Abstract
V(D)J recombination is the assembly of gene segments at the antigen receptor loci to generate antigen receptor diversity in T and B lymphocytes. This process is regulated, according to defined developmental programs, by the action of a single specific recombinase complex formed by the recombination antigen gene (RAG-1/2) proteins that are expressed in immature lymphocytes. V(D)J recombination is strictly controlled by RAG-1/2 accessibility to specific recombination signal sequences in chromatin at several levels: cellular lineage, temporal regulation, gene segment order, and allelic exclusion. DNA cleavage by RAG-1/2 is regulated by the chromatin structure, transcriptional elongation, and three-dimensional architecture and position of the antigen receptor loci in the nucleus. Cis-elements specifically direct transcription and V(D)J recombination at these loci through interactions with transacting factors that form molecular machines that mediate a sequence of structural events. These events open chromatin to activate transcriptional elongation and to permit the access of RAG-1/2 to their recombination signal sequences to drive the juxtaposition of the V, D, and J segments and the recombination reaction itself. This chapter summarizes the advances in this area and the important role of the structure and position of antigen receptor loci within the nucleus to control this process.
1. Introduction
The immune system is considered one of the best models to study the molecular mechanisms of epigenetic control of cellular differentiation in vivo. Development of B and T lymphocytes occurs through a series of well-defined differentiation stages initiating from a common hematopoietic stem cell. Each of these differentiation stages involves the activation and repression of antigen receptor loci that influence cellular identity. Expression of these loci in immature lymphocytes requires the activation and silencing of genomic DNA rearrangements that are regulated through the accessibility of chromatin, transcriptional elongation, three-dimensional structure, and nuclear positioning during cell development [1, 2]. In fact, lymphocytes are the only vertebrate cells that use genomic DNA rearrangements as an integral component of their developmental program. This DNA rearrangement process is known as V(D)J recombination and consists of the assembly of the genomically dispersed gene segments V (variable), D (diversity), and J (joining) to generate the functional variable region of antigen receptors: immunoglobulins (Igs) in B lymphocytes and T-cell receptors (TCRs) in T lymphocytes (Figure 1). This process results in the expression of a unique antigen receptor in each developed lymphocyte and is therefore responsible for the generation of antigen receptor diversity in T and B lymphocytes that defines the vertebrate adaptive immune responses.
Figure 1.
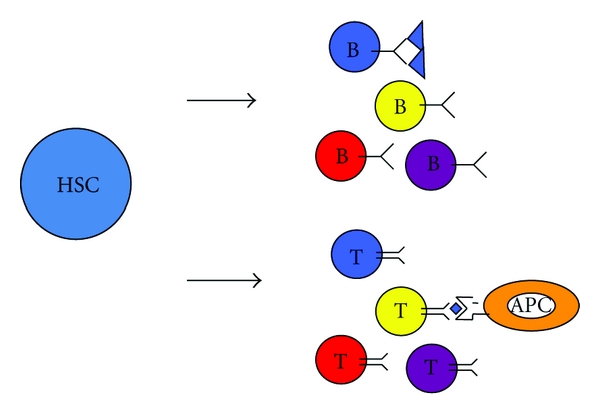
Lymphocyte maturation and expression of clonotypic antigen receptors. Scheme represents lymphocyte maturation from hematopoietic stem cells (HSCs). Each lymphocyte expresses a specific antigen receptor during cell development. B lymphocyte antigen receptors and immunoglobulins (Ig) recognize soluble antigens, whereas T lymphocyte antigen receptors (TCRs) recognize antigenic peptides presented by antigen presenting cells (APCs).
V(D)J recombination is initiated through the action of the protein products of the recombination activating gene (RAG) 1 and 2; together, RAG-1 and RAG-2 form a specific endonuclease in immature lymphoid cells. The RAG-1/2 complex is responsible for the double-strand DNA cleavage between the segments that allows recombination through the recognition of specific recombination signal sequences (RSSs) that flank them (Figure 2); these RSSs consist of conserved heptamer and nonamer elements that are separated by a less conserved spacer region of 12 or 23 base pairs. The 12–23 rule limits recombination to segments between a 12-bp RSS and a 23-bp RSS (here, we will refer to RSSs with 12-bp and 23-bp spacers as compatible RSSs) [3]. Proteins responsible for the joining of the nonhomologous ends then process the double-strand broken ends to generate the coding DNA and the extrachromosomal circles containing the RSSs and the deleted internal gene regions.
Figure 2.
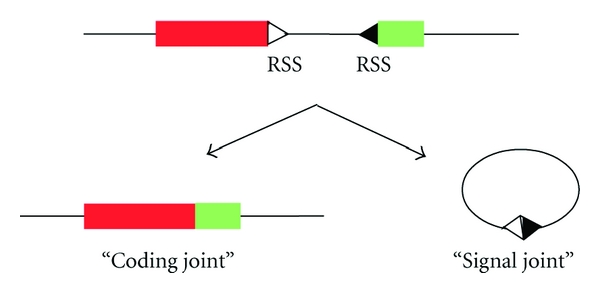
V(D)J recombination process. Gene segments are represented as red and green rectangles, and a pair of compatible RSSs is represented as white and black triangles.
