Abstract
The malaria vaccine RTS,S/AS01, based on immunogenic regions of the Plasmodium falciparum circumsporozoite protein (CSP), has partial efficacy against clinical malaria in African children. Understanding how sequence diversity in CSP T- and B-cell epitopes relates to naturally acquired and vaccine-induced immunity may be useful in efforts to improve the efficacy of CSP-based vaccines. However, limitations in sequencing technology have precluded thorough evaluation of diversity in the immunogenic regions of this protein. In this study, 454, a next generation sequencing technology, was evaluated as a method for assessing diversity in these regions. Portions of the circumsporozoite gene (cs) were sequenced both by 454 and Sanger sequencing from samples collected in a study in Bandiagara, Mali. 454 detected more single nucleotide polymorphisms and haplotypes in the T-cell epitopes than Sanger sequencing, and it was better able to resolve genetic diversity in samples with multiple infections; however, it failed to generate sequence for the B-cell epitopes.
Introduction
Substantial resources are being invested in clinical trials to evaluate the potential of vaccines targeting specific immunogenic antigens of Plasmodium falciparum, including the circumsporozoite protein (CSP) encoded by the circumsporozoite gene (cs). However, malaria vaccine development and testing has generally not been informed by molecular epidemiologic evaluations of how genetic diversity in vaccine antigens in parasite populations may affect vaccine efficacy. For example, vaccines that confer strain-specific protection may not be effective in a parasite population in which the vaccine strain is rare. Furthermore, vaccines may create a selective advantage for non-vaccine strain types, compromising vaccine efficacy.1
The cs gene is polymorphic, with diversity in regions that code for epitopes recognized by the human immune system. The central repeat region of the cs gene contains tetrameric repeats that vary in both the sequence and number of tetramers.2 This region codes for epitopes recognized by anti-CSP antibodies.3,4 The 3′ regions of the cs gene, Th2R and Th3R, encode epitopes that are recognized by CD8+ and CD4+ T cells.5 The diversity in these regions, which occurs in the form of non-synonymous SNPs, increases as malaria transmission increases across distinct geographic areas,6,7 with the highest diversity occurring in Africa. Molecular surveys in Sierra Leone and the Gambia found 42 haplotypes in 99 samples and 24 haplotypes in 44 samples for the regions containing Th2R and Th3R, respectively.7,8 The current leading malaria vaccine candidate RTS,S/AS01, which consists of the CSP repeat region and Th regions fused to the hepatitis B surface antigen, has shown modest efficacy in phase 2 trials9–12 and is currently being evaluated in a multicenter phase 3 trial in 11 countries in Africa.13
A follow-up study to a phase 2 trial of the vaccine concluded that there was no selection of non-vaccine strains in vaccinated children versus non-vaccinated children.14 However, this study, which used Sanger sequencing to detect polymorphism in the regions coding for the T-cell epitopes Th2R and Th3R, excluded samples that could not be resolved into predominant alleles from the analysis. Furthermore, diversity in the central repeat region of the cs gene, which codes for the B-cell epitopes of CSP and is also included in the vaccine, was not considered, presumably owing to the limitations in Sanger sequencing technology.
Sanger sequencing is limited in its ability to detect multiple parasite types in a mixed infection, because this method depends on reading major and minor peaks on a chromatogram to determine allele presence or absence in a sample. The proportion of these peaks may not correlate well with the actual proportion of parasite DNA in a sample, because incorporation of dye-labeled dideoxynucleotide can vary within a sample and be influenced by flanking sequence.15 Moreover, complete haplotypes for each unique parasite clone that is in a sample cannot be determined by Sanger sequencing. It is also impossible to resolve diversity with respect to repetitive DNA sequences in mixed infections using this sequencing method.
New more powerful sequencing technologies may have the potential to address some of these limitations. 454, a next generation sequencing platform, generates massively parallel DNA sequences from polymerase chain reaction (PCR) products, potentially making it possible to resolve diversity in complex infections. With longer read lengths than other next generation sequencing platforms, 454 might permit sequencing of the variable-length central repeat region of cs that has defied other sequencing methods on field samples. Furthermore, by providing massively parallel sequences of the target region that permits quantification of reads with different variants, this technology may help determine the alleles that are predominant at polymorphic sites more reliably than Sanger sequencing.
