset of the Wechsler Memory Scale-Revised and immediate and delayed recall of the East Boston Story), eight instruments in ADNI (word list memory, recognition and recall from CERAD, immediate and delayed recall of Story A from the logical memory subset of the Wechsler Memory Scale-Revised and the Rey Auditory Verbal Learning Trials 1–6 totals, 30 min delay total and recognition score) and two instruments in CHAP (immediate and delayed recall of the East Boston Story). For each cohort, we used previously described methods (12,40–42) to create a normalized summary measure relative to the baseline population. General linear mixed models were used to model episodic memory decline over time within each cohort, adjusted for age at enrollment, years of education and sex. From these models, we obtained estimated residual slopes for each individual with at least two recorded measures of episodic memory.
Clinical diagnosis of probable AD was based on the NINCDS–ADRDA (National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association) criteria (43) in ROS, MAP, ADNI and CHAP, or clinically characterized and neuropathologically defined in TGEN. We created standardized summary measures of neurofibrillary tangles and neuritic plaques in ROS and MAP by dividing each person's count by SD for that particular count and taking the square root of this value to normalize the outcome, as previously reported (13,20). More phenotype information is presented in Supplementary Material, Methods, and previously published literature (12,39,40,44–46).
Genotypic data
Genetic information included genotyped and imputed SNPs. Detailed genotyping and QC methods for the discovery and replication cohorts are described in detail in Supplementary Material, Methods, and elsewhere (2,12,47–49). Imputation in all cohorts was performed using the MACH software (version 1.0.16a) and HapMap release 22 CEU (build 36) or the 1000 Genomes Project as a reference (38). The Rsq quality measure for the index CR1 SNP rs6656401 was 0.9997 in ROS and MAP, 0.9270 in ADNI, 0.9091 in CHAP and 0.9890 in TGEN. The Rsq for the nscSNP rs4844609 was 0.7212 in ROS and MAP, 0.1426 in ADNI, 0.6868 in CHAP and 0.5570 in TGEN.
Statistical methods
Detailed statistical methods are presented in Supplementary Material, Methods. Briefly, within each cohort, general linear mixed models were used to obtain residual slope estimates of episodic memory decline for each individual with at least two recorded measures. Using these estimates as outcomes, we fit linear regression models within our discovery cohort, examining associations with the SNPs in the CR1 LD block. Candidate functional variants were those SNPs that were highly associated with episodic memory and had evidence of functional significance. For secondary AD outcomes, we examined the association with candidate functional variants in each sample, using linear regression for continuous AD pathology measures and logistic regression for AD diagnoses. Where appropriate, we conducted a fixed effects meta-analysis across the replication data sets and a joint meta-analysis, incorporating ROS and MAP. Models in ROS and MAP, ADNI and CHAP were adjusted for the first three principal components, age at enrollment, years of education and sex. Models in ROS and MAP were additionally adjusted for cohort. Models in TGEN were adjusted for sex and age at death. To determine whether the candidate causal variant accounted for the observed effect of the index CR1 SNP, we additionally included both SNPs as covariates and examined the changes in effect sizes compared with the single-SNP models. To assess our hypothesized causal chains of intermediate phenotypes, we further adjusted for AD pathology outcomes associated with our functional variants of interest. We assessed whether there was evidence of interaction between APOE-ɛ4 and identified functional variants by including both the main effects for each variant and a product term (APOE-ɛ4 × SNP) in the regression models. If we observed a P value of <0.05 for interaction, we further examined the effect of our candidate functional variant in stratified subsets. Statistical analyses were performed using SAS software, version 9.1 (SAS Institute, Cary, NC, USA), R, version 2.13 (www.r-project.org) and the PLINK toolkit (http://pngu.mgh.harvard.edu/~purcell/plink/). The meta-analysis was conducted using R, version 2.13 (www.r-project.org) and METAL (50).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
ACKNOWLEDGEMENTS
The authors are grateful to the participants in the Religious Orders Study, the Rush Memory and Aging Project, the Alzheimer's Disease Neuroimaging Initiative, the Chicago Health and Aging Project and the Translational Genetics Research Institute, and thank Alessandro Biffi, Christopher Anderson and Jonathan Rosand for sharing the quality-controlled genotyping data set from ADNI.
Conflict of Interest statement. The authors have no conflicts of interest in regard to this manuscript.
