Abstract
Proteins represent the most sophisticated building blocks available to an organism or the laboratory chemist. Yet, in contrast to nearly all other types of molecular building blocks, the designed self-assembly of proteins has been largely inaccessible owing to the chemical and structural heterogeneity of protein surfaces. To circumvent the challenge of programming extensive non-covalent interactions for controlling protein self-assembly, we had previously exploited the directionality and strength of metal coordination interactions to guide the formation of closed, homoligomeric protein assemblies. Here, we extend this strategy to the generation of periodic protein arrays. We show that a monomeric protein with properly oriented coordination motifs on its surface can arrange upon metal binding into one-dimensional nanotubes, and two-or three-dimensional crystalline arrays whose dimensions collectively span nearly the entire nano- and micrometer length scale. The assembly of these arrays is predictably tuned by external stimuli, such as metal concentration and pH.
Introduction
Proteins are nature’s most versatile building blocks, programmed at the genetic level to perform myriad functions and largely responsible for the complexity of an organism. The exquisite, error-free self-assembly of proteins into ordered yet dynamic nano- and microscale architectures, such as 1D microtubules, 2D bacterial surface layers (S-layers), and 3D virus capsids is a cornerstone of life1, and has served as a major inspiration for synthetic supramolecular chemistry2 and nanoscience3. In fact, such self-assembled protein arrays themselves have been widely exploited as well-ordered and genetically/chemically modifiable structural templates in numerous nanotechnological applications4-6. Significantly, crystalline protein arrays form the basis of diffraction-based methods for structure determination, and their acquisition generally represents the rate-limiting step in this process. Thus, a bottom-up strategy for arranging proteins into periodic structures would have wide-ranging applications in crystallography and in nanotechnology, where it would allow access to novel biological polymers that incorporate the wide range of chemical functionalities afforded by the proteins.
In contrast to simpler building blocks such as DNA7, 8, RNA9, 10, and peptides11, 12, the designed self-assembly of proteins has largely been inaccessible, plagued by the chemical heterogeneity and size of the molecular surfaces required for protein-protein interactions13. Previous efforts in designed protein self-assembly have had limited success: they have generally required building blocks with high preexisting symmetry, allowed little or no external control over self-assembly, and yielded static protein architectures that did not display long-range order beyond a few hundred nanometers14-18.
Inspired by efforts in Supramolecular Coordination Chemistry (SCC)19-21, we have adopted the strategy of using directional metal coordination bonds to control protein self-assembly, which led to the facile generation of discrete superprotein architectures22, 23. In this study, we further extend into the protein realm the concepts of SCC to create extended 1, 2, and 3D molecular arrays. The premise of our approach is that metal-bonding interactions can capture all salient features of protein-protein interactions (stability, specificity, directionality, symmetry, reversibility) on a much smaller surface than is needed by non-covalent interactions, and thus require “little design”. The reversibility of metal coordination allows for self-healing that is necessary to produce assemblies with long-range order. Importantly, chemical control and tunability are inherent in this approach: metal-directed protein self-assembly is, by default, dependent on external stimuli (i.e., metal binding or any stimulus that affects metal coordination), and all defining features of protein self-assembly listed above can be dictated by the choice of the metal ion and the coordination motif.
Using this approach, we show here that a monomeric protein (cytochrome cb562) with properly oriented metal coordinating motifs on its surface can self-assemble upon Zn2+ binding into 1D helical nanotubes, and 2- or 3D crystalline arrays. The dimensions of these arrays collectively span nearly the entire nano- and micrometer scale (5 nm-200 μm). In-depth characterization by X-ray crystallography, cryo-electron microscopy and image reconstruction revealed the structural basis of cyt cb562 self-assembly in atomic detail, and guided the design of a variant whose 3D assembly was further controlled by surface-attached functional groups.
Results and discussion
Design of a self-assembling building block, RIDC3
Previously, we focused on the use of the four-helix-bundle heme protein, cyt cb562, as a building block, because it is stable, monomeric at mM concentrations, and has a compact, cylindrical shape22. We showed that the cyt cb562 variant, MBPC1, with just two metal-chelating bis-histidine motifs (His59/His63 and His73/His77) on its surface, assembled into discrete oligomeric states upon stoichiometric Zn2+, Cu2+ or Ni2+ binding24, 25. When Zn2+ was added in greater than 1.5-fold molar excess, MBPC1 formed large, heterogeneous aggregates, which we attributed to multiple and indiscriminate modes of Zn-mediated interprotein coordination. Combining the symmetry- and directional bonding-based self-assembly strategies of others17, 19, 20, we posited that, if two or more selective Zn coordination sites on the MBPC1 surface could be stabilized in appropriate orientations relative to each other, it should be possible to dictate the self-assembly of the protein into regular, extended arrays through metal coordination.
