Abstract
Environmental conditions can alter the form, function, and behavior of organisms over short and long timescales, and even over generations. Aphid females respond to specific environmental cues by transmitting signals that have the effect of altering the development of their offspring. These epigenetic phenomena have positioned aphids as a model for the study of phenotypic plasticity. The molecular basis for this epigenetic inheritance in aphids and how this type of inheritance system could have evolved are still unanswered questions. With the availability of the pea aphid genome sequence, new genomics technologies, and ongoing genomics projects in aphids, these questions can now be addressed. Here, we review epigenetic phenomena in aphids and recent progress toward elucidating the molecular basis of epigenetics in aphids. The discovery of a functional DNA methylation system, functional small RNA system, and expanded set of chromatin modifying genes provides a platform for analyzing these pathways in the context of aphid plasticity. With these tools and further research, aphids are an emerging model system for studying the molecular epigenetics of polyphenisms.
1. Introduction
While the genome has been portrayed as a “blueprint” instructing the development of an adult organism, the articulation of genotype into phenotype is a more complex phenomenon. Context-dependent development and environment-dependent phenotypic variation have been observed for decades [1]. Like the changes in gene expression that intrinsically occur in development, environment can affect gene expression and alter developmental trajectories [2]. If these developmental responses to the environment, and plasticity itself, can increase fitness and are heritable, then morphology, physiology, behavior, or life history strategies can evolve elements of adaptive phenotypic plasticity [1, 3]. This can result in the production of continuous or discrete phenotypic variation (polyphenism). The possibility for nongenetic heritable effects of environment on development raises doubts about the “blueprint” view of the genome [4].
Waddington originally defined “epigenetics” as the study of phenomena that act to produce phenotype from genotype all within in a framework of evolutionary biology [5–7]. Waddington's view of epigenetics now largely encompasses the fields of developmental biology and evolutionary developmental biology, which describe, in part, how patterns of gene expression change during ontogeny and through evolution [8]. The modern field of epigenetics examines how patterns of gene expression, instructed by extrinsic biotic or abiotic factors, can be passed to offspring through means other than the inheritance of DNA sequence. Examples of inherited epigenetic phenomena include stable cell fate specification during pluripotent stem cell divisions, dosage compensation and X chromosome inactivation, imprinting, and position effect variegation in Drosophila [9]. Models for seemingly disparate phenomena have converged on common mechanisms for establishing heritable gene expression patterns: changes in chromatin architecture due to the effects of DNA methylation, small RNAs, and chromatin modifying enzymes [10, 11].
2. Predictive Adaptive Developmental Plasticity through the Aphid Life Cycle
Aphids, soft-bodied insects that feed on the phloem sap of plants, have long been a model for studying the causes and consequences of phenotypic plasticity. They exhibit both a wing polyphenism (consisting of winged and unwinged females) and a reproductive polyphenism (consisting of asexual and sexual individuals). The production of alternative morphs by genetically identical individuals by definition involves epigenetic mechanisms. Here, we describe these two polyphenisms within the context of the aphid life cycle. We then discuss the environmental cues that trigger the polyphenisms. Finally, we discuss what is known about epigenetic mechanisms in the pea aphid.
The life cycle of a model aphid species, the pea aphid Acyrthosiphon pisum, begins as a “foundress”—a female aphid that hatches in the spring from an overwintering egg. The foundress produces, via live birth (viviparity), a population of female unwinged aphids through asexual reproduction (apomictic parthenogenesis) that continues to reproduce asexually over several generations. This population is genetically identical, aside from spontaneous mutations [12], and lacks males during the spring and summer months. Environmental factors such as high aphid density, host plant quality, and predation can induce unwinged females to produce winged offspring. Winged asexual females disperse and colonize new host plants, founding new colonies via parthenogenesis. The parthenogenetic production of winged and unwinged female aphids continues during the spring and summer.
In fall, a change from asexual to sexual reproductive modes occurs. Asexual females sense the changing photoperiod and temperature and respond by parthenogenetically producing sexual females and males. Males are produced genetically by the loss of one X chromosome during parthenogenetic oocyte division and can be winged or unwinged. Since only sperm containing an X chromosome are viable, sexual females lay only female eggs on the host plant. The egg must “overwinter” for three to four months at cold temperatures in order to complete development and hatch as a foundress in the spring [13, 14]. Other aphid species switch host plants, produce winged sexual females or produce males earlier than pea aphids. These adaptations (sexual versus asexual, winged versus unwinged) have evolved in response to environmental changes that are predictable (seasons) and unpredictable but common (population density, host plant quality, and predation).
3. Experimental Evidence for Epigenetic Phenomena in Aphids
The wing and reproductive polyphenisms are examples of how the maternal environment affects the development of the offspring as a “predictive adaptive response” [15]. Several groups have described the triggering environmental cues and aphid responses. Though the cues differ for the reproductive and wing polyphenisms, the developmental response for both is separated by at least one generation from the triggering cue. Additionally, the resulting morphs are discrete forms and not simply continuous differences along a phenotypic gradient. This binary phenotypic output from an inductive signal gives the aphid experimental system an advantage for studying the epigenetic contribution to phenotypic plasticity.
3.1. Induction of Winged Aphids
Winged offspring can be induced by tactile stimulation of unwinged asexual aphids, either by interactions with other aphids, interactions with nonpredator insects, or experimental stimulation [16–18]. Unwinged mothers produce both unwinged and winged offspring; winged aphid mothers rarely produce winged offspring [16]. Other factors, such as the age of the mothers and temperature, can also modulate the degree of wing induction [18, 19]. The production of winged offspring can also be induced by the presence of aphid predators [20–22]. However, this effect may be driven by increased aphid walking, and thus increased inter-aphid interactions, in response to predator presence [23]. The environmental changes listed above are unpredictable but generally common, and aphids facultatively express the wing phenotype to limit predation and competition for resources.
In some aphids, wing induction occurs prenatally [24] while other species can be induced postnatally [16]. In prenatal determination, the environmental cue perceived by the mother must be transmitted to its embryos in utero, and the daughter embryos respond to this maternal signal. The precise nature of this maternal signal or its response is not known, though some studies implicate the juvenile hormone (JH) pathway (but see [16]). However, wing development itself does not occur until the second to third larval stage and is accompanied by the development of wing musculature, increased sclerotization of the cuticle, changes in eyes, antennal sensory rhinaria, and reproductive output [25, 26]. Thus, several days and presumably several rounds of cell division separate induction and resulting developmental response.
3.2. Induction of Sexual Aphids
The production of sexual morphs and the resulting overwintering egg coincides with predictable, seasonal changes in photoperiod and temperature. Sexual aphid morphs are observed in temperate zones during the fall and winter but not in the spring or summer, and aphids were the first animals shown to respond to changes in photoperiod [27]. Later studies defined the lengths of light and dark phases necessary for the induction of sexual aphids (reviewed in [28]). An embryo that developed under experimentally controlled long-day “summer” conditions (16 hours of light, 8 hours of darkness), and shifted to short day “fall/winter” conditions (12 hours light, 12 hours darkness) upon birth, can produce sexuals-producing mothers that consequently give birth to sexual offspring [28]. Experimental manipulations of temperature can modulate the degree of sexual morph production [29], and high temperatures can override the effect of short days on sexual induction [29, 30]. Based on the timing of sexual offspring birth, determination of embryos destined to become sexual morphs is thought to occur after embryonic germ cell cluster formation and migration, roughly corresponding to stage 17 of asexual embryo development [13, 31]. Induction of sexual-producing aphids and their sexual offspring requires at least 10 consecutive days of fall/winter conditions [32], which appears to prevent aphids born prior to the vernal equinox from undergoing sexual induction. Some strains of aphids also produce sexual aphids followed by asexual females, possibly to hedge bets against a harsh winter and the lack of host plants [33, 34].