Materials and Methods
Standardized mixed infections.
Mixtures of PCR product containing Th2R and Th3R amplified from laboratory strains (3D7, Hb3, and Dd2) for which the sequences are known were created, quantified, and diluted to concentrations of 100, 50, 25, 12.5, and 6.25 ng/μL. 3D7 comprised 60% of each mixture, Hb3 comprised 30%, and Dd2 comprised 10%. The PCR products for each strain were generated in triplicate, and three mixtures were made and serially diluted in parallel. Each of the mixtures was sequenced by both 454 and Sanger sequencing to test the ability of each technology to quantitate the different alleles in a mixture. The sequence output for each dilution was combined for both 454 and Sanger sequencing. The observed allele frequencies for the 454 sequencing method were determined by calculating the percentage of reads that contained each type of allele at each of seven polymorphic sites for each concentration from the three parallel dilutions. The observed allele frequencies for Sanger sequencing were determined by calculating the relative peak heights of the major and minor alleles at each polymorphic site at each concentration from the three parallel dilutions. These frequencies were subtracted from the expected frequency for each allele, and the sums of the absolute value of these differences were averaged for each concentration.
Sensitivity analyses were performed on sequence output generated from each concentration of the standardized mixed infections from each technology to determine a threshold for inclusion of minor alleles from the clinical samples. Each dilution yielded the same inflection points for both technologies, except for the highest concentration, which yielded a slightly higher error rate. At this concentration, erroneous single nucleotide polymorphisms (SNPs) were found at minor allele frequencies of 0.08 and 0.25 for 454 and Sanger sequencing, respectively (data not shown). However, because this concentration was much higher than the concentration of PCR products generated for clinical samples, the results of the middle (25 ng/μL) concentration were used to generate the curves in Figure 1. The largest numbers of erroneous SNPs were found between the frequencies of 0.025 and 0.01 for 454 and between 0.2 and 0.15 for Sanger sequencing. However, because of the fact that erroneous SNPs were still present at a frequency of 0.05 and 0.075, a conservative minor allele frequency threshold of 0.1 for calling SNPs was selected for 454. A minor allele frequency threshold of 0.2 was selected for calling SNPs by Sanger sequencing.
Figure 1.
Determination of a minor allele frequency (MAF) threshold for calling SNPs from the 25 ng/μL concentration of standardized mixed infections. In 454 output (Left), the largest increase in number of SNPs occurred below the MAF of 0.025 (broken line). The solid line marks the MAF chosen for inclusion in this study. In Sanger sequencing output (Right), the largest increase in number of SNPs occurred below the MAF of 0.2 (solid line).
Clinical sample selection.
DNA extracted from 45 parasite-positive filter paper blood samples was used to compare the ability of 454 and Sanger sequencing to detect CSP diversity in field samples. Samples were randomly selected from among participants in a prospective cohort study of malaria incidence conducted in Bandiagara, Mali, from 1999 to 2001,16 and they represent both clinical and asymptomatic infections detected through passive and active surveillance.
Sanger sequencing.
PCR amplification
Two nested PCR assays were designed to amplify a region of the cs gene containing both Th2R and Th3R and the central repeat region. The primary PCR was designed to amplify the region of the gene that contains both regions, and the secondary PCRs amplify Th2R and Th3R as well as the central repeat region individually. The primary forward and reverse PCR primers were GTTGAGGCCTTTTCCAGGAATACCAG and GTACAACTCAAACTAAGATGTGTTC. Primary PCR conditions were as follows: 30 cycles of 95°C for 30 seconds, 52°C for 30 seconds, and 72°C for 1 minute. Secondary PCR conditions for the repeat regions and ThRs were 30 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1.5 minutes and 25 cycles of 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 1 minute, respectively. Secondary primers were the same as the sequence-specific primers shown in Figure 2. Expected product sizes for the Th region and repeat region were 214 and 516 base pairs, respectively, based on the 3D7 strain of P. falciparum. PCR products were amplified using HotStar Taq (Qiagen, Venlo, The Netherlands). PCR products were loaded on a 2% agarose gel, stained with ethidium bromide, and run at 100 V for 1 hour. Bands were detected using geneSNAP (Synoptics LTD, Cambridge, United Kingdom) gel imaging software.