FUNDING
This work is supported by the National Institutes of Health (NIH) (R01 AG30146, R01 AG179917, R01 AG15819, K08 AG034290, P30 AG10161 and R01 AG11101) and the Illinois Department of Public Health. J.M.S. was additionally supported by the Burroughs Wellcome Fund and the Clinical Investigator Training Program (Beth Israel Deaconess Medical Center—Harvard/MIT Health Sciences and Technology in collaboration with Pfizer, Inc. and Merck & Co.). M.J.H., J.J.C. and E.M.R. are supported by grants from the NIH (R01NS059873), Arizona Alzheimer's Disease Center P30 AG19610, RO1 AG023193, Mayo Clinic Alzheimer's Disease Center P50 AG16574 and Intramural Research Program, Kronos Life Sciences Laboratories, the National Alzheimer's Coordinating Center (U01 AG016976), the NIH Neuroscience Blueprint (U24NS051872), the ENDGAME Consortium (UO1HL084744), NIH grants to Carl Cotman (University of California, Irvine, P50 AG16573) and K01AG024079, and the state of Arizona. A.J.M. is supported by the NIH grant R01 AG034504 and the Johnnie B. Byrd Institute. J.A.H. would like to thank the Verum Foundation. Data collection and sharing for ADNI was funded by the NIH Grant U01 AG024904. ADNI is funded by the NIH, the National Institute of Biomedical Imaging and Bioengineering, through generous contributions from Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer, Inc., F. Hoffman-La Roche, Schering-Plough, Synarc, Inc. and non-profit partners the Alzheimer's Association and Alzheimer's Drug Discovery Foundation, with participation from the US Food and Drug Administration. Private sector contributions are facilitated by the Foundation for the NIH (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Alzheimer's Association (2010) Alzheimer's disease facts and figures. Alzheimers Dement. 6:158–194. doi: 10.1016/j.jalz.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Corneveaux J.J., Myers A.J., Allen A.N., Pruzin J.J., Ramirez M., Engel A., Nalls M.A., Chen K., Lee W., Chewning K., et al. Association of CR1, CLU and PICALM with Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum. Mol. Genet. 2010;19:3295–3301. doi: 10.1093/hmg/ddq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A., et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert J.C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M., Combarros O., Zelenika D., Bullido M.J., Tavernier B., et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 5.Seshadri S., Fitzpatrick A.L., Ikram M.A., DeStefano A.L., Gudnason V., Boada M., Bis J.C., Smith A.V., Carassquillo M.M., Lambert J.C., et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J.C., Carrasquillo M.M., Abraham R., Hamshere M.L., Pahwa J.S., Moskvina V., et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K., et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krych-Goldberg M., Atkinson J.P. Structure-function relationships of complement receptor type 1. Immunol. Rev. 2001;180:112–122. doi: 10.1034/j.1600-065x.2001.1800110.x. [DOI] [PubMed] [Google Scholar]
- 9.Liu D., Niu Z.X. The structure, genetic polymorphisms, expression and biological functions of complement receptor type 1 (CR1/CD35) Immunopharmacol. Immunotoxicol. 2009;31:524–535. doi: 10.3109/08923970902845768. [DOI] [PubMed] [Google Scholar]
- 10.Fallman M., Andersson R., Andersson T. Signaling properties of CR3 (CD11b/CD18) and CR1 (CD35) in relation to phagocytosis of complement-opsonized particles. J. Immunol. 1993;151:330–338. [PubMed] [Google Scholar]
- 11.Tas S.W., Klickstein L.B., Barbashov S.F., Nicholson-Weller A. C1q and C4b bind simultaneously to CR1 and additively support erythrocyte adhesion. J. Immunol. 1999;163:5056–5063. [PubMed] [Google Scholar]
- 12.Chibnik L., Shulman J., Leurgans S., Schneider J., Wilson R., Tran D., Aubin C., Buchman A., Heward C., Myers A., et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Annals of Neurology. 2011;69:560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett D.A., De Jager P.L., Leurgans S.E., Schneider J.A. Neuropathologic intermediate phenotypes enhance association to Alzheimer susceptibility alleles. Neurology. 2009;72:1495–1503. doi: 10.1212/WNL.0b013e3181a2e87d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiman E.M., Chen K., Liu X., Bandy D., Yu M., Lee W., Ayutyanont N., Keppler J., Reeder S.A., Langbaum J.B., et al. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer's disease. Proc. Natl Acad. Sci. USA. 2009;106:6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Querfurth H.W., LaFerla F.M. Alzheimer's disease. N. Engl. J. Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 16.De Jager P.L., Shulman J.M., Chibnik L.B., Keenan B.T., Raj T., Wilson R.S., Yu L., Leurgans S.E., Tran D., Aubin C., et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiol. Aging. 2011 doi: 10.1016/j.neurobiolaging.2011.09.033. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brouwers N., Van Cauwenberghe C., Engelborghs S., Lambert J.C., Bettens K., Le Bastard N., Pasquier F., Montoya A.G., Peeters K., Mattheijssens M., et al. Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Mol. Psychiatry. 2012;17:223–233. doi: 10.1038/mp.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett D.A., Schneider J.A., Wilson R.S., Bienias J.L., Arnold S.E. Neurofibrillary tangles mediate the association of amyloid load with clinical Alzheimer disease and level of cognitive function. Arch. Neurol. 2004;61:378–384. doi: 10.1001/archneur.61.3.378. [DOI] [PubMed] [Google Scholar]
- 19.Bennett D.A., Schneider J.A., Wilson R.S., Bienias J.L., Berry-Kravis E., Arnold S.E. Amyloid mediates the association of apolipoprotein E e4 allele to cognitive function in older people. J. Neurol. Neurosurg. Psychiatry. 2005;76:1194–1199. doi: 10.1136/jnnp.2004.054445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett D.A., Wilson R.S., Schneider J.A., Evans D.A., Aggarwal N.T., Arnold S.E., Cochran E.J., Berry-Kravis E., Bienias J.L. Apolipoprotein E epsilon4 allele, AD pathology, and the clinical expression of Alzheimer's disease. Neurology. 2003;60:246–252. doi: 10.1212/01.wnl.0000042478.08543.f7. [DOI] [PubMed] [Google Scholar]
- 21.Bowness P., Davies K.A., Norsworthy P.J., Athanassiou P., Taylor-Wiedeman J., Borysiewicz L.K., Meyer P.A., Walport M.J. Hereditary C1q deficiency and systemic lupus erythematosus. QJM. 1994;87:455–464. [PubMed] [Google Scholar]
- 22.Ghiran I., Barbashov S.F., Klickstein L.B., Tas S.W., Jensenius J.C., Nicholson-Weller A. Complement receptor 1/CD35 is a receptor for mannan-binding lectin. J. Exp. Med. 2000;192:1797–1808. doi: 10.1084/jem.192.12.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klickstein L.B., Barbashov S.F., Liu T., Jack R.M., Nicholson-Weller A. Complement receptor type 1 (CR1, CD35) is a receptor for C1q. Immunity. 1997;7:345–355. doi: 10.1016/s1074-7613(00)80356-8. [DOI] [PubMed] [Google Scholar]
- 24.Moulds J.M., Zimmerman P.A., Doumbo O.K., Diallo D.A., Atkinson J.P., Krych-Goldberg M., Hourcade D.E., Moulds J.J. Expansion of the Knops blood group system and subdivision of Sl(a) Transfusion. 2002;42:251–256. doi: 10.1046/j.1537-2995.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- 25.Lanzrein A.S., Jobst K.A., Thiel S., Jensenius J.C., Sim R.B., Perry V.H., Sim E. Mannan-binding lectin in human serum, cerebrospinal fluid and brain tissue and its role in Alzheimer's disease. Neuroreport. 1998;9:1491–1495. doi: 10.1097/00001756-199805110-00045. [DOI] [PubMed] [Google Scholar]
- 26.Yasojima K., Schwab C., McGeer E.G., McGeer P.L. Up-regulated production and activation of the complement system in Alzheimer's disease brain. Am. J. Pathol. 1999;154:927–936. doi: 10.1016/S0002-9440(10)65340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonseca M.I., Zhou J., Botto M., Tenner A.J. Absence of C1q leads to less neuropathology in transgenic mouse models of Alzheimer's disease. J. Neurosci. 2004;24:6457–6465. doi: 10.1523/JNEUROSCI.0901-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fonseca M.I., Kawas C.H., Troncoso J.C., Tenner A.J. Neuronal localization of C1q in preclinical Alzheimer's disease. Neurobiol. Dis. 2004;15:40–46. doi: 10.1016/j.nbd.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Zanjani H., Finch C.E., Kemper C., Atkinson J., McKeel D., Morris J.C., Price J.L. Complement activation in very early Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2005;19:55–66. doi: 10.1097/01.wad.0000165506.60370.94. [DOI] [PubMed] [Google Scholar]
- 30.Tooyama I., Sato H., Yasuhara O., Kimura H., Konishi Y., Shen Y., Walker D.G., Beach T.G., Sue L.I., Rogers J. Correlation of the expression level of C1q mRNA and the number of C1q-positive plaques in the Alzheimer Disease temporal cortex. Analysis of C1q mRNA and its protein using adjacent or nearby sections. Dement. Geriatr. Cogn. Disord. 2001;12:237–242. doi: 10.1159/000051265. [DOI] [PubMed] [Google Scholar]
