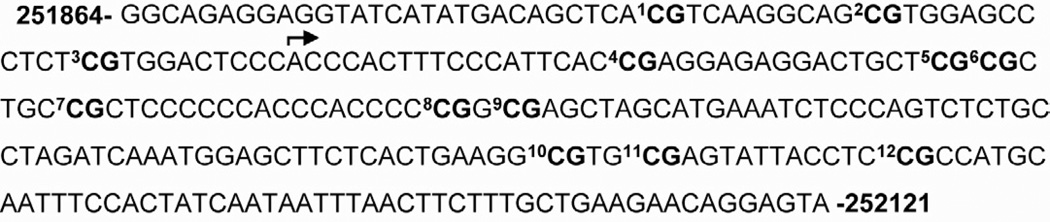Abstract
Epigenetic alterations of the brain-derived neurotrophic factor (Bdnf) gene have been linked with memory, stress, and neuropsychiatric disorders. Here we examined whether there was a link between an established rat model of post-traumatic stress disorder (PTSD) and BdnfDNA methylation. Adult male Sprague-Dawley rats were given psychosocial stress composed of two acute cat exposures in conjunction with 31 days of daily social instability. These manipulations have been shown previously to produce physiological and behavioral sequelae in rats that are comparable to symptoms observed in traumatized people with PTSD. We then assessed BdnfDNA methylation patterns (at exon IV) and gene expression. We have found here that the psychosocial stress regimen significantly increased BdnfDNA methylation in the dorsal hippocampus, with the most robust hypermethylation detected in the dorsal CA1 subregion. Conversely, the psychosocial stress regimen significantly decreased methylation in the ventral hippocampus (CA3). No changes in BdnfDNA methylation were detected in the medial prefrontal cortex or basolateral amygdala. In addition, there were decreased levels of BdnfmRNA in both the dorsal and ventral CA1. These results provide evidence that traumatic stress occurring in adulthood can induce CNS gene methylation, and specifically, support the hypothesis that epigenetic marking of the Bdnfgene may underlie hippocampal dysfunction in response to traumatic stress. Furthermore, this work provides support for the speculative notion that altered hippocampal BdnfDNA methylation is a cellular mechanism underlying the persistent cognitive deficits which are prominent features of the pathophysiology of PTSD.
Keywords: Animal model, post-traumatic stress disorder (PTSD), epigenetic, DNA methylation, brain-derived neurotrophic factor (Bdnf) gene, hippocampus
1. Introduction
Post-traumatic stress disorder (PTSD) is a debilitating anxiety disorder which develops in a subset of people after they experience emotional trauma (American Psychiatric Association, 1994). PTSD patients are commonly plagued by recurrent frightening thoughts and memories of the experience and suffer from a host of persistent physiological and behavioral sequelae that include altered sympathetic and hypothalamic-pituitary-adrenal (HPA) axis responsivity, chronic anxiety, exaggerated startle, and cognitive dysfunction (Johnsen & Asbjornsen, 2008; Mittal et al., 2001; Moore, 2009; Nemeroff et al., 2006; Yehuda & LeDoux, 2007). The biological basis of the development of PTSD is not fully understood, but the symptoms of PTSD have been linked to abnormalities in the functioning of the hippocampus, medial prefrontal cortex and amygdala (Bremner, 2007; Bremner et al., 2008; Heim & Nemeroff, 2009).
As PTSD shows moderate heritability, the etiology of PTSD has been hypothesized to be a product of complex gene-environment interplay (Afifi et al., 2010; Koenen et al., 2008; Yehuda & Bierer, 2009). One way traumatic events could affect genes is through DNA methylation, an epigenetic mechanism that produces functional changes in the genome without changes to the DNA sequence. Whereas DNA methylation has long been recognized for its role in establishing gene expression patterns during cellular development and differentiation, recent work has provided compelling evidence that DNA methylation remains an active process in the developing, as well as mature, CNS (Champagne & Curley, 2009; Graff & Mansuy, 2008; Roth & Sweatt, 2009; Zhang & Meaney, 2010). The susceptibility of the genome to epigenetic modifications provides a layer of genetic regulation that is potentially sensitive to a lifetime of experiential and environmental factors. Thus, an epigenetic explanation for environmental contributions to cognitive health and risk for neuropsychiatric disorders continues to gain traction (Costa et al., 2009; McGowan & Szyf, 2010; Roth et al., 2009; Tsankova et al., 2007).
The involvement of DNA methylation specifically in emotional trauma is suggested from a broad range of work on stress, memory and animal models of PTSD. For example, Chertkow-Deutsher et al. (2010) recently provided evidence of methylation of the Disks Large-Associated Protein (DIgap2) gene in the hippocampus of rats briefly exposed to predator-related stimuli. Related work has focused on the brain-derived neurotrophic factor (Bdnf) gene, which is seen as a central player in mediating the effects of stress on CNS function and psychopathology, including PTSD and major depressive disorder (Calabrese et al., 2009; Casey et al., 2009; Duman & Monteggia, 2006; Kauer-Sant’Anna et al., 2007; McEwen, 2008). For example, rats exhibiting a PTSD-like response to trauma have been shown to exhibit a significant down-regulation of BdnfmRNA in the hippocampus which correlated with PTSD-like behavioral stress responses (Kozlovsky et al., 2007). A recent study has documented changes in histone acetylation at individual Bdnf promoters following fear conditioning in animals previously subjected to a bout of prolonged stress (Takei et al., 2010). Moreover, a series of studies has demonstrated isoform-specific epigenetic marking, including methylation of Bdnf DNA, in rats exposed to either early developmental stress or fear conditioning in adulthood (Lubin et al., 2009; Roth et al., 2009). Finally, hippocampus-specific deletion of the Bdnfgene impaired learning, and more importantly, reduced extinction of conditioned fear (Heldt et al., 2007), two findings that resonate with cognitive and emotional abnormalities commonly observed in people with PTSD. Taken together, there is strong support for the hypothesis that an ideal genetic target linking clinical PTSD symptoms with animal models of PTSD is the Bdnfgene in the hippocampus.
