Abstract
This project aims at investigating the link between individual epigenetic variability (not related to genetic variability) and the variation of natural environmental conditions. We studied DNA methylation polymorphisms of individuals belonging to a single genetic lineage of the clonal diploid fish Chrosomus eos-neogaeus sampled in seven geographically distant lakes. In spite of a low number of informative fragments obtained from an MSAP analysis, individuals of a given lake are epigenetically similar, and methylation profiles allow the clustering of individuals in two distinct groups of populations among lakes. More importantly, we observed a significant pH variation that is consistent with the two epigenetic groups. It thus seems that the genotype studied has the potential to respond differentially via epigenetic modifications under variable environmental conditions, making epigenetic processes a relevant molecular mechanism contributing to phenotypic plasticity over variable environments in accordance with the GPG model.
1. Introduction
Over the years, the debate about the evolutionary advantage of sexual over asexual reproduction has focused in part on the higher adaptive potential of populations with standing genetic variation [1] (and references therein). Each generation, the reproduction of amphimictic organisms results in genetic mixing, thus creating a multitude of new genotypes (and potentially novel phenotypes) in natural populations. While in sexually reproducing organisms each individual possesses a different genotype, asexually reproducing individuals from the same clonal lineage are presumed to be genetically identical.
On the other hand, asexuality has some advantages of its own; there is no need to produce males, and asexual populations can double their size each generation [2]. This twofold advantage of asexual reproduction is thought to be constrained by their limitation in colonizing new environments and/or when living in temporally unstable or heterogeneous environments. In such conditions, the survival, flexibility, and adaptive potential of asexual lineages are aspects that are not well understood. The general-purpose genotype (GPG) model [3] (Figure 1(a)) proposed that evolutionary success of asexual organisms could be possible via generalist lineages selected for their flexible phenotypes utilizing wide ecological niches. Such phenotypic flexibility enables a given genotype to be successful in many different and variable environments [4, 5]. Other models, such as the frozen niche variation (FNV) model [6], rely on the existence of genetic diversity among multiple highly specialized clonal lineages within a population each having respective narrow ecological subniches to explain the maintenance of asexual lineages. Each specialist lineage persists through time by partitioning of available ecological space so as to avoid clonal competition. However, microniche models do not provide explanations for how single clonal lineages can be successful across different and temporally variable environmental conditions.
Figure 1.
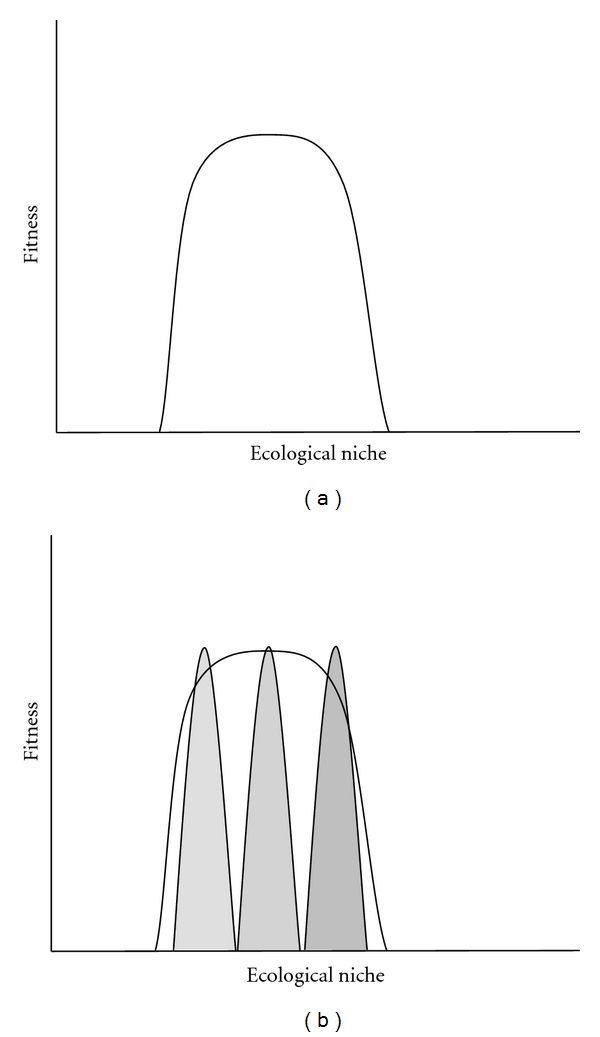
Graphic representation of the general-purpose genotype (GPG) model and the flexibility hypothesis. (a) GPG model, a flexible genetic lineage (unfilled distribution) with a wide ecological niche and a high fitness under variable environmental conditions. (b) Epigenetic as a mechanism extending the flexibility of a genome, environmentally induced epigenotypes (grey distributions) from a single genetic lineage (unfilled distribution from (a)).