Similar to winged aphid induction, the induction of sexual aphids is a complex process that involves multiple tissues and extends over several days of development. Though external morphological differences between asexual and sexual females are few, the difference in internal morphology is striking. Aphid ovaries consist of 12–16 ovarioles, each of which contains germ cells housed in an anterior germarium. In sexual ovaries, germaria are connected to oocytes [31, 35]. The sexual haploid oocyte will fill with yolk contributed by nurse cells in the germarium, grow in size, and pass through the uterus to undergo fertilization. However, in asexual ovaries, the germarium is connected to a posterior string of successively older asexual embryos progressing through development, from one-celled embryos to fully developed embryos ready for parturition.
Both aphid polyphenisms are examples of the maternal epigenetic determination of offspring phenotype. The maternal inducing signal, received by the offspring as embryos, is translated into an expansive suite of developmental changes well after birth. Over 90 years ago, Ewing [36] reviewed several studies on wing induction and postulated a transgenerational “physiological inheritance” that is “not dependent on the germplasm (or at least the chromosomes) but which modifies the expression of somatic characters.” Sutherland [37] also hypothesized a nongenetic “intrinsic factor” that delayed production of winged offspring from mothers born early from winged grandmothers. The transgenerational response to changing environmental conditions in aphids in some cases may involve juvenile hormone (JH). Application of JH or JH analogs to aphid mothers can prevent sexual induction under fall/winter conditions [38, 39]. Neurosecretory cells within the mother's brain likely perceive light and dark and transduce the photoperiod signal to the progeny directly or indirectly through JH [40, 41]. Thus, in the reproductive polyphenism, this “physiological inheritance” may be due to maternal hormonal signals that establish heritable epigenetic information that sets gene expression patterns in the developing embryo. Below, we discuss how genomics technologies and bioinformatics have invigorated investigation of the molecular basis of this epigenetic phenomenon.
4. The Aphid Genome: A Model for Plasticity
The genome of the pea aphid A. pisum is distinctive among insect and even animal genomes for several reasons [42]. With its large size (~517 Mbp) and large number of predicted genes (~35,000 genes, many well-supported by homology, EST, or RNA-seq data), the pea aphid possesses one of the largest gene repertoires among animals, rivaling that of Daphnia pulex, another polyphenic arthropod [43]. Repetitive elements (REs) account for a large fraction of the assembled genome (38%) [42, 44]. The large number of genes is due to a large number of gene duplications: 2,459 gene families of various functions have undergone duplication, with many families containing more than 5 paralogs. Indeed, paralogs account for nearly half of the total aphid genes, similar to that of Daphnia [43]. Notable among these are duplications of genes involved in DNA methylation, small RNA pathway, and chromatin modifications and remodeling (discussed in detail below). Furthermore, the aphid genome has the lowest G/C content among sequenced insects at 29.6%. The pea aphid community now has an impressive set of genomic data and tools: a draft genome sequence, expressed sequence tags (EST), full-length cDNA sequences, microarrays, and RNAi [42, 45–60].
This genome information can be leveraged toward understanding the basis of aphid plasticity and the role of epigenetics in that plasticity. For example, aphid-specific gene duplications may have facilitated the evolution of developmental plasticity, as greater phenotypic space can be explored through the differential expression of diverged paralogs in response to environmental variation. Indeed, reports of differential paralogous gene expression between different aphid morphs lend support to this hypothesis [51, 61–65]. The molecular basis for the differential expression of aphid paralogs is thus far unknown. We speculate that, in a manner similar to other arthropods [66], environmentally sensitive expression of maternal hormones helps establish heritable patterns of chromatin architecture in the embryo that affect gene expression patterns during development. This could involve DNA methylation, which can regulate gene expression in arthropods [67, 68], small RNAs, and chromatin modifications. Below, we discuss recent results lending support to a functional epigenetic system in aphids that may underlie polyphenic aphid development.
5. DNA Methylation
Several epigenetic processes rely on DNA methylation, which involves the addition of a methyl group (–CH3) to the 5-carbon of cytosine in genomic DNA to form 5-methylcytosine. Methylation modifications are most commonly found on cytosines at CG dinucleotides, resulting in a symmetrical double-stranded pattern. They are less commonly found in a CHG or CHH context, where H = A, G, or T [69]. These methyl groups act as a “memory” at particular genes and function during the normal growth and differentiation of many organisms [70, 71]. DNA methylation can negatively affect transcription by either physically interfering with the binding of proteins that activate transcription, or recruiting other proteins that affect chromatin structure (see Chromatin Remodeling section). They also silence the activity of transposons and inactive genes [72].
In aphids, DNA methylation was originally observed at the E4 esterase gene in insecticide-resistant green peach aphids, Myzus persicae [73–75]. Contrary to the generally understood role of DNA methylation in negatively regulating transcription, the E4 esterase gene was only expressed when it was methylated [76]. At the time, few studies had investigated the functional consequences of the observed low levels of methylation in insects [67]; thus, few conclusions could be made about the role of methylation in vertebrates versus invertebrates. Mandrioli and Borsatti [77] reported the presence of DNA methylation in the heterochromatic regions of pea aphid DNA, although they did not identify specific regions that were methylated.
DNA methyltransferases (Dnmts) are the enzymes that add methyl groups to nucleotides in DNA, using S-adenosyl methionine as the methyl donor. Animals use three classes of Dnmts [69, 78]. Dnmt1 acts as a maintenance methyltransferase, attaching methyl tags to newly synthesized DNA strands; Dnmt3 typically methylates DNA de novo; Dntmt2, an RNA cytosine methyltransferase, is no longer considered a true DNA methyltransferase [79, 80]. However, current evidence suggests that all three active Dnmts (Dnmt1, Dnmt3a, and Dnmt3b) may be involved in the maintenance of DNA methylation [81]. Considerable variation across taxa exists as to the presence or absence of each category of Dnmt [82]. For example, the honey bee (Apis mellifera) has two copies of Dnmt1, one of Dnmt2 and one of Dnmt3 [83], while C. elegans has lost all Dnmts and seems to lack DNA methylation [70, 84] (Figure 1). Clearly, some organisms develop and reproduce successfully without methylation enzymes and thus without methylated DNA.
Figure 1.
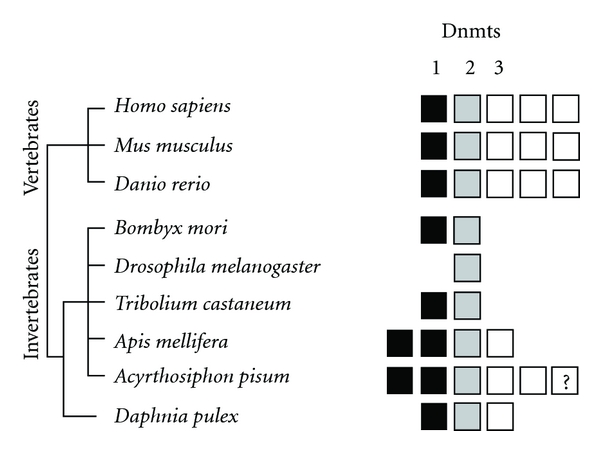
Vertebrates and invertebrates vary in Dnmt subfamily enzyme copy number. The number of boxes in each color (black, grey, white) indicates the number of paralogs of each type of Dnmt.
