Abstract
In the bacterial chemotaxis network, receptor clusters process input1–3, and flagellar motors generate output4. Receptor and motor complexes are coupled by the diffusible protein CheY-P. Receptor output (the steady-state concentration of CheY-P) varies from cell to cell5. However, the motor is ultrasensitive, with a narrow [CheY-P] operating range6. How might the match between receptor output and motor input be optimized? Here we show that the motor can shift its operating range by changing its composition. The number of FliM subunits in the C-ring increases in response to a decrement in the concentration of CheY-P, increasing motor sensitivity. This shift in sensitivity explains the slow partial adaptation observed in mutants that lack the receptor methyltransferase and methylesterase7–8 and why motors exhibit signal-dependent FliM turnover9. Adaptive remodelling is likely to be a common feature in the operation of many molecular machines.
The chemotaxis signalling pathway allows bacterial cells to sense and respond to changes in concentrations of chemical attractants or repellents1–2. Binding of chemicals by receptors modulates the activity of an associated histidine kinase, CheA, thereby changing the level of phosphorylation of the response regulator, CheY. CheY-P binds to FliM, a component of the switch complex at the base of the flagellar motor and modulates the direction of motor rotation. A phosphatase, CheZ, dephosphorylates CheY-P. The chemotaxis pathway is well known for its high gain8,10–11, wide dynamic range11–12, and robust adaptation5,13, mediated by receptor methylation and demethylation (by CheR and CheB).
The output of the chemotaxis pathway, the flagellar motor, is ultrasensitive to the intracellular concentration of CheY-P, with a Hill coefficient of about 10, imposing a narrow operational range for [CheY-P]6. While precise adaptation is a robust property of the chemotaxis pathway for certain attractants, e.g., aspartate, the steady-state concentration of CheY-P is not5. Given cell-to-cell variations in the concentration of CheY-P and the fact that different cells can maintain their chemotactic sensitivity14, it has been suggested that cells might have additional molecular mechanisms to adjust the CheY-P concentration around the operational value of ~3 μM6. One possibility is a feedback mechanism that allows a cell to adjust its kinase activity in response to motor output. This mechanism would increase the kinase activity if cells only ran, and would decrease the kinase activity if cells only tumbled. In earlier work, we looked for such a mechanism by monitoring the kinase activity with a FRET technique15 while jamming flagellar bundles with an anti-filament antibody. Stopping motors had no effect on kinase activity16.
Here, we report that the motor itself adapts, shifting its response function according to the steady-state concentration of CheY-P. It does this by increasing the complement of FliM when the concentration of CheY-P is low. Motor remodeling is well known for the stator elements MotA and MotB, which if defective, can be replaced by wild-type protein, as evidenced by stepwise increments in motor torque17–18. Such exchange also has been visualized by TIRF microscopy of GFP-labeled protein19, and a similar technique has been used to demonstrate FliM9 and FliN20 exchange in cells containing CheY-P. The present work addresses the functional consequences of FliM exchange. We studied cheR cheB cells, which are defective in methylation and demethylation, and monitored motor and kinase responses to step-addition of the non-metabolisable attractant α-methylaspartate (MeAsp), using bead21 and FRET15 assays. One cannot do these experiments with wild-type cells, because their adaptation to aspartate is robust, so that the steady-state concentration of CheY-P does not change. Motor adaptation occurs on a minute rather than on a second timescale and does not play a direct role in sensing temporal gradients. Instead, it helps to match the operating point of the motor to the output of the chemotaxis receptor complex, obviating the requirement for fine tuning of that output.