One of the process underlying the GPG model is the concept of phenotypic plasticity, an environmentally induced phenotypic difference that occurs within an organism's life-time in the absence of genetic variation [7] (but see [8]). Epigenetic variation potentially represents a molecular mechanism that can generate phenotypic plasticity under natural environmental conditions [9]. The modification of the epigenome of an organism by variable methylation of DNA sequences has been shown to play a role in the regulation of some genes expression [10]. There are now numerous examples of epigenetically driven phenotypic variations that are not related to DNA sequence encoded genetic polymorphisms [11–14]. Such phenotypic variation can also be caused by an inability to maintain the original epigenetic state during embryogenesis [15]. Environmental cues (extrinsic signal) such as the diet [11, 16], temperature [17], maternal behaviour [14] and chemicals exposure [18], have been shown to influence the epigenetic profile of individuals.
The fact that the genome is able to integrate extrinsic signals from the environment to vary gene expression is a potentially important mechanism for producing phenotypic plasticity. This stands in sharp contrast with better understood mechanisms which are based on sequence encoded genetic variation. More importantly, some epigenetic variation has been shown not to be related to genetic polymorphism in natural populations [19]. While the genome provides the material to work upon, it is the epigenetic regulation that in part enables genomic flexibility. Finally, recent studies have argued that some naturally occurring epimutations can be adaptive [11, 20].
This project aims at investigating the link between individual epigenetic variability (not related to genetic variability) and the variation of natural environmental conditions. In accordance with the general-purpose genotype (GPG) model, a flexible genotype under different environmental conditions would exhibit distinct methylation patterns due to alternate gene expression profiles necessary to produce flexible phenotypes (Figure 1(b)). As a result, DNA methylation would represent a molecular mechanism extending the plasticity and flexibility of phenotypes produced by a given genotype. As a model, we used the clonal fish hybrid Chrosomus eos-neogaeus (Cyprinidea and Pisces). We chose this system because a given clonal lineage of C. eos-neogaeus can be present over a large geographic distribution [21], is found in many different types of habitats [22], is thought to be generalist [23, 24], and, more importantly, has been shown to be epigenetically variable [19].
2. Materials and Methods
2.1. Biological Model and Sampling
The all-female C. eos-neogaeus taxon resulted from hybridization events between female finescale dace (C. neogaeus) and male northern redbelly dace (C. eos) [25]. The diploid hybrids reproduce clonally via gynogenesis [26, 27]. Sperm from one of the parental species is thus required but only to trigger embryogenesis: the resulting offspring are generally genetically identical to the mother [26]. In this complex, the paternal genome can be incorporated into the zygote [22, 26, 28] resulting in triploid or mosaic hybrids which differ in the proportion of diploid-triploid cell lineages [25].
Fish from seven lakes belonging to different watersheds of the St. Lawrence River, QC, Canada (Table 1; Figure 2(a)) were sampled in the reproduction season and over a short period of approximately two weeks. Total DNA from muscle tissue of parental species, three C. eos and three C. neogaeus, and 26 gynogenetic hybrids belonging to seven different lakes were extracted by proteinase K digestion followed by phenol-chloroform purification and ethanol precipitation [30]. The lakes sampled were each classified as one of the four different types of environment according to a characterization previously used to describe C. eos-neogaeus populations [29], water pH, and temperature were also measured. Total body length, total body weight, and gonads weight were measured for each individual in order to estimate the gonadosomatic index (GSI) and the Fulton's K condition factor index (K) [31]. The lakes sampled are known to contain either one or both parental species (C. eos and C. neogaeus) as well as gynogenetic and triploids hybrids [21, 28].