The previous reports of methylated aphid DNA indicated the presence of Dnmts in the aphid genome. However, given variation among taxa in Dnmt occurrence, it was not obvious a priori that an aphid genome would contain all of the DNA methylation enzymes. By searching the pea aphid genome sequence [42], Walsh et al. [85] found two copies of Dnmt1, a Dnmt2 a Dnmt3, and a gene distantly related to the other Dnmts that they called Dnmt3X. Dnmt3X lacks key amino acids thought to be necessary for Dnmt function. It may, therefore, be a pseudogene. Additional proteins involved in DNA methylation are present in the pea aphid genome: the methylated-CpG binding proteins MECP2 (one copy) and NP95 (three copies), and Dnmt1 associated protein that associates with Dnmt1 to recruit histone deacetylases [85, 86]. Walsh et al. [85] also quantified overall methylcytosine levels, finding that 0.69% (±0.25%) of all of the cytosines were methylated. This low percentage closely matches the low methylation levels observed in other insect genomes [82]. Further, twelve pea aphid genes are methylated in their coding regions, but not in their introns [85]. Three of those genes are juvenile hormone (JH) associated genes, chosen for analysis because JH has previously been shown to be involved in phenotypic plasticity in aphids [28]. Further investigation of the gene for JH binding protein revealed one methylated site that had a marginally significant higher level of methylation in winged relative to wingless asexual females [85]. Overall, these data indicated that the pea aphid has a functional DNA methylation system.
6. Aphid Genome Methylation Patterns
With the pea aphid genome sequence, patterns of DNA methylation could be investigated using an indirect measure that utilizes the observed versus expected levels of CpG methylation (CpGO/E). This method is based on the fact that methylated cytosines are hypermutable, resulting in a loss of CpGs in methylated regions. Regions of DNA with low CpGO/E are inferred to have been historically methylated and thus are considered areas of dense methylation [87].
Walsh et al. [85] used this method to examine the coding regions of all predicted genes in the pea aphid genome. The resulting histogram of gene frequency by CpGO/E exhibited a clear bimodal distribution, indicating two gene classes: genes with and without a history of DNA methylation. This same pattern was observed in another polyphenic species, the honey bee, whereas it was not observed in nonpolyphenic species like the red flour beetle (Tribolium castaneum), Anopheles gambiae, and Drosophila melanogaster [88]. These data began to approach the intriguing question of whether methylation levels associate with aphid alternative phenotypes, but to take this question a step further required gene expression data.
Brisson et al. [89] used a pea aphid microarray to identify significantly differentially expressed (DE) genes between fourth instars and adults, males and females, and wing morphs within each sex (wing morphology in asexual females is polyphenic, while in males it is genetically determined). Using these data, Hunt et al. [90] asked whether gene methylation density associated with patterns of DE genes among the different phenotypic groups. Overall, genes with condition-specific expression (i.e., genes with DE among categories) showed higher CpGO/E levels than genes that were more ubiquitously expressed. They concluded that morph-biased genes have sparse levels of methylation while non-morph-biased genes have dense levels of methylation. In a similar study, Elango et al. [88] showed that genes with DE between honey bee queens and workers had higher CpGO/E levels. These studies, along with others [68, 91], suggest that ubiquitously expressed genes in insects are the most likely to be densely methylated.
What are the gene categories with low and high CpGO/E values? The highly methylated class includes gene ontology (GO) terms associated with general organismic functions such as metabolic processes. In contrast, genes with sparser methylation encompass a wider variety of functions such as signal transduction, cognition, and behavior [90]. Given the putative role for methylation in alternative morphologies, these patterns are counterintuitive since morph-biased genes would be presumed to be the most highly methylated. One way to reconcile this contradiction is to modify the hypothesis: if morph-biased genes have sparser CpG methylation, their CpG sites are available for the action of de novo methylation. These genes could then acquire differential methylation states, and thus different expression states, on a generation-by-generation basis, induced by relevant environmental circumstances. In support of this, RNAi of the Dnmt3 de novo methyltransferase in honey bees led to changes in reproductive morph specification [92].
These previous studies relied on indirect measures of methylation specifically focused on methylation at CG dinucleotides. A catalog of all base positions in the genome that exhibit methylation, known as a “methylome,” would allow for global comparative analyses of DNA methylation. This has been achieved in other organisms (e.g., [91, 93, 94]), and indeed a pea aphid methylome is currently being pursued (O. Edwards, D. Tagu, J. A. B., S. Jaubert-Possamai, unpublished data). With the methylome, it will be possible to answer the following questions: Are there differences in CG, CHG, or CHH methylation patterns between winged and wingless or sexual and asexual females? If so, what specific genes exhibit methylation differences between morphs? Does methylation associate with alternative splicing? Does methylation have a role in regulating the abundant paralogs in the pea aphid genome? Does methylation correlate with expression levels?
7. Chromatin Modification and Remodeling Pathway
The production of a cell fate relies on stable gene expression patterns specified by intrinsic and/or external factors during development. Current models propose that DNA methylation and chromatin architecture set stable, yet modifiable, patterns of gene expression. An array of different DNA-bound proteins, largely consisting of histones, acts in concert to create higher-order structures that alter chromatin shape from local to global scales. Histones H2A, H2B, H3, and H4 form an octamer on which DNA is wrapped, forming a structure known as a nucleosome, that can make DNA locally inaccessible to DNA-binding factors. Histone tails extend from the core octamer and are available for modification such as acetylation, ADP ribosylation, methylation, phosphorylation, SUMOylation, and ubiquitylation. These modifications affect local chromatin function by adjusting its accessibility and attractiveness to regulatory complexes [95]. Variant histones can replace core octamer subunits, endowing the local chromatin environment with unique structural and functional properties [96]. Nucleosomes themselves can be repositioned to allow local access to DNA by nucleosome remodeling complexes [97]. This large array of activities is thought collectively to establish a “code” of chromatin characteristics, which reflects the functional and structural state of the underlying chromosomal DNA. Histone modifications, nucleosome remodeling, DNA methylation, and even small RNA pathways may be functionally linked and interdependent in a context-dependent manner [98–100].
Increasing evidence shows that a simple model of “open” and “closed” chromatin is insufficient to explain functional and structural differences among different regions of the genome. Instead, chromatin structure can be viewed as a composite of structural and functional domains with unique combinations of histone post-translational modifications, DNA methylation patterns, variant histone members, nucleosome position and chromosome territory within the nucleus [101, 102]. Chromatin structure is maintained across mitotic divisions, although theoretical and experimental evidences have not yet converged on a mechanism for that transmission.
A survey of aphid chromatin genes is the first step in understanding how heritable chromatin structure may be associated with aphid polyphenisms. The current draft of the aphid genome indicates expansions of antagonistic chromatin modifying and remodeling pathways [61]. Histones and histone variants are conserved in the aphid genome at numbers similar to Drosophila melanogaster, though histone variants such as Cenp-A and protamines appear absent [61]. The major chromatin remodeling complexes (SWI/SNF, CHD1, ISWI, and NURD) are represented in the aphid genome. The most striking observation is that expansions of gene families involved in histone acetylation are mirrored by expansions of genes involved in histone deacetylation. A similar situation is seen for genes involved in histone methylation and histone demethylation [61, 103]. Since the effect of acetylation and methylation on chromatin state and gene expression is context-dependent, these multiple antagonistic activities could contribute towards a complex regulation of chromatin state in aphids.
Evidence thus far for morph-associated chromatin architecture in aphids is in its early stages. The holocentric structure of aphid chromosomes (which presumably have diffuse kinetochores) could have effects on higher-order chromatin structure. Stainings of pea aphid chromatin detected several histone modifications, such as methylation of histone H3 on lysine 4 and lysine 9 [61, 77, 104]. In particular, largely overlapping differential histone methylation of these residues was observed in specific regions of chromatin [61]. Duplications of antagonistic histone modifying genes could be interpreted as a “need” for a balance of chromatin modifying activities. Alternatively, these duplications could be merely coincident with the general level of gene duplication in the aphid genome and may not have biological relevance for any specific trait. Chromatin immunoprecipitation (ChIP), expression analysis, and evolutionary analysis of these genes should help distinguish between these hypotheses. Additionally, next-generation sequencing technologies can be used to survey morph-specific chromatin modifications [105].