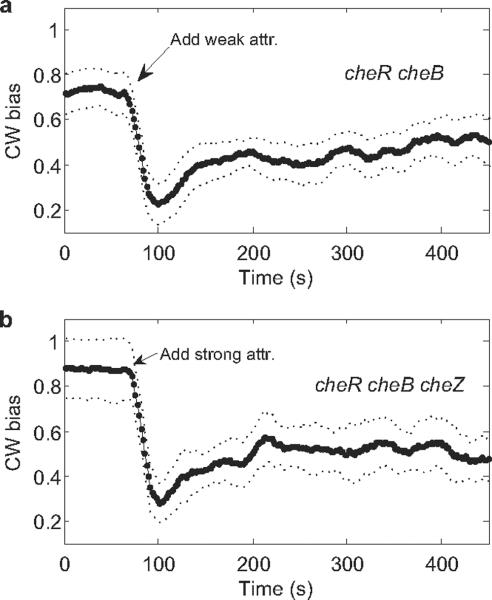
Using a bead assay, we found partial adaptation in cheR cheB cells within 1 min following the initial response, Fig. 1 a, which shows the averaged responses of 7 motors on different cells to stepwise addition of 1 mM MeAsp. These results are similar to those obtained previously with tethered cells7–8. A recent model suggests that partial adaptation might be due to dynamic localization of CheZ22. To test this hypothesis, we repeated the bead experiments using cheR cheB cheZ cells. The results were essentially the same, Fig. 1b, which shows the averaged responses of 4 motors on different cells of a cheR cheB cheZ strain to stepwise addition of 2 mM MeAsp + 0.5 mM L-serine (a stronger stimulus needed because of the lower sensitivity of cheZ strains). So CheZ is not required for this partial adaptation.
Fig. 1.
Motor responses to stepwise addition of chemical attractants monitored by the bead assay. The attractants were applied at the times indicated by the arrows. Error bounds for standard errors of the mean are shown as dotted lines. (a) Averaged responses of 7 cheR cheB cells (JY35 carrying pKAF131) to 1 mM MeAsp (weak attractant). (b) Averaged responses of 4 cheR cheB cheZ cells (JY32 carrying pVS7 and pKAF131) to 2 mM MeAsp + 0.5 mM L-serine (strong attractant).
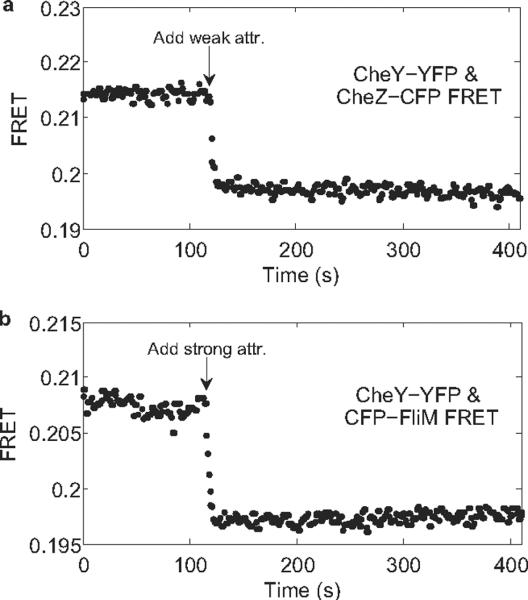
CheY-P concentrations were monitored by measuring FRET between CheZ-CFP and CheY-YFP. We measured responses in cheR cheB cells to stepwise addition of 1mM MeAsp, Fig. 2a. The response shown in Fig. 2a is similar to that obtained previously11. No adaptation is apparent. To rule out possible complications due to CheZ oligomerization23, we also measured CFP-FliM/CheY-YFP FRET24 in cheR cheB cells following stepwise addition of 2 mM MeAsp + 0.5 mM L-serine (a stronger stimulus needed because of the lower sensitivity of CFP-FliM/CheY-YFP FRET), as shown in Fig. 2b. No adaptation is apparent in either panel of Fig. 2, so the partial adaptation shown in Fig. 1 does not occur upstream of CheY-P. It must occur at the level of the flagellar motor.
Fig. 2.
FRET responses (Y/C ratio) of cheR cheB cells to stepwise addition of chemical attractants. The attractants were added at the times indicated by the arrows. (a) CheY-YFP/CheZ-CFP FRET responses to 1 mM MeAsp (weak attractant). (b) CheY-YFP/CFP-FliM FRET responses to 2 mM MeAsp + 0.5 mM L-serine (strong attractant).
