Abstract
Background
The growth inhibitory effect of tamoxifen is used for the treatment of breast cancer. Tamoxifen efficacy is mediated by its biotransformation, predominantly via the cytochrome P450 2D6 (CYP2D6) isoenzyme, to the active metabolite endoxifen. We investigated the relationship of CYP2D6 genotypes to the metabolism of dextromethorphan (DM), which is frequently used as a surrogate marker for the formation of endoxifen.
Methods
The CYP2D6 genotype was determined by polymerase chain reaction (PCR) in previously untreated patients with hormone receptor-positive invasive breast cancer considered to receive antihormonal therapy. The DM/dextrorphan (DX) urinary excretion ratios were obtained in a subset of patients by high-pressure liquid chromatography (HPLC)-mediated urine analysis after intake of 25 mg DM. The relationships of genotype and corresponding phenotype were statistically analyzed for association.
Results
From 151 patients predicted based on their genotype data for the ‘traditional’ CYP2D6 phenotype classes poor, intermediate, extensive and ultrarapid, 83 patients were examined for their DM/DX urinary ratios. The genotype-based poor metabolizer status correlated with the DM/DX ratios, whereas the intermediate, extensive and ultrarapid genotypes could not be distinguished based on their phenotype. Citalopram intake did not significantly influence the phenotype.
Conclusions
The DM metabolism can be reliably used to assess the CYP2D6 enzyme activity. The correlation with the genotype can be incomplete and the metabolic ratios do not seem to be compromised by citalopram. DM phenotyping may provide a standardized tool to better assess the CYP2D6 metabolic capacity.
Keywords: CYP2D6, Phenotype, Genotype, Dextromethorphan/Dextrorphan, Breast cancer
Abstract
Hintergrund
Das Wachstum von Brustkrebs kann durch Tamoxifen gehemmt werden. Tamoxifen ist ein Prodrug das in einer ersten Reaktion vom Cytochrom P450 2D6 (CYP2D6)-Isoenzym in das aktive Endoxifen umgewandelt wird. Wir haben das Verhältnis zwischen CYP2D6-Genotypen und dem Metabolismus von Dextromethorphan (DM), das häufig als Surrogat für die Bildung von Endoxifen verwendet wird, untersucht.
Material und Methoden
Der CYP2D6-Genotyp wurde mittels Polymerase-Kettenreaktion (PCR) bei vorgängig unbehandelten Frauen mit Hormonrezeptor-positivem Brustkrebs untersucht, die für eine antihormonelle Therapie in Betracht kamen. Die renalen DM/Dextrorphan (DX)-Exkretionsquotienten wurden nach Einnahme von 25 mg DM über Hochdruck-Flüssigchromatographie (HPLC)-vermittelte Urin-Analyse bestimmt. Das Verhältnis von Genotyp und Phänotyp wurde statistisch analysiert.
Ergebnisse
Von 151 Patientinnen, die über den CYP2D6-Genotyp den traditionellen” Phänotypklassen poor, intermediate, extensive und ultrarapid zugeordnet wurden, konnten 83 anhand ihrer renalen DM/DX-Exkretionsratio phänotypisiert werden. Der Genotyp poor metabolizer-Status korrelierte dabei mit der DM/DX-Exkretionsratio, während intermediate, extensive und ultrarapide Genotypen aufgrund ihres Phänotyps nicht voneinander unterschieden werden konnten. Die Einnahme von Citalopram hatte keinen signifikanten Einfluss auf den Phänotyp.
Schlussfolgerungen
Mittels DM-Metabolismus kann die CYP2D6-Aktivität zuverlässig bestimmt werden. Die Korrelation mit dem Genotyp kann inkomplett sein und scheint durch Citalopram unbeeinträchtigt. Die DM-Phänotypisierung könnte zur Bestimmung der metabolischen Kapazität von CYP2D6 standardisiert werden.