Figure 2.
Primers for amplification of 454 PCR products.
Sequencing
After amplification was verified by gel electrophoresis, PCR products were purified by vacuum filtration in Excela Pure (Edge Biosystems, Gaithersburg, MD) 96-well plates. Purified PCR product was then amplified and sequenced on a ABI3730 xl at the University of Maryland School of Medicine Biopolymer Laboratory.
Sequence analysis
Sequences were aligned to the 3D7 reference genome using Sequencher (Gene Codes Corp, Ann Arbor, MI) software. For samples containing more than one allele at a polymorphic site, a predominant allele was designated if the secondary peak height was less than or equal to 40% of the height of the primary peak on the chromatogram for that sample. If the secondary peak was greater than 40% of the height of the primary peak, the polymorphic site and sample were designated as mixed, and therefore, haplotypes were not constructed for these samples. Minor alleles that were not represented by a peak that was at least 20% of the primary peak height were not included in the total number of SNPs discovered in Sanger sequencing output.
454 sequencing.
PCR amplification
The primary PCR used for 454 sequencing was identical to the PCR used for Sanger sequencing. Secondary PCR primers contained specific adapters necessary for the emulsion PCR17 step of 454 sequencing as well as unique barcodes to identify sequences from individual samples (Figure 2).
Multiplexing
The concentration of each PCR product was determined by band intensity compared with a standard of similar molecular weight using geneSNAP software, and 100 ng of each product were then pooled. PCR products for each region were pooled separately.
Sequencing
Pooled PCR products were sequenced at the University of Maryland School of Medicine Genomic Resource Center on the GS FLX Titanium 454 Platform (Roche Diagnostics, Branford, CT).
Sequence analysis
Sequences were aligned using gsAmplicon (Roche Diagnostics, Branford, CT) software. For samples containing more than one allele at a polymorphic site, predominance was determined if the majority allele was present in 71% or more of all reads obtained for that sample. If a majority allele could not be determined, that polymorphic site was considered mixed. Haplotype information, however, was still obtained for samples with mixed polymorphic sites.
Results
Detection of allele frequencies in standardized mixed infections.
The average difference between observed and expected allele frequencies for 454 was less than 0.1 for each concentration. The highest difference was 9%, which occurred at a concentration of 100 ng/μL and decreased with decreasing concentration, leveling off at 12.5 ng/μL. The average difference between observed and expected allele frequencies for Sanger sequencing also decreased from high to low concentrations, with the highest (0.38) occurring at 100 ng/μL and the lowest (0.14) occurring at 6.25 ng/μL. Overall, the difference between observed and expected allele frequencies was lower at each concentration for 454 than Sanger sequencing (Figure 3). The proportion of correctly identified predominant alleles was 91% and 75% for 454 and Sanger sequencing, respectively (data not shown).
Figure 3.
Mean difference between observed allele frequency in sequence output for 454 and Sanger sequencing and expected allele frequency in five dilutions of standardized mixed infections. The average difference between observed and expected allele frequencies in 454 output was less than 0.1 for each concentration (100, 50, 25, 12.5, and 6.25 ng/μL) and greater than 0.1 for each concentration in Sanger sequencing output. The largest differences occurred at the highest concentration for both technologies. Statistical significance was calculated using a Student t test and is denoted by an asterisk.
SNP detection.
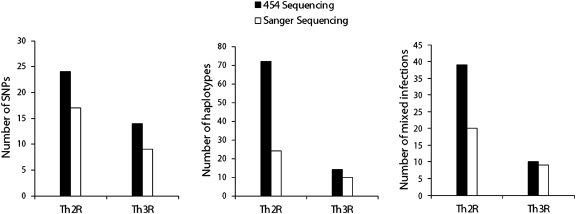
A total of 17 and 9 SNPs were detected by Sanger sequencing (two times coverage, forward and reverse) in Th2R and Th3R, respectively, from the 45 samples selected for sequencing. A total of 24 and 14 SNPs were detected by 454 in Th2R and Th3R, respectively (Figure 4). The average coverage of the Th regions in 454 sequence output was ∼500×, with a range of ∼200–1,000×.