Clockwise (CW) biases of motors were measured before addition of attractant, immediately after addition of attractant, and after time was allowed for partial adaptation. We focused on motors with pre-stimulus CW biases around 0.8 (ranging from 0.70 to 0.95). Due to cell-to-cell variation, the lowest biases following stimulation ranged from 0 to 0.75. The concentration of CheY-P in a given cell was estimated from its CW bias at the time of the lowest bias, using the response curve measured previously by Cluzel et al.6, shown by the red line in Fig. 3 (Hill coefficient NH=10.3, CheY-P dissociation constant K=3.1 μM). Then the CW bias found after that cell had adapted was plotted as a function of this concentration, as shown by the blue data points in Fig. 3. The measurements were carried out for 49 motors on different cheR cheB cells with stepwise addition of 0.5 or 1 mM MeAsp. Following adaptation, the relationship between the CW bias and the concentration of CheY-P shifted to lower concentrations of CheY-P, increasing motor sensitivity to CheY-P.
Fig. 3.
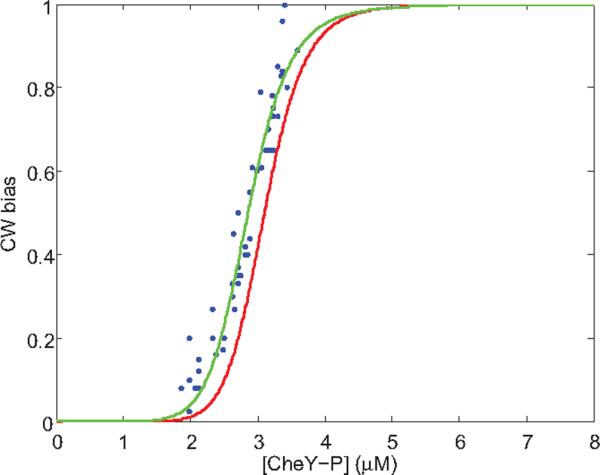
CW bias as a function of CheY-P concentration. The red curve is for pre-stimulus wild-type motors, as measured by Cluzel et al.6 and fit with an MWC model with 34 FliM units26. The blue dots are data for motors that have partially adapted to stepwise addition of attractant, with a two-parameter fit to the MWC model with Nav = 36 FliM units shown by the green curve; see text.
How the motor accomplishes this shift is intriguing. We sought to explain the shift in the motor response curve by using an MWC type model25–26, which has been used previously to explain the motor switching kinetics26. In this model, the C-ring is considered to be an allosteric switch, stochastically switching between two conformational states, CCW and CW, with N independent binding sites for CheY-P, corresponding to N units of FliM in the C-ring. The CW state has a higher affinity to CheY-P than the CCW state. The CW bias of the motor is given by: BCW=(1+[CheY-P]/K)N/((1+[CheY-P]/K)N+L(1+[CheY-P]/(KC))N), where L is the ratio of the probability that the motor is in the CCW state to the probability that it is in the CW state in the absence of CheY-P, K is the CheY-P dissociation constant for the CW state, and C is the ratio of dissociation constants for the CCW and CW states, respectively26. With reasonable values for the parameters, e.g., N=34, L=107, and K=3.1 μM6,27, the model can be fit to the ultrasensitivity data of Cluzel et al. with a best-fit value of C of 4.1, as shown by the red curve in Fig. 3. With these values for the parameters L, K, and C, and assuming that the number of FliM units N varies with [CheY-P], we can fit the data measured for the adapted motor using the MWC model with N = Nav + α([CheY-P] − 2.7), where N is written as a Taylor expansion about the average value of [CheY-P], 2.7 μM. We obtain a two-parameter fit with Nav = 36 and α = −1.2 μM−1, shown by the green curve in Fig. 3. The average value of N has increased from 34 to 36. Sensitivity of an MWC complex is known to increase with N for fixed values of L, K, and C28. Equivalently, the motor bias versus [CheY-P] curve shifts to smaller [CheY-P] with larger N as shown in Fig. 3. Intuitively, the fact that increasing the number of FliM units causes an increase in CW bias can be understood by considering the energetics of the switch. Each CheY-P binding decreases the energy level of CW state by a specific amount. With the values for parameters L, K and C fixed, increasing the number of FliM, i.e. CheY-P binding sites, increases the number of CheY-P bound to the motor. This decreases the energy level of the CW state, thereby increasing the CW bias.