Introduction
Tamoxifen is a selective estrogen reuptake inhibitor that is used in the neo-adjuvant and adjuvant treatment of pre- and postmenopausal patients with estrogen receptor (ER)-positive breast cancer, palliatively as well as in chemoprevention. Previous studies indicate that the tamoxifen efficacy depends on its biotransformation, predominantly via the cytochrome P450 2D6 (CYP2D6) isoenzyme, to the active metabolite endoxifen [1]. Levels of endoxifen may vary with the number of mutant CYP2D6 alleles or concurrent use of drug inhibitors of CYP2D6 [2, 3, 4, 5, 6]. In the adjuvant setting, non-functional and severely impaired CYP2D6 genetic variants and concomitant CYP2D6-inhibitory drug intake seem to affect the disease-free survival and have even been associated with overall survival [7, 8, 9, 10, 11, 12] as well as with a lower incidence of hot flashes in patients taking tamoxifen [13, 14]. So far, methodological problems such as lack of interstudy comparability, sample sizes and the growing number of complex allelic variants (more than 80 CYP2D6 variants are defined) make the prediction of the CYP2D6 phenotype from genotype data particularly challenging [1]. Meanwhile, more than 10 epidemiologic studies have focused on the association between inheriting a variant CYP2D6 allele and breast cancer recurrence on tamoxifen and have reported highly heterogeneous relative risks [15] (for studies in which previous experiences are presented and summarized, please refer to table 1). In addition, a modeling analysis to calculate the disease-free survival of tamoxifen-treated patients who are wild type for CYP2D6 in comparison to aromatase inhibitors [16] and scoring systems were proposed that incorporate the impact of concomitant CYP2D6-inhibiting medications [17]. Also the combination of CYP2D6*4 and/or SULT1A1*1/*1 genotypes and incorporation of the ABCC2 gene to create a prediction for the prognosis of patients treated with tamoxifen and testing the CYP2D6 gene dose effect on plasma concentrations of endoxifen has been explored [18, 19]. However, subjects with identical genotypes may exhibit considerable interindividual variability in metabolic ratio-based phenotypes, which often cover 1–2 orders of magnitude, and the range of the observed values may not always fall within one of the ‘traditional’ phenotype classes [15, 20, 21]. While there is no widespread testing for CYP2D6 gene mutations in breast cancer patients, of particular interest in clinical practice is how pharmacogenomics can reliably determine a patient's individual phenotype for tailoring treatment [22]. Gaedigk et al. [23] established phenotype scores that have been derived from urinary ratios of dextromethorphan (DM), a frequent probe drug for CYP2D6 functional assessment. DM is O-demethylated into dextrorphan (DX) in humans by CYP2D6. Clinically, DM has been successfully used as an index of CYP2D6, and analytical data have validated the urinary molar ratio DM/DX to assess the CYP2D6 activity [24, 25].
Table 1.
Studies in which previous experiences are presented and summarized
| Authors | Okishiro et al. [11] | Nowell et al. [33] | Wegmann et al. [18] | Goetz et al. [38] | Newmann et al. [8] | Schroth et al. [7] | Ramón y Cajal et al. [9] | Bijl et al. [10] | Xu et al. [12] | Kyotani et al. [19] |
|---|---|---|---|---|---|---|---|---|---|---|
| Patients, n | 173 | 337 | 677 | 190 | 115 | 1325 | 91 | 85 | 293 | 282 |
| Tamoxifen dose | 20 mg/day, median 52 months | not reported | 40 mg/day for 2 or 5 years; 20 mg/day for 5 years | 20 mg/day for 5 years | 20 mg/day, median duration > 4 years | not reported | not reported | average 33.7 mg/day | 20 mg/day for 5 years | 20 mg/day for 5 years |
| Age group | 78% postmeno-pausal | 60% > 50 years | postmeno-pausal | postmeno-pausal | 27-68 years | 96% post-menopausal | 40% postmeno-pausal | 55 years | 75% > 50 years | median 50 years |
| Adjustments treatment | adjuvant therapy | none | none | no chemo-therapy | none | no chemo-therapy | none | none | no chemo-therapy | no chemo-therapy |
| Adjustments CYP2D6 inhibitors | excluding paroxetine | none | none | exposure definition | exposure definition | none | none | exposure definition | none | restricted to SSRI |
| Adjustments tamoxifen adherence | none | none | tamoxifen duration | none | none | tamoxifen duration | none | tamoxifen duration | none | 5 years completed |
| Adjustments prognostic markers | tumor size, node status, histologic grade, ER, PR, HER2 | age, stage, race, ER, PR | age, tumor size, node status | age, tumor size, node status | node status | tumor size, grade, ER, PR, node status, retrospective versus prospective | none | age, calendar time | age, tumor size, node status, adjuvant therapy, ER, PR, c-erb2 | age, tumor size, node status, menopausal status, nuclear grade, ER, PR, HER2 |
| Impact | no clinically significant impact on prognosis | no significant impact on survival | clinically significant impact of CYP3A5 | clinically significant impact on prognosis | significant impact on survival | significant impact on event-free and disease-free survival | significant impact on disease-free survival | increased risk of breast cancer mortality | significant impact on disease-free survival | significant impact on recurrence-free survival |
All studies (except for [7]) were retrospectively evaluated. Sample size varied widely between 91 and 1325. Data were derived from clinical series and trials [18, 38 during the years 1975-2006 ([8], data collection not indicated). Follow-up duration was heterogeneous and varied between 4.7 [11] and 11.4 years [38]. ER testing was either performed by pathology laboratory protocol or trial protocol [18, 38. Tamoxifen dose was not reported in 3 studies. Postmenopausal status was indicated in 4 studies, 2 studies reported 40-78% postmenopausal status, while 4 studies did not clearly indicate the menopause status. Selection of study subjects was performed either by frozen tumor tissue (4 studies), successful genotyping (3 studies) or patient survival follow-up (4 studies). 5 studies performed no adjustments to treatment; 4 studies allowed no chemotherapy. Scattered adjustments were performed with respect to CYP2D6 inhibitors, tamoxifen adherence and prognostic markers.
Here, we investigate the relationship between 5 CYP2D6 variants in previously untreated early-stage breast cancer patients with hormone receptor-positive tumors considered to receive antihormonal therapy and their functional capacity to metabolize DM as a surrogate predictive phenotypic marker of tamoxifen activation. The primary objective of this study was to determine whether the CYP2D6 genotype would correlate with the phenotype of DM metabolism based on the measurement of the DM/DX urinary excretion quotient.
