Abstract
Objectives:
To characterize clinically and genetically a family with autosomal dominant lateral temporal epilepsy (ADLTE) negative to LGI1 exon sequencing test.
Methods:
All participants were personally interviewed and underwent neurologic examination. Most affected subjects underwent EEG and neuroradiologic examinations (CT/MRI). Available family members were genotyped with the HumanOmni1-Quad v1.0 single nucleotide polymorphism (SNP) array beadchip and copy number variations (CNVs) were analyzed in each subject. LGI1 gene dosage was performed by real-time quantitative PCR (qPCR).
Results:
The family had 8 affected members (2 deceased) over 3 generations. All of them showed GTC seizures, with focal onset in 6 and unknown onset in 2. Four patients had focal seizures with auditory features. EEG showed only minor sharp abnormalities in 3 patients and MRI was unremarkable in all the patients examined. Three family members presented major depression and anxiety symptoms. Routine LGI1 exon sequencing revealed no point mutation. High-density SNP array CNV analysis identified a genomic microdeletion about 81 kb in size encompassing the first 4 exons of LGI1 in all available affected members and in 2 nonaffected carriers, which was confirmed by qPCR analysis.
Conclusions:
This is the first microdeletion affecting LGI1 identified in ADLTE. Families with ADLTE in which no point mutations are revealed by direct exon sequencing should be screened for possible genomic deletion mutations by CNV analysis or other appropriate methods. Overall, CNV analysis of multiplex families may be useful for identifying microdeletions in novel disease genes.
Autosomal dominant lateral temporal epilepsy (ADLTE; OMIM 600512), or autosomal dominant partial epilepsy with auditory features (ADPEAF), is characterized by lateral temporal seizures with prominent auditory or aphasic auras, onset in adolescence or early adulthood, and normal MRI.1,2 Mutations in the leucine rich, glioma inactivated 1 (LGI1) gene are responsible for ADLTE in up to 50% of the families.2–4 Over 25 LGI1 mutations have been identified so far, all of which are point mutations, either missense or splice-site mutations or short indels.5,6
We report the first genomic microdeletion involving LGI1 identified in an ADLTE family by high-density single nucleotide polymorphism (SNP) array copy number variation (CNV) analysis.
METHODS
Standard protocol approvals, registrations, and patient consents.
All participants signed informed consent approved by the local ethics committee. Neurologic examination, video-EEG recordings, and brain MRI were performed in each subject. LGI1 exon sequencing was performed as described.2 Genotyping was performed using the HumanOmni1-Quad v1.0 BeadChip (Illumina, San Diego, CA). The PennCNV algorithm7 was used to infer CNVs from the signal intensity data. Real time quantitative PCR (qPCR) was performed using the TaqMan Gene Exp Assay (Applied Biosystems). For detailed methods see e-Methods on the Neurology® Web site at www.neurology.org.
RESULTS
Clinical descriptions.
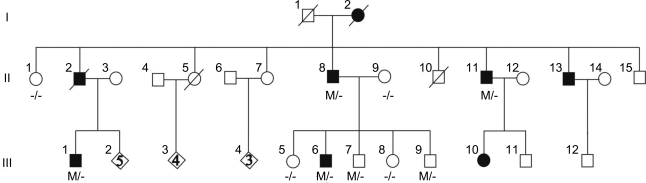
The pedigree structure is shown in figure 1 and the clinical features of the affected family members are summarized in the table. The proband (III-1), a 42-year-old right-handed man, reported monthly episodes of high-pitched sounds and unformed voices, usually accompanied by feeling of confusion, from age 15 years. At the same age, he also experienced a tonic-clonic seizure (TCS). His EEG recording showed independent bilateral (left > right) runs of slow waves in the frontotemporal areas (figure e-1a). Neurologic examination, brain MRI, and auditory evoked potentials were unremarkable. At age 30 years, oxcarbazepine (600 mg/day) was started with good results. Patient III-6 is a 35-year-old, left-handed man who had a TCS at age 34 years and monthly focal seizures starting with unformed noises or screaming and commonly followed by déjà vu, epigastric aura, or dysmnesia. Bilateral temporal slow waves were observed only at 1 EEG. He was started on valproate (1,000 mg/day) with benefit. Patient II-11 is a 47-year-old, right-handed man with a history of focal seizures with retained awareness that started at age 14 years characterized by unformed voices or sounds in both ears progressively getting louder; a secondarily generalized TCS could sometimes ensue. Sleep deprivation and stress facilitated seizures. His neurologic examination, prolonged EEG monitoring, and brain MRI were normal. Therapy with phenobarbital (75 mg/day) in adult age allowed almost complete seizure control. However, at age 42 years, voluntary withdrawal of therapy resulted in a TCS. No further seizures occurred after resumption of the treatment. Individual II-8 also had similar episodes beginning in his adolescence. Neurologic examination, EEGs, and brain MRI were unremarkable. He was taking phenobarbital (50 mg/day) and phenytoin (300 mg/day). Subjects I-2 and II-2 experienced repeated TCS from adolescence or adult age but insufficient clinical data were available for these cases. The other family members did not report any auras or symptoms and had normal neurologic examination and EEG, except for individual III-9 who showed borderline intelligence and rare left anterior temporal theta activity (figure e-1b). In addition, major depression and anxiety symptoms were present in individuals II-11, III-1, and III-6.