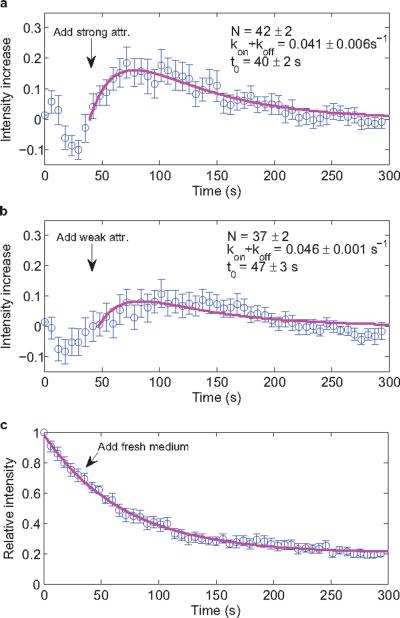
To directly test for this increase of the number of FliM units, we fused YFP to the C-terminus of FliM and monitored the fluorescence intensity of single motors using total internal reflection fluorescence (TIRF) microscopy. To minimize shifts in motor position, we tethered a cheR cheB strain that lacks the flagellar filament to glass via single hooks with anti-hook antibody. Changes in fluorescence were measured upon addition of 2 mM MeAsp + 0.5 mM L-serine, which should saturate the chemoreceptors and eliminate CheY-P, or of 1 mM MeAsp, which should simply reduce the concentration of CheY-P. The results are shown in Fig. 4. Fig. 4a is the averaged response of 20 motors to the strong attractant, and Fig. 4b is the averaged response of 22 motors to the weak attractant. In either case, the fluorescence intensity increased following the addition of attractant on a time scale consistent with partial adaptation, by a larger amount for the stronger attractant. We compensated for fluorescence bleaching by subtracting a control curve (Fig. 4c) and fitting the results to a model in which the FliM off rate decreases when the CheY-P concentration decreases; see Methods. Steady state is reached when Nkoff =(M− N)kon, where N is the number of FliM molecules in the motor, and M is the maximum number of FliM binding sites in the motor. The fits are shown in magenta and the final values for N are given in each panel (assuming an initial value of 34). These values agree with those presented in Fig. 3. So the motor increases the number of FliM units as it partially adapts to a decrement in the concentration of CheY-P. By doing so, it increases its dynamic range.
Fig. 4.
Changes in single-motor FliM-YFP fluorescence intensities in cheR cheB cells tethered by hooks and stimulated by addition of attractant (at time t0, arrow). The intensity for a given motor was normalized by its intensity at time 0, intensities for different motors were averaged, and the fit to the control of panel c was subtracted. Fits with the model are shown in magenta and the parameters for these fits are given. Error bars are standard errors of the mean. (a) Responses of 20 motors to addition of 2 mM MeAsp + 0.5 mM L-serine. (b) Responses of 22 motors to addition of 1 mM MeAsp. (c) Responses of 15 motors without addition of attractant, fit with an exponential decay function plus a constant.
We eliminated the concern that binding of CheY-P to FliM-YFP might be different than binding to wild-type FliM by using the bead assay to compare the biases, switching rates, and speeds for motors of cheR cheB cells expressing FliM-YFP or wild-type FliM: the biases were 61±16% or 58±19%, the switching rates were 3.8±1.2 or 3.4±1.0 s−1, and the speeds were 50.4±8.4 or 51.5±8.2 Hz, respectively.