Methods
For genotyping, 151 patients with primary hormone receptor-positive early-stage breast cancer were recruited from July 2009 to September 2010. All patients were Caucasian women pathologically diagnosed with ER- and/or progesterone receptor (PR)-positive, invasive cancer, who were considered to receive antihormonal therapy. Patients had neither received prior chemotherapy nor antihormonal treatment and were not included in this study in case of concomitant inhibitory drug intake other than antidepressants. Data on the stage of primary breast cancer diagnosis or recurrence were confirmed from the patients’ medical records. For the DM/DX phenotype study, a subset of 83 patients, including 7 patients with intake of the selective serotonin reuptake inhibitor (SSRI) citalopram, were permitted. This observational study was approved by the local ethical committee of Zürich, Switzerland, and written informed consent was obtained from all patients. Due to the relatively small number of patients, no survival analysis was planned or performed in this cohort.
Genotyping and Genotype Classification
Genomic DNA was extracted from 200 μl ethylenediaminetetraacetic acid (EDTA)-whole blood samples in a fully automated manner on the MagNa Pure Compact instrument from Roche Applied Science using the MagNA Pure Compact Nucleic Acid Isolation Kit®. All samples were genotyped for the polymorphic CYP2D6 gene. The definition of CYP2D6 allelic variants is in accordance with the Cytochrome P450 (CYP) Allele Nomenclature Committee (www.cypalleles.ki.se). We focused on the *3, *4, *5, and *6 alleles because these 4 variant alleles account for approximately 97% of the non-functional CYP2D6 variants in white populations [23]. The single-nucleotide polymorphisms (SNPs) CYP2D6*3, CYP2D6*4 and CYP2D6*6 were analyzed using tetra-primer polymerase chain reaction (PCR), and the CYP2D6*5 deletion allele using multiplex long PCR, followed by subsequent agarose gel electrophoresis according to methods previously described by Hersberger et al. [26]. We also looked for the presence of a CYP2D6 gene duplication or gene amplification using a long-range PCR-based method [27].
All investigated polymorphisms in the CYP2D6 gene, with the exception of the CYP2D6 gene duplication or amplification, give rise to null alleles that either code for an inactive enzyme or do not code for any enzyme protein at all. To enable the result evaluation, we stratified our patient phenotypes according to genotype as follows: Patients with 2 functional copies of the CYP2D6 gene were considered to be ‘extensive metabolizers’ (EM) for CYP2D6. These were subjects in whom we did not detect any of the investigated polymorphisms or variants and who, according to the analysis, were wild-type carriers and expected to exhibit normal CYP2D6 enzyme activity. Patients with 1 wild-type and 1 deficient allele were considered to be ‘intermediate metabolizers’ (IM) with impaired enzyme activity, whereas patients with 2 deficient alleles were classified as ‘poor metabolizers’ (PM) lacking enzyme activity. Carriers of more than 2 functional CYP2D6 gene copies were defined as CYP2D6 ‘ultrarapid metabolizers’ (UM) with increased CYP2D6 enzyme activity. This phenotype definition is compatible with the classification suggested from previous work and is based on the assumption of a gene dosage effect [1, 7].
DM Phenotyping
DM undergoes polymorphic metabolism depending on variations in the cytochrome P450 enzyme phenotype, with the prime specific enzyme catalyzing the DM metabolism being CYP2D6 [28]. Formation of the major active tamoxifen metabolites is primarily catalyzed by CYP2D6 and CYP3A4/5 [1]. The CYP2D6 activity was determined using DM as the phenotyping probe by high-pressure liquid chromatography (HPLC) analysis as described in detail by Abdel-Rahman et al. [29], and subsequently modified as described by Blake et al. [30]. In essence, 6 h after intake of 25 mg of DM, the urinary concentrations of DM and its O-demethylated metabolite, DX, were determined by reversed-phase HPLC with fluorescence detection. For this, 1.25 ml of every urine sample was deglucuronized by adding 500 µl phosphate buffer and 25 µl glucuronidase. After incubating the solution at 50 °C in a water bath and subsequent cooling of the samples, an aliquot of every sample was centrifuged and proceeded for further analysis. Upon determination of the peak height concentrations of DM and its O-demethylated metabolite, DX, the quotient of DM/DX was calculated.
Statistical Analysis
DM/DX phenotype quotients are reported as median with range and were logarithmically transformed to obtain initial information on the approximately normal distribution. Phenotype expression in each defined genotype group was reported as median ± standard deviation (SD). Genotypes were compared using simple analysis of variance with Bonferroni-post hoc tests. The statistical program SPSS 17 (SPSS Inc., Chicago, IL, USA) was used for analyses. A receiver operating characteristic (ROC) analysis was employed to evaluate a cut-off for the DM/DX quotient and to obtain maximum sensitivity (= 1) and specificity (< 0.9) for all phenotyping results. The 2-way analysis of variance was used to evaluate the effect of antidepressants on the DM/DX ratios; p < 0.05 was considered statistically significant.