Figure 4.
The number of SNPs (Left), haplotypes (Center), and mixed infections (Right) detected in P. falciparum circumsporozoite genes by 454 and Sanger sequencing in 45 dried blood spot field samples from Mali, West Africa.
Haplotype detection.
The total number of haplotypes found in Th2R and Th3R, respectively, was 24 and 10 in Sanger sequencing output and 72 and 14 in 454 output (Figure 4). Only haplotypes representing at least 10% of all 454 reads obtained for a sample were included (Genbank accession numbers JN849502–JN849573). Samples that contained polymorphic sites with more than one allele in Sanger sequencing output could not be resolved into haplotypes and therefore, were excluded from haplotype analyses. The proportion of unique haplotypes to the total number of haplotypes detected was 0.53 (24/45) for Sanger sequencing and 0.49 (72/147) for 454.
Mixed infections.
Most samples (39 of 45) contained more than one distinct parasite type based on 454 data, whereas only 20 samples had more than one haplotype detected by Sanger sequencing with respect to Th2R, the Th region with the most diversity (Figure 4).
Determination of majority alleles.
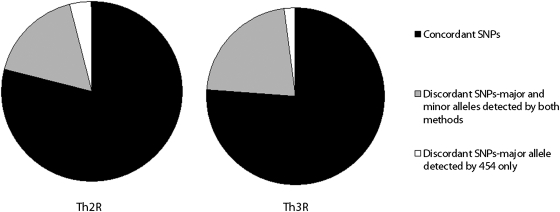
Of the SNPs identified as majority alleles by either 454 or Sanger sequencing, approximately 74% were identified as majority alleles by both technologies, 24% were identified as a majority allele by one technology and not the other but were detected by both, and 2% were identified as the majority allele by 454 but not detected by Sanger sequencing with respect to Th2R. In the case of Th3R, approximately 77% were identified as majority alleles by both technologies, 18% were identified as a majority allele by one technology and not the other but were detected by both, and 4% were identified as the majority allele by 454 but not detected by Sanger sequencing (Figure 5). There were no samples in which an allele was identified as predominant in Sanger sequencing output and not detected by 454.
Figure 5.
Concordance between Sanger sequencing and 454 in determination of majority alleles in the Th2R and Th3R regions of the circumsporozoite gene. Concordant SNPs are those SNPs that are determined to be majority alleles in an infection by both technologies. Discordant SNPs are those SNPS on which the two technologies disagree on the majority allele.
Haplotype diversity.
The number of distinct haplotypes found within each sample with respect to Th2R and Th3R was explored in 454 data. An average of 3.5 parasite types were found with respect to Th2R (range = 1–8), and an average of 2.5 parasite types were found with respect to Th3R (range = 1–4) (Table 1).
Table 1.
Haplotype diversity with respect to the Th2R and Th3R T-cell epitope regions of the P. falciparum circumsporozoite gene as measured by 454 sequencing
| Th2R | Th3R | |
|---|---|---|
| Mean | 3.5 | 1.6 |
| Median | 4.5 | 2.5 |
| Range | 1–8 | 1–4 |
Sequencing the repeat region.
Complete sequences for the region of the cs coding for B-cell epitopes were not obtained, and therefore, diversity and haplotype data could not be generated for this region.
Discussion
454 was the more sensitive method for assessing diversity in cs, detecting approximately 41% more SNPs in Th2R and 64% more SNPs in Th3R than Sanger sequencing. 454 was also more accurate at identifying diversity in a known mixture of parasite clones. A previous study has shown that adjustments can be made to peak height in chromatograms to diminish inaccuracies caused by dye effects in Sanger sequencing when sequencing genomic DNA.18 However, it is unclear how well these methods would work for sequencing field samples, particularly filter paper blood samples, which tend to have DNA of poorer quality and variable quantity.