The motor adaptation mechanism observed here is related to the turnover of motor C-ring components discovered recently 9,20, where exchange of FliM was found to be signal dependent and exhibited a similar timescale 9. The detailed mechanism should involve changes in FliM on/off rates dependent upon either CheY-P binding or rates of motor switching. As noted earlier, the timescale for motor adaptation (1 min) is much slower than that for receptor methylation/demethylation (1 s), which enables cells to make rapid temporal comparisons; thus, motor adaptation does not play a critical role in that aspect. Instead, it helps match the operating point of the motor to the output of the chemotaxis receptor complex.
Methods summary
All strains used in this study were derivatives of E. coli K12 strain RP437. Cells were grown at 33 °C in 10 ml T-broth supplemented with the appropriate antibiotics and inducers to an OD600 of 0.45 to 0.50. Cells were collected by centrifugation (10 min at 1300 × g), washed twice in 10 ml of motility medium (10 mM potassium phosphate/0.1 mM EDTA/1 μM methionine/10 mM lactic acid, pH 7.0), and resuspended in 10 ml of this medium. They were used immediately for experiments or stored at 4 °C for up to 2 h. All experiments were carried out with a custom-made flow chamber at room temperature.
For the bead assay, cells were sheared to truncate flagella, and 1.0-μm-diameter polystyrene latex beads were attached to the filament stubs. Rotation of the bead was monitored with a laser dark-field setup described previously 29. Rotational velocity as a function of time was determined for each motor and smoothed with a 25-point running average. CW bias was calculated over a 20-s interval every 2 s, leading to a plot of CW bias versus time.
FRET measurements of bacterial populations were carried out as described previously 30.
For TIRF measurements, cells were tethered to the bottom window of the flow chamber by single hooks using anti-FlgE antibody, following a protocol adapted from Yuan & Berg 21. The fluorescence intensity of the motors was monitored with a TIRF microscope (Nikon Eclipse Ti-U), and images were recorded with a back-illuminated, cooled (−55 °C), electron-multiplying CCD camera (DV887ECS-BV, Andor Technology). Image analysis of the motor spots was carried out using a Gaussian mask method described previously 19.
Methods
Strains and plasmids
All strains used in this study are derivatives of E. coli K12 strain RP437 31: JY32 [cheR cheB cheY cheZ fliC], JY35 [cheR cheB fliC], RP2893 [Δ2206(tap-cheZ)] 11, JY37 [cheR cheB cheY fliM], and JY40 [cheR cheB fliM fliC]. The fliM-eyfpA206K fusion with a 3× glycine linker was cloned into pTrc99A 32 under an IPTG-inducible promoter, yielding pRWB7. pDFB72 carrying wild-type fliM on pTrc99A was a gift from David Blair. pVS7 carrying wild-type cheY on a pBAD18-Kan 33 vector was a gift from Victor Sourjik. pVS18 carrying cheY-eyfp on pTrc99A, pVS31 carrying ecfp-fliM on pBAD33 33, and pVS54 carrying cheZ-ecfp on pBAD33, were described previously 11,24. pKAF131 carrying the sticky fliC allele under control of the native fliC promoter, was described previously 34. For studies of cheR cheB cells with the bead assay, JY35 carrying pKAF131 was used. For studies of cheR cheB cheZ cells with the bead assay, JY32 carrying pVS7 and pKAF131 was used. For CheZ-CFP/CheY-YFP FRET studies of cheR cheB cells, RP2893 carrying pVS18 and pVS54 was used. For CFP-FliM/CheY-YFP FRET studies of cheR cheB cells, JY37 carrying pVS18 and pVS31 was used. For the TIRF studies of single motors, JY40 carrying pRWB7 was used. For comparison of motors with wild-type FliM and FliM-YFP, JY40 carrying pDFB72 and pKAF131, and JY40 carrying pRWB7 and pKAF131 were used. Cells were grown at 33 °C in 10 ml T-broth (1% tryptone and 0.5% NaCl) supplemented with the appropriate antibiotics (Amp: 100 μg/ml, Kan: 50 μg/ml, Cam: 34 μg /ml) and inducers (0.005% arabinose for the bead assay, 0.01% arabinose and 50 μM IPTG for the FRET studies, 100 μM IPTG for the TIRF studies) to an OD600 of 0.45 to 0.50. Cells were collected by centrifugation (10 min at 1300 × g), washed twice in 10 ml of motility medium (10 mM potassium phosphate/0.1 mM EDTA/1 μM methionine/10 mM lactic acid, pH 7.0), and resuspended in 10 ml of this medium. They were used immediately for experiments or stored at 4 °C for up to 2 h.