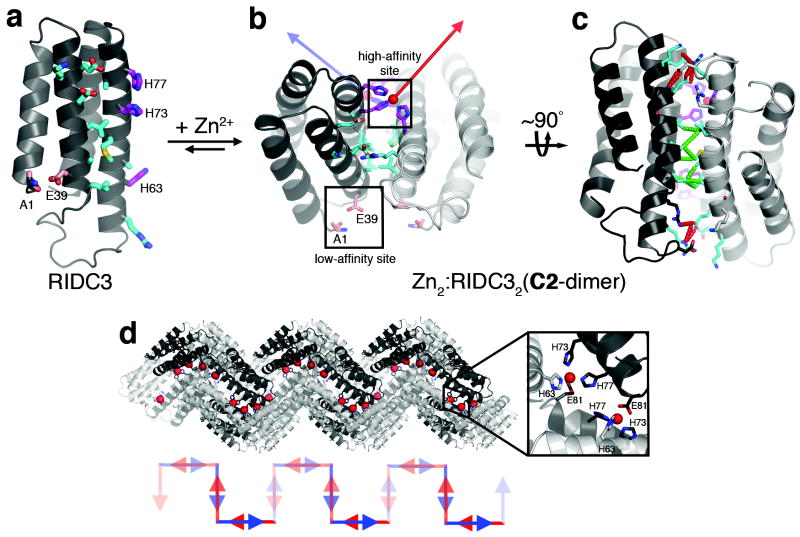
Here we accomplished this using RIDC3 (RosettaInterfaceDesignedCytochrome3) (Fig. 1a), which incorporates ten Rosetta26-prescribed surface mutations (K27E, D28K, T31E, R34L, L38A, Q41L, H59R, D66A, V69M, L76A) into MBPC1. The mutations stabilize a C2-symmetric dimer (the “C2-dimer”) that forms upon Zn2+ binding by His73 and His77 from one monomer and His63 from the other (the “high-affinity site”, Fig. 1b); this particular Zn coordination mode and the resulting C2-dimer geometry were derived from the structure of the Zn-directed tetramer of MBPC124 (Supplementary Fig. S1). Each RIDC3 monomer also possesses a weaker metal binding site (the “low-affinity site”) formed by the N-terminal alanine and Asp39, which we have seen in various cyt cb562 structures (PDB IDs: 3QVY, 3QVZ, 3DE9). C2-dimers contain three key design elements: 1) The tricoordinate high- and low-affinity sites provide stable and selective, yet labile, Zn2+ binding motifs that allow metal-directed self-assembly to proceed under thermodynamic control. 2) Each metal-binding motif leaves one Zn coordination site open for binding to another C2-dimer. Coordination vectors originating from the high-affinity sites are nearly orthogonal to one another, suggesting that they could promote the self-assembly of C2-dimers in two perpendicular directions. 3) Small, hydrophobic residues are incorporated into the center of the dimeric interface for enhanced stability, and polar residues are incorporated onto the periphery for geometric specificity (Fig. 1c), generating a pattern commonly observed in transient, but specific protein-protein interactions27.
Figure 1. Design and initial crystallographic characterization of RIDC3.
a, Cartoon representation of the RIDC3 monomer. Surface residues predicted by RosettaDesign to stabilize the Zn-induced RIDC3 dimer, the high-affinity and the low-affinity Zn binding sites are highlighted as cyan, magenta and pink sticks, respectively. b, Zn binding by RIDC3 produces a C2-symmetric dimer (Zn2:RIDC32 or C2-dimer) with two orthogonal coordination vectors (red and blue arrows) originating from the two high-affinity Zn sites. The C2-dimer geometry is derived from the structure of the Zn-mediated tetramer of MBPC1 (Zn4:MBPC14) as illustrated in Supplementary Fig. 1. c, Rotated view of the C2-dimer showing the crystallographically observed hydrophobic (dashed green lines) and polar (dashed red lines) interactions in the dimeric interface. d, Pairwise Zn coordination interactions between neighboring C2-dimers involving the high-affinity sites and E81 (inset) in the lattice leads to a helical 1D chain. Shown below is a cartoon representation of the C2-dimer units in this chain, illustrated as pairs of blue and red arrows that approximately represent the direction of Zn coordination vectors.
Sedimentation velocity experiments showed that RIDC3 was monomeric, but that it fully converted into a dimeric form upon binding equimolar Zn2+ at 5 μM concentration (Supplementary Fig. S2). The 2.3-Å crystal structure of the Zn-mediated dimer (Zn2:RIDC32), obtained using polyethylene glycol (PEG)-precipitated crystals, confirmed that it possessed the desired C2-dimer geometry. The overall root mean square deviation (rmsd) in the α-C positions between the crystallographically-determined and the Rosetta-predicted C2-dimer conformations is <0.6 Å (Supplementary Fig. S3). In the lattice, C2-dimers formed 1D coordination polymers, in which each high-affinity-site-bound Zn is ligated to E81 from a neighboring dimer (Fig. 1d). This arrangement provided the first evidence that C2-dimers might indeed self-assemble into extended arrays in solution, particularly in the presence of excess Zn2+.
Zn-directed self-assembly of RIDC3 in solution
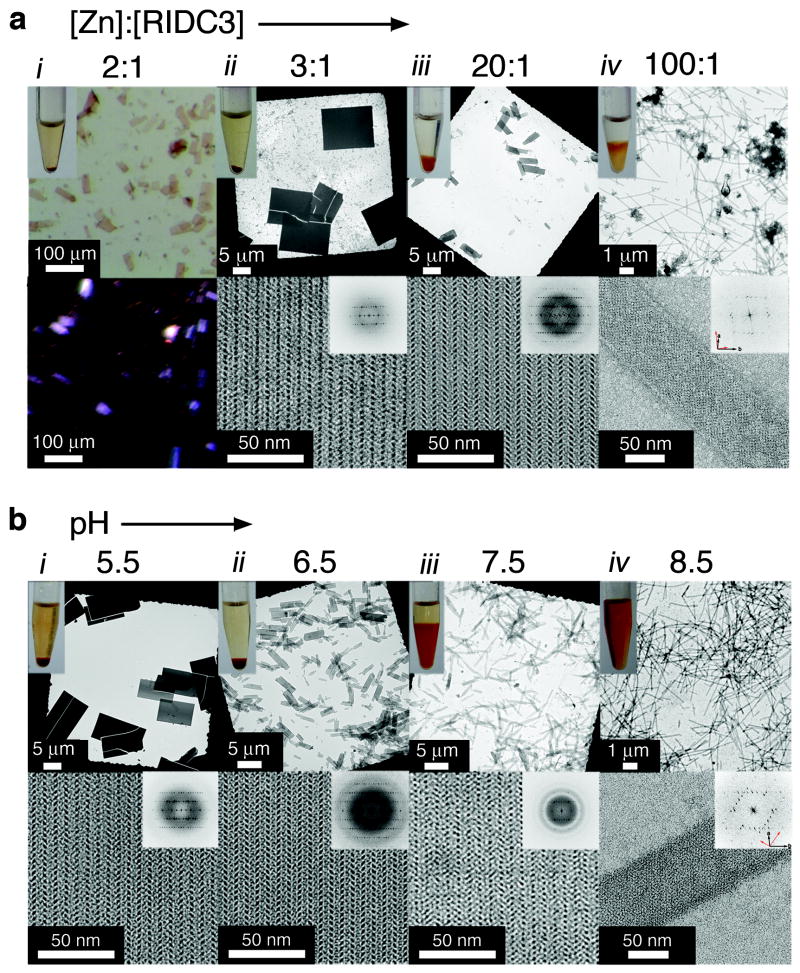