8. Small RNA Pathway
Work over the last 15 years has implicated small noncoding RNAs as a layer of epigenetic control. Small RNAs direct the transcriptional or post-transcriptional repression of gene activity in a gene-specific manner. Classes of small noncoding RNAs include endogenous microRNAs (miRNAs); exogenous and endogenous short interfering RNAs (siRNAs and esiRNAs); and Piwi protein-associated small RNAs (piRNAs) [106–108]. This dizzying array of small RNAs is generated by transcription either of endogenous miRNA- and siRNA-encoding genes, or of repetitive elements, transposons and noncoding regions [109]. These classes differ in their biogenesis, processing, function, and partner proteins [110]. Here, we discuss progress in studying aphid miRNA and piRNA pathways.
The miRNA and siRNA pathways provide animals and plants a means of attenuating the activities of viruses and selfish genetic elements [111]. Additionally, miRNAs post-transcriptionally regulate the expression of many endogenous genes [112, 113]. Primary miRNA transcripts in the form of a stem-loop are processed by the Drosha/Pasha complex [114] into pre-miRNAs which are exported from the nucleus via Exportin 5 [115–117]. The Dicer1/Loquacious complex then pares the pre-miRNA down to a 21-nucleotide miRNA duplex [118–120]. Mature miRNAs or endogenous siRNAs are then loaded onto a RNA-induced silencing complex (RISC), which contains an Argonaute (Ago) family member protein, of which there are five in Drosophila (Ago1–3, Piwi and Aubergine) [121]. In Drosophila, Ago1-containing RISCs bind miRNAs while Ago2 RISCs contain siRNAs [122]. One strand of the duplex is retained in this complex as the single-strand miRNA or “guide” siRNA [123, 124]. RISC facilitates annealing of the single-strand miRNA to 3′ UTRs of target mRNAs to either block protein translation and promote target mRNA degradation [109], or, if the miRNA is nearly fully complementary to the target, direct cleavage of the target mRNA by RISC, similar to a siRNA (Figure 2).
Figure 2.
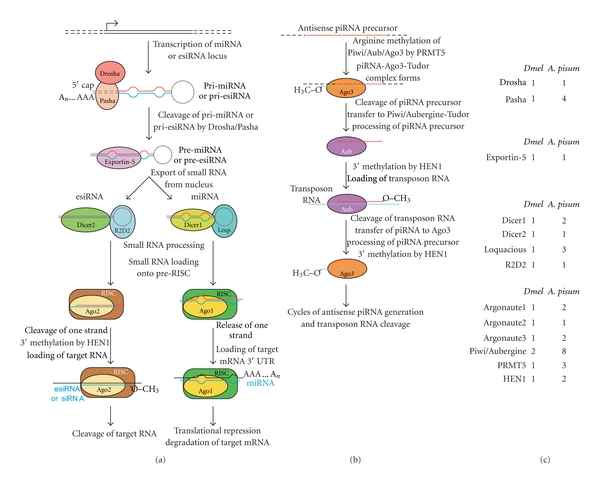
Small RNA pathways are conserved between Drosophila melanogaster and A. pisum. (a) The esiRNA and siRNA pathway is initiated typically with nearly perfectly complementary dsRNA produced endogenously or introduced exogenously, respectively. miRNAs are endogenously transcribed and processed by a parallel pathway in Drosophila, arises from imperfectly complementary dsRNAs, and repress translation of endogenous genes. (b) piRNAs are generated from piRNA clusters in the genome and are processed by a different set of Argonaute family proteins to repress transposon activity. (c) Comparison of small RNA pathway gene copy number between D. melanogaster and A. pisum reveals aphid-specific duplications. Dmel: D. melanogaster.
In Drosophila, Ago3 and the germline-specific Piwi and Aubergine Argonaute subfamily members associate with Piwi-associated piRNAs [125–127]. This class of small RNAs arises from “piRNA clusters” in heterochromatin in a manner distinct from siRNAs [107]. Piwi and Aubergine proteins exhibit “Slicer” activity when bound to piRNAs and cleave their piRNA's cognate RNA [128, 129]. Tudor domain proteins and arginine methylation of Piwi/Aubergine by the PRMT5 methyltransferase modulate Piwi/Aubergine association with piRNAs [130–133]. In addition, Piwi subfamily members may regulate the translation of germline transcripts [134, 135] and affect chromatin architecture to promote silencing [69, 136–138].
Analysis of the aphid genome sequence has revealed that the miRNA pathway has expanded in aphids [139]. Drosophila contains two Dicer genes, Dicer1 and Dicer2, while mammals and C. elegans possess only one Dicer [140]. Jaubert-Possamai et al. [139] showed that the aphid genome, however, contains single copies of the Dicer2 and Ago2 siRNA pathway components and duplicates of Pasha, Dicer1, Loquacious and Ago1 miRNA pathway genes relative to Drosophila (Figure 2). Aphid Ago1b and two of the four Pasha paralogs have undergone rapid evolution since duplication. The aphid dcr-1a and dcr-1b genes are lineage-specific duplications distinguished by a 44-amino acid insertion in the first RNAse III domain of DCR-1B. The aphid-specific duplication of Loquacious, a partner protein of Dicer1 that binds to precursor miRNAs and esiRNAs, complements the Dicer1 duplication. These potential binding partners could form an array of complexes that regulate gene expression.
The identification of miRNAs encoded by the aphid genome firmly establishes the presence of active small RNA pathways in aphids [141]. Legeai et al. [141] used homology, deep sequencing, and predictive methods to converge on 149 pea aphid miRNAs, of which 55 are conserved among insects and 94 are thus far aphid-specific. Seventeen miRNAs showed differential expression among asexual, sexuals-producing asexual, and sexual females. Polyphenic locusts [142] and honeybees [143] also express small RNAs in morph-specific patterns. Aphid miRNAs can now be tested for their roles in aphid plasticity. As of yet, no aphid esiRNAs or piRNAs have been identified, but these small RNAs could be identified by prediction or by empirical methods.
The piRNA pathway also expanded in aphids. Within aphids, the Piwi/Aubergine subfamily has undergone extensive gene duplications, with eight Piwi paralogs and three Ago3 paralogs found in the genome ([144], and Figure 2). Parallel to the expansion of aphid Piwi/Aubergine members, the aphid genome contains three PRMT5 methyltransferase paralogs (compared to one in Drosophila) [61], at least three Tudor-domain containing proteins and two copies of the HEN1 2′-OH RNA methyltransferase (D. G. Srinivasan, unpublished results). Similarly, in C. elegans, 27 Argonaute family proteins have been identified—some without Slicer activity—that act in different aspects of the small RNA pathway [145]. This may be the case in aphids as aphid Piwi paralogs do show differential expression between different aphid morphs [144]. The high number of repetitive and mobile genetic elements in the aphid genome mirrors the expansion of the Argonaute protein family in aphids [42, 44, 146]. This suggests morph-specific regulation of transposons and mRNAs in a Piwi-dependent manner.