Table 1.
Summary of ecological and molecular data. Lake environmental characteristics, individual morphometric characteristics, sampling size, genetic diversity (number of genotypes), and epigenetic diversity (number of epigenotypes).
| Lakes | Geographic coordinates | Date of sampling | Habitat type* | Drainage | Altitude (m) | T (°C) | pH | Weight (g) | Length (cm) | K | GSI | Sampling size | Genotype | Epigenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Richer | 45°50′35′′ N 74°11′39′′ W |
2007-05-29 | C | Nord | 360 | 24 | 6.4 | 3.32 ± 0.94 | 6.93 ± 0.71 | 0.97 ± 0.05 | 7.33 ± 2.27 | 4 | 1 | 2 |
| Merde | 45°57′55.9′′ N 74°1′41,8′′ W |
2007-05-28 | B | L'Assomption | 360 | 23 | 6.2 | 3.44 ± 1.36 | 6.78 ± 0.77 | 1.04 ± 0.07 | 11.37 ± 2.7 | 4 | 3 | 4 |
| Barbotte | 46°5′36′′ N 73°52′7′′ W |
2007-05-30 | C | L'Assomption | 280 | 22 | 6.5 | 2.16 ± 0.4 | 6.04 ± 0.41 | 0.97 ± 0.05 | 7.19 ± 2.5 | 8 | 6 | 2 |
| Jonction | 45°46′37′′ N 74°34′29′′ W |
2007-06-01 | C | Rouge | 340 | 24 | 7.1 | 2.38 ± 0.56 | 6.08 ± 0.54 | 1.05 ± 0.07 | 9.74 ± 0.71 | 4 | 1 | 1 |
| Dépotoir | 45°50′41.6′′ N 74°33′20.9′′ W |
2007-05-31 | B | Rouge | 320 | 25 | 7.1 | 1.85 ± 0.38 | 5.65 ± 0.45 | 1.01 ± 0.03 | 10.73 ± 0.53 | 2 | 1 | 1 |
| Saumons | 45°59′38′′ N 74°18′21′′ W |
2007-06-16 | A | Nord | 490 | 22 | 7.0 | 3.2 ± 0.26 | 6.8 ± 0 | 1.01 ± 0.08 | 6.51 ± 1.31 | 2 | 1 | 2 |
| Saad | 45°54′51.4′′ N 74°1′41.3′′ W |
2007-06-16 | D | L'Assomption | 320 | 24 | 6.8 | 1.89 ± 0.18 | 5.9 ± 0.2 | 0.92 ± 0.008 | 4.48 ± 2.54 | 2 | 1 | 1 |
*Habitats characterization according to Schlosser et al. [29]: A: pond of moderate depth, B: a shallow beaver pond, C: a moderately deep area of open water upstream from a beaver dam, and D: pond of moderate depth with flooded standing and fallen tree.
Figure 2.