We have therefore investigated whether there are changes in BdnfDNA methylation and mRNA expression in an animal model of PTSD which incorporates psychosocial factors known to contribute to the development and expression of PTSD in humans (Zoladz et al., 2008). Specifically, rats were immobilized while they were inescapably exposed to a cat, which elicits intense fear in rats (Blanchard et al., 1990, 1998; Hubbard et al., 2004) and inhibits hippocampal functioning and enhances amygdaloid activity (Adamec et al., 2005; Diamond et al., 1999, 2007; Mesches et al., 1999; Park et al., 2008; Vouimba et al., 2006), all of which are clinical features of PTSD (Bremner, 2007; Bremner et al., 2008). Cat exposure, in conjunction with immobilization, models the lack of control and sense of horror and helplessness during trauma that are commonly reported by PTSD patients (American Psychiatric Association, 1994). As a core symptom of PTSD is the repeated re-experiencing of the traumatic event, such that patients often feel as if the trauma is actually happening at the present as opposed to a mere recollection (Ehlers et al., 2004; Reynolds & Brewin, 1999), rats were given a second inescapable cat exposure episode 10 days after the first to provide them with a reminder of their traumatic experience. We acknowledge there is a difference in internal re-experiencing, as can be seen in many PTSD patients following a single traumatic exposure, and the actual re-experiencingcomponent that we use on rats.Thus,this aspect of our model may be most relevant to those people who develop PTSD as a result of multiple traumatic experiences.Finally, since a lack of social support and stability have been shown to contribute to PTSD (Andrews et al., 2003; Boscarino, 1995; Ullman & Filipas, 2001), housing conditions of rats were continually disturbed and randomized over the entire psychosocial stress period. We have shown that this combination of a life-threatening stressor, re-experiencing of the trauma, and chronic social instability produces a broad array of physiological and behavioral sequelae in rats, such as increases in cardiovascular and corticosteroid reactivity, increased anxiety, exaggerated startle, and cognitive impairments, all of which are remarkably similar to symptoms expressed by PTSD patients (Zoladz et al., 2008; Diamond & Zoladz, 2010a).
In summary, an extensive body of research has identified the hippocampus and Bdnfas vulnerable targets to traumatic stress in clinical and preclinical studies on PTSD. The current work extends this research to provide the first assessment of epigenetic modifications of the Bdnfgene in the hippocampus of adult rats in a well-established psychosocial stress animal model of PTSD.
2. Methods and Materials
2.1 Animals
Experimentally naïve adult male Sprague-Dawley rats (225–250 g upon delivery) obtained from Charles River laboratories (Wilmington, MA) were used for the present experiment. The rats were pair-housed on a 12-hr light/dark schedule (lights on at 0700) in standard Plexiglas cages (two per cage) with free access to food and water. The colony room temperature and humidity were maintained at 20±1°C and 60±3%, respectively. Upon arrival, all rats were given 1 week to acclimate to the housing room environment, as well as cage changing procedures, before any experimental manipulations took place. All procedures were approved by the Institutional Animal Care and Use Committee at the University of South Florida.
2.2 Stress manipulations
Following the 1-week acclimation phase, rats were randomly assigned to “stress” or “no stress” groups (n=10 rats/group). Rats in the stress group were immobilized in plastic DecapiCones (Braintree Scientific; Braintree, MA) and placed in a perforated wedge-shaped Plexiglas enclosure (Braintree Scientific; Braintree, MA; 20 × 20 × 8 cm). Then, the rats, still immobilized in the plastic DecapiCones within the Plexiglas enclosure, were taken to the cat housing room where they were placed in a metal cage (24 × 21 × 20 in) with an adult female cat for 1 hr. The Plexiglas enclosure prevented any contact between the cat and rats, but the rats were still exposed to all non-tactile sensory stimuli associated with the cat. Canned cat food was smeared on top of the Plexiglas enclosure to direct cat gustatory activity toward the rats. One hour later, the rats were returned to the laboratory. Rats in the no stress group remained in their home cages in the laboratory for the 1-hr stress period. Rats in the stress group were exposed to two of the acute stress sessions, separated by 10 days. As perceived control and predictability are important factors in PTSD development and expression (Gidron et al., 2004), we minimized the predictability of when cat exposure might occur with the first stress session taking place during the light cycle (between 0800–1300), and the second stress session taking place during the dark cycle (between 1900–2200).
Beginning on the day of the first stress session, rats in the stress groups were exposed to unstable housing conditions, which continued for the entire 31 days of psychosocial stress. Specifically, rats in the non-stress group were pair-housed with the same cage cohorts, and rats in the stress group were pair-housed with a different cohort on a daily basis. That is, the 10 rats in the stress group were pseudorandomly arranged in 5 different cage-mate pairs on a daily basis. Therefore, no rat in the stress group had the same cage-mate on two consecutive days during the entire 31-day stress period. Unstable housing was an important factor in producing the full PTSD-like behavioral and physiological profile (Zoladz et al., 2008; Diamond & Zoladz, 2010a).
2.3 Methylation analysis
On Day 32, rats were taken from the housing room, weighed and immediately decapitated. The rat brains were extracted from the skull, and the hippocampal subregions (dentate gyrus, CA3, CA1), medial prefrontal cortex, and basolateral amygdala were dissected on ice and frozen at −80°C. We also removed and weighed each rat’s adrenal glands and thymus. The frozen brain tissue was shipped to the University of Alabama at Birmingham where it was assayed by one of the authors (TLR) who was blind to the behavioral manipulations.
Bdnf gene expression is controlled by a complex regulatory region that in rodents includes at least nine promoters (Aid et al., 2007). Bdnf promoter IV is known to contribute significantly to environmentally-induced BDNF gene transcription, and the DNA methylation state of CpG sites within promoter IV has been shown to be correlated with gene expression during development (Dennis & Levitt, 2005), learning and memory (Aid et al., 2007; Bredy et al., 2007; Lubin et al., 2008), depression (Tsankova et al., 2006), early-life stress (Roth et al., 2009), and suicide (Keller et al., 2010). To this end, we focused our efforts on characterizing methylation of Bdnf exon IV.
DNA was extracted from the brain tissue using an AllPrep DNA/RNA kit (Qiagen). Methylation status was assessed via direct bisulfite DNA sequencing PCR (BSP) on bisulfite-modified DNA (Qiagen). Bisulfite-treated samples were amplified by primers that targeted a CpG-rich region within Bdnfexon IV (NW_047673.1: 251864–252121). The primer sequences were as follows: 5’-GGTAGAGGAGGTATTATATGATAGTTTA-3’ (forward) and 5’-TACTCCTATTCTTCAACAAAAAAATTAAAT-3’ (reverse). As depicted by Figure 1, the target amplicon spans the transcription start site, as well a cAMP response element site (TCACGTCA) for transcription factor CAMP response element binding protein (at CpG site 1).
Figure 1.
Schematic of target Bdnf exon IV amplicon and its 12 CG dinucleotides relative to the transcription initiation site (bent arrow).
PCR products were purified using a gel extraction kit (Qiagen), and sequenced using the reverse primer at the University of Alabama at Birmingham Genomics Core Facility of the Heflin Center for Human Genetics. The percent methylation of each CpG site within the region amplified was determined by the ratio between peaks values of G and A (G/[G+A]), and these levels on the electropherogram were determined using Chromas software. To confirm that direct bisulfite sequencing is adequately sensitive to detect methylation, universally unmethylated and methylated standards (EpigenDx) were run in parallel with samples and analyzed as described above (Table 1). Analysis of data generated from the standards indicated that the ratio of cytosine methylation increased proportionately with expected methylation rates (R2=0.947) and that there was small sample deviation (slope=0.963). Others have similarly shown the sensitivity of direct bisulfite sequencing (Jiang et al., 2009; Niculescu et al., 2006; Zhao et al., 2005).
Table 1.