We examined RIDC3 self-assembly in solution with respect to Zn and RIDC3 concentrations and pH, the parameters expected to govern Zn-RIDC3 interactions. The macroscopic and microscopic characteristics of RIDC3 self-assembly as a function of the [Zn]:[RIDC3] ratio at [RIDC3]=50 μM and pH=5.5 were monitored by light and electron microscopy (Fig. 2a). It is important to note that RIDC3 concentrations used here are significantly lower than those (>2 mM) needed during conventional cyt cb562 crystallization procedures that require precipitants like PEG. Upon Zn addition, RIDC3 rapidly formed precipitates, which evolved into planar, rectangular crystalline arrays over the course of several days (Supplementary Fig. S4). We refer to these as “arrays” rather than “crystals” to emphasize that they are obtained by bottom-up self-assembly. These 2D arrays decreased in size as the [Zn]:[RIDC3] ratio was increased (Fig. 2a). The lattice parameters for all 2D arrays were identical within experimental error (a=37±1 Å, b=137±1 Å, α=90±1°), indicating they share the same underlying 2D arrangement of RIDC3 molecules. At 100-fold excess Zn, the arrays converted into uniformly thin, unbranched tubes up to 15 μm long and with widths distributed around 80 nm (Fig. 2a and Supplementary Fig. S5). Fourier transforms computed from transmission electron microscopy (TEM) images of these tubes indicated the presence of two, mirror-symmetric lattices (interlattice angle ~85°). Such overlapping lattices are typical of hollow, helical tubes flattened during specimen preparation for negative-stain TEM 28.
Figure 2. Zn-induced RIDC3 self-assembly in solution characterized by light and electron microscopy.
a, Dependence of the morphology of self-assembled RIDC3 arrays on the [Zn]:[RIDC3] ratio at [RIDC3]=50 μM and pH=5.5. b, Dependence of the morphology of self-assembled RIDC3 arrays on pH at [RIDC3]=100 μM and [Zn]=300 μM. These microscopy images illustrate a trend of decreasing array size with either an increase in the [Zn]:[RIDC3] ratio (a) or an increase in pH (b). For [Zn]:[RIDC3]=2 at pH=5.5 (column i in a), only macroscopic crystalline arrays were observed and were viewed by light microscopy with (top) and without (bottom) polarizers. The birefringence of the RIDC3 arrays is indicative of crystalline order. In all other experiments, the top and the bottom rows show low and high magnification TEM images, respectively, with the computed Fourier transforms of the high-magnification images shown as insets. Discrete reflections in the Fourier transforms with identical spacings under all conditions demonstrate that all RIDC3 arrays are crystalline and contain the same underlying two-dimensional lattice. The overlapping lattice axes in the tubular structures (column iv) are highlighted with black and red arrows (bottom right of transform).
The same morphological trend was observed at a higher [RIDC3] (400 μM), except that the transitions occurred at lower [Zn]:[RIDC3] ratios (Supplementary Fig. S6). Similarly, when pH was raised from 5.5 to 8.5 at a fixed [Zn]:[RIDC3] ratio, the morphology of the assembled arrays gradually changed from 2D plates into helical nanotubes (Fig. 2b). Under these conditions, the lattice constants remained the same as those at pH=5.5. However, the nanotubes obtained at pH=8.5 featured a different interlattice angle (~60°) and greater width (~90 nm, Supplementary Fig. S5), indicating pH-dependent polymorphism. All RIDC3 assemblies were stable for at least six months at room temperature and remained intact upon exchange into a solution with no Zn, but could be dissolved by adding ethylenediaminetetraacetic acid (EDTA). Nanotubes obtained at pH=8.5 fully converted into 2D arrays upon lowering the pH to 5.5, but this transition was not reversible.
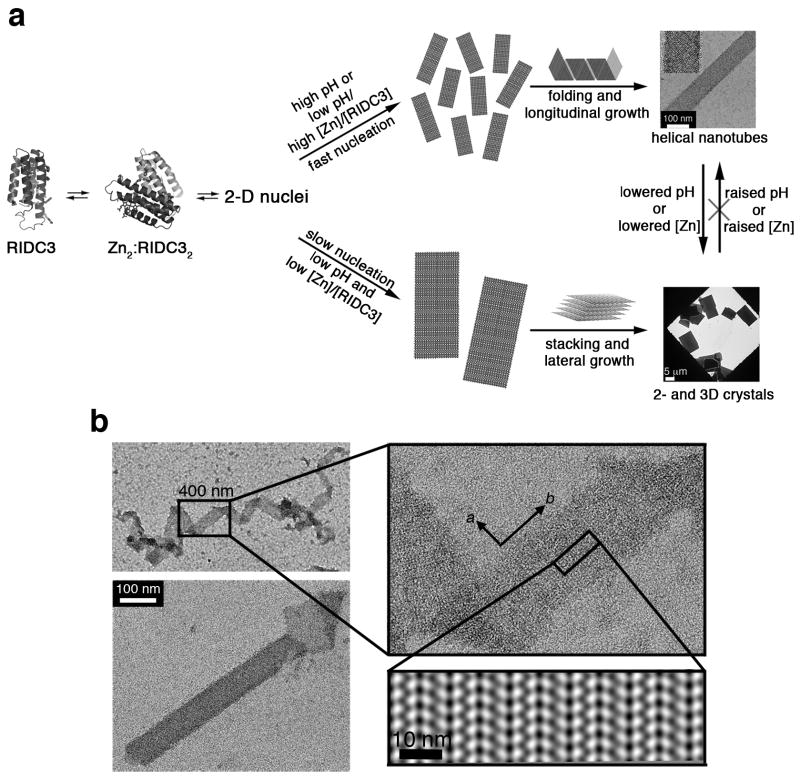
Based on these results, we propose a mechanism for the Zn-mediated self-assembly of RIDC3 (Fig. 3a), in which all observed arrays originate from 2D nuclei that have the same arrangement of RIDC3 molecules. At high [Zn]:[RIDC3] ratios or at high pH, where the high-affinity sites are fully deprotonated, Zn-RIDC3 interactions are favored and nucleation is rapid, resulting in a large number of small nuclei that “roll up” into helical nanotubes. This hypothesis is supported by the observation of monolayered, 2D sheets at intermediate maturation times at pH=8.5 and near the frayed ends of the nanotubes (Fig. 3b). At pH=5.5, where the high-affinity sites are partially protonated, and at low [Zn]:[RIDC3] ratios, nucleation is slow, yielding a smaller number of large 2D sheets. Because RIDC3 (pI = 5.3) is essentially charge neutral at this pH, repulsion between RIDC3 molecules is minimized, which encourages the stacking of the 2D sheets and additional lateral growth. As clearly demonstrated by the structural studies described below, the crystalline RIDC3 arrays are products of Zn-directed self-assembly and do not result from chemically-indiscriminate protein precipitation caused by changes in pH or [Zn].