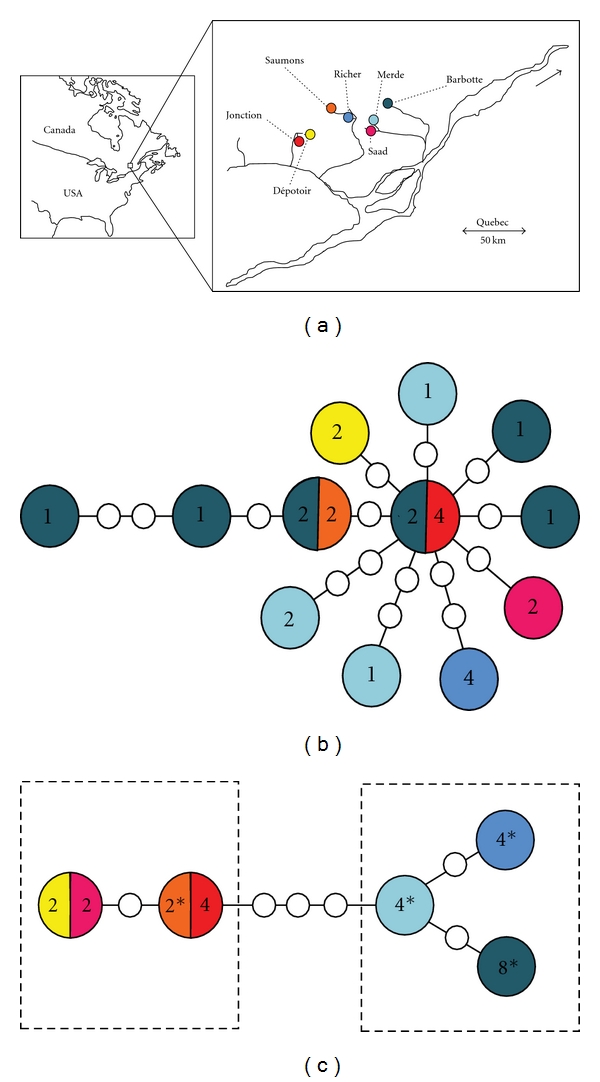
Details of sampling, genotypes, and epigenotypes diversity. (a) Sampled lakes in the Laurentian region, QC, Canada. (b) Minimum spanning network of the 12 genotypes identified by scoring nine microsatellite loci. The number of gynogenetic hybrids of each genotype per lake is indicated. (c) Minimum spanning network of the five main epigenotypes and two epigenetic groups (dash boxes) identified by the MSAP analysis. The number of gynogens of each epigenotype per lake is indicated. *refers to intrapopulation variation. The colour code of the sampled lakes from panel (a) is maintained throughout the rest of the figure.
2.2. Genetic Identification
The gynogenetic hybrids were identified according to Binet and Angers [28]. Briefly, C. eos-neogaeus hybrids were identified using diagnostic markers designed on two genes. Primers of each marker were designed to provide PCR products of different sizes for C. eos and C. neogaeus, allowing chromosome identification. Individuals that displayed alleles of both parental species were classified as gynogenetic hybrids.
Gynogenetic hybrids (diploid) were then discriminated from triploid hybrids according to the ploidy level of the nuclear genome by using nine hypervariable microsatellites as detailed in Binet and Angers [28] and Angers and Schlosser [21]. Gynogens are expected to be hemizygous at every species-specific locus, while triploid hybrids (C. eos-neogaeus x eos) are expected to be heterozygous at loci specific for C. eos species. The microsatellites analysis also enabled the identification of the clonal lineage [21] and the discrimination of derived mutations. Only gynogenetic hybrids (diploid) were used for further analysis.
2.3. MSAP Analysis
We investigated epigenetic polymorphism at CCGG motif via an MSAP analysis [32] performed on parental species, three C. eos and three C. neogaeus, and the 26 C. eos-neogaeus gynogenetic hybrids identified in the procedure mentioned above. Each DNA sample was, respectively, digested with MseI/HpaII and MseI/MspI to allow the detection of differentially methylated sequences. Aliquots (4 μL) of each sample for each primer combinations were loaded on 6% polyacrylamide gels (19 : 1 acrylamide to bisacrylamide) containing 8 M urea and 1X TBE. Fragments that displayed methylation polymorphism among samples at restriction sites were identified by the presence/absence banding pattern between the two treatments. Full methylation of both cytosines and hemimethylation of the internal cytosines cannot be investigated by MSAP. As a consequence, it was impossible to distinguish these fragments from unmethylated sequences.