2. V(D)J Recombination Control by Chromatin Structure
The 5′ region of the Ig and Tcr loci (which encode the variable regions responsible for antigen recognition) contains the V, D (only in some loci), and J gene segments (Figure 3) that are assembled through the action of RAG-1/2 proteins in lymphocyte precursors. The restricted expression of RAG-1 and RAG-2 in immature lymphocytes explains the specificity of the V(D)J recombination process in these cells. However, antigen receptor loci (Igh, Igk, Igl, Tcra, Tcrb, Tcrg, or Tcrd) and lineage (B or T) specific regulation is defined by controlled RAG-1/2 accessibility to the specific locus chromatin in B and T lymphocytes precursors [6]. This control establishes that the immunoglobulin loci (Igh, Igk, and Igl) only rearrange in B-lymphocyte precursors, and the T-cell receptor loci (Tcra, Tcrb, Tcrd, and Tcrg) rearrange exclusively in T lymphocyte precursors. Additionally, there is temporal control of V(D)J recombination that ensures that this process occurs in a developmental stage-specific manner during lymphocyte development. During T-lymphocyte development, Tcrb, Tcrd, and Tcrg loci rearrange earlier than Tcra locus. Similarly, during B lymphocyte development, Igh rearranges earlier than the Igk and Igl loci. Furthermore, there is an additional developmental control imposed on sets of gene segments within each antigen receptor locus. For example, D-to-J rearrangements precede V-to-DJ rearrangements at the Tcrb and Igh loci. This locus-, lineage-, temporal-, and gene segment order-specific regulation of V(D)J recombination is mediated through the control of RSS accessibility to the RAG-1/2 proteins. Hence, the chromatin imposes a barrier to RAG-1/2 accessibility that is controlled through strict epigenetic control, which is dependent on the specific antigen receptor locus, gene segment, cellular lineage, and developmental stage. This is the basis for the accessibility model proposed 25 years ago by Yancopoulos and Alt [7]. These investigators observed that the developmental activation of VH gene segment recombination at the Igh locus coincided with VH germline transcription (the process of transcription of sterile transcripts at an un-rearranged locus originating from V-associated promoters) during B lymphocyte development [7]. Based on these results, they proposed that the transcription of the VH gene segments reflects an increase in the accessibility of the VH gene segments to both the transcriptional and recombinational machineries (RNA polymerase II (RNAPII) and RAG-1/2 proteins, resp.). Since then, germline transcripts initiating at V, D, and J gene segments have been found to developmentally coincide with the activation of V(D)J recombination at each antigen receptor locus [7–9]. In addition to reports of sense transcription, developmentally regulated antisense intergenic transcription across the VH gene segments that correlates with VH to DJH recombination has also been reported [10]. In agreement with this model, it has been proven that the barrier that the chromatin imposes on RAG-1/2 accessibility is eliminated through the activation of cis-transcriptional elements present at these loci during lymphocyte development [1]. Each Ig or Tcr locus is equipped with at least one transcriptional enhancer in the vicinity of the constant region and numerous promoters associated with V, D, and J gene segments (Figure 3). The essential role of each of these cis-elements in controlling the accessibility to the RAG-1/2 proteins was demonstrated in numerous studies using transgenic mini-loci as recombination reporters and directed mutagenesis at the endogenous loci [1]. These studies clearly established that the enhancers are the elements that are responsible for specific lineage determination and temporal control of V(D)J recombination through the general regulation of locus chromatin structure; thus, enhancers control the accessibility of the RAG-1/2 proteins to multiple gene segments separated by large distances, whereas promoters are the elements that mediate the accessibility of the RAG-1/2 proteins to regions located at the proximal regions of the specific gene segments [1]. The accessibility model was reinforced by observations demonstrating a direct correlation between V(D)J recombination and activating epigenetic modifications such as histone H3 and H4 acetylation (H3ac and H4ac), methylation of lysine 4 of histone H3 (H3K4me), nuclease accessibility and DNA hypomethylation [1, 11–14], and changes in nucleosomal structure [15]. Furthermore, establishment of inactive chromatin suppresses V(D)J recombination [16]. Additional in vitro studies have demonstrated that assembly of RSSs into nucleosomes inhibits V(D)J recombination [17–20], supporting the notion that nucleosomes impede RAG1/2 binding or function. The barrier for V(D)J recombination imposed by nucleosomes can be surmounted by ATP-dependent chromatin remodeling complexes, such as SWI/SNF [17, 18, 21–23]. Recently, it has been directly demonstrated that chromatin accessibility to RAG-1/2 is indeed mediated by enhancers and promoters [24]. In this study, it was proven that the enhancers control global RAG-1 binding, whereas promoters direct local RAG-1 binding at the antigen receptor loci. RAG-1 binding to accessible RSSs can be targeted in the absence of RAG-2, which is recruited directly to trimethylated H3K4 (H3K4me3), a mark of open and active chromatin [25–27]. Hence, both the enhancer and promoter elements are necessary to confer RAG-1/2 accessibility to specific RSSs within a given locus and to facilitate the recombination synapse between the RSSs (Figure 4). To date, the precise molecular mechanisms by which distal enhancers control the transcriptional activation from promoters separated by large distances and V(D)J recombination are not known. Chromatin immunoprecipitation and chromosomal conformation capture experiments have demonstrated that V(D)J recombination involves physical interactions between distal transcriptional regulatory elements such as promoters and enhancers to permit RAG-1/2 accessibility to their target RSSs [28–30].
Figure 3.
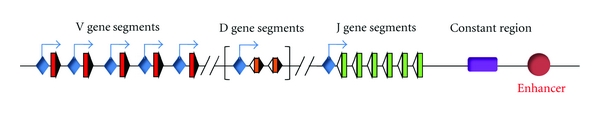
Representation of an antigen receptor locus. V, D, and J gene segments are represented as red, orange, and green rectangles, respectively. RSSs are represented as white and black triangles. The constant region is represented as a purple rectangle. Promoters are represented as blue diamonds, and arrows indicate transcription. The enhancer is represented as a red circle.
Figure 4.