Summary of results from methylated standardized control samples.
| Expected | Observed (mean ± SEM) |
|---|---|
| Standard, 0% Methylation | 3.75% ± 0.46 |
| Standard, 5% Methylation | 6.90% ± 1.36 |
| Standard, 10% Methylation | 13.91% ± 2.93 |
| Standard, 50% Methylation | 68.98% ± 2.74 |
| Standard, 75% Methylation | 83.83% ± 2.79 |
| Standard, 100% Methylation | 92.98% ± 0.27 |
Linear regression y=0.963x + 6.827, R2=0.947
2.4 mRNA analysis
RNA was extracted from the same tissue using an AllPrep DNA/RNA kit (Qiagen). Reverse transcription was performed using a cDNA synthesis kit (Qiagen). cDNA was amplified by real-time PCR (Bio-Rad iQ5 system) using Taqman probes (Applied Biosystems), with tubulin serving as a reference gene. Ct values were chosen within the linear range, and the comparative Ct method was used to calculate differences in gene expression between samples (Livak & Schmittgen, 2001; Pfaffl, 2001).
2.5 Statistics
Differences in BSP data were analyzed by analysis of variance (ANOVA) tests with Bonferroni’s post hoc tests. Differences in growth rate, adrenal gland weight, thymus weight and mRNA levels were analyzed by two-tailed, independent samples t-tests. Statistical significance was set to p ≤ 0.05.
3. Results
3.1 Growth rate and organ weights
Growth rates, expressed as grams per day (g/day), were calculated for all rats by dividing their total body weight gained during the course of the experiment by the total number of days in the experiment (i.e., 31 days). Rats in the stress group exhibited significantly lower growth rates (4.47 ± 0.22 g/day) than rats in the no stress group (5.65 ± 0.47 g/day), t = 2.29, p< 0.05. There was no significant difference between the adrenal gland weights of the two groups (raw weight: t = 0.14, p = 0.888; relative to body weight: t = 1.41, p = 0.176). The stress group did, however, exhibit significantly smaller thymus glands than the no stress group, particularly in terms of raw organ weight (raw weight: t = 2.78, p< 0.05; relative to body weight: t = 1.97, p = 0.065).
3.2 DNA methylation in hippocampus
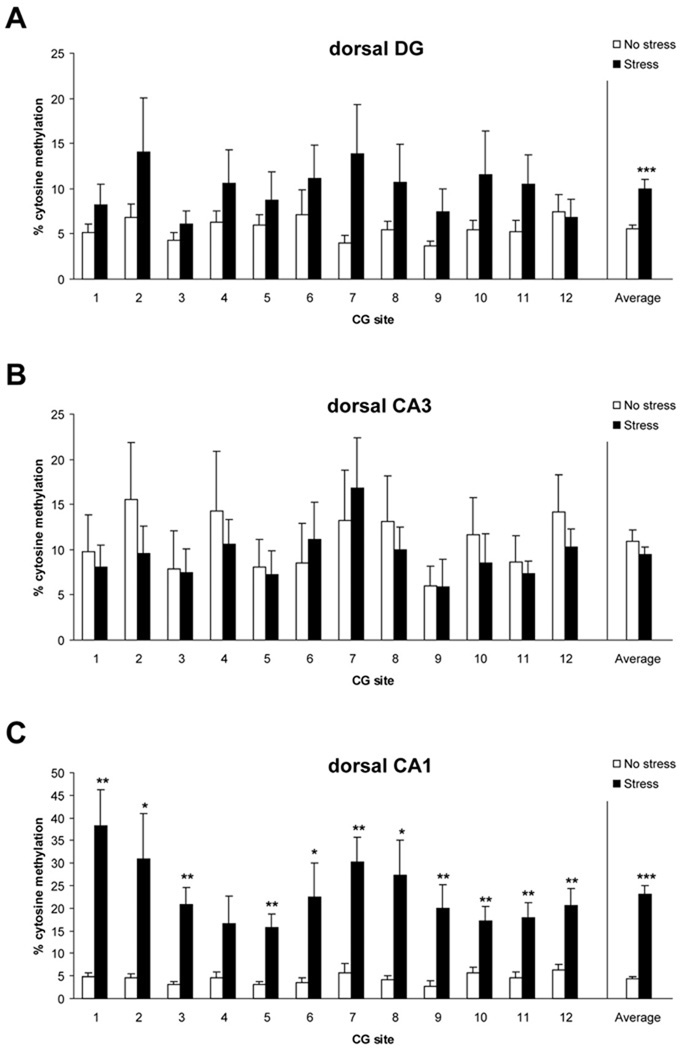
Studies have demonstrated that there are epigenetic changes in the hippocampus associated with stress and memory (Collins et al., 2009; Hunter et al., 2009; Lubin et al., 2008). We thus hypothesized that our PTSD model would likewise produce epigenetic changes in the hippocampus. First, we examined the methylation status of BdnfDNA in the dorsal hippocampus (Figure 2). We found a significant main effect of stress in the dorsal dentate gyrus, indicating that the psychosocial stress regimen significantly increased DNA methylation across the targeted exon IV region (Fig. 2A, F = 12.70, p< 0.001). While there was no significant effect detected for the dorsal CA3 subregion (Fig. 2B, p = 0.353), cytosine methylation within dorsal CA1 was strongly affected by psychosocial stress (Fig. 2C, F = 112.10, p< 0.0001).
Figure 2.
Levels of methylated Bdnfexon IV DNA in dorsal dentate (A), CA3 (B), and CA1 (C) in control (no stress) and stress-exposed rats. *** p< 0.001 vs. no stress controls. DG n=7–8/group, CA3 n=8/group, CA1 n=6/group. Data are mean ± SEM.