- 31.Head E., Azizeh B.Y., Lott I.T., Tenner A.J., Cotman C.W., Cribbs D.H. Complement association with neurons and beta-amyloid deposition in the brains of aged individuals with Down Syndrome. Neurobiol. Dis. 2001;8:252–265. doi: 10.1006/nbdi.2000.0380. [DOI] [PubMed] [Google Scholar]
- 32.Fan R., DeFilippis K., Van Nostrand W.E. Induction of complement proteins in a mouse model for cerebral microvascular A beta deposition. J. Neuroinflammation. 2007;4:22. doi: 10.1186/1742-2094-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichwald J., Danner S., Wiederhold K.H., Staufenbiel M. Expression of complement system components during aging and amyloid deposition in APP transgenic mice. J. Neuroinflammation. 2009;6:35. doi: 10.1186/1742-2094-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuoka Y., Picciano M., Malester B., LaFrancois J., Zehr C., Daeschner J.M., Olschowka J.A., Fonseca M.I., O'Banion M.K., Tenner A.J., et al. Inflammatory responses to amyloidosis in a transgenic mouse model of Alzheimer's disease. Am. J. Pathol. 2001;158:1345–1354. doi: 10.1016/S0002-9440(10)64085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fonseca M.I., Chu S.H., Berci A.M., Benoit M.E., Peters D.G., Kimura Y., Tenner A.J. Contribution of complement activation pathways to neuropathology differs among mouse models of Alzheimer's disease. J. Neuroinflammation. 2011;8:4. doi: 10.1186/1742-2094-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Changelian P.S., Fearon D.T. Tissue-specific phosphorylation of complement receptors CR1 and CR2. J. Exp. Med. 1986;163:101–115. doi: 10.1084/jem.163.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jozsi M., Prechl J., Bajtay Z., Erdei A. Complement receptor type 1 (CD35) mediates inhibitory signals in human B lymphocytes. J. Immunol. 2002;168:2782–2788. doi: 10.4049/jimmunol.168.6.2782. [DOI] [PubMed] [Google Scholar]
- 38.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennett D.A., Schneider J.A., Arvanitakis Z., Kelly J.F., Aggarwal N.T., Shah R.C., Wilson R.S. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology. 2006;66:1837–1844. doi: 10.1212/01.wnl.0000219668.47116.e6. [DOI] [PubMed] [Google Scholar]
- 40.Bennett D.A., Schneider J.A., Buchman A.S., Mendes de Leon C., Bienias J.L., Wilson R.S. The Rush Memory and Aging Project: study design and baseline characteristics of the study cohort. Neuroepidemiology. 2005;25:163–175. doi: 10.1159/000087446. [DOI] [PubMed] [Google Scholar]
- 41.Wilson R.S., Barnes L.L., Krueger K.R., Hoganson G., Bienias J.L., Bennett D.A. Early and late life cognitive activity and cognitive systems in old age. J. Int. Neuropsychol. Soc. 2005;11:400–407. [PubMed] [Google Scholar]
- 42.Wilson R.S., Beckett L.A., Barnes L.L., Schneider J.A., Bach J., Evans D.A., Bennett D.A. Individual differences in rates of change in cognitive abilities of older persons. Psychol. Aging. 2002;17:179–193. [PubMed] [Google Scholar]
- 43.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 44.Bennett D.A., Schneider J.A., Bienias J.L., Evans D.A., Wilson R.S. Mild cognitive impairment is related to Alzheimer disease pathology and cerebral infarctions. Neurology. 2005;64:834–841. doi: 10.1212/01.WNL.0000152982.47274.9E. [DOI] [PubMed] [Google Scholar]
- 45.Schneider J.A., Wilson R.S., Bienias J.L., Evans D.A., Bennett D.A. Cerebral infarctions and the likelihood of dementia from Alzheimer disease pathology. Neurology. 2004;62:1148–1155. doi: 10.1212/01.wnl.0000118211.78503.f5. [DOI] [PubMed] [Google Scholar]
- 46.Bennett D.A., Wilson R.S., Schneider J.A., Evans D.A., Beckett L.A., Aggarwal N.T., Barnes L.L., Fox J.H., Bach J. Natural history of mild cognitive impairment in older persons. Neurology. 2002;59:198–205. doi: 10.1212/wnl.59.2.198. [DOI] [PubMed] [Google Scholar]
- 47.Bienias J.L., Beckett L.A., Bennett D.A., Wilson R.S., Evans D.A. Design of the Chicago Health and Aging Project (CHAP) J. Alzheimers Dis. 2003;5:349–355. doi: 10.3233/jad-2003-5501. [DOI] [PubMed] [Google Scholar]
- 48.Biffi A., Anderson C.D., Desikan R.S., Sabuncu M., Cortellini L., Schmansky N., Salat D., Rosand J. Genetic variation and neuroimaging measures in Alzheimer disease. Arch. Neurol. 2010;67:677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 50.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.