3. Results
3.1. Genetic Polymorphism: Microsatellite Loci Analysis
The analysis of nine highly variable microsatellite loci indicates that all samples belong to the same clonal lineage (lineage B6, [21]). Survey of microsatellite variation detected 14 mutations over nine loci and twelve multilocus mutant genotypes were identified within the clonal lineage (Figure 2(b)). These genotypes display very little divergence, since all but one genotype differ by only one or two mutations from the putative ancestral clone, with an average of 2.3 mutations among genotypes. The number of sublineages carrying derived mutations per lake varied from one to six (Table 1).
3.2. Epigenetic Polymorphism: MSAP Analysis
A total of 257 reproducible fragments detected between 150 and 600 bp were assessed with a set of six primer pairs. Over the 257 fragments detected in C. eos-neogaeus hybrids, 60 were exclusive to C. neogaeus, 67 to C. eos, and 114 were present in both parental species genomes. The remaining 16 fragments detected could not be associated to either of the parental species genomes. Eight fragments (3.11%) revealed informative methylation polymorphism among populations. Three fragments exclusive to C. eos, three fragments exclusive to C. neogaeus, and two fragments that were present in both parental species genome were differently methylated for some C. eos-neogaeus hybrids. The number of epigenotypes per lake varied from one to four (Table 1) and is not correlated with the number of samples (R 2 = 0.07, P = 0.56).
Two of the eight fragments are variable within populations, while the others are only variable among populations. For the 6 fragments that varied among populations, five main epigenotypes were detected. Although the sample size is low for some populations, individuals from a given population consistently shared the same methylation profile (Figure 2(c)). In most instances, individuals could be regrouped according to the lake of origin on the basis of their unique methylation profile.
Contrasting with genetic relationships among clones where variants are descendents of an ancestral genotype (Figure 2(b)), populations clustered in two distinct epigenetic groups separated by three epimutations (Figure 2(c)). No significant relationship was detected between genetic and epigenetic variation (Figure 3). For instance, individuals from two distinct lakes and harbouring the same genotype clustered in distinct epigenetic groups. Similarly, there is no relationship between genetic intrapopulation variability and epigenotypes. As an example, the six different genotypes from Barbotte Lake clustered into the same epigenetic group (Figure 2(c)).
Figure 3.

Relationship between the genotypes (genetic variation and microsatellite analysis) and the epigenotypes (methylation profiles difference and MSAP analysis).
There is no indication that epigenetic profile is related to geographic position, hydrologic network (Figure 2(a)), or date of sampling (Table 1). Also, no difference in individual body size length (P = 0.26), body weight (P = 0.28), Fulton's K (P = 0.91), and GSI (P = 0.72) were detected among populations. In addition, the shared epigenetic profiles among populations are not correlated with the habitats characterization of lakes (Table 1). While there is no important temperature fluctuation among lakes, we observed a significant pH variation that is consistent with the two epigenetic groups (Table 1). This is a particularly important result, since it correlates the clustering of populations in two epigenetic groups to the variation of a local environmental condition.
4. Discussion
The present study report an effect of the local environmental conditions on the variation of the methylation profile among genetically identical individuals belonging to different natural populations. This is a particularly important result, considering that most studies investigating the influence of the integration of the extrinsic signal of the environment on epigenetic variation were performed under control conditions (e.g., [14, 16, 17]) (but see [18]). This indicates that the variation of natural environmental conditions can lead to DNA methylation polymorphism at the population level.
4.1. A Successful Generalist Lineage
The C. eos-neogaeus hybrid lineage studied here (lineage B6) is widespread in the south-western part of Quebec and is abundant in many populations from numerous watersheds [21]. The seven lakes under investigation are thought to be characterized by different environmental conditions of a variety of abiotic and biotic conditions (e.g., the oxygen concentration, the diets, the predation level, and the presence of competitors) [29]. Accordingly, each of the different lakes can be thought of as a different ecological niche. As a result, clonal lineage B6 can be characterized as a generalist lineage that is able to adjust in order to persist among many ecological niches. This situation has already been reported in northern Minnesota lakes (USA) and Algonquin Park lakes (Ontario) [23, 29]. Interestingly, C. eos-neogaeus hybrids from a single clonal lineage have been shown to present a high level of phenotypic variation [22]. Such variation of the phenotype in the absence of genetic variation has also been observed among C. eos-neogaeus hybrids from Quebec populations (B. Angers, unpublished data).