Figure 3. Model for RIDC3 self-assembly.
a, Hypothetical model for Zn-mediated RIDC3 self-assembly under fast and slow nucleation conditions. Fast nucleation/growth conditions (high pH or high [Zn]:[RIDC3] ratio) promote the formation of many 2D nuclei, which “roll up” into helical nanotubes. Under slow nucleation conditions (low pH and low [Zn]:[RIDC3]), a smaller number of large 2D nuclei form, which can stack up in the third dimension due to the lack of repulsive interactions near the isoelectric point (pI = 5.3) of RIDC3. b, TEM images of a 2D RIDC3 ribbon and a tubular structure with frayed ends, illustrating the interconversion between tubular and sheet-like morphologies. Shown in the right bottom corner is the 2D image reconstruction of a single-layered portion of the ribbon, indicating the molecular arrangement of RIDC3 molecules.
Structural basis of RIDC3 self-assembly
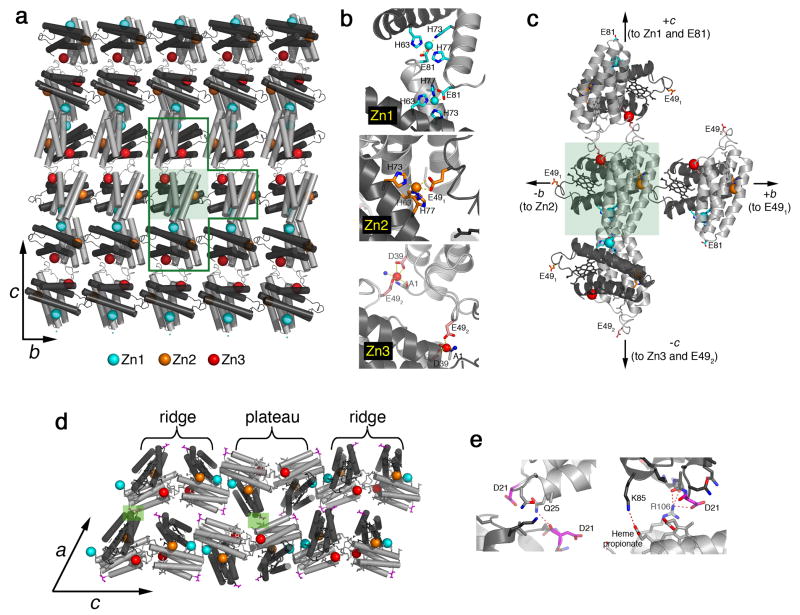
To help determine the structural basis of Zn-mediated RIDC3 self-assembly, we set out to grow 3D arrays large enough for X-ray diffraction analysis. According to our model above, this could be achieved by further retarding the nucleation of 2D RIDC3 arrays. Large, 3D RIDC3 crystalline arrays (≥200 × 20 × 5 μm3) (Supplementary Fig. S7) were obtained in a pH=6.0 solution that contained 2.5 mM RIDC3, 8 mM Zn2+, and 200 mM bis-Tris as a dual pH and Zn buffer. Given the pKa (6.46) and Zn2+ association constant KZn (240 M-1) of bis-Tris at 25°C 29, we calculate [Zn]free to be <1 mM in the solution. Thus, bis-Tris can act as an internal “drip hose” for Zn delivery and promote growth of 3D arrays under slow nucleation conditions. The X-ray crystal structure of 3D Zn-RIDC3 arrays, determined at 2.3-Å resolution (Supplementary Table 1), corroborated our self-assembly model and revealed the central role of Zn coordination. The b and c axes of these 3D arrays (C2 spacegroup, a=95.6 Å, b=37.8 Å, c=138.5 Å, β= 112.6°) match the lattice constants determined for the 2D arrays. We can conclude that the 3D arrays indeed form by the stacking of the 2D RIDC3 layers.
Intermolecular associations between RIDC3 molecules in the bc-plane are mediated entirely through three different types of Zn coordination at the previously described high- and low-affinity sites and involve no other surface contacts (Figs. 4a and 4b). The layers are built through the repetition of the stabilized C2-dimers (depicted as a pair of dark and light grey monomers). The observed arrangement of C2-dimers perfectly matches the pattern seen in monolayered, 2D sheets (Fig. 3b, see Supplementary Fig. S8 for an overlay). Geometric relationships between the Zn-mediated protein interactions in the bc-layer are best understood by considering a single C2-dimer as the origin of nucleation (green shaded box in Figs. 4a and 4c). The orthogonal Zn1 and Zn2 coordination vectors that originate from the two high-affinity sites are the key determinants of crystal growth along the c and b cell axes, respectively, and yield the 90° angle between them. Along the c axis, two C2-dimers interconnect through pairwise Zn1-Glu81 interactions (same mode of interaction as in Fig. 1d). Along the b axis, neighboring C2-dimers connect in a head-to-tail fashion through Zn2 and Glu49 (“E491”), giving rise to an infinite chain. Finally, a second set of pairwise interactions mediated by Zn3 bound to a low-affinity site and a second Glu49 (“E492”) introduce 2-fold symmetry normal to the bc-plane, allowing propagation along c and formation of the 2D array. As opposed to these strong Zn-mediated interactions, growth along the a axis involves two small (<250 Å2) surface patches of polar interactions between the 2D layers (Figs. 4d and e). These only promote crystal growth along a when the interlayer repulsions are minimized at pH=5.5.