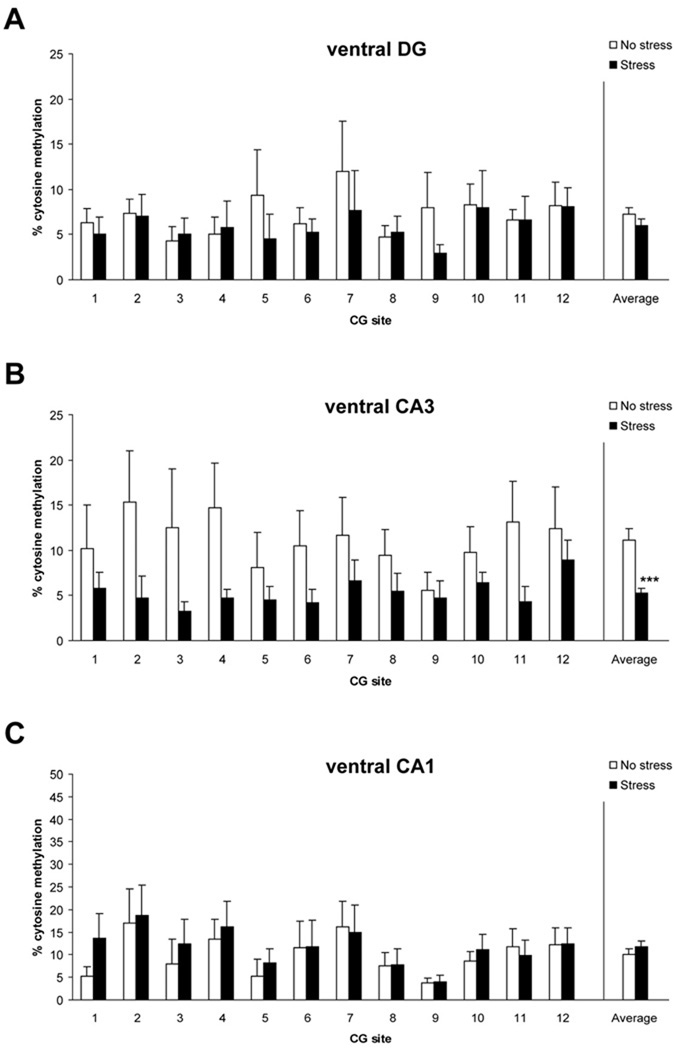
We also examined methylation within the ventral hippocampus (Figure 3). Average methylation levels in the ventral CA3 subregion were lower in the stress group, relative to the no stress group (Fig. 3B, F = 16.2, p< 0.0001). No effect was detected for the ventral DG (Fig. 3A, p = 0.267) or CA1 (Fig. 3C, p = 0.360). Overall, these results indicate that our psychosocial stress regimen differentially affected BdnfDNA methylation in the dorsal versus ventral hippocampus.
Figure 3.
Levels of methylated Bdnfexon IV DNA in ventral dentate (A), CA3 (B), and CA1 (C) in control (no stress) and stress-exposed rats. *** p< 0.001 vs. no stress controls. DG n=8/group, CA3 n=7–8/group, CA1 n=8/group. Data are mean ± SEM.
3.3 Gene expression in hippocampus
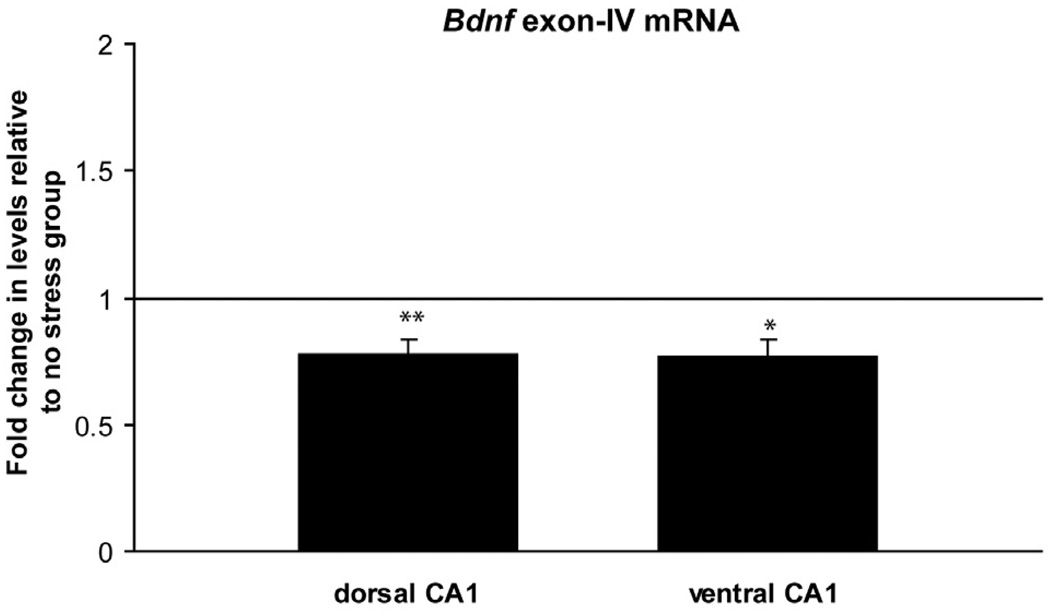
DNA methylation is a known regulator of gene expression. Since our methylation analysis suggested that area CA1 was one of the hippocampal subregions most strongly affected by the psychosocial stress regimen, we examined whether this area exhibited corresponding changes in BDNF gene expression. Bdnfexon-IV containing transcripts were significantly lower in the stress group in both dorsal (Fig. 4, t = 3.8, p< 0.01) and ventral CA1 (Fig. 4, t = 3.3, p< 0.05) relative to the non-stress group. These data support the idea that increased DNA methylation is one mechanism by which transcriptional suppression of the Bdnfgene following the psychosocial stress regimen is accomplished. The finding that BdnfmRNA was suppressed in the ventral CA1, as well, suggests that there are multiple levels of regulation of BdnfmRNA which extend beyond methylation of BdnfDNA.
Figure 4.
Exon IV mRNA levels in PTSD-like rats. Dorsal CA1, ** p< 0.01 vs. no stress controls, n=8/group. Ventral CA1, * p< 0.05 vs. no stress controls, n=8/group. Data are mean ± SEM.