4.2. Environmentally Induced Epigenotypes
First, we did not detect any relationship between genotype and epigenotype. This is in accordance with a previous study that demonstrated pure (or facilitated) epigenetic variation in natural populations of C. eos-neogaeus hybrids [19]. More importantly, the genomic mutations detected are restricted to highly variable microsatellites loci, there is no mutation at mtDNA [21], and very few mutations were detected on AFLP loci [19]. This supports that the fragment variation detected with the MSAP analysis is due to difference in methylation not to DNA mutation.
Interestingly, the epigenetic polymorphism observed is shared among individuals of the same population in most instances. This suggests an influence of common environmental factors on the resulting epigenetic profiles or a long-term inheritance of epigenetic variation (modifications that could have been acquired before postglacial colonization). Considering the low probability of the inheritance of epigenetic variation across generations [33] and the absence of correlation between genetic and epigenetic polymorphism, the long-term heritability hypothesis can be ruled out. Accordingly, the observation of among lakes epigenetic variation suggests that current environmental conditions have an influence on the DNA methylation profiles among genetically identical individuals from different populations as opposed to hard-wired or germline dependent [34, 35]. In contrast with previous observations, the detection of the same epigenotype in different lakes indicates that the epigenetic polymorphisms observed are not the result of random variation [19]. More importantly, the correlation between the two epigenetic groups and the pH variation strongly support an effect of the local environmental conditions on the variation of methylation profile. Such pH variation may be caused by and/or will result in the variation of many other environmental factors potentially having respective or conjoint effects on the methylation polymorphism.
4.3. Revisiting the Importance of Heritability for Epigenetic Variation
Previous reports in the literature suggests that in order to be of importance in evolution, epigenetic changes must be heritable across generations [36–38]. In the situation for which an epimutation leading to a beneficial phenotypic modification appears in one generation and that the environmental conditions do not change in subsequent generations, the heritability of the new epigenetic mark may represent a transient step leading to genetic assimilation [39]. Although epimutations potentially represent a fast pathway toward adaptation [38], we do not believe that the main interest of epigenetic mechanisms is to mimic what is occurring at the adaptive genomic level. If heritable, both genetic and epigenetic polymorphisms are frozen. In temporally unstable or heterogeneous environments, such canalization of the phenotype does not seem beneficial [40]. Furthermore, heritability of epigenetic changes in vertebrates is not expected to be frequent considering the two phases of erasure prior to the initiation of zygote development [33]. Angers and coauthors [9] have recently identified some of the beneficial aspects of epigenetic mechanisms in that these processes may enable rapid and reversible changes in response to environmental perturbations. For instance, such is observed for the influence of the maternal behaviour on a glucocorticoid receptor gene promoter in the rat hippocampus [14]. Rather than passing on to the next-generation epimutations that may not be adaptive under new environmental regimes, selection might favour individuals with a plastic genome that easily adjusts epigenetically to environmental variables. Thus, the hard-wired genetic variation and the flexible epigenetic variation may be complementing each other by, respectively, leading to long-term and short-term adaptation.
5. Conclusion
While preliminary, these results appear to confirm that response of the genome when under variable environmental conditions leads to the formation of different epigenotypes. Each population presenting different epigenetic profiles can be seen as an acclimated epigenotype from a single flexible genetic lineage. It thus seems that this lineage has the potential to respond via epigenetic modifications such as DNA methylation when under variable environmental conditions. Even more importantly, this lineage potentially has the capacity to colonize different environments and/or the ability to adjust following a perturbation in the environment as expected from the long-term maintenance of multiple populations in this lineage. Thus, epigenetic processes may represent a molecular mechanism sustaining the GPG model.
Acknowledgments
The authors are particularly grateful to J. Boizard, E. Castonguay, M. Laporte, C.-O. Silva-Beaudry, and F. Vallières for their invaluable field and laboratory assistance. This research was supported by a research grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) to BA and from the Fonds Québécois pour la Recherche sur la Nature et les Technologies (FQRNT) and Étienne-Magnin scholarships at Université de Montréal to RM.