Figure 4. Structural basis of Zn-mediated RIDC3 assembly.
a, Crystallographically-determined molecular arrangement in 2D Zn-RIDC3 sheets, viewed normal to the bc plane of 3D crystalline arrays. A single C2-dimer is highlighted in the shaded green box. b, Close-up view of the three different Zn coordination environments that enable RIDC3 self-assembly in two dimensions. Zn1 and Zn2 sites are formed by the high affinity coordination motif described in Fig. 1, whereas Zn3 is formed by the low affinity coordination motif. c, Close-up view of the T-shaped boxed area in (a). d, Contacts between each 2D RIDC3 layer, responsible for growth in the third dimension, are highlighted in shaded green boxes and detailed in e.
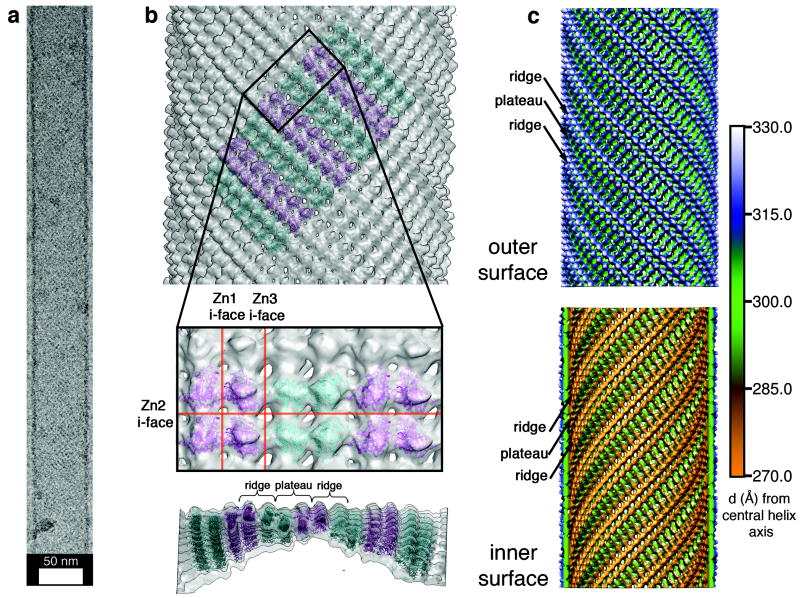
To complete our characterization of RIDC3 arrays, we determined the structure of non-flattened RIDC3 nanotubes using cryo-TEM. Images of the pH-5.5 nanotubes (Fig. 5a) that have C9 helical symmetry were processed using helical, real-space reconstruction methods30, which yielded a final map at near-nanometer resolution (Figs. 5b and 5c). Each subunit of the tube contains four discrete densities, each of whose volume is consistent with a C2-dimer (Supplementary Fig. S9). The outer and inner surfaces of the nanotubes display the same pattern of ridges and plateaus (Figs. 5b and c) seen in each 2D layer of the 3D RIDC3 arrays (Fig. 4). This finding, again, is in accord with our model that both nanotubes and 3D arrays emerge from the same 2D arrangement of RIDC3 molecules.
Figure 5. RIDC3 nanotube structure and assembly.
a, A typical vitrified, unstained nanotube used for helical cryo-EM reconstruction; see Supplementary Fig. S14 for collective views of RIDC3 nanotubes. b, Tetrameric RIDC3 units (Zn1-linked dimer of C2-dimers) modeled into the reconstructed tube density map. (top) View of the outer surface showing the alternating orientations (cyan and magenta) of tetrameric units; (middle) closeup view of tube surface with modeled tetramers; (bottom) lateral view of the tube highlighting its curvature. c, Radially color-coded representations of the outer (top) and inner (bottom) surfaces of the reconstructed nanotube revealing ridges and plateaus consistent with the X-ray crystal structure.
Conversion of a 2D crystalline sheet into a helical tube arises when curvature is induced along both lattice vectors of the sheet. Using the Zn1-linked dimer of C2-dimers as a rigid unit (see Supplementary Methods for details), we built a pseudo-atomic model of the tube to help identify the structural origins of this curvature (Fig. 5b). In this model, the crystallographically observed Zn2 coordination geometry appeared to be maintained (Supplementary Figs. S10 and S11). In contrast, the Zn3 interface must simultaneously bend and twist to enable the transformation of the 2D sheet into a hollow tube (Supplementary Figs. S10 and S12). Such a significant conformational change precludes the Zn3-Glu492 coordination observed in crystals. Instead, the EM model indicates that other nearby acidic residues (Asp2 and Asp50) may participate in an alternative mode of Zn coordination (Supplementary Fig. S12).
Directed stacking of RIDC3 sheets in the third dimension through chemical functionalization
Because RIDC3 self-assembly is mediated by easily identifiable intermolecular interactions in all three dimensions (Fig. 4), we can correlate crystal morphology with the strength of these interactions. Strongest are the pairwise interactions between C2-dimers mediated by Zn1 and Zn3 along c, followed by a singular Zn2 interaction between adjacent C2-dimers along b. Consequently, the 2D sheets observed under fast nucleation conditions are rectangular (Fig. 3b), with their long edges parallel to c (b in 2D designation) and short edges parallel to b (a in 2D designation). 3D crystalline arrays are thinnest along a, which is consistent with the weakest interactions occurring in this direction.