Figure 1. Family pedigree and mutation.
Circles denote females; squares denote males; blackened symbols denote affected subjects. Individuals carrying 1 mutant and 1 normal allele are denoted by M/−; those with no mutations by −/−.
Table.
Clinical details of the affected family members
Abbreviations: AED = antiepileptic drug; N = normal; NA = not available; ND = not done; OXC = oxcarbazepine; PB = phenobarbital; PHT = phenytoin; VPA = valproate.
Deceased
LGI1 sequencing and microsatellite analysis.
Earlier routine sequencing of LGI1 exons excluded point mutations in the proband III-1, and linkage analysis of microsatellite markers flanking LGI1 gave a maximum lod score (0.79) considerably lower than the expected lod score (1.84) at θ = 0.0, assuming 70% penetrance and disease allele frequency 0.0001. We therefore considered this family unlikely to have LGI1 mutations and further investigated it to search for novel ADLTE genes.
SNP array analysis.
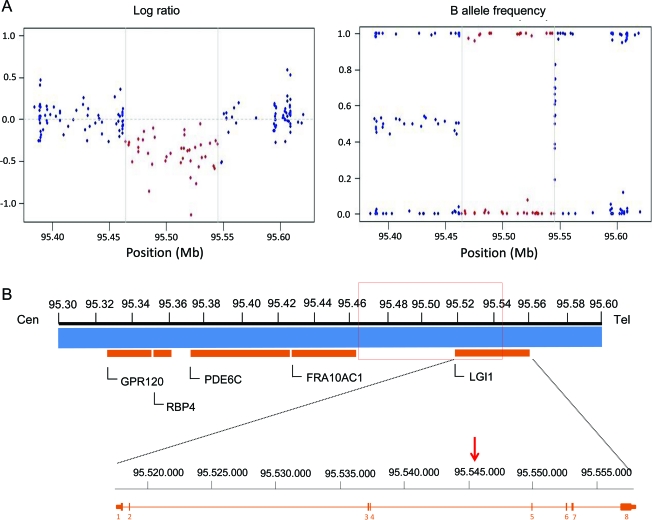
The available family members were genotyped using the HumanOmni1-Quad v1.0 platform (Illumina), which contains over 1 million SNPs. The whole genotype dataset was analyzed to evaluate the amount of structural variation in this family and to examine the effect of CNVs on the affection status. Overall we identified 710 CNVs corresponding to 383 unique CNV regions with sizes ranging from 10,041 to 724,099 bp (mean = 66,880; SD = 93,350). The number of CNVs per individual varied from 51 to 106 (mean = 71; SD = 19.9). Among all the CNVs identified, we focused on those that were common to all the available affected individuals, in order to recognize a possible cause of the syndrome. We found a deletion spanning 81,245 bp on chromosome 10q23.33 (figure 2) in 4 patients (II-8, II-11; III-1, III-6) and 2 nonaffected individuals (III-7 and III-9) aged 24 and 17 years. The deletion encompasses the region between SNPs rs11187602 (chromosome position 95464132) and rs7099034 (95545377) and includes 40 SNPs; it spans part of the LGI1 gene, resulting in the loss of the first 4 exons and disruption of the gene. This deletion is not present in any of the available databases: 1000 Genomes Project (http://www.1000genomes.org), Database of Genome Variants (http://projects.tcag.ca/variation), and Decipher (http://decipher.sanger.ac.uk). Gene dosage performed by qPCR with primers specific for LGI1 exon 1 confirmed the deletion in the 6 affected and unaffected family members as predicted by CNV analysis (figure e-2).
Figure 2. Illustration of the genomic microdeletion detected at 10q23.33.
(A) Results of copy number analysis using high-density single nucleotide polymorphism (SNP) arrays. Genomic profiles are shown for chromosome 10 (95, 384, 353–95, 620, 059). Log R ratios and B allele frequencies are plotted against chromosome 10 position. The length of the deletion is displayed as distance between the 2 gray vertical lines and included SNPs indicated by red dots. Hemizygosity is shown by a drop in log R ratio (red dots below 0.0) and missing heterozygote SNP calls (lack of dots at 0.5). (B) Schematic representation of the deleted genomic region on chromosome 10. The deletion (red rectangle) encompasses exons 1–4 of LGI1 (genes are represented by horizontal orange bars). The zoomed region displays the exon–intron organization of LGI1 and the red arrow indicates the telomeric end point of the deletion inside the gene. Cen = centromere; tel = telomere.