Results
For genetic analysis, all 151 breast cancer patients were screened for 5 different CYP2D6 allele variants, including multiple copies of the gene, gene deletion, and 3 null alleles. The frequencies of the individual CYP2D6 genotypes are presented in table 2. CYP2D6 variants that predicted for the ‘traditional’ phenotype classes PM, IM, EM and UM [6] were present in 16 (11%), 53 (35%), 71 (47%) and 11 (7%) individuals, respectively. These results correspond to frequencies previously observed in Caucasians populations [1, 31]. Genotype analysis revealed no statistically significant association between the CYP2D6 mutation status and tumor size, nodal status, or histological grade in the women tested (data not shown), as was indicated in previous work [7].
Table 2.
Numbers and percentages from 151 patients tested for CYP2D6 variants and predicted for the 'traditional' phenotype classes in the whole cohort of patientsa
| Genotype | All patients | Paired | Phenotype | ||
|---|---|---|---|---|---|
| n | % | n | Median | Range | |
| Ultrarapid | 11 | 7 | 8 | 0.014 | 0.01–0.1 |
| Extensive | 71 | 47 | 23 | 0.019 | 0.01–0.3 |
| Intermediate | 53 | 35 | 38 | 0.098 | 0.01–2.5 |
| Poor | 16 | 11 | 14 | 1.774 | 0.8–5.3 |
Results of CYP2D6 testing in patients where paired genotype and phenotype were available, and associated results of the DM test (median, range).
Next, we studied the magnitude of difference between the analyzed CYP2D6 genotype and the corresponding DM metabolic ratios in patients where both genotype and phenotype data were available (23 patients with wild-type genotype and 60 patients with mutant alleles). According to their genotype, the subset of 83 patients available for DM/DX ratios included 14 PM, 38 IM, 23 EM and 8 UM, respectively.
According to phenotyping, the values for the DM/DX quotient ranged from less than 0.1 to 5.3. The mean value for all phenotypically evaluated patients was 0.50, with a median of 0.06. Patients in the UM group exhibited a median of 0.014 for the DM/DX quotient (range 0.01–0.1) and a mean of 0.02. According to the DM test, patients of the EM group exhibited a median of 0.019 (range 0.01–0.3) and a mean of 0.04. All further results are listed in table 2.
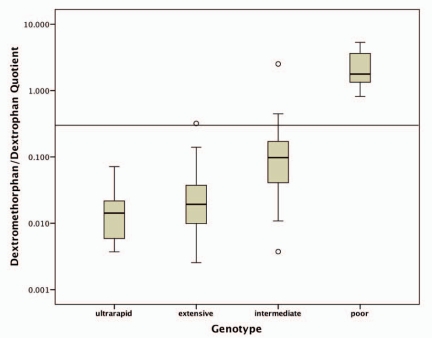
The DM/DX ratios differed significantly between the PM CYP2D6 genotypes and all other genotypes (p < 0.0001, after Bonferroni correction). In contrast, the extensive and ultrarapid genotypes could not be distinguished based on their metabolic DM/DX ratio (p = 1.0). The associations of the DM/DX ratios with the CYP2D6 genotypes are shown in figure 1. Overall, according to the obtained phenotypes, the genotype was predictive in distinguishing between poor and non-poor metabolizer status. When applying a cut-off of 0.3 for the DM/DX ratio, the CYP2D6 genotype-based prediction of phenotype failed in 5 (6%) patients (genotypically 1 extensive and 4 intermediate) who were phenotypically stratified as PM. In contrast, by using a metabolic ratio cut-off of 0.45, we achieved a phenotype-genotype concordance, in stratifying patients into poor and non-poor metabolizers, for all but 1 patient. This genotypically intermediate patient had a drug history of polymedication that could not be specified more precisely while an occasional use of antidepressants was also reported (table 3).
Fig. 1.
Graphical representation of the 4 classical genotypes when correlated with the results of DM testing.
Table 3.
Patients with discordant genotype and phenotype
| Patient | Genotype | Metabolic ratio |
|---|---|---|
| 1 | extensive wild type | 0.32 |
| 2 | intermediate CYP2D6*4, CYP2D6*2×2 | 0.43 |
| 3 | intermediate CYP2D6*1/*3 | 0.44 |
| 4 | intermediate CYP2D6*1/*4 | 0.45 |
| 5 | intermediate CYP2D6*1/*4 | 2.52 |
Of note, none of the 8 patients genotyped as UM in the subset where both phenotypic and genotypic data were available were carriers of the screened null alleles (CYP2D6*3, *4 and *6) nor of the deletion (CYP2D6*5). On the basis of the investigated polymorphisms and their higher frequency in the general population, one can assume with a high degree of certainty that a common functional allele was amplified (for example, allele *1 or *2). We cannot exclude the rather small possibility of the presence of further non-investigated polymorphisms; however, the possibility that this rare allele would also be amplified is even smaller.
In addition, we examined a panel of 7 patients with concomitant intake of the antidepressant citalopram, which supposedly does not reduce the efficacy of CYP2D6 metabolism [3]. Genotypically, 3 patients were classified as EM, another 3 as IM, and 1 as PM. Except for 1 patient (genotypically classified as IM) who phenotypically demonstrated poor metabolizing features (ratio 0.45), all 6 remaining patients were not detected to have an impaired DM/DX metabolic urinary ratio. As it was expected from the literature [32], overall the use of citalopram antidepressant did not exhibit a significant effect (p = 0.23) on the DM/DX ratios.