Given these considerations, we asked whether RIDC3 crystal morphology can be further directed through designed interactions. To test this possibility, we quantitatively labeled the D21C variant of RIDC3 with Rhodamine Red C2 maleimide at the surface position 21, which is involved in interlayer interactions in 3D arrays (Figs. 4e and 6a). Rhodamine was chosen for functionalization because its derivatives are known to dimerize with sub-mM affinities31, which should be strong enough to promote interlayer interactions and 3D growth, but weak enough to permit self-healing during self-assembly such that crystallinity is achieved. Rhodamine has the added benefit of exhibiting distinct spectral changes upon dimerization32, which can be used to monitor self-assembly.
Figure 6. Rhodamine-directed stacking of 2D RIDC3 arrays.
a, Structural model of a 2D RIDC3 sheet uniformly labeled with Rhodamine Red C2 (green sticks) at Cys21. b and c, TEM images of fully matured, negatively stained rhodamine-C21RIDC3 crystals obtained at pH 8.5 (b) and pH 5.5 (c). The formation of 3D structures at pH 8.5 rather than nanotubes evidences that rhodamine labels promote favorable interactions between 2D, Zn-mediated RIDC3 layers. d, UV-visible absorption spectrum of a 50 μM rhodamine-C21RIDC3 (1-mm pathlength) sample obtained prior to Zn addition (black), after crystal maturation (red) and upon dissolution of crystals by addition of EDTA (green). The intense band at 415 nm is due to the haem Soret absorption. e, CD spectra of the same rhodamine-C21RIDC3 sample. The increase in absorption at 534 nm upon Zn addition (d) indicates the formation of rhodamine dimers, while the emergence of a CD signal in the 520-630 nm region (e) during the maturation of rhodamine-C21RIDC3 crystals suggests that the rhodamine dimers are in discrete, rigid environments.
As a critical test, we examined the Zn-mediated assembly of rhodamine-D21CRIDC3 (R-C21RIDC3) under fast nucleation conditions (pH=8.5 and 10-fold excess [Zn]), which normally yield nanotubes with unmodified RIDC3. As desired, R-C21RIDC3 instead assembled into multilayered crystalline arrays with the previously observed 2D lattice parameters (a=36±1 Å, b=137±1 Å, α=90±1°) (Fig. 6b). At pH=5.5, which is near the isoelectric point of RIDC3, the 3D growth of R-C21RIDC3 was further amplified (Fig.6c). The UV-visible spectrum of a suspension of Zn-mediated R-C21RIDC3 crystals showed the rhodamine dimer band emerging at 534 nm (Fig. 6d), which is not pronounced when the protein is monomeric in solution. The CD spectrum of the same crystal suspension featured a distinct signature in the 520-630 nm region associated with the formation of a rigid environment around the rhodamine dimers (Fig. 6e), and this signature only emerged upon full crystal maturation (Supplementary Fig. S13). These observations suggest that the R-C21RIDC3 arrays display order along the third dimension as well.
Conclusions
Our results demonstrate that the self-assembly of a protein can be directed in all three dimensions to yield periodic arrays with nano- and microscale long-range order. The 1- and 2D RIDC3 arrays described here resemble natural assemblies like microtubules33, helical viruses34 and S-layers35 in terms of their dimensions and structural uniformity. More importantly, they recapitulate the ability of these macromolecules to assume distinct conformational states in response to environmental cues such as pH and metal concentrations36. A particular advantage of metal-directed protein self-assembly (MDPSA) lies in its inherently chemically-tunable and modular nature. Self-assembling proteins can be chosen based on their built-in functions (e.g., electron transfer for cyt cb562), and/or be functionally augmented through derivatization (e.g., by rhodamine) or through the interfacial metals themselves. Together, these features create a versatile platform for engineering functional biomaterials and for studying the fundamentals of protein self-assembly and crystallization under highly controllable conditions.
The practical challenge in the general application of MDPSA stems from the broad and relatively smooth energy landscape of metal-protein interactions, particularly when exchange-labile metal ions like Zn2+ are utilized. That is, owing to the abundance of coordinating residues on any protein’s surface, there are potentially many metal-mediated aggregation states that lie close in energy with small barriers separating them22. The breadth of the MDPSA landscape would make it difficult to design from scratch or reengineer a protein that self-assembles into discrete supramolecular arrays rather than a heterogeneous mixture of metal-crosslinked aggregates. Our findings suggest that the search space for finding discrete protein assemblies can be significantly reduced by a) constructing symmetrical protein building blocks, and b) installing properly oriented metal coordination motifs on the protein surface that selectively, stably and reversibly bind metal ions. Once these design criteria are met, the smoothness of the MDPSA landscape can help ensure that protein self-assembly proceeds under thermodynamic control to yield the lowest-energy conformational states, which can interconvert in response to external stimuli. Indeed, such stimulus-responsive switching between nearly isoenergetic states is a hallmark of biological self-assembly.
Methods
Protein Biochemistry
Generation of plasmids, expression of RIDC3 and C21RIDC3 and SV analysis were performed according to previously published procedures24. After purification, mutations were verified by MALDI mass spectrometry (RIDC3: MWcalc=12,235 Da, MWmeasured=12241 Da; C21RIDC3: MWcalc=12,209 Da, MWmeasured=12210 Da).
Prior to labeling with Rhodamine Red C2 maleimide, C21RIDC3 was reduced with 10 mM DTT and exchanged into a buffer containing 20 mM HEPES (pH 7.5). A mixture containing 95 μM C21RIDC3 and 330 μM Rhodamine Red C2 maleimide was then let to react in the dark for one hour at room temperature with gentle stirring. After quenching the reaction with 10 mM DTT, singly-labeled protein was purified by anion exchange chromatography, and quantitative labeling of the single free cysteine was verified by MALDI mass spectrometry (MWcalc=12890 Da, MWmeasured=12889 Da).