A relatively high minor allele frequency was used for defining SNPs (at least 10% of all reads) to strengthen confidence that the SNPs included in the analyses were genuine and not products of PCR or 454 sequencing error. The 10% limit is very conservative, being well above both the lower minor allele frequency thresholds typically used for SNP discovery and the error rates reported by the manufacturers; 0.4% error is reported for HotStar Taq polymerase, and for GS FLX Titanium, a 1% error rate for read lengths of 400 base pairs and better for smaller read lengths is reported. Also, it was two times the frequency at which erroneous SNPs were discovered in the sensitivity analysis performed on the standardized mixed infections. Additionally, one base pair indels were excluded from the analysis, because one of the limitations of pyrosequencing is that it has a higher error rate when sequencing homopolymers (stretches of a single base such as AAAA). This exclusion was used to help ensure that erroneous SNPs were excluded from the analysis and also to increase confidence in our main conclusion about the comparative ability of the sequencing platforms to detect diversity; however, it does raise the possibility that some real SNPs were also excluded.
Because this study was not focused on detecting rare variants, the conservative threshold for SNP calling would not have affected the conclusions drawn; however, using a lower threshold would have compromised our goal of comparing the ability of the two sequencing methods to detect genetic diversity in polymorphic genes in the same samples, because a seemingly more sensitive method might simply have detected more false-positive SNPs if too low of a minor allele threshold was used for SNP calling. For 454 applications where detecting infrequent alleles is important, a lower minor allele frequency threshold could likely be identified using more rigorous SNP-calling algorithms. The proportion of unique haplotypes to total haplotypes detected was reassuringly similar for Sanger sequencing (0.53) and 454 (0.49); however, the data from this study showed that using 454 more than tripled the number of evaluable haplotypes that were generated from a sample set. In 45 samples, 72 haplotypes were found in Th2R alone, whereas in earlier studies using Sanger sequencing on samples from other West African settings with similar malaria epidemiology, only 24 haplotypes were found in 44 Gambian samples7; also, just 42 haplotypes were found8 for Th2R and Th3R combined in 99 samples from Sierra Leone.
The high number of SNPs and unique haplotypes in our study is consistent with this region of the cs gene being under diversifying selection pressure.19 Although no evidence has yet been reported of selection of non-vaccine variants of CSP after immunization with CSP-based vaccines, this evidence of diversifying selection supports the notion that genetic variation in CSP may be driven by the human immune system and could be important in naturally acquired and vaccine-induced immunity.
The results of the sensitivity analysis revealed different error rates for the highest DNA concentrations for both technologies. A possible explanation for this finding may be that the high concentration of DNA results in signal interference. In output from Sanger sequencing, base ambiguities may result from overlap between peaks. In 454 output, light signals that occur when bases are incorporated can bleed into signals from surrounding reactions, which may result in errors.
A few alleles were determined to be majority alleles by 454 but were not detected by Sanger sequencing. Two possible explanations for this finding are (1) the majority alleles were actually not majority alleles and were the result of PCR bias or (2) these alleles were on the lower end of the cutoff for a majority allele by 454 and were detected as a minority allele in Sanger sequencing but excluded because of the minority allele cutoff for this method. Given the variability found in the results from the sensitivity analysis performed on laboratory strains for direct sequencing, the second explanation is more likely.
Despite several attempts and extensive troubleshooting, complete sequence data could not be generated for the cs repeat region, which seems to not be amenable to sequencing in filter paper samples using current 454 technology. Longer read lengths are required to get full coverage across this ∼450- to 550-base pair region. Although read lengths in this range are possible on the GS FLX Titanium 454 Platform, they are at the upper limit of what is routinely obtained. In addition, a known limitation of pyrosequencing is difficulty in reliably generating data on long repetitive DNA sequences because of nucleotide exhaustion resulting in premature termination of read synthesis (Tallon L, personal communication). It was initially thought that, because the cs gene repeats with 12 nucleotides are longer than many short tandem repeats, 454 could still be a viable option for this region. However, despite the longer length of the repeat, premature termination still occurred. Because diversity in the cs repeat region may be an important driver of allele-specific natural and vaccine-induced immunity, technology development efforts that will enable sequencing this region are warranted.