9. Current Hypotheses and Comparisons with Other Arthropod and Nonarthropod Systems
Most of what is known about the patterns and processes associated with DNA methylation come from studies in noninsect systems, primarily in mammals and plants. From these studies, a view emerged that methylation levels are high in CpG contexts, with transposons, other repeats, promoters, and gene bodies exhibiting methylation [78, 93, 147]. Promoter methylation is associated with a downregulation of transcription. More recent studies have shown that gene body methylation is ancestral to eukaryotes, but other methylation patterns, such as methylation of transposons, are taxon specific [148, 149].
Methylation in insects has traditionally been understudied due to the finding that Drosophila melanogaster, the most well-developed insect model, has almost no detectable DNA methylation [150]. It was therefore assumed that DNA methylation does not play an integral role in insect biology as it does in mammals and plants. Recent efforts have changed this impression. Whole-genome bisulfite sequencing of Apis mellifera [151] and Bombyx mori [91] has shown that insect genomes are, indeed, methylated. However, these studies have also shown that there are key differences between insect methylomes and vertebrate or plant methylomes. First, less than one percent of cytosines in insects is methylated compared with 20–80% in plants and mammals. Second, as mentioned above, insects exhibit variable numbers of each of the Dnmt enzymes. Third, methylation in insects is highest in gene bodies. And finally, transposable elements and other repetitive elements do not appear to be methylated at high levels. One pattern is shared among insects, plants, and mammals: genes with the highest and lowest expression levels show the least gene body methylation, while those with moderate levels of expression are the most highly methylated [68, 88, 148].
One intriguing idea that insect methylation studies have raised is the possibility that gene body methylation controls alternative splicing of transcripts. In fact, methylation in A. mellifera is enriched near alternatively spliced exons, and alternative transcripts of at least one gene are expressed in workers versus queens [68, 151]. Thus, methylation could control alternative splicing, with alternative transcripts being deployed to achieve alternative phenotypes. In general, because of their smaller genomes, accessibility as study organisms, and gene body methylation, insects may emerge as valuable systems for understanding the causes and consequences of DNA methylation [82].
How can DNA methylation be coupled to other epigenetic pathways in aphids? The measurement of relative methylation and accompanying chromatin states is a clear first step to test the connections between aphid gene duplications, gene expression, and chromatin structure. The interplay between chromatin modifications and DNA methylation may converge on differential expression and/or splicing of morph-specific genes. Additionally, small RNAs are expressed in morph-specific expression patterns in polyphenic locusts [142] and honeybees [143], and loss of piwi in Drosophila is associated with the loss of heterochromatic histone modifications and of HP1 association with chromatin in somatic cells [138]. Interestingly, the piRNA pathway in Drosophila has been associated with the suppression of phenotypic variation through the Hsp90 pathway [152] and with de novo DNA methylation of an imprinted locus in mice [153]. Additionally, Drosophila piRNAs can be epigenetically transmitted from mother to egg and affect the suppression of transposons in the next generation [154]. Identification, characterization, and correlation of small RNAs, DNA methylation, and chromatin structure to polyphenic aphid traits will help resolve the epigenetics underlying aphid life cycles.
Acknowledgments
The authors thank the reviewers for helpful comments on this paper. D. G. Srinivasan acknowledges the support of Rowan University and Jenna Lewis, Ahmed Abdelhady, and Ruthsabel Cortes for comments on the paper. J. A. Brisson acknowledges the support of the National Institute of Environmental Health Sciences (R00ES017367) and the School of Biological Sciences, University of Nebraska-Lincoln.
References
- 1.DeWitt TJ, Scheiner SM, Ebrary Inc. Phenotypic Plasticity Functional and Conceptual Approaches. New York, NY, USA: Oxford University Press; 2004. [Google Scholar]
- 2.Gilbert SF. Mechanisms for the environmental regulation of gene expression. Birth Defects Research. Part C. 2004;72(4):291–299. doi: 10.1002/bdrc.20026. [DOI] [PubMed] [Google Scholar]
- 3.West-Eberhard MJ. Developmental Plasticity and Evolution. New York, NY, USA: Oxford University Press; 2003. [Google Scholar]
- 4.Pigliucci M. Genotype-phenotype mapping and the end of the “genes as blueprint” metaphor. Philosophical Transactions of the Royal Society B. 2010;365(1540):557–566. doi: 10.1098/rstb.2009.0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 6.Jamniczky HA, Boughner JC, Rolian C, et al. Rediscovering Waddington in the post-genomic age: operationalising Waddington’s epigenetics reveals new ways to investigate the generation and modulation of phenotypic variation. BioEssays. 2010;32(7):553–558. doi: 10.1002/bies.200900189. [DOI] [PubMed] [Google Scholar]
- 7.Waddington CH. The Strategy of the Genes; A Discussion of Some Aspects of Theoretical Biology. London, UK: Allen & Unwin; 1957. [Google Scholar]
- 8.Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity : Molecular Genetics and the Evolution of Animal Design. Malden, Mass, USA: Blackwell; 2001. [Google Scholar]
- 9.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nature Reviews Molecular Cell Biology. 2009;10(3):192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 10.Blomen VA, Boonstra J. Stable transmission of reversible modifications: maintenance of epigenetic information through the cell cycle. Cellular and Molecular Life Sciences. 2010;68:27–44. doi: 10.1007/s00018-010-0505-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bollati V, Baccarelli A. Environmental epigenetics. Heredity. 2010;105(1):105–112. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lushai G, Loxdale HD, Brookes CP, Von Mende N, Harrington R, Hardie J. Genotypic variation among different phenotypes within aphid clones. Proceedings of the Royal Society B. 1997;264(1382):725–730. doi: 10.1098/rspb.1997.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miura T, Braendle C, Shingleton A, Sisk G, Kambhampati S, Stern DL. A Comparison of parthenogenetic and sexual embryogenesis of the pea Aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea) Journal of Experimental Zoology Part B. 2003;295(1):59–81. doi: 10.1002/jez.b.3. [DOI] [PubMed] [Google Scholar]
- 14.Shingleton AW, Sisk GC, Stern DL. Diapause in the pea aphid (Acyrthosiphon pisum) is a slowing but not a cessation of development. BMC Developmental Biology. 2003;3:p. 7. doi: 10.1186/1471-213X-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gluckman PD, Hanson MA, Bateson P, et al. Towards a new developmental synthesis: adaptive developmental plasticity and human disease. The Lancet. 2009;373(9675):1654–1657. doi: 10.1016/S0140-6736(09)60234-8. [DOI] [PubMed] [Google Scholar]
- 16.Braendle C, Davis GK, Brisson JA, Stern DL. Wing dimorphism in aphids. Heredity. 2006;97(3):192–199. doi: 10.1038/sj.hdy.6800863. [DOI] [PubMed] [Google Scholar]
- 17.Lees AD. The production of the apterous and alate forms in the aphid Megoura viciae Buckton, with special reference to the rôle of crowding. Journal of Insect Physiology. 1967;13(2):289–318. [Google Scholar]
- 18.Müller CB, Williams IS, Hardie J. The role of nutrition, crowding and interspecific interactions in the development of winged aphids. Ecological Entomology. 2001;26(3):330–340. [Google Scholar]
- 19.MacKay PA, Wellington WG. Maternal age as a source of variation in the ability of an aphid to produce dispersing forms. Researches on Population Ecology. 1976;18(1):195–209. [Google Scholar]
- 20.Mondor EB, Rosenheim JA, Addicott JF. Predator-induced transgenerational phenotypic plasticity in the cotton aphid. Oecologia. 2005;142(1):104–108. doi: 10.1007/s00442-004-1710-4. [DOI] [PubMed] [Google Scholar]
- 21.Dixon AFG, Agarwala BK. Ladybird-induced life-history changes in aphids. Proceedings of the Royal Society B. 1999;266(1428):1549–1553. [Google Scholar]