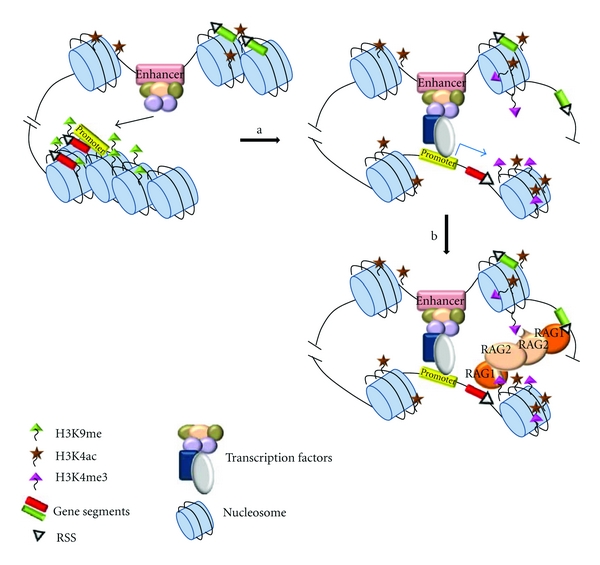
Representation of the molecular mechanism for activation of V(D)J recombination by enhancers, promoters, and transcription. Shown is a depiction of the physical interaction between the enhancer and the promoter within an antigen receptor locus. This interaction is mediated through protein-protein interactions among transcription factors and triggers the processes of transcription and V(D)J recombination that is derived from chromatin opening and subsequent accessibility of the RSSs to the RNAPII and RAG-1/2 proteins. Gene segments are represented as red and green rectangles and RSSs as white triangles. (a) Enhancer activation by the assembly of a functional multiprotein complex on the enhancer mediates the recruitment of the RNAPII to the promoter. This activates germline transcription (blue arrow) and opens the chromatin structure by repositioning nucleosomes, evicting nucleosomes, and/or changing the covalent modifications of histones (e.g., changing H3K9me, which is indicative of repressive chromatin, to H3K4me and H3/H4ac, which are indicative of activated chromatin). (b) The new chromatin configuration allows recruitment of the RAG-2 protein through H3K4me3 and recruitment of the RAG-1 protein to accessible RSSs. Recruitment of a RAG-1/2 complex to two compatible RSSs allows initiation of V(D)J recombination.
It is interesting that the antigen receptor loci undergo a process of gene contraction (juxtaposition of V and D-J regions), which is strictly regulated during development. As demonstrated by three-dimensional fluorescence in situ hybridization (3D-FISH) experiments using distal DNA probes, this contraction correlates with transcription and V(D)J recombination [29, 31–33]. In fact, gene contraction occurs at the same moment that a particular locus is transcribed and is ready to recombine even in the absence of recombinase activity in Rag −/− mice. These contractions could be mediated by interactions between regulatory regions and/or by specific nuclear structures. Enhancer-promoter interactions are thought to direct the long-distance communications (Figure 4). Comparative analysis of Tcra/Tcrd locus contraction in wild-type, Tcra locus enhancer (Eα)−/− and Tcrd locus enhancer (Eδ)−/− thymocytes revealed no significant differences between the cell types [33]. Hence, enhancer-promoter interaction is not necessarily sufficient to mediate Tcra locus contraction and RSSs synapse, but locus contraction might facilitate both enhancer-promoter interactions and RSSs synapse that are required for transcription and V(D)J recombination. These data suggest that a preexisting conformation of the locus that is mediated by an enhancer-independent mechanism may promote the enhancer-promoter interactions necessary to activate transcription and V(D)J recombination of the distant gene segments [32, 33]. Hence, enhancer-promoter interactions could establish molecular bridges over long distances but only after they have been brought into proximity by locus contraction mediated by chromatin-organizing proteins. The mechanism for locus contraction itself is not known. Deficiencies in specific transcription factors such as Pax5, Ikaros, and YY1 disrupt the contracted Igh locus configuration, but it is not known how they do this [34–38]. Additionally, locus contraction could be mediated by chromatin-organizing proteins such as SATB1, CCCTC-binding factor (CTCF), and CTCF-associated cohesin that have been shown to promote long-distance looping interactions at other loci [39–44]. In fact, CTCF and cohesin have been shown to colocalize at multiple sites within the Igh and Igk loci in immature B lymphocytes [45, 46]. Consistent with this, very recent experiments have functionally demonstrated that CTCF and cohesin influence the genomic structure of the Igh locus in developing B lymphocytes [47]. The precise molecular mechanisms involved in how locus-specific conformational changes can regulate the enhancer-promoter interactions to subsequently direct the different V(D)J programs during lymphocyte development is an issue of intense research in the field.
3. V(D)J Recombination Control by Transcriptional Elongation
The finding that germline transcription at a given antigen receptor locus occurs concomitantly with its recombination [7] suggests a linkage between transcription and V(D)J recombination. Based on this evidence, it is accepted that both transcription and V(D)J recombination are consequences of locus accessibility that is mediated through the activation of promoters and enhancers during lymphocyte development [1]. These regulatory elements serve as docking elements to recruit transcription factors that initiate and help to propagate changes in chromatin structure that are essential for the accessibility of the RNAPII and RAG-1/2 proteins (Figure 4). Consistent with this, several transcription factors have also been shown to coordinately regulate both transcription and V(D)J recombination. For example, overexpression of E2A in nonlymphoid cells that express RAG-1 and RAG-2 proteins induced germline transcription and V(D)J recombination at the Igk, Tcrg, and Tcrd loci [48, 49]; OcaB−/− mice displayed defective transcription and recombination of a subset of Vκ gene segments [50]; Stat5 is required for the transcription and VγJγ recombination at the Tcrg locus in response to IL-7 [51, 52]; and deletion of the enhancers and promoters at Tcr and Ig loci or inclusion of mutations at motifs for required transcription factors within these cis-elements inhibit both transcription and V(D)J recombination at each locus [1].
For many years, it was not clear whether germline transcription was merely a side effect of chromatin accessibility generated by the activation of enhancers and promoters or whether it was causal in the V(D)J recombination process itself. In an elegant and definitive study, Abarrategui and Krangel have proven that germline transcription is a key developmental regulator of accessibility for Vα-to-Jα recombination at the Tcra locus [4]. This locus spans 1.5 Mb and contains around 100 Vα/δ gene segments in a 1 Mb region at the 5′ end of the locus and 61 Jα gene segments in a 65 kb region at the 3′ end of the locus [53] (Figure 5). Vα-to-Jα recombination events depend on both Eα, located at the 3′ end of the locus [54], and the promoters associated with the Jα gene segments [55]. These investigators introduced a strong transcription terminator downstream of either the TEA promoter (TEA-T) or the Jα56 gene segment (56R) in the endogenous mouse locus to block transcription originating at the upstream TEA promoters [4, 5] (Figure 5). The terminator sequence they used consists of four polyadenylation sites followed by an array of twelve bacterial lac operons that are thought to function as strong pause sites for RNAPII. The introduced terminator was able to impose an effective block to RNAPII passage. Interestingly, the transcriptional block in both TEA-T and 56R mice caused a strong reduction in recombination at the Jα gene segments located immediately downstream of the terminator sequence. Thus, these experiments clearly demonstrated that transcriptional elongation by RNAPII is necessary for creating accessible chromatin for the RAG-1/2 proteins to initiate Tcra recombination.