Discussion
The effect of CYP2D6 genotypic variations on the tamoxifen metabolism is one of the best characterized and clinically important examples of pharmacogenomics in cancer. Wide interpatient variations in circulating levels of both tamoxifen and metabolites seem to be explained by combinations of several mechanisms, e.g., those responsible for resistance and ‘at-risk’ alleles predictive for the response towards tamoxifen [18, 33]. Despite this potential link between the CYP2D6 mutation status and altered clinical outcomes among mutation carriers on tamoxifen intake, the discussion how this would affect the type of adjuvant endocrine therapy remains highly controversial [15, 19]. Moreover, dose-setting studies with clinical and biomarker outcomes and models of receptor binding suggest that tamoxifen and its metabolites might potentially reach concentrations sufficient to achieve the therapeutic effect regardless of CYP2D6 inhibition. In addition, the absence of both functional CYP2D6 alleles does not preclude distinct concentrations of the tamoxifen metabolite endoxifen (of which the required amount is not known) in sera of pre-and postmenopausal women [15]. Finally, the above-mentioned considerations seem to be complicated by suggestions that the density of ERs on the surface of breast cancer cells might even be a surrogate marker for the efficacy of tamoxifen [34].
Analyses of retrospective data from 2 large trials (Arimidex, Tamoxifen Alone or in Combination (ATAC) and Breast International Group (BIG) 1–98 presented at the San Antonio Breast Cancer Symposium (SABCS) 2010) found, in contrast to several previous studies, no effect of the CYP2D6 genotype in predicting breast cancer recurrence [35, 36]. While further animating the substantial controversy regarding the pharmacokinetics of tamoxifen biotransformation and CYP2D6 enzyme activity, these retrospective data may also reflect the poorly understood adherence in breast cancer patients undergoing antihormonal treatment [37, 38, 39]. Thus, wide inter-individual variations in serum levels of tamoxifen metabolites seem to become more relevant, and additional therapeutic drug monitoring might link different genotypes to clinical outcome [40]. However, standardized endoxifen measurements require attainment of steady state concentrations and interpatient variability in the endoxifen concentration may occur even after correcting for CYP2D6 status [17], possibly due to differences in tamoxifen metabolite elimination half-life, distribution volume, and formation rate [3]. While prospective trials currently explore whether the in vivo assay measuring CYP2D6 enzyme activity provides more accuracy in identifying patients with low endoxifen concentrations, caregivers exert caution and probably avoid potent CYP2D6 inhibitors in women treated with tamoxifen.
The results in our study provide further support for the surrogate functional testing of the tamoxifen metabolism. Beside a wide interpatient variability, we found that 5 (6%) of the 83 patients tested phenotypically, due to their functional capacity and/or being compromised by inhibitor drugs, did not fall within one of the ‘traditional’ genotype classes [15, 21]. Our findings are in line with results from Jin et al. [3], who demonstrated a wide range of efficacy in these subsets. Our model assumed a ratio of < 0.30 as threshold for sufficient metabolism, which was derived from ROC analyses to determine a cut-off with maximum sensitivity and specificity and was deemed comparable to ratios from previously published data [25]. Accordingly, all patients determined to be PM by genotyping also phenotypically proved to be PM and thus were identified as presumably producing inferior amounts of endoxifen (ratio > 0.30). Except for 5 patients, all wild-type homozygotes for CYP2D6 (EM) and all patients with loss of 1 allele (IM) demonstrated adequate phenotypes with respect to the measured DM/DX metabolism. However, further analysis of these patients (table 2) disclosed that a slightly higher metabolic ratio cut-off of 0.45 would have identified them in an analogous way to genotyping. Finally, the traditional CYP2D6 genotypes of UM, EM and IM did not correlate with the DM/DX phenotypes in these women. Although statistically not significant, in addition, 1 of 7 patients with concomitant intake of citalopram exhibited a poor CYP2D6 metabolism despite genotypically being tested as IM, and 1 additional EM also exhibited an insufficient ratio in the DM testing, indicating the constraints of genotyping (fig. 1).
There are several limitations of our study. Patients were only genotyped for the most frequent null variants in the Caucasian population by omitting further null and reduced-function alleles. Thus, a difference between the genotype and the phenotype-based classification might become less apparent upon a broader CYP2D6 allele coverage through extended genotyping. Even so, by screening for the most frequent known CYP2D6 polymorphisms, the effects of novel or rare polymorphisms might still remain unidentified [41].
Further, a relatively small number of patients with both phenotypic and genotypic data were available. However, mainly the frequencies observed in the genotyped cohort approximate those described in the literature for larger study samples (www.imm.ki.se/cypalleles/cyp2d6.htm).