- 22.Kunert G, Weisser WW. The interplay between density- and trait-mediated effects in predator-prey interactions: a case study in aphid wing polymorphism. Oecologia. 2003;135(2):304–312. doi: 10.1007/s00442-003-1185-8. [DOI] [PubMed] [Google Scholar]
- 23.Kunert G, Otto S, Röse USR, Gershenzon J, Weisser WW. Alarm pheromone mediates production of winged dispersal morphs in aphids. Ecology Letters. 2005;8(6):596–603. [Google Scholar]
- 24.Sutherland ORW. The role of crowding in the production of winged forms by two strains of the pea aphid, Acyrthosiphon pisum . Journal of Insect Physiology. 1969;15(8):1385–1410. [Google Scholar]
- 25.Ishikawa A, Miura T. Differential regulations of wing and ovarian development and heterochronic changes of embryogenesis between morphs in wing polyphenism of the vetch aphid. Evolution and Development. 2009;11(6):680–688. doi: 10.1111/j.1525-142X.2009.00375.x. [DOI] [PubMed] [Google Scholar]
- 26.Brisson JA. Aphid wing dimorphisms: linking environmental and genetic control of trait variation. Philosophical Transactions of the Royal Society B. 2010;365(1540):605–616. doi: 10.1098/rstb.2009.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcovitch S. The migration of the Aphididae and the appearance of the sexual forms as affected by the relative length of daily light exposure. Journal of Agricultural Research. 1924;27:513–522. [Google Scholar]
- 28.Le Trionnaire G, Hardiet J, Jaubert-Possamai S, Simon JC, Tagu D. Shifting from clonal to sexual reproduction in aphids: physiological and developmental aspects. Biology of the Cell. 2008;100(8):441–451. doi: 10.1042/BC20070135. [DOI] [PubMed] [Google Scholar]
- 29.Lees AD. The role of photoperiod and temperature in the determination of parthenogenetic and sexual forms in the aphid Megoura viciae Buckton-III. Further properties of the maternal switching mechanism in apterous aphids. Journal of Insect Physiology. 1963;9(2):153–164. [Google Scholar]
- 30.Lees AD. The role of photoperiod and temperature in the determination of parthenogenetic and sexual forms in the aphid Megoura viciae Buckton-I. The influence of these factors on apterous virginoparae and their progeny. Journal of Insect Physiology. 1959;3(2):92–117. [Google Scholar]
- 31.Blackman RL. Reproduction, cytogenetics and development. In: Minks A, Harrewijn KP, editors. In Aphids: Their Biology, Natural Enemies and Control. Amsterdam, The Netherlands: Elsevier; 1987. pp. 163–195. [Google Scholar]
- 32.Hardie J, Vaz Nunes M. Aphid photoperiodic clocks. Journal of Insect Physiology. 2001;47(8):821–832. doi: 10.1016/s0022-1910(99)00161-4. [DOI] [PubMed] [Google Scholar]
- 33.Halkett F, Harrington R, Hullé M, et al. Dynamics of production of sexual forms in aphids: theoretical and experimental evidence for adaptive "coin-flipping" plasticity. The American Naturalist. 2004;163(6):E112–125. doi: 10.1086/383618. [DOI] [PubMed] [Google Scholar]
- 34.Dedryver CA, Le Gallic JF, Gauthier JP, Simon JC. Life cycle of the cereal aphid Sitobion avenae F.: polymorphism and comparison of life history traits associated with sexuality. Ecological Entomology. 1998;23(2):123–132. [Google Scholar]
- 35.Büning J. Morphology, ultrastructure, and germ cell cluster formation in ovarioles of aphids. Journal of Morphology. 1985;186(2):209–221. doi: 10.1002/jmor.1051860206. [DOI] [PubMed] [Google Scholar]
- 36.Ewing HE. The factors of inheritance and parentage as affecting the ratio of alate to apterous individuals in aphids. The American Naturalist. 1925;59:311–326. [Google Scholar]
- 37.Sutherland ORW. An intrinsic factor influencing alate production by two strains of the pea aphid, Acyrthosiphon pisum . Journal of Insect Physiology. 1970;16(7):1349–1354. [Google Scholar]
- 38.Mittler TE, Nassar SG, Staal GB. Wing development and parthenogenesis induced in progenies of kinoprene-treated gynoparae of Aphis fabae and Myzus persicae . Journal of Insect Physiology. 1976;22(12):1717–1725. [Google Scholar]
- 39.Mittler TE, Eisenbach J, Searle JB, Matsuka M, Nassar SG. Inhibition by kinoprene of photoperiod-induced male production by apterous and alate viviparae of the aphid Myzus persicae . Journal of Insect Physiology. 1979;25(3):219–226. [Google Scholar]
- 40.Lees AD. The location of the photoperiodic receptors in the aphid Megoura viciae buckton. The Journal of Experimental Biology. 1964;41:119–133. doi: 10.1242/jeb.41.1.119. [DOI] [PubMed] [Google Scholar]
- 41.Steel CG, Lees AD. The role of neurosecretion in the photoperiodic control of polymorphism in the aphid Megoura viciae . Journal of Experimental Biology. 1977;67:117–135. doi: 10.1242/jeb.67.1.117. [DOI] [PubMed] [Google Scholar]
- 42.Richards S, Gibbs RA, Gerardo NM, et al. Genome sequence of the pea aphid Acyrthosiphon pisum . PLoS Biology. 2010;8(2) doi: 10.1371/journal.pbio.1000313. Article ID e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colbourne JK, Pfrender ME, Gilbert D, et al. The ecoresponsive genome of Daphnia pulex . Science. 2011;331(6017):555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernet GP, Munoz-Pomer A, Dominguez-Escriba L, et al. GyDB mobilomics: LTR retroelements and integrase-related transposons of the pea aphid Acyrthosiphonpisum genome. Mobile Genetic Elements. 2011;1:97–102. doi: 10.4161/mge.1.2.17635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabater-Muñoz B, Legeai F, Rispe C, et al. Large-scale gene discovery in the pea aphid Acyrthosiphon pisum (Hemiptera) Genome Biology. 2006;7(3, article no. R21) doi: 10.1186/gb-2006-7-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tagu D, Prunier-Leterme N, Legeai F, et al. Annotated expressed sequence tags for studies of the regulation of reproductive modes in aphids. Insect Biochemistry and Molecular Biology. 2004;34(8):809–822. doi: 10.1016/j.ibmb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 47.Hunter WB, Dang PM, Bausher MG, et al. Aphid biology: expressed genes from alate Toxoptera citricida, the brown citrus aphid. Journal of Insect Science. 2003;3:p. 23. doi: 10.1093/jis/3.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shigenobu S, Richards S, Cree AG, et al. A full-length cDNA resource for the pea aphid, Acyrthosiphon pisum . Insect Molecular Biology. 2010;19(2, supplement 2):23–31. doi: 10.1111/j.1365-2583.2009.00946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakabachi A, Shigenobu S, Sakazume N, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(15):5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramsey JS, Wilson ACC, de Vos M, et al. Genomic resources for Myzus persicae: EST sequencing, SNP identification, and microarray design. BMC Genomics. 2007;8, article no. 423 doi: 10.1186/1471-2164-8-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortés T, Tagu D, Simon JC, Moya A, Martínez-Torres D. Sex versus parthenogenesis: a transcriptomic approach of photoperiod response in the model aphid Acyrthosiphon pisum (Hemiptera: Aphididae) Gene. 2008;408(1-2):146–156. doi: 10.1016/j.gene.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 52.Altincicek B, Gross J, Vilcinskas A. Wounding-mediated gene expression and accelerated viviparous reproduction of the pea aphid Acyrthosiphon pisum . Insect Molecular Biology. 2008;17(6):711–716. doi: 10.1111/j.1365-2583.2008.00835.x. [DOI] [PubMed] [Google Scholar]
- 53.Carolan JC, Fitzroy CIJ, Ashton PD, Douglas AE, Wilkinsonl TL. The secreted salivary proteome of the pea aphid Acyrthosiphon pisum characterised by mass spectrometry. Proteomics. 2009;9(9):2457–2467. doi: 10.1002/pmic.200800692. [DOI] [PubMed] [Google Scholar]
- 54.Ollivier M, Legeai F, Rispe C. Comparative analysis of the Acyrthosiphon pisum genome and expressed sequence tag-based gene sets from other aphid species. Insect Molecular Biology. 2010;19(2, supplement 2):33–45. doi: 10.1111/j.1365-2583.2009.00976.x. [DOI] [PubMed] [Google Scholar]