3.4 DNA methylation in other brain regions
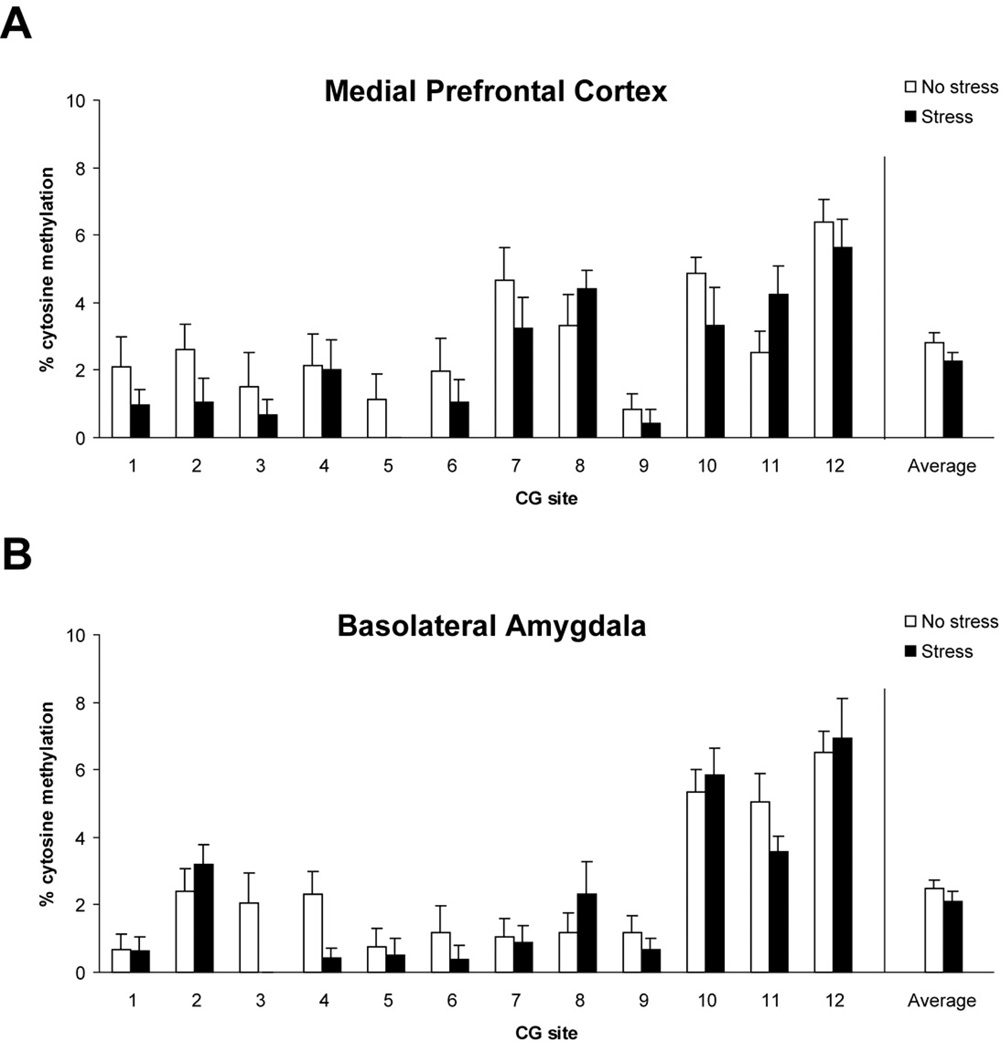
To examine the regional specificity of our epigenetic observations, we also examined whether DNA methylation in the psychosocial stress group was altered in two other PTSD-related brain regions that share connections with the hippocampus – the medial prefrontal cortex and basolateral amygdala. As shown in Figure 5, there was no significant effect of psychosocial stress on exon IV methylation in either the medial prefrontal cortex (panel A, p = 0.0610) or basolateral amygdala (panel B, p = 0.446). It is important to note that a recent study has shown opposing effects of BDNF within the infralimbic and prelimbic regions of the medical prefrontal cortex in regards to fear behavior (Choi et al., 2010). These regions were combined in our analysis, thus our lack of a significant finding in our medial prefrontal cortex could reflect such opposing effects.
Figure 5.
Levels of methylated Bdnfexon IV DNA in medial prefrontal cortex (A), and basolateral amygdala (B) in control (no stress) and stress-exposed rats. Medial prefrontal cortex n=9–10/group, basolateral amygdala n=10/group. Data are mean ± SEM.
4. Discussion
The role of neurotrophins, particularly BDNF, in the neurobiological mechanisms underlying stress-related and memory disorders has been widely emphasized in a broad range of clinical and preclinical research (Calabrese et al., 2009; Casey et al., 2009; Duman & Monteggia, 2006; Kauer-Sant’Anna et al., 2006; McEwen, 2008). In the present study, we explored the influence of a chronic psychosocial stress regimen which has been shown to produce PTSD-like effects in rats on methylation of the Bdnfgene. We first showed that psychosocial stress produced significant changes in hippocampal DNA methylation, most notably a robust hypermethylation within dorsal CA1. Second, we showed that this combination of acute cat exposures and social instability produced a significant down-regulation of BdnfmRNA in the dorsal and ventral CA1. Finally, we demonstrated regional specificity of the Bdnfepigenetic modifications, as no detectable changes were found in the medial prefrontal cortex or basolateral amygdala, two other PTSD-relevant brain regions.
These are the first findings we are aware of that document dramatic effects of traumatic stressors in adulthood on BdnfDNA methylation in the hippocampus. Likewise, our results are the first to demonstrate differential DNA methylation within specific hippocampal subfields in response to stress. These observations reinforce the view that DNA methylation remains an active process in the mature CNS that is modifiable by the environment, and indicate that changes in BdnfDNA methylation are an important aspect in potentially understanding how traumatic stress affects the hippocampus.
As this is the first study addressing this issue, we were cautious to obtain brain tissue while rats were in a baseline (non-stress) behavioral state. Any disturbance of the animals, such as conducting a behavioral or physiological test just prior to brain removal, could affect DNA methylation and make it difficult to interpret epigenetic changes specific to the PTSD manipulations as compared to those produced by behavioral testing. For example, exploration of a novel environment or contextual fear conditioning has been shown to evoke rapid and transient changes in DNA methylation in the hippocampus of behaving animals (Lubin et al., 2008; Penner et al., 2010).Therefore, in the current work brain tissue was obtained 3 weeks after the second cat exposure with minimal disturbance of the animals at the time of tissue harvesting. The findings we have described here suggest that the Bdnf gene was in a persistent hypermethylated (dorsal DG and CA1) and hypomethylated (ventral CA3) state under baseline conditions.
These documented epigenetic changes presumably reflect the combination of the psychosocial stress manipulations (cat exposure and unstable housing) the animals experienced. Whether the epigenetic changes we have found reflect general changes in baseline activity, or specific learning (such as fear conditioning) effects produced by the re-experiencing component of our model cannot be ruled out since methylation was examined after both exposures had occurred. We make this point to acknowledge that there may be a difference between epigenetic changes associated with internal re-experiencing following a single traumatic exposure, and those associated with an actual re-experiencing of the traumatic event, as used in our model. Although in this study our methylation findings cannot be linked to specific behavioral changes, it is important to note the strength of our model in reliably producing a broad range of PTSD-like effects in psychosocially stressed rats (Zoladz et al., 2008). Indeed, we have replicated the PTSD-like effects in other samples and have even shown the persistence of the behavioralchangesin animals tested 4 months after termination of the stress treatments (Diamond and Zoladz, 2010b; Zoladz et al., 2009). It is also important to note that in this study we do report significant changes in growth rate and organ weight of the stressed animals, an effect that we have previously documented in animals displaying PTSD-like effects (Zoladz et al., 2008).
Even with taking these limitations into account, our results are an important step in the direction of understanding the myriad of symptoms in PTSD, and suggest that such changes in BdnfDNA methylation specifically in the hippocampus could contribute to the persistent impairment of hippocampal functioning which can last for years after emotional trauma in PTSD patients (Johnsen & Asbjornsen, 2008; Mittal et al., 2001; Moore, 2009). In related work, we have shown that shock avoidance conditioning in rats can exert a powerful intrusive effect on new hippocampal memory processing as much as 1 year after the original traumatic experience (Zoladz et al., 2010). Hence, the basis of such long-lasting adverse effects of emotional trauma on new memory processing in people may involve hippocampus-specific DNA methylation changes of the Bdnfgene.
The current findings resonate with electrophysiological studies of stress and hippocampal functioning. Our work here indicates the dorsal CA1 region as the primary site of cognitive-related processing which is most vulnerable to psychosocial stress-induced hypermethylation of BdnfDNA. CA1 is also the most sensitive area of the hippocampus to exhibit a suppression of synaptic plasticity in response to stress. That is, studies of long-term potentiation (LTP), the primary physiological model of memory, have demonstrated that prolonged stress reliably suppresses the induction of LTP in CA1, with less consistent findings in other hippocampal regions (Diamond et al., 2007; Joels, 2008; Kim & Diamond, 2002).