Figure 5.
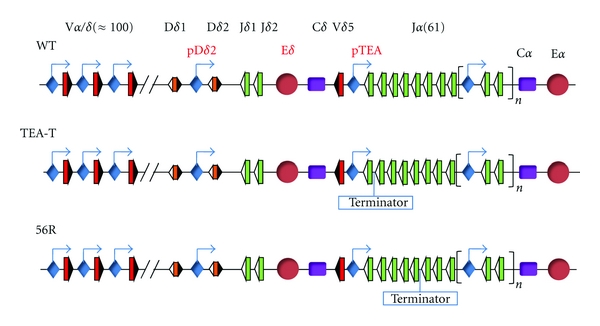
Representation of WT Tcra locus and mutant versions containing a terminator sequence [4, 5]. V, D, and J gene segments are represented as red, orange, and green rectangles, respectively. RSSs are represented as white and black triangles. Constant regions are represented as purple rectangles. Promoters, including the TEA promoter, are represented as blue diamonds, and arrows indicate transcription. Eδ and Eα are represented as red circles. The position of the terminator sequence in mutant TEA-T and 56R Tcra is indicated.
Although instances of recombination in the apparent absence of transcription have also been reported, credible explanations for each of them can be found. For example, isolated nuclei of lymphocytes from Rag −/− mice can rearrange their receptor antigen loci in vitro by addition of RAG-1 and RAG-2 proteins in the absence of ongoing transcription [6]; however, the chromatin structure of the receptor antigen loci could remain accessible during nuclei isolation. Additionally, it has been demonstrated that the VH gene segments rearrange in pro-B lymphocytes with no detectable transcription of these segments [56]; however, the state of transcription of these segments at the time of recombination could not be analyzed. Furthermore, the requirement for transcription itself might not be necessary for RSSs that are located near a cis-element; this is the case for RSSs that are positioned adjacent to a promoter. Several examples of this phenomenon include the inducible mouse mammary tumor virus long terminal repeat that can confer accessibility to a tightly associated RSS [57], the endogenous Tcrb locus that has a Tcrb enhancer-(Eβ-) dependent promoter tightly associated with the Dβ1 segment to confer accessibility to this segment [58, 59], and the physical interaction of the Dβ1 promoter with Eβ to deliver SWI/SNF chromatin remodeling complexes that results in a decrease of nucleosome occupancy at the Dβ1 gene segment [28, 30, 60]. These results argue that transcription is not required to confer accessibility to RAG-1/2 when the RSS is tightly associated with a promoter. However, this is not the situation for many RSSs within the antigen receptor loci in vivo because they are positioned far away from the promoters.
More recent experiments with the TEA-T and 56R mouse models have definitively demonstrated that chromatin accessibility to the RAG-1 protein is mediated by transcriptional elongation itself [24]. In this study, it was proven that the transcriptional terminator introduced within the Jα cluster in TEA-T and 56R mice (Figure 5) inhibited the recruitment of the RAG-1 protein to downstream chromatin. Hence, transcriptional elongation itself confers accessibility to the RAG-1/2 proteins to specific RSSs. Although the pattern of RAG-2 binding was not assessed in this study, it is expected that it would closely resemble that of RAG-1 binding because the pattern of H3K4me3 (which accurately predicts RAG-2 binding) was similar to that of RAG-1 [4, 5, 24, 27]. Hence, H3K4me3 recruits the RAG-2 protein [25–27] and directly contributes to the opening of the chromatin by either repositioning or evicting nucleosomes, which allows free access of the RAG-1 protein to RSSs. Although it has not been directly demonstrated, it is expected that this is likely to be the case for the RSSs that are distantly located from the promoters in the other Ig and Tcr loci. However, a direct role for transcripts in directing V(D)J recombination, as has been shown for class switching at the Igh locus, is not expected because the RAG-1/2 substrate is double strand DNA.
Thus, Abarrategui and Krangel's studies [4, 5] have demonstrated that V(D)J recombination at the Tcra locus requires that the elongation machinery travels or has traveled through the RSS's DNA to allow the RAG-1/2 proteins to access to the RSS's chromatin. However, the precise molecular mechanism by which transcriptional elongation directs the process of VαJα recombination is not known. Chromatin structure imposes significant obstacles on the passage of the RNAPII through the DNA. Because the elongation of transcription is associated with the transient disruption of nucleosome structure [61] and because of the transcription-dependent reduction of nucleosome density at the coding region [62], it is possible that RAG-1 is recruited to the RSSs due to either a transient disruption of the chromatin structure or to less compacted chromatin derived from RNAPII transit. Consistent with the role of transcriptional elongation in transient disrupting of nucleosomal structure, it has been shown using the TEA-T mouse model (Figure 5) that germline transcription originating from the TEA promoter at the Tcra locus causes covalent histone modifications related to opening of chromatin [5], as well as repositioning and loss of the nucleosomes at a 600 base-pair region including the TEA promoter itself and its closest 3′ Jα gene segment, Jα61 [15].
Gene transcription is initiated by the binding of transcription factors to promoters and enhancers (Figure 4). The binding of transcription factors to the enhancer recruits histone acetyltransferases (HATs) that acetylate the N-terminal tails of histones H3 and H4. These acetylated histones provide binding sites for the bromodomains present in other chromatin remodeling complexes and histone-modifying enzymes. Binding of these complexes and enzymes results in nucleosome displacement or disassembly and thus frees promoters for binding by RNAPII thereby allowing transcriptional initiation [63–65]. The RNAPII complexes assembled on the promoter subsequently transit through the chromatin to mediate transcriptional elongation. Clearance of the RNAPII from the promoter requires the phosphorylation of its carboxy-terminal domain (CTD) [66], which is a molecular platform that can recruit a variety of histone modifier complexes, chromatin remodeling complexes, histone chaperones, and elongation factors that are associated and travel with the elongating form of RNAPII; these cofactors are required for efficient transcription through the chromatin [65, 67–72]. Among the most important chromatin modifications associated with transcriptional elongation are the methylation of lysines 4, 36, and 79 of histone H3 and the monoubiquitination of histone H2A.