The bead assay and data analysis
Cells were sheared to truncate flagella by passing 1 ml of the washed-cell suspension 50 times between two syringes equipped with 23-gauge needles and connected by a 7-cm length of polyethylene tubing (0.58 mm i.d., catalog no. 427411; Becton Dickinson, Waltham, MA). The sheared cell suspension was centrifuged and resuspended in 0.5 ml of motility medium. 50 μl of this suspension was placed on a glass coverslip coated with poly-L-lysine (0.01%, catalog no. P4707; Sigma, St. Louis, MO) and allowed to stand for 2 min, then 5 μl of 1.0-μm-diameter polystyrene latex beads (2.69%, catalog no. 07310; Polysciences, Warrington, PA) was added, mixed by gentle pipetting, and allowed to stand for 2 min. The coverslip was installed as the top window of a flow chamber 35 and rinsed with motility medium. The chamber was kept under a constant flow of buffer (400 μl/min) by a syringe pump (Harvard Apparatus, Holliston, MA). Rotation of the bead was monitored with a laser dark-field setup described previously 36. Outputs from the photomultiplier tubes were DC-coupled to an 8-pole low-pass Bessel filter (3384, Krohn-Hite) with a cutoff frequency of 200 Hz and sampled at 500 Hz using LabView. For each experiment, the rotation of the bead was monitored for 70 s, then the medium was switched to attractants, and the rotation was monitored further for about 400 s.
Rotational velocity as a function of time was determined for each motor as described previously 36 and smoothed with a 25-point running average. CW bias was calculated over a 20-s interval every 2 s, leading to a plot of CW bias versus time.
FRET measurements
FRET measurements of bacterial populations were carried out as described previously 15,30, except that the epifluorescent illumination was provided by a LED white light source (MCWHL2-C3, Thorlabs) through an excitation bandpass filter (FF01-438/24–25, Semrock). For each experiment, 4 ml of the washed-cell suspension was centrifuged and resuspended in 55 μl of motility medium, which was placed on a glass coverslip coated with poly-L-lysine (0.1% , catalog no. P8920, Sigma) and allowed to stand for 5 min. The coverslip was installed as the top window of a flow chamber 35 and rinsed with motility medium. The chamber was kept under a constant flow of buffer (500 μl/min). Epifluorescent emission was split into donor (cyan, C) and acceptor (yellow, Y) channels and collected by photon-counting photomultipliers (H7421-40, Hamamatsu). Signal intensities of these two channels were recorded by a computer running LabView, and the ratio between them (R=Y/C) provided an indicator of FRET activity. The FRET traces were smoothed with a median filter of rank 3.
TIRF measurements, data analysis, and fits to the model
Cells were tethered to glass by hooks using anti-FlgE antibody, following a protocol adapted from Yuan & Berg 21: 350 μl of the washed-cell suspension was centrifuged and resuspended in 100 μl of motility medium; 10 μl of anti-FlgE antibody (0.1 mg/ml) was added, and the mixture was incubated at 23 °C for 25 min. The antibody-treated cells were washed twice with 350 μl of motility medium and gently resuspended in 55 μl of motility medium. This cell suspension was placed on the bottom coverslip of a flow chamber (which was washed earlier with ethanol and dH2O and air-dried for 2 h) and allowed to stand for 15 min. The top coverslip of the flow chamber was then installed and rinsed with motility medium. The chamber was kept under a constant flow of buffer (100 μl/min). For each experiment, the flow was switched to attractants at 1 min before the start. It took about 1 min and 30 s for the attractant to reach the flow chamber and about 30 s to replace the medium, so effectively the medium reached the cells at between 30 s to 60 s after the start of each experiment. Only stably rotating and switching motors were monitored. The motors observed usually started with high CW bias; upon addition of strong attractant, they changed to exclusively CCW and remained 100% CCW throughout the observation time; upon addition of weak attractant, their CW bias reduced and later partially recovered; upon addition of motility medium, their CW bias did not change. The fluorescent intensity of the motors was monitored with a TIRF microscope (Nikon Eclipse Ti-U), and images were recorded at 65 nm/pixel with a back-illuminated, cooled (−55 °C), electron-multiplying CCD camera (DV887ECS-BV, Andor Technology). The camera was controlled by Andor Solis software running on a desktop computer. Image acquisition was under Andor Solis `kinetic' mode, with 200 ms exposures every 6 s for 50 exposures for each motor. The laser illumination was blocked in between exposures.