References
- 1.Neiman M, Schwander T. Using parthenogenetic lineages to identify advantages of sex. Evolutionary Biology. 2011;38(2):115–123. [Google Scholar]
- 2.Maynard-Smith J. The Evolution of Sex. Cambridge, UK: Cambridge University Press; 1978. [Google Scholar]
- 3.Baker H. Characteristics and modes of origin of weeds. In: Stebbins G, editor. The Genetics of Colonizing Species. New York, NY, USA: Academic Press; 1965. pp. 147–168. [Google Scholar]
- 4.Vrijenhoek RC. Unisexual fish: model systems for studying ecology and evolution. Annual Review of Ecology and Systematics. 1994;25:71–96. [Google Scholar]
- 5.Schlutz RJ. Evolution and ecology of unisexual fishes. Evolutionary Ecology. 1977;10:231–277. [Google Scholar]
- 6.Vrijenhoek RC. Coexistence of clones in a heterogeneous environment. Science. 1978;199(4328):549–552. doi: 10.1126/science.199.4328.549. [DOI] [PubMed] [Google Scholar]
- 7.Stearns SC. The evolutionary significance of phenotypic plasticity—phenotypic sources of variation among organisms can be described by developmental switches and reaction norms. Bioscience. 1989;39:436–445. [Google Scholar]
- 8.Drown DM, Levri EP, Dybdahl MF. Invasive genotypes are opportunistic specialists not general purpose genotypes. Evolutionary Applications. 2011;4:132–143. doi: 10.1111/j.1752-4571.2010.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angers B, Castonguay E, Massicotte R. Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Molecular Ecology. 2010;19(7):1283–1295. doi: 10.1111/j.1365-294X.2010.04580.x. [DOI] [PubMed] [Google Scholar]
- 10.Bird A. DNA methylation patterns and epigenetic memory. Genes and Development. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 11.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319(5871):1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 12.Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends in Genetics. 2002;18(7):348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 13.Manning K, Tör M, Poole M, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature Genetics. 2006;38(8):948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 14.Weaver ICG, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nature Neuroscience. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 15.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genetics. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 16.Waterland RA, Jirtle RL. Maternal dietary methyl donor supplementation affects offspring phenotype by increasing cytosine methylation at the agouti locus in Avy mice. The FASEB Journal. 2002;16(4):p. A228. [Google Scholar]
- 17.Sheldon CC, Burn JE, Perez PP, et al. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11(3):445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crews D, Gore AC, Hsu TS, et al. Transgenerational epigenetic imprints on mate preference. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(14):5942–5946. doi: 10.1073/pnas.0610410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massicotte R, Whitelaw E, Angers B. DNA methylation: a source of random variation in natural populations. Epigenetics. 2011;6(4):422–428. doi: 10.4161/epi.6.4.14532. [DOI] [PubMed] [Google Scholar]
- 20.Martin A, Troadec C, Boualem A, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461(7267):1135–1138. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- 21.Angers B, Schlosser IJ. The origin of Phoxinus eos-neogaeus unisexual hybrids. Molecular Ecology. 2007;16(21):4562–4571. doi: 10.1111/j.1365-294X.2007.03511.x. [DOI] [PubMed] [Google Scholar]
- 22.Doeringsfeld MR, Schlosser IJ, Elder JF, Evenson DP. Phenotypic consequences of genetic variation in a gynogenetic complex of Phoxinus eos-neogaeus clonal fish (Pisces: Cyprinidae) inhabiting a heterogeneous environment. Evolution. 2004;58(6):1261–1273. doi: 10.1111/j.0014-3820.2004.tb01705.x. [DOI] [PubMed] [Google Scholar]