Abarrategui and Krangel observed that H3ac, H3K4me3, H3K4me2, and H3K36me3 were significantly reduced at the 3′ end of the terminator introduced immediately downstream of the TEA promoter at the Tcra locus [5] (Figure 5); these results suggest that the histone methyl transferases and HATs responsible for these histone covalent modifications might be involved in activating VαJα recombination through RAG-1/2 recruitment mediated by transcriptional elongation [70, 73]. In addition, H3K4me2/3 that recruits RAG-2 [25–27] also recruits other chromatin remodeling complexes such as the ATPase RSC, ISWI, or SWI/SNF that can reposition or evict nucleosomes to facilitate the passage of the RNAPII that can be involved in recruitment of RAG-1 [23, 74–76]. Currently, the identity of the specific chromatin modifying activities that are involved in facilitating the recruitment of RAG-1/2 proteins to RSSs is unknown. Additionally, chaperones that travel with the elongating RNAPII and the chromatin remodeling complexes that facilitate RNAPII's transit could also favor RAG-1 binding [61, 77]. It is also possible that RAG-1 recruitment to the RSSs might be facilitated by direct interaction with the elongating RNAPII through the RNAPII CTD. This would require a total coupling between transcription and V(D)J recombination; the coupling of transcription and RNA splicing is already accepted to occur at the RNAPII CTD [78]. Understanding how precisely transcription activates V(D)J recombination is an important goal for future research in this field.
4. Control of V(D)J Recombination by the Nuclear Position of the Antigen Receptor Loci
In spite of the above cited data, there is evidence that shows that transcription and RSS accessibility are not necessarily sufficient to activate V(D)J recombination in vivo. For example, in pre-T lymphocytes, a deletion within the Tcrb locus that placed the Vβ gene segments under the influence of the Eβ promoted high levels of Vβ transcription but not Vβ to DJβ rearrangement [79]. Additionally, in pre-T lymphocytes, ectopic introduction of Eα within the cluster of Vβ gene segments enhanced transcription of these segments but did not induce Vβ-to-DJβ rearrangement [80]. Furthermore, germline Vβ transcripts are detected from both alleles of the Tcrb locus even in the presence of allelic exclusion during Vβ-to DβJβ recombination [81, 82]. Finally, germline transcription similarly occurs at the Igk locus, which undergoes bi-allelic germline transcription in pre-B lymphocytes during allelic exclusion of Vκ-to-Jκ recombination [83–86]. These findings indicate that additional regulatory constraints on V(D)J recombination exist that operate beyond transcription and chromatin accessibility.
Over the last ten years, it has become clear that the position of Ig and Tcr loci in the nucleus has an essential role in directing V(D)J recombination between distantly located gene segments. It is now accepted that, in addition to transcriptional competence, a particular locus or allele must move away from repressive chromatin to allow distant RSSs to form a recombinational synapse through RAG-1/2 binding that initiates V(D)J recombination [87]. It is now evident that the Ig and Tcr loci move away from repressive compartments such as the nuclear periphery or pericentric heterochromatin when they undergo recombination; the loci then reassociate with them following recombination [85, 87]. Furthermore, associations of Igh, Igk, and Tcrb loci with repressive nuclear compartments seem to be responsible for the establishment of allelic exclusion [81, 86, 88]. Hence, there is a clear connection between the association with repressive nuclear compartments and the inhibition of V(D)J recombination, but germline transcription does not always correlate with this phenomenon.
The mechanism by which locus association with repressive compartments inhibits V(D)J recombination without inhibiting transcription remains unknown. RAG-1/2 binding to D and J gene segments is robust at the Igh and Tcrb loci in pro-B/pre-B and pro-T/pre-T lymphocytes, respectively, even when V-to-DJ recombination is inhibited and one or both alleles are associated with repressive compartments in both stages [24, 27, 31, 81]. It is known that the Igh locus is tethered to the nuclear membrane through the distal VH region cluster, whereas the DHJH region is located away [85]. Thus, persistent RAG-1/2 binding to Igh and Tcrb D and J gene segments in pro-B/T and pre-B/T lymphocytes might be consequence of RSS accessibility due to the spatial orientation of these loci within the nucleus [27, 85]. It is known that VH and Vβ gene segment transcription and accessibility are reduced in the transition from pro-B/T to pre-B/T lymphocytes, respectively, and hence both parameters do seem to correlate with allelic exclusion of Igh and Tcrb loci at pre-B/T lymphocytes [89–91]. These results support the current model that feedback inhibition of Igh and Tcrb loci in pre-B/T lymphocytes, but not in pro-B/T lymphocytes, operates primarily on VH and Vβ gene segment accessibility, respectively. The different chromatin structure at the VH and Vβ gene segments in the two stages indicates clear differences about what might be happening in pro-B/T versus pre-B/T lymphocytes [27, 81, 89–91]. Identification of cis-elements and transfactors that are involved in controlling the association of specific antigen receptor alleles with repressive nuclear compartments is required to elucidate the function of nuclear positioning in regulating V(D)J recombination and allelic exclusion. In the case of the Igk locus, which is also relocated during B lymphocyte development [85], a cis-element that binds the Ikaros transcriptional repressor targets Igk transgenes to centromeric chromatin and inhibits Vκ-to-Jκ recombination [92]; another candidate is IRF-4, which directs the Igk allele away from the pericentromeric heterochromatin [93]. In the case of the Tcrb locus, the helix-loop-helix protein, E47, is a good candidate to direct the interaction of this locus with pericentromeric heterochromatin since its dosage is rate-limiting with regard to V(D)J recombination and forced E47 expression interferes with pre-TCR-mediated feedback inhibition [94]. Additionally, it was also proposed that V(D)J recombination events occurring on one allele could activate signals that inhibit rearrangements on the second allele [95]. Consistent with this idea, it has been shown recently that homologous pairing of Ig alleles occurs during recombination and is mediated by RAG-1/2 binding [88, 96]. Furthermore, it has been demonstrated that RAG-mediated cleavage on one allele induces the other allele to relocate to pericentromeric heterochromatin by a mechanism related to the recognition of the cleaved allele by the DNA damage sensor ataxia telangiectasia mutated (ATM) protein [88]. Hence, activation of ATM by the cleaved allele acts in trans on the uncleaved allele to prevent recombination. Interallelic pairing has been proposed as a general mechanism for establishing the monoallelic gene expression that contributes to the maintenance of genomic integrity and suppresses oncogenic translocations during V(D)J recombination of antigen receptor loci.