The results of these initial studies show that there is more extensive polymorphism in the regions of the cs gene coding for T-cell epitopes than has been previously described in this geographic region. Additional examination of polymorphism in CSP in vaccine trials and epidemiological studies may elucidate the contribution of CSP immunity to clinical protection against malaria and inform the development of improved CSP-based vaccines. As read lengths continue to improve and costs continue to decline, 454 and other next and third generation sequencing platforms may be better suited to handle the long repeats of the cs gene; therefore, important diversity in this important region can be examined. Until the entire cs gene, including the repeat regions that are thought to drive protective humoral immunity, can be fully sequenced in a high-throughput fashion, the role, if any, of allelic diversity in limiting the efficacy of RTS,S and other CSP-based vaccines will remain uncertain. With RTS,S continuing to show modest protective efficacy in a large phase 3 trial as it moves to licensure,13 the ability to sequence the full cs gene in thousands of filter paper samples could be a critical technical advance that would aid the possible improvement of this first partially successful malaria vaccine.
ACKNOWLEDGMENTS
We would like to thank Jacques Ravel of the Institute for Genome Sciences at the University of Maryland, Baltimore, MD, for providing barcodes used in this project.
Footnotes
Financial support: This research was supported by Contract N01AI85346 and Cooperative Agreement U19AI065683 from the National Institute of Allergy and Infectious Diseases (NIAID), Grant D43TW001589 from the Fogarty International Center, National Institutes of Health, and Contract W81XWH-06-1-0427 from the US Department of Defense and the US Agency for International Development. S.T.-H. is supported by the University of Maryland Multidisciplinary Clinical Research Career Development Program (National Institutes of Health Grant K12RR023250). C.V.P. is supported by a Distinguished Clinical Scientist Award from the Doris Duke Charitable Foundation and by the Howard Hughes Medical Institute.
Authors' addresses: Kavita Gandhi, Shannon Takala-Harrison, and Christopher V. Plowe, Howard Hughes Medical Institute/Center for Vaccine Development, University of Maryland, Baltimore, MD, E-mails: Kavita.Gandhi@som.umaryland.edu, stakala@medicine.umaryland.edu, and cplowe@medicine.umaryland.edu. Mahamadou A. Thera, Drissa Coulibaly, Karim Traoré, Ando B. Guindo, and Ogobara K. Doumbo, Malaria Research and Training Center, University of Bamako, Bamako, Mali. E-mails: mthera@icermali.org, coulibalyd@icermali.org, karim@icermali.org, ando@icermali.org, and okd@icermali.org.
References
- 1.Takala SL, Plowe CV. Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming ‘vaccine resistant malaria’. Parasite Immunol. 2009;31:560–573. doi: 10.1111/j.1365-3024.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escalante AA, Grebert HM, Isea R, Goldman IF, Basco L, Magris M, Biswas S, Kariuki S, Lal AA. A study of genetic diversity in the gene encoding the circumsporozoite protein (CSP) of Plasmodium falciparum from different transmission areas–XVI. Asembo Bay Cohort Project. Mol Biochem Parasitol. 2002;125:83–90. doi: 10.1016/s0166-6851(02)00216-5. [DOI] [PubMed] [Google Scholar]