Another limitation of our study is the reliance on the model that equals the metabolism of tamoxifen with DM, since DM urinary ratios have some methodological requirements for assessing in vivo enzyme activity [25]. In addition, further enzymes are involved in the metabolism of tamoxifen. Certainly, CYP2D6 is the most important enzyme catalyzing the formation of the active metabolite endoxifen; however, contributions from other enzymes such as CYP3A4, CYP2C9, CYP2C19 have been described [24]. Moreover, sulfation and glucuronidation are mechanisms contributing to the further metabolism of tamoxifen's active metabolites [1], for which the effects of allelic variants of the genes encoding for the responsible enzymes, steroid sulfuryl transferase (SULT) and UDP-glucuronosyltransferase (UGT), on the steady-state concentrations in vivo is, however, not well understood [3].
In contrast, potentially underestimated issues like intake of comedications in the adjuvant setting and lifestyle are more likely to be reflected in a functional phenotype analysis. Certainly, a reliable classification into poor and good metabolizers of tamoxifen will be available using the DM/DX metabolic ratios [38]. Perhaps, in some pre- and menopausal breast cancer patients, this more personalized approach would allow an adjustment in therapeutic strategies in case of substantial side effects and risk of fragile drug adherence to tamoxifen and associated outcome [2, 15, 39].
In conclusion, DM phenotyping is an efficient and reproducible method (double testing was performed in 5 patients; data not shown) that would permit to encounter the interpatient variability of DM/DX ratios within a common genotype group as well as concomitant drug intake and lifestyle factors. Based on our analysis and results of recent work [42], DM phenotyping might provide a standardized tool to assess the CYP2D6 metabolic capacity, which may even better predict the tamoxifen efficacy and endoxifen formation than extensive CYP2D6 genotyping.
Whether this method may serve as a substitute for the increasingly complex, costly and protracted genotyping with poorly defined categories shall be explored in prospectively randomized studies, e.g. the CYPTAM trial guided by the University of Leuven and assisted by the Swiss Group for Clinical Cancer Research (SAKK), which is designed to also evaluate a larger panel of biomarkers and dosages of tamoxifen according to the phenotype.
Disclosure Statement
All authors and coauthors indicated no potential conflicts of interest. Acknowledgement
Acknowledgement
This work was supported by the Swiss Tumor Institute, Zurich, Switzerland.
References
- 1.Brauch H, Muerdter TE, Eichelbaum M, Schwab M. Pharmacogenetics of tamoxifen therapy. Clin Chem. 2009;55:1770–1782. doi: 10.1373/clinchem.2008.121756. [DOI] [PubMed] [Google Scholar]
- 2.Barrière J, Formento JL, Milano G, Ferrero JM. CYP2D6 polymorphisms and therapeutic perspectives in the management of hormone-dependent breast cancer patients. Bull Cancer. 2010;97:311–320. doi: 10.1684/bdc.2010.1038. [DOI] [PubMed] [Google Scholar]
- 3.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, Skaar T, Storniolo AM, Li L, Araba A, Blanchard R, Nguyen A, Ullmer L, Hayden J, Lemler S, Weinshilboum RM, Rae JM, Hayes DF, Flockhart DA. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 4.Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, Bhargava P, Hayes DF, Desta Z, Flockhart DA. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 5.Johnson MD, Zuo H, Lee KHJP, Rae JM, Weatherman RV, Desta Z, Flockhart DA, Skaar TC. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 6.Borges S, Desta Z, Li L, Skaar TC, Ward BA, Nguyen A, Jin Y, Storniolo AM, Nikoloff DM, Wu L, Hillman G, Hayes DF, Stearns V, Flockhart DA. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Schroth W, Goetz M, Hamann U, Fasching PA, Schmidt M, Winter S, Fritz P, Simon W, Suman VJ, Ames MM, Safgren SL, Kuffel MJ, Ulmer HU, Boländer J, Strick R, Beckmann MW, Koelbl H, Weinshilboum RM, Ingle JN, Eichelbaum M, Schwab M, Brauch H. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newman WG, Hadfield KD, Latif A, Roberts SA, Shenton A, McHague C, Lalloo F, Howell S, Evans DG. Impaired tamoxifen metabolism reduces survival in familial breast cancer patients. Clin Cancer Res. 2008;14:5913–5918. doi: 10.1158/1078-0432.CCR-07-5235. [DOI] [PubMed] [Google Scholar]
- 9.Ramón y Cajal T, Altés A, Paré L, del Rio E, Alonso C, Barnadas A, Baiget M. Impact of CYP2D6 polymorphisms in tamoxifen adjuvant breast cancer treatment. Breast Cancer Res Treat. 2010;119:33–38. doi: 10.1007/s10549-009-0328-y. [DOI] [PubMed] [Google Scholar]