5. Conclusions
V(D)J recombination is essential for the development of adaptive immune responses in vertebrates. In developing lymphocytes, V(D)J recombination is subjected to very tight spatial and temporal regulation. The regulation of this process is very complex and involves nuclear dynamics and changes in higher-order chromatin architecture to create gene segment accessibility to RAG-1/2 proteins. Active chromatin is bound by RAG-2 through interactions with specific H3K4me3, whereas RAG-1 binds to accessible RSSs derived from transcriptional elongation in large loci. The correlation between transcription and V(D)J recombination in both the recombined and allelic excluded antigen receptor loci led to the studies that established that the differential positioning of such loci at transcriptionally repressive nuclear regions might be responsible for allelic exclusion. Allelic association with the repressive nuclear compartments can inhibit V(D)J recombination by a mechanism other than transcription or RAG-1/2 accessibility to chromatin. Future experiments should be focused on identifying the cis-elements and trans-factors that regulate V(D)J recombination in vivo.
Acknowledgments
The authors are grateful for support from the Spanish Ministry of Science and Innovation Grant BFU2009-08796, Junta de Andalucía Grants CTS-6587 and CVI-4526, and CSIC Grants 200620I040 and 201020E060. The research in our lab is partly supported by European Region Development Funds (ERDF/FEDER). They are also grateful to an anonymous reviewer for comments on the paper.
References
- 1.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Advances in Immunology. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 2.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nature Reviews Immunology. 2011;11:252–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 3.Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280(5720):288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 4.Abarrategui I, Krangel MS. Regulation of T cell receptor-alpha gene recombination by transcription. Nature Immunology. 2006;7(10):1109–1115. doi: 10.1038/ni1379. [DOI] [PubMed] [Google Scholar]
- 5.Abarrategui I, Krangel MS. Noncoding transcription controls downstream promoters to regulate T-cell receptor α recombination. EMBO Journal. 2007;26(20):4380–4390. doi: 10.1038/sj.emboj.7601866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanhope-Baker P, Hudson KM, Shaffer AL, Constantinescu A, Schlissel MS. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85(6):887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- 7.Yancopoulos GD, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged V(H) gene segments. Cell. 1985;40(2):271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 8.Fondell JD, Marcu KB. Transcription of germ line Vα segments correlates with ongoing T-cell receptor α-chain rearrangement. Molecular and Cellular Biology. 1992;12(4):1480–1489. doi: 10.1128/mcb.12.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlissel MS, Corcoran LM, Baltimore D. Virus-transformed preB-cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. Journal of Experimental Medicine. 1991;173(3):711–720. doi: 10.1084/jem.173.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolland DJ, Wood AL, Johnston CM, et al. Antisense intergenic transcription in V(D)J recombination. Nature Immunology. 2004;5(6):630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 11.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annual Review of Immunology. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 12.Goldmit M, Bergman Y. Monoallelic gene expression: a repertoire of recurrent themes. Immunological Reviews. 2004;200:197–214. doi: 10.1111/j.0105-2896.2004.00158.x. [DOI] [PubMed] [Google Scholar]
- 13.Krangel MS. T cell development: better living through chromatin. Nature Immunology. 2007;8(7):687–694. doi: 10.1038/ni1484. [DOI] [PubMed] [Google Scholar]
- 14.Schlissel MS. Regulating antigen-receptor gene assembly. Nature Reviews Immunology. 2003;3(11):890–899. doi: 10.1038/nri1225. [DOI] [PubMed] [Google Scholar]
- 15.Kondilis-Mangum HD, Cobb RM, Osipovich O, Srivatsan S, Oltz EM, Krangel MS. Transcription-dependent mobilization of nucleosomes at accessible TCR gene segments in vivo. Journal of Immunology. 2010;184(12):6970–6977. doi: 10.4049/jimmunol.0903923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osipovich O, Milley R, Meade A, et al. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nature Immunology. 2004;5(3):309–316. doi: 10.1038/ni1042. [DOI] [PubMed] [Google Scholar]
- 17.Kwon J, Morshead KB, Guyon JR, Kingston RE, Oettinger MA. Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Molecular Cell. 2000;6(5):1037–1048. doi: 10.1016/s1097-2765(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 18.Kwon J, Imbalzano AN, Matthews A, Oettinger MA. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Molecular Cell. 1998;2(6):829–839. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- 19.Golding A, Chandler S, Ballestar E, Wolffe AP, Schlissel MS. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO Journal. 1999;18(13):3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBlane F, Boyes J. Stimulation of V(D)J recombination by histone acetylation. Current Biology. 2000;10(8):483–486. doi: 10.1016/s0960-9822(00)00449-8. [DOI] [PubMed] [Google Scholar]
- 21.Osipovich O, Cobb RM, Oestreich KJ, Pierce S, Ferrier P, Oltz EM. Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Tcrb genes. Nature Immunology. 2007;8(8):809–816. doi: 10.1038/ni1481. [DOI] [PubMed] [Google Scholar]
- 22.Patenge N, Elkin SK, Oettinger MA. ATP-dependent remodeling by SWI/SNF and ISWI proteins stimulates V(D)J cleavage of 5 S arrays. Journal of Biological Chemistry. 2004;279(34):35360–35367. doi: 10.1074/jbc.M405790200. [DOI] [PubMed] [Google Scholar]
- 23.Du H, Ishii H, Pazin MJ, Sen R. Activation of 12/23-RSS-dependent RAG cleavage by hSWI/SNF complex in the absence of transcription. Molecular Cell. 2008;31(5):641–649. doi: 10.1016/j.molcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji Y, Little AJ, Banerjee JK, et al. Promoters, enhancers, and transcription target RAG1 binding during V(D)J recombination. Journal of Experimental Medicine. 2010;207(13):2809–2816. doi: 10.1084/jem.20101136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27(4):561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramón-Maiques S, Kuo AJ, Carney D, et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(48):18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141(3):419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM. Regulation of TCRβ gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24(4):381–391. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, Busslinger M. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nature Immunology. 2007;8(4):378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 30.Spicuglia S, Kumar S, Yeh J-H, et al. Promoter activation by enhancer-dependent and -independent loading of activator and coactivator complexes. Molecular Cell. 2002;10(6):1479–1487. doi: 10.1016/s1097-2765(02)00791-8. [DOI] [PubMed] [Google Scholar]