- 55.Jaubert-Possamai S, Le Trionnaire G, Bonhomme J, Christophides GK, Rispe C, Tagu D. Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum . BMC Biotechnology. 2007;7, article no. 63 doi: 10.1186/1472-6750-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shakesby AJ, Wallace IS, Isaacs HV, Pritchard J, Roberts DM, Douglas AE. A water-specific aquaporin involved in aphid osmoregulation. Insect Biochemistry and Molecular Biology. 2009;39(1):1–10. doi: 10.1016/j.ibmb.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 57.Mutti NS, Louis J, Pappan LK, et al. A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(29):9965–9969. doi: 10.1073/pnas.0708958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mutti NS, Park Y, Reese JC, Reeck GR. RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum . Journal of Insect Science. 2006;6:1–7. doi: 10.1673/031.006.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whyard S, Singh AD, Wong S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochemistry and Molecular Biology. 2009;39(11):824–832. doi: 10.1016/j.ibmb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 60.Xu W, Han Z. Cloning and phylogenetic analysis of sid-1-like genes from aphids. Journal of Insect Science. 2008;8, article no. 30:1–6. doi: 10.1673/031.008.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rider SD, Srinivasan DG, Hilgarth RS. Chromatin-remodelling proteins of the pea aphid, Acyrthosiphon pisum (Harris) Insect Molecular Biology. 2010;19(2):201–214. doi: 10.1111/j.1365-2583.2009.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brisson JA, Ishikawa A, Miura T. Wing development genes of the pea aphid and differential gene expression between winged and unwinged morphs. Insect Molecular Biology. 2010;19(2, supplement):63–73. doi: 10.1111/j.1365-2583.2009.00935.x. [DOI] [PubMed] [Google Scholar]
- 63.Ramos S, Moya A, Martínez-Torres D. Identification of a gene overexpressed in aphids reared under short photoperiod. Insect Biochemistry and Molecular Biology. 2003;33(3):289–298. doi: 10.1016/s0965-1748(02)00243-6. [DOI] [PubMed] [Google Scholar]
- 64.Price DRG, Duncan RP, Shigenobu S, Wilson ACC. Genome expansion and differential expression of amino acid transporters at the aphid/Buchnera symbiotic interface. Molecular Biology and Evolution. 2011;28(11):3113–3126. doi: 10.1093/molbev/msr140. [DOI] [PubMed] [Google Scholar]
- 65.Le Trionnaire G, Francis F, Jaubert-Possamai S, et al. Transcriptomic and proteomic analyses of seasonal photoperiodism in the pea aphid. BMC Genomics. 2009;10, article no. 1471:p. 456. doi: 10.1186/1471-2164-10-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nijhout HF. Development and evolution of adaptive polyphenisms. Evolution and Development. 2003;5(1):9–18. doi: 10.1046/j.1525-142x.2003.03003.x. [DOI] [PubMed] [Google Scholar]
- 67.Field LM, Lyko F, Mandrioli M, Prantera G. DNA methylation in insects. Insect Molecular Biology. 2004;13(2):109–115. doi: 10.1111/j.0962-1075.2004.00470.x. [DOI] [PubMed] [Google Scholar]
- 68.Foret S, Kucharski R, Pittelkow Y, Lockett GA, Maleszka R. Epigenetic regulation of the honey bee transcriptome: unravelling the nature of methylated genes. BMC Genomics. 2009;10, article no. 1471:p. 472. doi: 10.1186/1471-2164-10-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews Genetics. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bird A. DNA methylation patterns and epigenetic memory. Genes and Development. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 71.Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286(5439):481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 72.Delcuve GP, Rastegar M, Davie JR. Epigenetic control. Journal of Cellular Physiology. 2009;219(2):243–250. doi: 10.1002/jcp.21678. [DOI] [PubMed] [Google Scholar]
- 73.Field LM, Blackman RL, Tyler-Smith C, Devonshire AL. Relationship between amount of esterase and gene copy number in insecticide-resistant Myzus persicae (Sulzer) Biochemical Journal. 1999;339(3):737–742. [PMC free article] [PubMed] [Google Scholar]
- 74.Hick CA, Field LM, Devonshire AL. Changes in the methylation of amplified esterase DNA during loss and reselection of insecticide resistance in peach-potato aphids, Myzus persicae . Insect Biochemistry and Molecular Biology. 1996;26(1):41–47. doi: 10.1016/0965-1748(95)00059-3. [DOI] [PubMed] [Google Scholar]
- 75.Field LM, Crick SE, Devonshire AL. Polymerase chain reaction-based identification of insecticide resistance genes and DNA methylation in the aphid Myzus persicae (Sulzer) Insect Molecular Biology. 1996;5(3):197–202. doi: 10.1111/j.1365-2583.1996.tb00054.x. [DOI] [PubMed] [Google Scholar]
- 76.Field LM. Methylation and expression of amplified esterase genes in the aphid Myzus persicae (Sulzer) Biochemical Journal. 2000;349(3):863–868. doi: 10.1042/bj3490863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mandrioli M, Borsatti F. Analysis of heterochromatic epigenetic markers in the holocentric chromosomes of the aphid Acyrthosiphon pisum . Chromosome Research. 2007;15(8):1015–1022. doi: 10.1007/s10577-007-1176-4. [DOI] [PubMed] [Google Scholar]
- 78.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annual Review of Biochemistry. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 79.Goll MG, Kirpekar F, Maggert KA, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311(5759):395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 80.Schaefer M, Lyko F. Lack of evidence for DNA methylation of Invader4 retroelements in Drosophila and implications for Dnmt2-mediated epigenetic regulation. Nature Genetics. 2010;42(11):920–921. doi: 10.1038/ng1110-920. [DOI] [PubMed] [Google Scholar]
- 81.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nature Reviews Genetics. 2009;10(11):805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lyko F, Maleszka R. Insects as innovative models for functional studies of DNA methylation. Trends in Genetics. 2011;27(4):127–131. doi: 10.1016/j.tig.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 83.Wang Y, Jorda M, Jones PL, et al. Functional CpG methylation system in a social insect. Science. 2006;314(5799):645–647. doi: 10.1126/science.1135213. [DOI] [PubMed] [Google Scholar]
- 84.Maleszka R. Epigenetic integration of environmental and genomic signals in honey bees. The critical interplay of nutritional, brain and reproductive networks. Epigenetics. 2008;3(4):188–192. doi: 10.4161/epi.3.4.6697. [DOI] [PubMed] [Google Scholar]
- 85.Walsh TK, Brisson JA, Robertson HM, et al. A functional DNA methylation system in the pea aphid, Acyrthosiphon pisum . Insect Molecular Biology. 2010;19(2, supplement 2):215–228. doi: 10.1111/j.1365-2583.2009.00974.x. [DOI] [PubMed] [Google Scholar]
- 86.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23(46):7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 87.Suzuki MM, Kerr ARW, de Sousa D, Bird A. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Research. 2007;17(5):625–631. doi: 10.1101/gr.6163007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Elango N, Hunt BG, Goodisman MAD, Yi SV. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera . Proceedings of the National Academy of Sciences of the United States of America. 2009;106(27):11206–11211. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brisson JA, Davis GK, Stern DL. Common genome-wide patterns of transcript accumulation underlying the wing polyphenism and polymorphism in the pea aphid (Acyrthosiphon pisum) Evolution and Development. 2007;9(4):338–346. doi: 10.1111/j.1525-142X.2007.00170.x. [DOI] [PubMed] [Google Scholar]
- 90.Hunt BG, Brisson JA, Yi SV, Goodisman MAD. Functional conservation of DNA methylation in the pea aphid and the honeybee. Genome Biology and Evolution. 2010;2(1):719–728. doi: 10.1093/gbe/evq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiang H, Zhu J, Chen Q, et al. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nature Biotechnology. 2010;28(7):516–520. doi: 10.1038/nbt.1626. [DOI] [PubMed] [Google Scholar]
- 92.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319(5871):1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 93.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lister R, O’Malley RC, Tonti-Filippini J, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis . Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 96.Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Developmental Cell. 2010;19(5):662–674. doi: 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463(7280):474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Steensel B. Chromatin: constructing the big picture. EMBO Journal. 2011;30(10):1885–1895. doi: 10.1038/emboj.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Iorio MV, Piovan C, Croce CM. Interplay between microRNAs and the epigenetic machinery: an intricate network. Biochimica et Biophysica Acta, Gene Regulatory Mechanisms. 2010;1799(10-12):694–701. doi: 10.1016/j.bbagrm.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 100.Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutation Research. 2008;659(1-2):40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 101.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nature Reviews Genetics. 2010;11(4):285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Filion GJ, van Bemmel JG, Braunschweig U, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143(2):212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krauss V, Fassl A, Fiebig P, Patties I, Sass H. The evolution of the histone methyltransferase gene Su(var)3-9 in metazoans includes a fusion with and a re-fission from a functionally unrelated gene. BMC Evolutionary Biology. 2006;6, article no. 18 doi: 10.1186/1471-2148-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mandrioli M, Azzoni P, Lombardo G, Manicardi GC. Composition and epigenetic markers of heterochromatin in the aphid Aphis nerii (Hemiptera: Aphididae) Cytogenetic and Genome Research. 2011;133(1):67–77. doi: 10.1159/000323510. [DOI] [PubMed] [Google Scholar]