Our results of altered Bdnfgene expression are in line with findings from other laboratories. Previous work using various animal models of stress (such as predator exposure, restraint, and animal isolation) has shown that stress-induced changes in hippocampal function and structure are typically associated with decreased BDNF activity, including down-regulation of BdnfmRNA and protein in hippocampal subregion CA1 (Bazak et al., 2009; Lippman et al., 2007; Nair et al., 2007; Nibuya et al., 1999; Rasmusson et al., 2002; Tsankova et al., 2006). Of particular relevance are the observations by Kozolovsky et al. (2007) who documented a down-regulation of BdnfmRNA in the CA1 region of animals exhibiting PTSD-like behavioral stress responses. Furthermore, clinical studies have also demonstrated that BDNF serum levels are altered in PTSD patients (Dell’Osso et al., 2009; Grassi-Oliveira et al., 2008; Hauck et al., 2009, 2010). Together, research efforts have made it clear that the hippocampal Bdnfgene is particularly susceptible to modulation by stress.
The mechanisms by which traumatic stress can affect the Bdnfgene are not currently understood. Here we examined whether there is a link between traumatic stress and alterations of BdnfDNA methylation. The predominant view in the literature is that methylation of DNA is associated with suppression of gene transcription, and in many cases extensive DNA methylation triggers complete silencing of the gene (Bird, 2002; Miranda & Jones, 2007). In this study, we show that PTSD-like rats have significant alterations in methylation of Bdnfexon IV DNA in their hippocampus, most notably increased methylation within CA1 that corresponded with decreased Bdnfgene expression. Increased cytosine methylation of exon IV has been shown to negatively regulate Bdnfgene expression (Aid et al., 2007; Chen et al., 2003; Martinowich et al., 2003), though there is evidence that decreased methylation could also be associated with decreased gene expression (Chahrour et al., 2008; Yasui et al., 2007). Our data are consistent with the likelihood that stress-evoked methylation of exon IV DNA (and perhaps even demethylation) contributed to transcriptional suppression of the Bdnfgene.
Our results also document that traumatic stress differentially affected Bdnfgene methylation within dorsal and ventral hippocampus. Specifically, we found that the predator-based psychosocial stress regimen evoked an increase in methylation in the dorsal hippocampus (dentate gyrus and CA1), but a decrease in the ventral hippocampus (CA3). These differences in methylation status of DNA in the two regions may be associated with the distinct functions of the dorsal versus ventral regions of the hippocampus. The dorsal hippocampus performs more cognitive-related functions, while the ventral hippocampus is activated more by strong emotions, such as fear and stress, and appears to be involved in mood disorders (Fanselow & Dong, 2010; Kjelstrup et al., 2002; Segal et al., 2010). This distinction between the dorsal and ventral hippocampal regions has been described at an electrophysiological level, with stress or corticosterone impairing LTP in the dorsal CA1 and enhancing LTP in the ventral CA1 (Maggio & Segal, 2009a,2009b).
Taken together, our findings advance our understanding of the functional distinction between the dorsal and ventral hippocampus by showing region-specific patterns of BdnfDNA methylation in the hippocampus. These observations may provide insight into the molecular mechanisms that maintain the memory of the traumatic experience for prolonged periods of time, thereby contributing to the cognitive impairments and increased anxiety that occur in our animal model (Zoladz et al., 2008), as well as in clinical PTSD (Johnsen & Asbjornsen, 2008; Mittal et al., 2001; Moore, 2009).
In summary, we have provided novel evidence linking epigenetic marking of BdnfDNA with a psychosocial stress model of PTSD. The issue remains as to whether these epigenetic alterations are causally related to the pathogenesis or maintenance of PTSD in humans. These findings provide us with important insight into the complex gene-by-environment interactions that are likely responsible for PTSD. Elucidation of the precise epigenetic mechanisms contributing to stress-induced brain pathology may contribute to useful therapeutic tools to ameliorate PTSD symptoms.
Acknowledgments
Support: Merit Review and Career Scientist Awards from the Veterans Affairs Department to DMD;National Alliance for Research on Schizophrenia and Depression and Civitan InternationalAwards to TLR;National Institutes of Health, the Rotary Clubs CART fund, and the Evelyn F. McKnight Brain Research Foundation to JDS
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamec RE, Blundell J, Burton P. Neural circuit changes mediating lasting brain and behavioral response to predator stress. Neuroscience & Biobehavioral Reviews. 2005;29:1225–1241. doi: 10.1016/j.neubiorev.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Afifi TO, Asmundson GJG, Taylor S, Jang KL. The role of genes and environment on trauma exposure and posttraumatic stress disorder symptoms: A review of twin studies. Clinical Psychology Review. 2010;30:101–112. doi: 10.1016/j.cpr.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNFgene structure and expression revisited. Journal of Neuroscience Research. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagonistic and statistical manual of mental disorders: DSM-IV. Washington DC: American Psychiatric Association; 1994. [Google Scholar]
- Andrews B, Brewin CR, Rose S. Gender, social support, and PTSD in victims of violent crime. Journal of Traumatic Stress. 2003;16:421–427. doi: 10.1023/A:1024478305142. [DOI] [PubMed] [Google Scholar]
- Bazak N, Kozlovsky N, Kaplan Z, et al. Pre-pubertal stress exposure affects adult behavioral response in association with changes in circulating corticosterone and brain-derived neurotrophic factor. Psychoneuroendocrinology. 2009;34:844–858. doi: 10.1016/j.psyneuen.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes &Development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Blanchard RJ, Blanchard DC, Rodgers J, Weiss SM. The characterization and modeling of antipredator defensive behavior. Neuroscience Biobehavioral Reviews. 1990;14:463–472. doi: 10.1016/s0149-7634(05)80069-7. [DOI] [PubMed] [Google Scholar]
- Blanchard RJ, Nikulina JN, Sakai RR, McKittrick C, McEwen B, Blanchard DC. Behavioral and endocrine change following chronic predatory stress. Physiology & Behaviour. 1998;63:561–569. doi: 10.1016/s0031-9384(97)00508-8. [DOI] [PubMed] [Google Scholar]