- 31.Roldan E, Fuxa M, Chong W, et al. Locus “decontraction” and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nature Immunology. 2005;6(1):31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shih HY, Hao B, Krangel MS. Orchestrating T-cell receptor α gene assembly through changes in chromatin structure and organization. Immunologic Research. 2011;49(1–3):192–201. doi: 10.1007/s12026-010-8181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih HY, Krangel MS. Distinct contracted conformations of the Tcra/Tcrd locus during Tcra and Tcrd recombination. Journal of Experimental Medicine. 2010;207(9):1835–1841. doi: 10.1084/jem.20100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldán E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes and Development. 2004;18(4):411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynaud D, Demarco IA, Reddy KL, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nature Immunology. 2008;9(8):927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Schmidt-Supprian M, Shi Y, et al. Yin Yang 1 is a critical regulator of B-cell development. Genes and Development. 2007;21(10):1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hesslein DGT, Pflugh DL, Chowdhury D, Bothwell ALM, Sen R, Schatz DG. Pax5 is required for recombination of transcribed, acetylated, 5′IgH V gene segments. Genes and Development. 2003;17(1):37–42. doi: 10.1101/gad.1031403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Espinoza CR, Yu Z, et al. Transcription factor Pax5 (BSAP) transactivates the RAG-mediated VH-to-DJVH rearrangement of immunoglobulin genes. Nature Immunology. 2006;7(6):616–624. doi: 10.1038/ni1339. [DOI] [PubMed] [Google Scholar]
- 39.Splinter E, Heath H, Kooren J, et al. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes and Development. 2006;20(17):2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han L, Lee DH, Szabo PE. CTCF is the master organizer of domain-wide allele-specific chromatin at the H19/Igf2 imprinted region. Molecular and Cellular Biology. 2008;28(3):1124–1135. doi: 10.1128/MCB.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hadjur S, Williams LM, Ryan NK, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460(7253):410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nature Genetics. 2006;38(11):1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 43.Nativio R, Wendt KS, Ito Y, et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genetics. 2009;5(11) doi: 10.1371/journal.pgen.1000739. Article ID e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galande S, Purbey PK, Notani D, Kumar PP. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Current Opinion in Genetics and Development. 2007;17(5):408–414. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Degner SC, Wong TP, Jankevicius G, Feeney AJ. Developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. Journal of Immunology. 2009;182(1):44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Degner-Leisso SC, Feeney AJ. Epigenetic and 3-dimensional regulation of V(D)J rearrangement of immunoglobulin genes. Seminars in Immunology. 2010;22(6):346–352. doi: 10.1016/j.smim.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Degner SC, Verma-Gaur J, Wong TP, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(23):9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romanow WJ, Langerak AW, Goebel P, et al. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Molecular Cell. 2000;5(2):343–353. doi: 10.1016/s1097-2765(00)80429-3. [DOI] [PubMed] [Google Scholar]
- 49.Ghosh JK, Romanow WJ, Murre C. Induction of a diverse T cell receptor γ/δ repertoire by the helix-loop-helix proteins E2A and HEB in nonlymphoid cells. Journal of Experimental Medicine. 2001;193(6):769–775. doi: 10.1084/jem.193.6.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casellas R, Jankovic M, Meyer G, et al. OcaB is required for normal transcription and V(D)J recombination of a subset of immunoglobulin κ genes. Cell. 2002;110(5):575–585. doi: 10.1016/s0092-8674(02)00911-x. [DOI] [PubMed] [Google Scholar]
- 51.Bertolino E, Reddy K, Medina KL, Parganas E, Ihle J, Singh H. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nature Immunology. 2005;6(8):836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- 52.Ye SK, Agata Y, Lee HC, et al. The IL-7 receptor controls the accessibility of the TCRγ locus by Stat5 and histone acetylation. Immunity. 2001;15(5):813–823. doi: 10.1016/s1074-7613(01)00230-8. [DOI] [PubMed] [Google Scholar]
- 53.Krangel MS, Carabana J, Abbarategui I, Schlimgen R, Hawwari A. Enforcing order within a complex locus: Current perspectives on the control of V(D)J recombination at the murine T-cell receptor α/δ locus. Immunological Reviews. 2004;200:224–232. doi: 10.1111/j.0105-2896.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- 54.Sleckman BP, Bardon CG, Ferrini R, Davidson L, Alt FW. Function of the TCR α enhancer in αβ and γδ T cells. Immunity. 1997;7(4):505–515. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- 55.Hawwari A, Bock C, Krangel MS. Regulation of T cell receptor α gene assembly by a complex hierarchy of germline Jα promoters. Nature Immunology. 2005;6(5):481–489. doi: 10.1038/ni1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Angelin-Duclos C, Calame K. Evidence that immunoglobulin VH-DJ recombination does not require germ line transcription of the recombining variable gene segment. Molecular and Cellular Biology. 1998;18(11):6253–6264. doi: 10.1128/mcb.18.11.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cherry SR, Baltimore D. Chromatin remodeling directly activates V(D)J recombination. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(19):10788–10793. doi: 10.1073/pnas.96.19.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehurst CE, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the Dβ1 gene segment at the TCRβ locus by a germline promoter. Immunity. 1999;10(3):313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- 59.Mathieu N, Hempel WM, Spicuglia S, Verthuy C, Ferrier P. Chromatin remodeling by the T cell receptor (TCR)-β gene enhancer during early T cell development: implications for the control of TCR-β locus recombination. Journal of Experimental Medicine. 2000;192(5):625–636. doi: 10.1084/jem.192.5.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sikes ML, Meade A, Tripathi R, Krangel MS, Oltz EM. Regulation of V(D)J recombination: a dominant role for promoter positioning in gene segment accessibility. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(19):12309–12314. doi: 10.1073/pnas.182166699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Molecular Cell. 2006;22(3):415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 62.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO Journal. 2004;23(21):4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nature Reviews Molecular Cell Biology. 2006;7(6):437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 64.Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Current Topics in Developmental Biology. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 65.Workman JL. Nucleosome displacement in transcription. Genes and Development. 2006;20(15):2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 66.Sims RJ, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes and Development. 2004;18(20):2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 67.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 68.Wittschieben BO, Otero G, de Bizemont T, et al. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Molecular Cell. 1999;4(1):123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 69.Krogan NJ, Dover J, Wood A, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Molecular Cell. 2003;11(3):721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 70.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Molecular Cell. 2003;11(3):709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 71.Xiao T, Hall H, Kizer KO, et al. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes and Development. 2003;17(5):654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao T, Kao CF, Krogan NJ, et al. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Molecular and Cellular Biology. 2005;25(2):637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Albert M, Helin K. Histone methyltransferases in cancer. Seminars in Cell and Developmental Biology. 2010;21(2):209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 74.Santos-Rosa H, Schneider R, Bernstein BE, et al. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Molecular Cell. 2003;12(5):1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- 75.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosmal block to RNA polymerase II elongation. Molecular Cell. 2006;24(3):481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Corey LL, Weirich CS, Benjamin IJ, Kingston RE. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes and Development. 2003;17(11):1392–1401. doi: 10.1101/gad.1071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301(5636):1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 78.Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Current Opinion in Cell Biology. 2008;20(3):260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Senoo M, Wang L, Suzuki D, Takeda N, Shinkai Y, Habu S. Increase of TCR Vβ accessibility within Eβ regulatory region influences its recombination frequency but not allelic exclusion. Journal of Immunology. 2003;171(2):829–835. doi: 10.4049/jimmunol.171.2.829. [DOI] [PubMed] [Google Scholar]
- 80.Jackson A, Kondilis HD, Khor B, Sleckman BP, Krangel MS. Regulation of T cell receptor β allelic exclusion at a level beyond accessibility. Nature Immunology. 2005;6(2):189–197. doi: 10.1038/ni1157. [DOI] [PubMed] [Google Scholar]
- 81.Schlimgen RJ, Reddy KL, Singh H, Krangel MS. Initiation of allelic exclusion by stochastic interaction of Tcrb alleles with repressive nuclear compartments. Nature Immunology. 2008;9(7):802–809. doi: 10.1038/ni.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jia J, Kondo M, Zhuang Y. Germline transcription from T-cell receptor Vβ gene is uncoupled from allelic exclusion. EMBO Journal. 2007;26(9):2387–2399. doi: 10.1038/sj.emboj.7601671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh N, Bergman Y, Cedar H, Chess A. Biallelic germline transcription at the κ immunoglobulin locus. Journal of Experimental Medicine. 2003;197(6):743–750. doi: 10.1084/jem.20021392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Amin RH, Cado D, Nolla H, et al. Biallelic, ubiquitous transcription from the distal germline Igk locus promoter during B cell development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(2):522–527. doi: 10.1073/pnas.0808895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kosak ST, Skok JA, Medina KL, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296(5565):158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 86.Goldmit M, Ji Y, Skok J, et al. Epigenetic ontogeny of the Igk locus during B cell development. Nature Immunology. 2005;6(2):198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 87.Jhunjhunwala S, van Zelm MC, Peak MM, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell. 2009;138(3):435–448. doi: 10.1016/j.cell.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hewitt SL, Yin B, Chaumeil J, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nature Immunology. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maes J, O’Neill LP, Cavelier P, Turner BM, Rougeon F, Goodhardt M. Chromatin remodeling at the Ig loci prior to V(D)J recombination. Journal of Immunology. 2001;167(2):866–874. doi: 10.4049/jimmunol.167.2.866. [DOI] [PubMed] [Google Scholar]
- 90.Tripathi R, Jackson A, Krangel MS. A change in the structure of Vβ chromatin associated with TCR β allelic exclusion. Journal of Immunology. 2002;168(5):2316–2324. doi: 10.4049/jimmunol.168.5.2316. [DOI] [PubMed] [Google Scholar]
- 91.Chowdhury D, Sen R. Transient IL-7/IL-7R signaling provides a mechanism for feedback inhibition of immunoglobulin heavy chain gene rearrangements. Immunity. 2003;18(2):229–241. doi: 10.1016/s1074-7613(03)00030-x. [DOI] [PubMed] [Google Scholar]
- 92.Liu Z, Widlak P, Zou Y, et al. A recombination silencer specifies heterochromatin positioning and Ikaros association in the immunoglobulin κ locus. Immunity. 2006;24(4):405–415. doi: 10.1016/j.immuni.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 93.Johnson K, Hashimshony T, Sawai CM, et al. Regulation of immunoglobulin light chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28(3):335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 94.Agata Y, Tamaki N, Sakamoto S, et al. Regulation of T cell receptor β gene rearrangement and allelic exclusion by the helix-loop-helix protein, E47. Immunity. 2007;27(6):871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 95.Alt FW, Enea V, Bothwell ALM, Baltimore D. Activity of multiple light chain genes in murine myeloma cells producing a single, functional light chain. Cell. 1980;21(1):1–12. doi: 10.1016/0092-8674(80)90109-9. [DOI] [PubMed] [Google Scholar]
- 96.Brandt VL, Hewitt SL, Skok JA. It takes two. Communication between homologous alleles preserves genomic stability during V(D)J recombination. Nucleus. 2010;1(1):23–29. doi: 10.4161/nucl.1.1.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]