Images of the motor spots showed radially symmetric and approximately Gaussian intensity profiles. The width of these spots was about 5 pixels (325 nm). The fluorescent intensity centroid for each motor was calculated using a Gaussian mask method described previously19,37. Specifically, an initial estimate was made based on the peak pixel intensity, a 9×9 pixel region of interest (ROI) was defined centring on the initial motor centroid, and the motor centroid was calculated as follows: Firstly, a circular motor mask of diameter 300 nm was applied to the ROI centring on the current motor centroid. Secondly, pixel intensities within the motor mask were multiplied by a radially symmetric 2-dimensional Gaussian mask of fixed half-width 170 nm, and a revised estimate for the motor centroid was calculated using a weighted average. Lastly, the previous two steps were iterated either 150 times or until the motor mask began clipping the side of the ROI. We also calculated the motor centroid with a 2-dimensional Gaussian fitting, and both methods yielded comparable results. After the centroid was calculated, the background intensity was defined as the mean pixel intensity within the ROI but external to the final motor mask, and the motor intensity was calculated as the sum of all pixel intensities within the motor mask after subtraction of the background intensity from each pixel value.
The model assumes that CheY-P binding destabilizes FliM, so that when [CheY-P] suddenly decreases due to addition of attractant, koff (the off rate of each FliM unit) decreases, while kon remains the same. When the number of FliM units (N) in the C-ring reaches a new steady state, Nkoff=(M−N)kon, where M is the maximum number of FliM binding sites in a motor. The pre-stimulus N is assumed to be 34. During the response to the attractant step, the increment of N satisfies dn=((M−(n+34))kon−(n+34)koff)dt, while the increment of the normalized motor intensity satisfies df=dn/a−λfdt, where a is the normalization factor that converts the number of FliM units to fluorescence intensity, and λ is the fluorescence bleaching rate obtained by fitting the control curve (Fig. 4c). Solving these two differential equations with the initial conditions: n(0)=0, f(0)=0 leads to:
If the time of arrival of the attractant at the cell is t0 instead of 0, change t in the above equations to t−t0.
Acknowledgements
This work was supported by National Institutes of Health Grant AI016478.
Footnotes
Author Contributions J.Y. and H.C.B. planned the work and wrote the first draft of the paper. J.Y. performed the research with help on the MWC model and TIRF experiment and analysis from R.W.B. and on TIRF analysis from B.G.H..