- 3.Egan JE, Hoffman SL, Haynes JD, Sadoff JC, Schneider I, Grau GE, Hollingdale MR, Ballou WR, Gordon DM. Humoral immune responses in volunteers immunized with irradiated Plasmodium falciparum sporozoites. Am J Trop Med Hyg. 1993;49:166–173. doi: 10.4269/ajtmh.1993.49.166. [DOI] [PubMed] [Google Scholar]
- 4.Nardin EH, Nussenzweig RS, McGregor IA, Bryan JH. Antibodies to sporozoites: their frequent occurrence in individuals living in an area of hyperendemic malaria. Science. 1979;206:597–599. doi: 10.1126/science.386511. [DOI] [PubMed] [Google Scholar]
- 5.de Groot AS, Johnson AH, Maloy WL, Quakyi IA, Riley EM, Menon A, Banks SM, Berzofsky JA, Good MF. Human T cell recognition of polymorphic epitopes from malaria circumsporozoite protein. J Immunol. 1989;142:4000–4005. [PubMed] [Google Scholar]
- 6.Chenet SM, Branch OH, Escalante AA, Lucas CM, Bacon DJ. Genetic diversity of vaccine candidate antigens in Plasmodium falciparum isolates from the amazon basin of peru. Malar J. 2008;7:93. doi: 10.1186/1475-2875-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weedall GD, Preston BM, Thomas AW, Sutherland CJ, Conway DJ. Differential evidence of natural selection on two leading sporozoite stage malaria vaccine candidate antigens. Int J Parasitol. 2007;37:77–85. doi: 10.1016/j.ijpara.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Jalloh A, Jalloh M, Matsuoka H. T-cell epitope polymorphisms of the Plasmodium falciparum circumsporozoite protein among field isolates from sierra leone: age-dependent haplotype distribution? Malar J. 2009;8:120. doi: 10.1186/1475-2875-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bojang KA, Milligan PJ, Pinder M, Vigneron L, Alloueche A, Kester KE, Ballou WR, Conway DJ, Reece WH, Gothard P, Yamuah L, Delchambre M, Voss G, Greenwood BM, Hill A, McAdam KP, Tornieporth N, Cohen JD, Doherty T, RTS, S Malaria Vaccine Trial Team Efficacy of RTS,S/AS02 malaria vaccine against Plasmodium falciparum infection in semi-immune adult men in the Gambia: a randomised trial. Lancet. 2001;358:1927–1934. doi: 10.1016/S0140-6736(01)06957-4. [DOI] [PubMed] [Google Scholar]
- 10.Alonso PL, Sacarlal J, Aponte JJ, Leach A, Macete E, Milman J, Mandomando I, Spiessens B, Guinovart C, Espasa M, Bassat Q, Aide P, Ofori-Anyinam O, Navia MM, Corachan S, Ceuppens M, Dubois MC, Demoitié MA, Dubovsky F, Menéndez C, Tornieporth N, Ballou WR, Thompson R, Cohen J. Efficacy of the RTS,S/AS02A vaccine against Plasmodium falciparum infection and disease in young african children: randomised controlled trial. Lancet. 2004;364:1411–1420. doi: 10.1016/S0140-6736(04)17223-1. [DOI] [PubMed] [Google Scholar]
- 11.Aponte JJ, Aide P, Renom M, Mandomando I, Bassat Q, Sacarlal J, Manaca MN, Lafuente S, Barbosa A, Leach A, Lievens M, Vekemans J, Sigauque B, Dubois MC, Demoitié MA, Sillman M, Savarese B, McNeil JG, Macete E, Ballou WR, Cohen J, Alonso PL. Safety of the RTS,S/AS02D candidate malaria vaccine in infants living in a highly endemic area of mozambique: a double blind randomised controlled phase I/IIb trial. Lancet. 2007;370:1543–1551. doi: 10.1016/S0140-6736(07)61542-6. [DOI] [PubMed] [Google Scholar]