- 10.Bijl MJ, van Schaik RH, Lammers LA, Hofman A, Vulto AG, van Gelder T, Stricker BH, Visser LE. The CYP2D6*4 polymorphism affects breast cancer survival in tamoxifen users. Breast Cancer Res Treat. 2009;118:125–130. doi: 10.1007/s10549-008-0272-2. [DOI] [PubMed] [Google Scholar]
- 11.Okishiro M, Taguchi T, Jin KS, Shimazu K, Tamaki Y, Noguchi S. Genetic polymorphisms of CYP2D6*10 and CYP2C19*2,*3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifen. Cancer. 2009;115:952–961. doi: 10.1002/cncr.24111. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y, Sun Y, Yao L, Shi L, Wu Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, He L, Li P, Xie Y. Association between CYP2D6 *10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. 2008;19:1423–1429. doi: 10.1093/annonc/mdn155. [DOI] [PubMed] [Google Scholar]
- 13.Mortimer JE, Flatt SW, Parker BA, Gold EB, Wasserman L, Natarajan L, Pierce JP. Tamoxifen, hot flashes and recurrence in breast cancer. Breast Cancer Res Treat. 2008;108:421–426. doi: 10.1007/s10549-007-9612-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Flockhart DA, Desta Z, Perez EA, Ingle JN. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 15.Lash TL, Lien EA, S0rensen HT, Hamilton-Dutoit S. Genotype-guided tamoxifen therapy: time to pause for reflection? Lancet Oncol. 2009;10:825–833. doi: 10.1016/S1470-2045(09)70030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Punglia RS, Burstein HJ, Winer EP, Weeks JC. Pharmacogenomic variation of CYP2D6 and the choice of optimal adjuvant endocrine therapy for postmenopausal breast cancer: a modeling analysis. J Natl Cancer Inst. 2008;100:642–648. doi: 10.1093/jnci/djn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borges S, Desta Z, Jin Y, Faouzi A, Robarge JD, Philips S, Nguyen A, Stearns V, Hayes D, Rae JM, Skaar TC, Flockhart DA, Li L. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J Clin Pharmacol. 2010;50:450–458. doi: 10.1177/0091270009359182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wegman P, Elingarami S, Carstensen J, Stál O, Nordenskjöld B, Wingren S. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9:R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiyotani K, Mushiroda T, Imamura CK, Hosono N, Tsunoda T, Kubo M, Tanigawara Y, Flockhart DA, Desta Z, Skaar TC, Aki F, Hirata K, Takatsuka Y, Okazaki M, Ohsumi S, Yamakawa T, Sasa M, Nakamura Y, Zembutsu H. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol. 2010;28:1287–1293. doi: 10.1200/JCO.2009.25.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langan-Fahey SM, Tormey DC, Jordan VC. Tamoxifen metabolites in patients on long-term adjuvant therapy for breast cancer. Eur J Cancer. 1990;26:883–888. doi: 10.1016/0277-5379(90)90191-u. [DOI] [PubMed] [Google Scholar]
- 21.Ratliff B, Dietze EC, Bean GR, Moore C, Wanko S, Seewaldt VL. Re: Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2004;96:883–885. doi: 10.1093/jnci/djh170. [DOI] [PubMed] [Google Scholar]
- 22.Bock KW, Schrenk D, Forster A, Griese EU, Mörike K, Brockmeier D, Eichelbaum M. The influence of environmental and genetic factors on CYP2D6, CYP1A2 and UDP-glucuronosyltrans-ferases in man using sparteine, caffeine, and paracetamol as probes. Pharmacogenetics. 1994;4:209–218. doi: 10.1097/00008571-199408000-00005. [DOI] [PubMed] [Google Scholar]
- 23.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83:234–242. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
- 24.Streetman DS, Bleakley JF, Kim JS, Nafoger AN, Leeder JS, Gaedigk A, Gotschall R, Kearns GL, Bertino JS., Jr Combined phenotypic assessment of CYP1A2, CYP2C19, CYP2D6, CYP3A, Nacetyltransferase-2, and xanthine oxidase with the ‘Cooperstown cocktail’. Clin Pharmacol Ther. 2000;68:375–383. doi: 10.1067/mcp.2000.109519. [DOI] [PubMed] [Google Scholar]
- 25.Ducharme J, Abdullah S, Wainer IW. Dextromethorphan as an in vivo probe for the simultaneous determination of CYP2D6 and CYP3A activity. J Chromatogr B Biomed Appl. 1996;678:113–128. doi: 10.1016/0378-4347(95)00574-9. [DOI] [PubMed] [Google Scholar]
- 26.Hersberger M, Marti-Jaun J, Rentsch K, Hänseler E. Rapid detection of the CYP2D6*3, CYP2D6*4 and CYP2D6*6 alleles by tetra-primer PCR and of the CYP2D6*5 allele by multiplex long PCR. Clin Chem. 2000;46:1072–1077. [PubMed] [Google Scholar]
- 27.Steijns LS, Van der Weide J. Ultrarapid drug metabolism: PCR-based detection CYP2D6 gene duplication. Clin Chem. 1998;44:914–917. [PubMed] [Google Scholar]