- 23.Mee JA, Rowe L. Distribution of Phoxinus eos, Phoxinus neogaeus, and their asexually-reproducing hybrids (Pisces: Cyprinidae) in Algonquin provincial Park, Ontario. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0013185. Article ID e13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elder JF, Schlosser IJ. Extreme clonal uniformity of phoxinus eos/neogaeus gynogens (Pisces: Cyprinidae) among variable habitats in northern Minnesota beaver ponds. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(11):5001–5005. doi: 10.1073/pnas.92.11.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawley RM, Schultz RJ, Goddard KA. Clonal reproduction and polyploidy in unisexual hybrids of Phoxinus eos and Phoxinus neogaeus (Pisces, Cyprinidae) Copeia. 1987:275–283. [Google Scholar]
- 26.Goddard KA, Dawley RM. Clonal inheritance of a diploid nuclear genome by a hybrid fresh-water minnow (Phoxinus eos-neogaeus, Pisces, Cyprinidae) Evolution. 1990;44:1052–1065. doi: 10.1111/j.1558-5646.1990.tb03825.x. [DOI] [PubMed] [Google Scholar]
- 27.Goddard KA, Megwinoff O, Wessner LL, Giaimo F. Confirmation of gynogenesis in Phoxinus eos-neogaeus (Pisces: Cyprinidae) Journal of Heredity. 1998;89(2):151–157. [Google Scholar]
- 28.Binet MC, Angers B. Genetic identification of members of the Phoxinus eos-neogaeus hybrid complex. Journal of Fish Biology. 2005;67(4):1169–1177. [Google Scholar]
- 29.Schlosser IJ, Doeringsfeld MR, Elder JF, Arzayus LF. Niche relationships of clonal and sexual fish in a heterogeneous landscape. Ecology. 1998;79(3):953–968. [Google Scholar]
- 30.Orkin S. Molecular-cloning—a laboratory manual, 2nd edition—Sambrook, J, Fritsch, EF, Maniatis, T. Nature. 1990;343:604–605. [Google Scholar]
- 31.Lambert Y, Dutil JD. Can simple condition indices be used to monitor and quantify seasonal changes in the energy reserves of atlantic cod (Gadus morhua)? Canadian Journal of Fisheries and Aquatic Sciences. 1997;54:104–112. [Google Scholar]
- 32.Xiong LZ, Xu CG, Maroof MAS, Zhang QF. Patterns of cytosine methylation in an elicite hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Molecular General Genetics. 1999;26:439–446. doi: 10.1007/s004380050986. [DOI] [PubMed] [Google Scholar]
- 33.Niemitz EL, Feinberg AP. Epigenetics and assisted reproductive technology: a call for investigation. American Journal of Human Genetics. 2004;74(4):599–609. doi: 10.1086/382897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crews D. Epigenetics and its implications for behavioral neuroendocrinology. Frontiers in Neuroendocrinology. 2008;29(3):344–357. doi: 10.1016/j.yfrne.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jablonka EVA, Raz GAL. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Quarterly Review of Biology. 2009;84(2):131–176. doi: 10.1086/598822. [DOI] [PubMed] [Google Scholar]
- 36.Bossdorf O, Richards CL, Pigliucci M. Epigenetics for ecologists. Ecology Letters. 2008;11(2):106–115. doi: 10.1111/j.1461-0248.2007.01130.x. [DOI] [PubMed] [Google Scholar]
- 37.Richards EJ. Inherited epigenetic variation—revisiting soft inheritance. Nature Reviews Genetics. 2006;7(5):395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 38.Richards EJ. Population epigenetics. Current Opinion in Genetics and Development. 2008;18(2):221–226. doi: 10.1016/j.gde.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 39.Pál C, Miklós I. Epigenetic inheritance, genetic assimilation and speciation. Journal of Theoretical Biology. 1999;200(1):19–37. doi: 10.1006/jtbi.1999.0974. [DOI] [PubMed] [Google Scholar]
- 40.Young RL, Badyaev AV. Evolution of ontogeny: linking epigenetic remodeling and genetic adaptation in skeletal structures. Integrative and Comparative Biology. 2007;47(2):234–244. doi: 10.1093/icb/icm025. [DOI] [PubMed] [Google Scholar]