- Boscarino JA. Post-traumatic stress and associated disorders among Vietnam veterans: the significance of combat exposure and social support. Journal of Traumatic Stress. 1995;8:317–336. doi: 10.1007/BF02109567. [DOI] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learning and Memory. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner JD. Neuroimaging in posttraumatic stress disorder and other stress-related disorders. Neuroimaging Clinics of North America. 2007;17:523–538. doi: 10.1016/j.nic.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner JD, Elzinga B, Schmahl C, Vermetten EE. Structural and functional plasticity of the human brain in posttraumatic stress disorder. Progress in Brain Research. 2008;167:171–186. doi: 10.1016/S0079-6123(07)67012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese F, Molteni R, Racagni G, Riva MA. Neuronal plasticity: A link between stress and mood disorders. Psychoneuroendocrinology. 2009;34:S208–S216. doi: 10.1016/j.psyneuen.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Casey BJ, Glatt CE, Tottenham N, et al. Brain-derived neurotrophic factor as a model system for examining gene by environment interactions across development. Neuroscience. 2009;164:108–120. doi: 10.1016/j.neuroscience.2009.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA, Curley JP. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neuroscience & Biobehavioral Reviews. 2009;33:593–600. doi: 10.1016/j.neubiorev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, et al. Depression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Chertkow-Deutsher Y, Cohen H, Klein E, Ben-Shachar D. DNA methylation in vulnerability to post-traumatic stress in rats: evidence for the role of the post-synaptic density protein Dlgap2. The International Journal of Neuropsychopharmacology. 2010;13:347–359. doi: 10.1017/S146114570999071X. [DOI] [PubMed] [Google Scholar]
- Choi DC, Maguschak KA, Ye K, Jang SW, Myers KM, Ressler KJ. Prelimbic cortical BDNF is required for memory of learned fear but not extinction or innate fear. Proceeings National Academy of Science. 2010;107:2675–2680. doi: 10.1073/pnas.0909359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Hill LE, Chandramohan Y, Whitcomb D, Droste SK, Reul JMHM. Exercise improves cognitive responses to psychological stress through enhancement of epigenetic mechanisms and gene expression in the dentate gyrus. PLoS ONE. 2009;4:e4330. doi: 10.1371/journal.pone.0004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E, Chen Y, Dong E, et al. GABAergic promoter hypermethylation as a model to study the neurochemistry of schizophrenia vulnerability. Expert Review of Neurotherapeutics. 2009;9:87–98. doi: 10.1586/14737175.9.1.87. [DOI] [PubMed] [Google Scholar]
- Dell'Osso L, Carmassi C, Del Debbio A, et al. Brain-derived neurotrophic factor plasma levels in patients suffering from post-traumatic stress disorder. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2009;33:899–902. doi: 10.1016/j.pnpbp.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Dennis KE, Levitt P. Regional expression of brain derived neurotrophic factor (BDNF) is correlated with dynamic patterns of promoter methylation in the developing mouse forebrain. Molecular Brain Research. 2005;140:1–9. doi: 10.1016/j.molbrainres.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Diamond DM, Campbell AM, Park CR, Halonen J, Zoladz PR. The temporal dynamics model of emotional processing: a synthesis on the neurobiological basis of stress-induced amnesia, flashbulb and traumatic memories, and the Yerkes-Dodson law. Neural Plasticity. 2007:60803. doi: 10.1155/2007/60803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond DM, Park CR, Heman KL, Rose GM. Exposing rats to a predator impairs spatial working memory in the radial arm water maze. Hippocampus. 1999;9:542–552. doi: 10.1002/(SICI)1098-1063(1999)9:5<542::AID-HIPO8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Diamond DM, Zoladz PR. An animal model of PTSD which integrates inescapable predator exposure and social instability. Culture Psy Neurosciences. 2010a;15:6–7. [Google Scholar]
- Diamond DM, Zoladz PR. Chronic psychosocial stress in adult rats produces PTSD-like sequelae, including heightened anxiety and predator-based fear conditioning, 4 months following the initiation of stress Program No. 265.12. 2010 Neuroscience Meeting Planner; San Diego, CA. Society for Neuroscience; 2010b. Online. [Google Scholar]
- Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biological Psychiatry. 2006;59:1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Ehlers A, Hackmann A, Michael T. Intrusive re-experiencing in post-traumatic stress disorder: Phenomenology, theory, and therapy. Memory. 2004;12:403–415. doi: 10.1080/09658210444000025. [DOI] [PubMed] [Google Scholar]
- Fanselow MS, Dong H-W. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;65:7–19. doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidron Y, Kaplan Y, Velt A, Shalem R. Prevalence and moderators of terror-related post-traumatic stress disorder symptoms in Israeli citizens. Israel Medical Association Journal. 2004;6:387–391. [PubMed] [Google Scholar]
- Graff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behavioural Brain Research. 2008;192:70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Grassi-Oliveira R, Stein LM, Lopes RP, Teixeira AL, Bauer ME. Low plasma brain-derived neurotrophic factor and childhood physical neglect are associated with verbal memory impairment in major depression--a preliminary report. Biological Psychiatry. 2008;64:281–285. doi: 10.1016/j.biopsych.2008.02.023. [DOI] [PubMed] [Google Scholar]
- Hauck S, Gomes F, Silveira ÉdM, Júnior, Almeida E, Possa M, Ceitlin LHF. Serum levels of brain-derived neurotrophic factor in acute and posttraumatic stress disorder: a case report study. Revista Brasileira de Psiquiatria. 2009;31:48–51. doi: 10.1590/s1516-44462009000100012. [DOI] [PubMed] [Google Scholar]
- Hauck S, Kapczinski F, Roesler R, et al. Serum brain-derived neurotrophic factor in patients with trauma psychopathology. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2010;34:459–462. doi: 10.1016/j.pnpbp.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Heim C, Nemeroff CB. Neurobiology of posttraumatic stress disorder. CNS Spectrums. 2009;14:13–24. [PubMed] [Google Scholar]
- Heldt SA, Stanek L, Chhatwal JP, Ressler KJ. Hippocampus-specific deletion of BDNF in adult mice impairs spatial memory and extinction of aversive memories. Molecular Psychiatry. 2007;12:656–670. doi: 10.1038/sj.mp.4001957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard DT, Blanchard DC, Yang M, et al. Development of defensive behavior and conditioning to cat odor in the rat. Physiology & behavior. 2004;80:525–530. doi: 10.1016/j.physbeh.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proceedings of the National Academy of Sciences. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Zhang Y, Fei J, et al. Rapid quantification of DNA methylation by measuring relative peak heights in direct bisulfite-PCR sequencing traces. Laboratory Investigation. 2009;90:282–290. doi: 10.1038/labinvest.2009.132. [DOI] [PubMed] [Google Scholar]
- Joëls M. Functional actions of corticosteroids in the hippocampus. European Journal of Pharmacology. 2008;583:312–321. doi: 10.1016/j.ejphar.2007.11.064. [DOI] [PubMed] [Google Scholar]
- Johnsen GE, Asbjørnsen AE. Consistent impaired verbal memory in PTSD: A meta-analysis. Journal of Affective Disorders. 2008;111:74–82. doi: 10.1016/j.jad.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Kauer-Sant'Anna M, Tramontina J, Andreazza AC, et al. Traumatic life events in bipolar disorder: impact on BDNF levels and psychopathology. Bipolar Disorders. 2007;9:128–135. doi: 10.1111/j.1399-5618.2007.00478.x. [DOI] [PubMed] [Google Scholar]