DISCUSSION
We identified the first ADLTE-causing microdeletion in the LGI1 gene locus by SNP array CNV analysis. It spans about 81 kb of genomic DNA (40 SNPs) and includes the first 4 LGI1 exons. The deletion was found in all the available patients and in 2 unaffected family members, who are still at risk of developing ADLTE; its occurrence in the family was confirmed by qPCR; and, finally, it is not present in public CNV databases. Altogether, these data support the pathogenic nature of this deletion.
Recognition of ADTLE is clinically important, as this form of focal epilepsy generally has a good prognosis. Since the clinical diagnosis of ADLTE is based mainly on the presence of an auditory aura, which may be elusive in some patients or families, testing for mutations in LGI1 is important to confirm diagnosis. Moreover, detection of LGI1 mutations in familial cases with lateral temporal epilepsy could contribute—together with the clinical data—to avoiding long presurgical studies and preventing unnecessary surgery. To ensure detection or exclusion of all possible LGI1 mutations, ADLTE families without DNA sequence-based mutations should be screened for possible LGI1 microdeletions by CNV analysis or other suitable methods.
The microdeletion described in this article encompasses about 60 kb upstream of exon 1 and very likely deletes most of or perhaps all the regulatory sequences of LGI1. Therefore, expression of the truncated allele is very unlikely, suggesting a loss-of-function effect of the deletion. Its proximal (telomeric) end lies less than 2 kb apart from the FRA10AC1 gene, which contains the folate-sensitive fragile site FRA10A, whose instability depends on variable numbers of a CGG repeat.8 Although FRA10A is deemed benign, its intrinsic instability may give rise to genomic microdeletions extending to the LGI1 gene, which may account for a proportion of ADLTE-causing mutations.
Common CNVs, that is, genomic microdeletions and duplications occurring in the general population with frequency >1%, have been shown to alter susceptibility to various neurologic disorders, including epilepsy.9 Conversely, rare CNVs have not yet been studied extensively. These latter include pathogenic deletions underlying Mendelian disorders, which altogether represent about 20% of all Mendelian mutations.10 Our results have shown that disease-causing microdeletions can be identified by high-density SNP-array CNV analysis of multiplex families. This approach may generally be useful for identifying microdeletions in novel disease genes, particularly those disease-related genes lying in relatively unstable genomic regions.
Supplementary Material
GLOSSARY
- ADLTE
autosomal dominant lateral temporal epilepsy
- ADPEAF
autosomal dominant partial epilepsy with auditory features
- CNV
copy number variation
- LGI1
leucine rich, glioma inactivated 1
- qPCR
quantitative PCR
- SNP
single nucleotide polymorphism
- TCS
tonic-clonic seizure
Footnotes
Supplemental data at www.neurology.org
AUTHOR CONTRIBUTIONS
Drafting/revising the manuscript for content, including medical writing for content: M.F., P.S., C.N. Study concept or design: A.R., R.M., S.U., S.S., F.A.d.F., P.S., C.N. Analysis or interpretation of data: M.F., L.S., L.E., C.B., L.T., T.C., L.R., A.d.F. Contribution of vital reagents/tools/patients: A.d.F., F.A.d.F. Acquisition of data: L.S., L.E., C.B., L.R., T.C. Statistical analysis: M.F., T.C. Study supervision or coordination: C.N. Obtaining funding: A.R., R.M., C.N.
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1. Ottman R, Risch N, Hauser WA, et al. Localization of a gene for partial epilepsy to chromosome 10q. Nat Genet 1995;10:56–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Michelucci R, Poza JJ, Sofia V, et al. Autosomal dominant lateral temporal epilepsy: clinical spectrum, new epitempin mutations, and genetic heterogeneity in seven European families. Epilepsia 2003;44:1289–1297 [DOI] [PubMed] [Google Scholar]
- 3. Kalachikov S, Evgrafov O, Ross B, et al. Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat Genet 2002;30:335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ottman R, Winawer MR, Kalachikov S, et al. LGI1 mutations in autosomal dominant partial epilepsy with auditory features. Neurology 2004;62:1120– 1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nobile C, Michelucci R, Andreazza S, et al. LGI1 mutations in autosomal dominant and sporadic lateral temporal epilepsy. Hum Mutat 2009;30:530–536 [DOI] [PubMed] [Google Scholar]
- 6. Heiman GA, Kamberakis K, Gill R, et al. Evaluation of depression risk in LGI1 mutation carriers. Epilepsia 2010;51:1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007;17:1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sarafidou T, Kahl C, Martinez-Garay I, et al. Folate-sensitive fragile site FRA10Ais due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein Genomics 2004;84:69–81 [DOI] [PubMed] [Google Scholar]
- 9. Helbig I, Mefford HC, Sharp AJ, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet 2009;41:160–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet 2003;33(suppl):228–237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.