- 12.Bejon P, Lusingu J, Olotu A, Leach A, Lievens M, Vekemans J, Mshamu S, Lang T, Gould J, Dubois MC, Demoitié MA, Stallaert JF, Vansadia P, Carter T, Njuguna P, Awuondo KO, Malabeja A, Abdul O, Gesase S, Mturi N, Drakeley CJ, Savarese B, Villafana T, Ballou WR, Cohen J, Riley EM, Lemnge MM, Marsh K, von Seidlein L. Efficacy of RTS,S/AS01E vaccine against malaria in children 5 to 17 months of age. N Engl J Med. 2008;359:2521–2532. doi: 10.1056/NEJMoa0807381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agnandji ST, Lell B, Soulanoudjingar SS, Fernandes JF, Abossolo BP, Conzelmann C, Methogo BG, Doucka Y, Flamen A, Mordmüller B, Issifou S, Kremsner PG, Sacarlal J, Aide P, Lanaspa M, Aponte JJ, Nhamuave A, Quelhas D, Bassat Q, Mandjate S, Macete E, Alonso P, Abdulla S, Salim N, Juma O, Shomari M, Shubis K, Machera F, Hamad AS, Minja R, Mtoro A, Sykes A, Ahmed S, Urassa AM, Ali AM, Mwangoka G, Tanner M, Tinto H, D'Alessandro U, Sorgho H, Valea I, Tahita MC, Kaboré W, Ouédraogo S, Sandrine Y, Guiguemdé RT, Ouédraogo JB, Hamel MJ, Kariuki S, Odero C, Oneko M, Otieno K, Awino N, Omoto J, Williamson J, Muturi-Kioi V, Laserson KF, Slutsker L, Otieno W, Otieno L, Nekoye O, Gondi S, Otieno A, Ogutu B, Wasuna R, Owira V, Jones D, Onyango AA, Njuguna P, Chilengi R, Akoo P, Kerubo C, Gitaka J, Maingi C, Lang T, Olotu A, Tsofa B, Bejon P, Peshu N, Marsh K, Owusu-Agyei S, Asante KP, Osei-Kwakye K, Boahen O, Ayamba S, Kayan K, Owusu-Ofori R, Dosoo D, Asante I, Adjei G, Adjei G, Chandramohan D, Greenwood B, Lusingu J, Gesase S, Malabeja A, Abdul O, Kilavo H, Mahende C, Liheluka E, Lemnge M, Theander T, Drakeley C, Ansong D, Agbenyega T, Adjei S, Boateng HO, Rettig T, Bawa J, Sylverken J, Sambian D, Agyekum A, Owusu L, Martinson F, Hoffman I, Mvalo T, Kamthunzi P, Nkomo R, Msika A, Jumbe A, Chome N, Nyakuipa D, Chintedza J, Ballou WR, Bruls M, Cohen J, Guerra Y, Jongert E, Lapierre D, Leach A, Lievens M, Ofori-Anyinam O, Vekemans J, Carter T, Leboulleux D, Loucq C, Radford A, Savarese B, Schellenberg D, Sillman M, Vansadia P, RTS,S Clinical Trials Partnership First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N Engl J Med. 2011;365:1863–1875. doi: 10.1056/NEJMoa1102287. [DOI] [PubMed] [Google Scholar]
- 14.Alloueche A, Milligan P, Conway DJ, Pinder M, Bojang K, Doherty T, Tornieporth N, Cohen J, Greenwood BM. Protective efficacy of the RTS,S/AS02 Plasmodium falciparum malaria vaccine is not strain specific. Am J Trop Med Hyg. 2003;68:97–101. [PubMed] [Google Scholar]
- 15.Carr IM, Robinson JI, Dimitriou R, Markham AF, Morgan AW, Bonthron DT. Inferring relative proportions of DNA variants from sequencing electropherograms. Bioinformatics. 2009;25:3244–3250. doi: 10.1093/bioinformatics/btp583. [DOI] [PubMed] [Google Scholar]
- 16.Coulibaly D, Diallo DA, Thera MA, Dicko A, Guindo AB, Koné AK, Cissoko Y, Coulibaly S, Djimdé A, Lyke K, Doumbo OK, Plowe CV. Impact of preseason treatment on incidence of falciparum malaria and parasite density at a site for testing malaria vaccines in Bandiagara, Mali. Am J Trop Med Hyg. 2002;67:604–610. doi: 10.4269/ajtmh.2002.67.604. [DOI] [PubMed] [Google Scholar]
- 17.Williams R, Peisajovich SG, Miller OJ, Magdassi S, Tawfik DS, Griffiths AD. Amplification of complex gene libraries by emulsion PCR. Nat Methods. 2006;3:545–550. doi: 10.1038/nmeth896. [DOI] [PubMed] [Google Scholar]
- 18.Hunt P, Fawcett R, Carter R, Walliker D. Estimating SNP proportions in populations of malaria parasites by sequencing: validation and applications. Mol Biochem Parasitol. 2005;143:173–182. doi: 10.1016/j.molbiopara.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Ochola LI, Tetteh KK, Stewart LB, Riitho V, Marsh K, Conway DJ. Allele frequency-based and polymorphism-versus-divergence indices of balancing selection in a new filtered set of polymorphic genes in Plasmodium falciparum. Mol Biol Evol. 2010;27:2344–2351. doi: 10.1093/molbev/msq119. [DOI] [PMC free article] [PubMed] [Google Scholar]