Macromolecular Crystallography
The PEG-precipitated RIDC3 crystals (Type 1, Fig. 1) and those formed through Zn-directed self-assembly (Type 2, Fig. 4) were both obtained by sitting drop vapor diffusion at room temperature (see Supplementary Table S1 for solution composition). Crystals suitable for diffraction experiments were transferred to a solution of mother liquor containing 20% glycerol as the cryoprotectant and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at 100 K and subsequently integrated using MOSFLM and scaled with SCALA37. Structures were then determined by molecular replacement using PHASER38 and subjected to rigid-body, positional, thermal and TLS refinement in REFMAC39, along with manual rebuilding in COOT40. All figures of the resulting structures were produced using PYMOL41.
Preparation and imaging of RIDC3 nanostructures
2D RIDC3 arrays or 1D nanotubes were produced at the indicated concentrations in 1-mL or 200-μL volume solutions buffered with either 20 mM MES (pH 5.5 and 6.5), MOPS (pH 7.5) or CHES (pH 8.5). After incubating at room temperature, 3 μL aliquots were deposited on home-made, carbon-coated Cu grids and stained with uranyl acetate. After being allowed to dry, grids were imaged on an FEI Sphera transmission electron microscope equipped with an LaB6 electron gun operated at 200 keV. Images were recorded on a Gatan 2K2 CCD using objective lens underfocus settings ranging from 400-800 nm. Where indicated, 2D image reconstructions were generated using the 2dx software package42.
For cryo-EM samples, solutions containing 450 μM RIDC3 and 4.5 mM Zn were incubated at room temperature for one week in a 20 mM MES buffer (pH 5.5) to allow the formation of protein nanotubes. A 3 μL aliquot of this solution was then deposited onto a home-made, lacey carbon grid covered with a thin layer of continuous carbon, or a Quantifoil grid (see Supplementary Fig. S14 for views of collections of nanotubes), plunged into liquid ethane after blotting, and transferred into a precooled, FEI Polara multi-specimen holder. Images were then recorded on a Gatan 4K2 CCD camera on an FEI Polara microscope using low-dose conditions (~12 e-/Å2) at a nominal magnification of 39,000 X (calibrated pixel size = 2.883 Å), with objective lens underfocus settings ranging from 1.5-4 μm.
Image processing and 3D reconstruction
Image preprocessing was carried out essentially as previously described30. Briefly, helical particles were boxed using the helixboxer routine in EMAN43 and padded to 81922 pixels, after which IHRSR++ was used for the following steps. A diffraction pattern was calculated for each tube using “helixautomation”, and tubes with the presence of specific layer lines (Supplementary Fig. S15) were included in the reconstruction. Each tube was then cut into segments using “helixcutstk”, diameters were computed for each segment using “get_tube_diameters”, and segments with consistent diameters were selected for further processing (diameter range 518-635 Å). Finally, the images were corrected for the CTF (estimated using RobEM (http://cryoEM.ucsd.edu/programs.shtm)) and merged into a single image stack for the final helical reconstruction.
Initial parameters as input for reconstruction using IHRSR++ were estimated from images of negatively stained tubes based on the interlattice angle and tube width. Tubes yielding consistent reconstructions, as indicated by alternating dimers of C2-dimers in the 3D density map, convergence to the same refined helical parameters and a computed Fourier transform consistent with an averaged spectrum from the raw images, were chosen for inclusion in the final reconstruction. Ultimately, the starting values for ΔZ and Δϕ were varied to ensure the global minimum was reached in the final reconstruction.
Supplementary Material
Acknowledgments
This work was primarily supported by DOE (Division of Materials Sciences, Office of Basic Energy Sciences, Award DE-FG02-10ER46677 to F.A.T.). Additional support was provided by the Agouron Foundation, the Beckman Foundation, the Sloan Foundation, NSF (CHE-0908115 to F.A.T., protein design), NIH (EM characterization, R37GM-033050 and 1S10 RR.020016 to T.S.B. and F32 AI078624 to K. N. P.), and the University of California, San Diego. Portions of this research were carried out at SSRL, operated by Stanford University on behalf of DOE.
Footnotes
Author Contributions J.D.B. designed and performed most of the experiments and data analysis, and wrote the paper. X.I.A. performed computational interface design calculations. C.T. and K.N.P. provided guidance and assistance with electron microscopy data collection and analysis. T.S.B. guided electron microscopy data analysis and wrote the paper. F.A.T. initiated and directed the project, analyzed data and wrote the paper.
Additional Information Coordinates, structure factors and EM maps were deposited into the PDB and EM databanks under accession codes 3TOL, 3TOM and EMD-5341.
The authors declare no competing financial interests.
Reprints and permission information is available online at www.nature.com/reprints.
Supplementary Information accompanies this paper at www.nature.com/naturechemistry.