- 105.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature Reviews Genetics. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 106.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135(7):1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 107.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135(1):3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 108.Czech B, Malone CD, Zhou R, et al. An endogenous small interfering RNA pathway in Drosophila . Nature. 2008;453(7196):798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nature Reviews Genetics. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila . Molecular Cell. 2008;32(4):592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lau NC. Small RNAs in the animal gonad: guarding genomes and guiding development. International Journal of Biochemistry and Cell Biology. 2010;42(8):1334–1347. doi: 10.1016/j.biocel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nature Reviews Genetics. 2011;12(2):136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nature Reviews Molecular Cell Biology. 2010;11(4):252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 114.Tomari Y, Zamore PD. MicroRNA biogenesis: drosha can’t cut it without a partner. Current Biology. 2005;15(2):R61–R64. doi: 10.1016/j.cub.2004.12.057. [DOI] [PubMed] [Google Scholar]
- 115.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes and Development. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bohnsack MT, Czaplinski K, Görlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear Export of MicroRNA Precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 118.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biology. 2005;3(7):1202–1212. doi: 10.1371/journal.pbio.0030235. Article ID e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila . Genes and Development. 2005;19(14):1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Forstemann K, Tomari Y, Du T, et al. Normal microRNA maturation and germ-line stem cell maintenance requires loquacious, a double-stranded RNA-binding domain protein. PLoS Biology. 2005;3(7):1187–1201. doi: 10.1371/journal.pbio.0030236. Article ID e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nature Reviews Molecular Cell Biology. 2008;9(1):22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 122.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes and Development. 2004;18(14):1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tomari Y, Du T, Zamore PD. Sorting of Drosophila Small Silencing RNAs. Cell. 2007;130(2):299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by Dicer-1. Cell. 2007;130(2):287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Current Biology. 2001;11(13):1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 126.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 127.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136(4):656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hock J, Meister G. The Argonaute protein family. Genome Biology. 2008;9(2, article no. 210) doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saito K, Siomi MC. Small RNA-mediated quiescence of transposable elements in animals. Developmental Cell. 2010;19(5):687–697. doi: 10.1016/j.devcel.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 130.Kirino Y, Kim N, de Planell-Saguer M, et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nature Cell Biology. 2009;11(5):652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nishida KM, Okada TN, Kawamura T, et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO Journal. 2009;28(24):3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kirino Y, Vourekas A, Sayed N, et al. Arginine methylation of aubergine mediates tudor binding and germ plasm localization. RNA. 2010;16(1):70–78. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu H, Wang JYS, Huang Y, et al. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes and Development. 2010;24(17):1876–1881. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(36):13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kennerdell JR, Yamaguchi S, Carthew RW. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes and Development. 2002;16(15):1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Brower-Toland B, Findley SD, Jiang L, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes and Development. 2007;21(18):2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Klenov MS, Lavrov SA, Stolyarenko AD, et al. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Research. 2007;35(16):5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pal-Bhadra M, Leibovitch BA, Gandhi SG, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303(5658):669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 139.Jaubert-Possamai S, Rispe C, Tanguy S, et al. Expansion of the miRNA pathway in the hemipteran insect Acyrthosiphon pisum . Molecular Biology and Evolution. 2010;27(5):979–987. doi: 10.1093/molbev/msp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Current Topics in Microbiology and Immunology. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- 141.Legeai F, Rizk G, Walsh T, et al. Bioinformatic prediction, deep sequencing of microRNAs and expression analysis during phenotypic plasticity in the pea aphid, Acyrthosiphon pisum . BMC Genomics. 2010;11(1) doi: 10.1186/1471-2164-11-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wei Y, Chen S, Yang P, Ma Z, Kang L. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome biology. 2009;10(1):p. R6. doi: 10.1186/gb-2009-10-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Weaver DB, Anzola JM, Evans JD, et al. Computational and transcriptional evidence for microRNAs in the honey bee genome. Genome Biology. 2007;8(6, article no. R97) doi: 10.1186/gb-2007-8-6-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lu HL, Tanguy S, Rispe C, et al. Expansion of genes encoding piRNA-associated argonaute proteins in the pea aphid: Diversification of expression profiles in different plastic morphs. PLoS One. 2011;6(12, article e28051) doi: 10.1371/journal.pone.0028051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yigit E, Batista PJ, Bei Y, et al. Analysis of the C. elegans argonaute family reveals that distinct argonautes act sequentially during RNAi. Cell. 2006;127(4):747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 146.Llorens C, Futami R, Covelli L, et al. The Gypsy Database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Research. 2011;39:D70–74. doi: 10.1093/nar/gkq1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cokus SJ, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 149.Feng S, Cokus SJ, Zhang X, et al. Conservation and divergence of methylation patterning in plants and animals. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(19):8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408(6812):538–540. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- 151.Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biology. 2010;8(11) doi: 10.1371/journal.pbio.1000506. Article ID e1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gangaraju VK, Yin H, Weiner MM, Wang J, Huang XA, Lin H. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nature Genetics. 2010;43:153–158. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Watanabe T, Tomizawa S, Mitsuya K, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332(6031):848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322(5906):1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]