- 28.Yu A, Haining RL. Comparative contribution to dextromethorphan metabolism by cytochrome P450 isoforms in vitro: Can dextromethorphan be used as a dual probe for both CYP2D6 and CYP3A activities. Drug Metab Dispos. 2001;29:1514–1520. [PubMed] [Google Scholar]
- 29.Abdel-Rahman SM, Leeder JS, Wilson JT, Gaedigk A, Gotschall RR, Medve R, Liao S, Spielberg SP, Kearns GL. Concordance between tramadol and dextromethorphan parent/metabolite ratios: the influence of CYP2D6 and non-CYP2D6 pathways on biotransformation. J Clin Pharmacol. 2002;42:24–29. doi: 10.1177/0091270002042001002. [DOI] [PubMed] [Google Scholar]
- 30.Blake MJ, Gaedigk A, Pearce RE, Bomgaars LR, Christensen ML, Stowe C, James LP, Wilson JT, Kearns GL, Leeder JS. Ontogeny of dextromethorphan O- and N-demethylation in the first year of life. Clin Pharmacol Ther. 2007;81:510–516. doi: 10.1038/sj.clpt.6100101. [DOI] [PubMed] [Google Scholar]
- 31.Brauch H, Schroth W. CYP2D6-Tamoxifen Pharmakogenetik beim frühen Mammakarzinom. Gynakologe. 2010;43:579–585. [Google Scholar]
- 32.Kelly CM, Juurlink DN, Gomes T, Duong-Hua M, Pritchard KI, Austin PC, Paszat LF. Selective serotonin reuptake inhibitors and breast cancer mortality in women receiving tamoxifen: a population based cohort study. BMJ. 2010;340:c693. doi: 10.1136/bmj.c693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowell SA, Ahn J, Rae JM, Scheys JO, Trovato A, Sweeney C, MacLeod SL, Kadlubar FF, Ambrosone CB. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91:249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 34.Pagani O, Gelber S, Simoncini E, Castiglione-Gertsch M, Price KN, Gelber RD, Holmberg SB, Crivellari D, Collins J, Lindtner J, Thürlimann B, Fey MF, Murray E, Forbes JF, Coates AS, Goldhirsch A. Is adjuvant chemotherapy of benefit for postmenopausal women who receive endocrine treatment for highly endocrine-responsive, node-positive breast cancer? International Breast Cancer Study Group Trials VII and 12–93. Breast Cancer Res Treat. 2009;116:491–500. doi: 10.1007/s10549-008-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rae JM, Drury S, Hayes DF, Stearns V, Thibert JN, Haynes BP, Salter J, Pineda S, Cuzick J, Dowsett M: Lack of correlation between gene variants in tamoxifen metabolizing enzymes with primary endpoints in the ATAC trial. San Antonio Breast Cancer Symposium (SABCS) 2010;abstr S1–7, www.abstracts2view.com/sabcs10/view.php?nu=SABCS10L_1093
- 36.Leyland-Jones B, Regan MM, Bouzyk M, Kammler R, Tang W, Pagani O, Maibach R, Dell'Orto P, Thurlimann B, Price KN, Viale G: Outcome according to CYP2D6 genotype among postmenopausal women with endocrine-responsive early invasive breast cancer randomized in the BIG 1–98 trial. San Antonio Breast Cancer Symposium (SABCS) 2010;abstr S1–8, www.abstracts2view.com/sabcs10/view.php?nu=SABCS10L_556
- 37.Rae JM, Sikora MJ, Henry NL, Li L, Kim S, Oesterreich S, Skaar TC, Nguyen AT, Desta Z, Storniolo AM, Flockhart DA, Hayes DF, Stearns V. Cytochrome P450 2D6 activity predicts discontinuation of tamoxifen therapy in breast cancer patients. Pharmacogenomics J. 2009;9:258–264. doi: 10.1038/tpj.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Weinshilboum RM, Fritcher EG, Nibbe AM, Desta Z, Nguyen A, Flockhart DA, Perez EA, Ingle JN. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 39.Thompson AM, Johnson A, Quinlan P, Hillman G, Fontecha M, Bray SE, Purdie CA, Jordan LB, Ferraldeschi R, Latif A, Hadfield KD, Clarke RB, Ashcroft L, Evans DG, Howell A, Nikoloff M, Lawrence J, Newman WG. Comprehensive CYP2D6 genotype and adherence affect outcome in breast cancer patients treated with tamoxifen monotherapy. Breast Cancer Res Treat. 2011;125:279–287. doi: 10.1007/s10549-010-1139-x. [DOI] [PubMed] [Google Scholar]
- 40.Gjerde J, Hauglid M, Breilid H, Lundgren S, Varhaug JE, Kisanga ER, Mellgren G, Steen VM, Lien EA. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol. 2008;19:56–61. doi: 10.1093/annonc/mdm434. [DOI] [PubMed] [Google Scholar]
- 41.McElroy S, Sachse C, Brockmoller J, Richmond J, Lira M, Friedman D, Roots I, Silber BM, Milos PM. CYP2D6 genotyping as an alternative to phenotyping for determination of metabolic status in a clinical trial setting. AAPS PharmSci. 2000;2:E33. doi: 10.1208/ps020433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Graan AJ, Teunissen SF, de Vos FY, Loos WJ, van Schaik RH, de Jongh FE, de Vos AI, van Alphen RJ, van der Holt B, Verweij J, Seynaeve C, Beijnen JH, Mathijssen RH. Dextromethorphan as a phenotyping test to predict endoxifen exposure in patients on tamoxifen treatment. J Clin Oncol. 2011;29:3240–3246. doi: 10.1200/JCO.2010.32.9839. [DOI] [PubMed] [Google Scholar]