References
- 1.Mann S. Life as a nanoscale phenomenon. Angew Chem Int Ed Eng. 2008;47:5306–5320. doi: 10.1002/anie.200705538. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen ST, Gin DL, Hupp JT, Zhang X. Supramolecular chemistry: Functional structures on the mesoscale. Proc Natl Acad Sci USA. 2001;98:11849–11850. doi: 10.1073/pnas.201373898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seeman NC, Belcher AM. Emulating biology: Building nanostructures from the bottom up. Proc Natl Acad Sci USA. 2002;99:6451–6455. doi: 10.1073/pnas.221458298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shenton W, Pum D, Sleytr UB, Mann S. Synthesis of cadmium sulphide superlattices using self-assembled bacterial S-layers. Nature. 1997;389:585–587. [Google Scholar]
- 5.McMillan RA, Paavola CD, Howard J, Chan SL, Zaluzec NJ, Trent JD. Ordered nanoparticle arrays formed on engineered chaperonin protein templates. Nat Mater. 2002;1:247–252. doi: 10.1038/nmat775. [DOI] [PubMed] [Google Scholar]
- 6.Lee YJ, et al. Fabricating Genetically Engineered High-Power Lithium-Ion Batteries Using Multiple Virus Genes. Science. 2009;324:1051–1055. doi: 10.1126/science.1171541. [DOI] [PubMed] [Google Scholar]
- 7.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 8.Zheng JP, et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of Intracellular Reactions with Rationally Designed RNA Assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 10.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 11.Aggeli A, et al. Responsive gels formed by the spontaneous self-assembly of peptides into polymeric beta-sheet tapes. Nature. 1997;386:259–262. doi: 10.1038/386259a0. [DOI] [PubMed] [Google Scholar]
- 12.Banwell EF, et al. Rational design and application of responsive [alpha]-helical peptide hydrogels. Nat Mater. 2009;8:596–600. doi: 10.1038/nmat2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kortemme T, Baker D. Computational design of protein-protein interactions. Curr Opin Chem Biol. 2004;8:91–97. doi: 10.1016/j.cbpa.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Dotan N, Arad D, Frolow F, Freeman A. Self-assembly of a tetrahedral lectin into predesigned diamondlike protein crystals. Angew Chem Int Ed. 1999;38:2363–2366. [PubMed] [Google Scholar]
- 15.Ringler P, Schulz GE. Self-Assembly of Proteins into Designed Networks. Science. 2003;302:106–109. doi: 10.1126/science.1088074. [DOI] [PubMed] [Google Scholar]
- 16.Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. In vitro self-assembly from a simple protein of tailorable nanotubes building block. Proc Natl Acad Sci USA. 2008;105:3733–3738. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padilla JE, Colovos C, Yeates TO. Nanohedra: Using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc Natl Acad Sci USA. 2001;98:2217–2221. doi: 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinclair JC, Davies KM, Venien-Bryan C, Noble MEM. Generation of protein lattices by fusing proteins with matching rotational symmetry. Nature Nanotech. 2011;6:558–562. doi: 10.1038/nnano.2011.122. [DOI] [PubMed] [Google Scholar]
- 19.Caulder DL, Raymond KN. Supermolecules by design. Acc Chem Res. 1999;32:975–982. [Google Scholar]
- 20.Leininger S, Olenyuk B, Stang PJ. Self-assembly of discrete cyclic nanostructures mediated by transition metals. Chem Rev. 2000;100:853–907. doi: 10.1021/cr9601324. [DOI] [PubMed] [Google Scholar]
- 21.Holliday BJ, Mirkin CA. Strategies for the construction of supramolecular compounds through coordination chemistry. Angew Chem Int Ed. 2001;40:2022–2043. [PubMed] [Google Scholar]
- 22.Salgado EN, Radford RJ, Tezcan FA. Metal-Directed Protein Self-Assembly. Acc Chem Res. 2010;43:661–672. doi: 10.1021/ar900273t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radford RJ, Brodin JD, Salgado EN, Tezcan FA. Expanding the utility of proteins as platforms for coordination chemistry. Coord Chem Rev. 2011;255:790–803. [Google Scholar]
- 24.Salgado EN, Faraone-Mennella J, Tezcan FA. Controlling Protein-Protein Interactions through Metal Coordination: Assembly of a 16-Helix Bundle Protein. J Am Chem Soc. 2007;129:13374–13375. doi: 10.1021/ja075261o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salgado EN, Lewis RA, Mossin S, Rheingold AL, Tezcan FA. Control of Protein Oligomerization Symmetry by Metal Coordination: C2 and C3 Symmetrical Assemblies through CuII and NiII Coordination. Inorg Chem. 2009;48:2726–2728. doi: 10.1021/ic9001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Kuhlman B. RosettaDesign server for protein design. Nucl Acids Res. 2006;34:W235–238. doi: 10.1093/nar/gkl163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polyakov A, Severinova E, Darst SA. Three-dimensional structure of E. coli core RNA polymerase: Promoter binding and elongation conformations of the enzyme. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 29.Scheller KH, Abel THJ, Polanyi PE, Wenk PK, Fischer BE, Sigel H. Metal Ion/Buffer Interactions. Eur J Biochem. 1980;107:455–466. [PubMed] [Google Scholar]
- 30.Parent KN, Sinkovits RS, Suhanovsky MM, Teschke CM, Egelman EH, Baker TS. Cryo-reconstructions of P22 polyheads suggest that phage assembly is nucleated by trimeric interactions among coat proteins. Phys Biol. 2010;7 doi: 10.1088/1478-3975/7/4/045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamman BD, Oleinikov AV, Jokhadze GG, Bochkariov DE, Traut RR, Jameson DM. Tetramethylrhodamine Dimer Formation as a Spectroscopic Probe of the Conformation of Escherichia coli Ribosomal Protein L7/L12 Dimers. J Biol Chem. 1996;271:7568–7573. doi: 10.1074/jbc.271.13.7568. [DOI] [PubMed] [Google Scholar]
- 32.Chambers RW, Kajiwara T, Kearns DR. Effect of Dimer Formation of Electronic Absorption and Emission-Spectra of Ionic Dyes - Rhodamines and Other Common Dyes. J Phys Chem. 1974;78:380–387. [Google Scholar]
- 33.Li HL, DeRosier DJ, Nicholson WV, Nogales E, Downing KH. Microtubule structure at 8 Ångstrom Resolution. Structure. 2002;10:1317–1328. doi: 10.1016/s0969-2126(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 34.Glucksman MJ, Bhattacharjee S, Makowski L. Three-dimensional structure of a cloning vector: X-ray diffraction studies of filamentous bacteriophage M13 at 7 Å resolution. J Mol Biol. 1992;226:455–470. doi: 10.1016/0022-2836(92)90960-r. [DOI] [PubMed] [Google Scholar]
- 35.Sara M, Sleytr UB. S-Layer Proteins. J Bacteriol. 2000;182:859–868. doi: 10.1128/jb.182.4.859-868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge P, Zhou ZH. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc Natl Acad Sci USA. 2011;108:9637–9642. doi: 10.1073/pnas.1018104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Cryst D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murshudov G, Vagin A, Dodson E. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Cryst. 1996;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.DeLano WL. The PYMOL Molecular Graphics System. 2003 http://www.pymol.org.
- 42.Stahlberg H, Gipson B, Zeng X, Zhang ZY. 2dx - User-friendly image processing for 2D crystals. J Struct Biol. 2007;157:64–72. doi: 10.1016/j.jsb.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 43.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.