Author Information Preprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Hazelbauer GL, Falke JJ, Parkinson JS. Baterial chemoreceptors: high - performance signaling in networked arrays. Trends Biochem. Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenfield D, et al. Self-organization of the Escherichia coli chemotaxis network Imaged with Super-Resolution Light Microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg HC. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 5.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 6.Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 7.Stock J, Kersulis G, Koshland DE., Jr. Neither methylating nor demthylating enzymes are required for bacterial chemotaxis. Cell. 1985;42:683–690. doi: 10.1016/0092-8674(85)90125-4. [DOI] [PubMed] [Google Scholar]
- 8.Segall JE, Block SM, Berg HC. Temporal comparisons in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 1986;83:8987–8991. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delalez NJ, et al. Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc. Natl. Acad. Sci. USA. 2010;107:11347–11351. doi: 10.1073/pnas.1000284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duke TAJ, Bray D. Heightened sensitivity of a lattice of membrane of receptors. Proc. Natl. Acad. Sci. USA. 1999;96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mello B, Tu Y. Effects of adaptation in maintaining high sensitivity over a wide dynamic range of backgrounds for E. coli chemotaxis. Biophys. J. 2007;92:2329–2337. doi: 10.1529/biophysj.106.097808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen CH, Endres RG, Wingreen NS. Chemotaxis in Escherichia coli: a molecular model for robust precise adaptation. PLoS Comp. Biol. 2008;4:e1. doi: 10.1371/journal.pcbi.0040001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin MD, Morton-Firth CJ, Abouhamad WN, Bourret RB, Bray D. Origins of individual swimming behavior in bacteria. Biophys. J. 1998;74:175–181. doi: 10.1016/S0006-3495(98)77777-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sourjik V, Vaknin A, Shimizu TS, Berg HC. In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol. 2007;423:363–391. doi: 10.1016/S0076-6879(07)23017-4. [DOI] [PubMed] [Google Scholar]
- 16.Shimizu TS, Delalez N, Pichler K, Berg HC. Monitoring bacterial chemotaxis by using bioluminescence resonance energy transfer: absence of feedback from the flagellar motors. Proc. Natl. Acad. Sci. USA. 2006;103:2093–2097. doi: 10.1073/pnas.0510958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blair DF, Berg HC. Restoration of torque in defective flagellar motors. Science. 1988;242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 18.Reid SW, et al. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. USA. 2006;103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leake MC, et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 20.Fukuoka H, Inoue Y, Terasawa S, Takahashi H, Ishijima A. Exchange of rotor components in functioning bacterial flagellar motor. Biochem. Biophys. Res. Commun. 2010;394:130–135. doi: 10.1016/j.bbrc.2010.02.129. [DOI] [PubMed] [Google Scholar]
- 21.Yuan J, Berg HC. Resurrection of the flagellar motor near zero load. Proc. Natl. Acad. Sci. USA. 2008;105:1182–1185. doi: 10.1073/pnas.0711539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipkow K. Changing cellular localization of CheZ predicted by molecular simulations. PLoS Comp. Biol. 2006;2:e39. doi: 10.1371/journal.pcbi.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blat Y, Eisenbach M. Oligomerization of the phosphatase CheZ upon interaction with the phosphorylated form of CheY, the signal protein of bacterial chemotaxis. J. Biol. Chem. 1996;271:1226–1231. doi: 10.1074/jbc.271.2.1226. [DOI] [PubMed] [Google Scholar]
- 24.Sourjik V, Berg HC. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA. 2002;99:12669–12674. doi: 10.1073/pnas.192463199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 26.Alon U, et al. Response regulator output in bacterial chemotaxis. EMBO J. 1998;17:4238–4248. doi: 10.1093/emboj/17.15.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner L, Caplan SR, Berg HC. Temperature induced switching of the bacterial flagellar motor. Biophys. J. 1996;71:2227–2233. doi: 10.1016/S0006-3495(96)79425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 29.Yuan J, Fahrner KA, Berg HC. Switching of the bacterial flagellar motor near zero load. J. Mol. Biol. 2009;390:394–400. doi: 10.1016/j.jmb.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu TS, Tu Y, Berg HC. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol. Syst. Biol. 2010;6:382. doi: 10.1038/msb.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkinson JS. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J. Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amann E, Ochs B, Abel K-J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 33.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan J, Fahrner KA, Turner L, Berg HC. Asymmetry in the clockwise and counter-clockwise rotation of the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA. 2010;107:12846–12949. doi: 10.1073/pnas.1007333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg HC, Block SM. A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J. Gen. Microbiol. 1984;130:2915–2920. doi: 10.1099/00221287-130-11-2915. [DOI] [PubMed] [Google Scholar]
- 36.Yuan J, Fahrner KA, Berg HC. Switching of the bacterial flagellar motor near zero load. J. Mol. Biol. 2009;390:394–400. doi: 10.1016/j.jmb.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson RR, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]