- Keller S, Sarchiapone M, Zarrilli F, et al. Increased BDNF promoter methylation in the wernicke area of suicide subjects. Archives of General Psychiatry. 2010;67:258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Diamond DM. The stressed hippocampus, synaptic plasticity and lost memories. Nature Reviews Neuroscience. 2002;3:453–462. doi: 10.1038/nrn849. [DOI] [PubMed] [Google Scholar]
- Kjelstrup KG, Tuvnes FA, Steffenach H-A, Murison R, Moser EI, Moser M-B. Reduced fear expression after lesions of the ventral hippocampus. Proceedings of the National Academy of Sciences. 2002;99:10825–10830. doi: 10.1073/pnas.152112399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen K, Nugent N, Amstadter A. Gene-environment interaction in posttraumatic stress disorder. European Archives of Psychiatry and Clinical Neuroscience. 2008;258:82–96. doi: 10.1007/s00406-007-0787-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlovsky N, Matar MA, Kaplan Z, Kotler M, Zohar J, Cohen H. Long-term down-regulation of BDNF mRNA in rat hippocampal CA1 subregion correlates with PTSD-like behavioural stress response. The International Journal of Neuropsychopharmacology. 2007;10:741–758. doi: 10.1017/S1461145707007560. [DOI] [PubMed] [Google Scholar]
- Lippmann M, Bress A, Nemeroff CB, Plotsky PM, Monteggia LM. Long-term behavioural and molecular alterations associated with maternal separation in rats. European Journal of Neuroscience. 2007;25:3091–3098. doi: 10.1111/j.1460-9568.2007.05522.x. [DOI] [PubMed] [Google Scholar]
- Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of bdnf gene transcription in the consolidation of fear memory. Journal of Neuroscience. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio N, Segal M. Differential corticosteroid modulation of inhibitory synaptic currents in the dorsal and ventral hippocampus. Journal of Neuroscience. 2009;29:2857–2866. doi: 10.1523/JNEUROSCI.4399-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio N, Segal M. Differential modulation of long-term depression by acute stress in the rat dorsal and ventral hippocampus. Journal of Neuroscience. 2009;29:8633–8638. doi: 10.1523/JNEUROSCI.1901-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, et al. DNA methylation-related chromatin remodeling in activity-dependent Bdnf gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Central effects of stress hormones in health and disease: Understanding the protective and damaging effects of stress and stress mediators. European Journal of Pharmacology. 2008;583:174–185. doi: 10.1016/j.ejphar.2007.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Szyf M. The epigenetics of social adversity in early life: Implications for mental health outcomes. Neurobiology of Disease. 2010 doi: 10.1016/j.nbd.2009.12.026. In Press, Uncorrected Proof. [DOI] [PubMed] [Google Scholar]
- Mesches MH, Fleshner M, Heman KL, Rose GM, Diamond DM. Exposing rats to a predator blocks primed burst potentiation in the hippocampus in vitro. Journal of Neuroscience. 1999;19:RC18. doi: 10.1523/JNEUROSCI.19-14-j0004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda TB, Jones PA. DNA methylation: The nuts and bolts of repression. Journal of Cellular Physiology. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- Mittal D, Torres R, Abashidze A, Jimerson N. Worsening of post-traumatic stress disorder symptoms with cognitive decline: case series. Journal of Geriatric Psychiatry and Neurology. 2001;14:17–20. doi: 10.1177/089198870101400105. [DOI] [PubMed] [Google Scholar]
- Moore SA. Cognitive abnormalities in posttraumatic stress disorder. Current Opinion in Psychiatry. 2009;22:19–24. doi: 10.1097/YCO.0b013e328314e3bb. [DOI] [PubMed] [Google Scholar]
- Nair A, Vadodaria KC, Banerjee SB, et al. Stressor-specific regulation of distinct brain-derived neurotrophic factor transcripts and cyclic AMP response element-binding protein expression in the postnatal and adult rat hippocampus. Neuropsychopharmacology. 2007;32:1504–1519. doi: 10.1038/sj.npp.1301276. [DOI] [PubMed] [Google Scholar]
- Nemeroff CB, Bremner JD, Foa EB, Mayberg HS, North CS, Stein MB. Posttraumatic stress disorder: A state-of-the-science review. Journal of Psychiatric Research. 2006;40:1–21. doi: 10.1016/j.jpsychires.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Nibuya M, Takahashi M, Russell DS, Duman RS. Repeated stress increases catalytic TrkB mRNA in rat hippocampus. Neuroscience Letters. 1999;267:81–84. doi: 10.1016/s0304-3940(99)00335-3. [DOI] [PubMed] [Google Scholar]
- Niculescu MD, Craciunescu CN, Zeisel SH. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. The Journal of the Federation of American Societies for Experimental Biology. 2006;20:43–49. doi: 10.1096/fj.05-4707com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CR, Zoladz PR, Conrad CD, Fleshner M, Diamond DM. Acute predator stress impairs the consolidation and retrieval of hippocampus-dependent memory in male and female rats. Learning and Memory. 2008;15:271–280. doi: 10.1101/lm.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiology of Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.01.009. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson AM, Shi L, Duman R. Downregulation of BDNF mRNA in the hippocampal dentate gyrus after re-exposure to cues previously associated with footshock. Neuropsychopharmacology. 2002;27:133–142. doi: 10.1016/S0893-133X(02)00286-5. [DOI] [PubMed] [Google Scholar]
- Reynolds M, Brewin CR. Intrusive memories in depression and posttraumatic stress disorder. Behaviour Research and Therapy. 1999;37:201–215. doi: 10.1016/s0005-7967(98)00132-6. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biological Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Sodhi M, Kleinman JE. Epigenetic mechanisms in schizophrenia. Biochimica et Biophysica Acta (BBA) - General Subjects. 2009;1790:869–877. doi: 10.1016/j.bbagen.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Sweatt JD. Regulation of chromatin structure in memory formation. Current Opinion in Neurobiology. 2009;19:336–342. doi: 10.1016/j.conb.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M, Richter-Levin G, Maggio N. Stress-induced dynamic routing of hippocampal connectivity: A hypothesis. Hippocampus. 2010 doi: 10.1002/hipo.20751. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Takei S, Morinobu S, Yamamoto S, Fuchikami M, Matsumoto T, Yamawaki S. Enhanced hippocampal BDNF/TrkB signaling in response to fear conditioning in an animal model of posttraumatic stress disorder. Journal of Psychiatric Research. 2010 doi: 10.1016/j.jpsychires.2010.08.009. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Berton O, Renthal W, Kumar A, Neve R, Nestler E. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nature Neuroscience. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nature Reviews Neuroscience. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Ullman SE, Filipas HH. Predictors of PTSD symptom severity and social reactions in sexual assault victims. Journal of Traumatic Stress. 2001;14:369–389. doi: 10.1023/A:1011125220522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vouimba R-M, Munoz C, Diamond DM. Differential effects of predator stress and the antidepressant tianeptine on physiological plasticity in the hippocampus and basolateral amygdala. Stress: The International Journal on the Biology of Stress. 2006;9:29–40. doi: 10.1080/10253890600610973. [DOI] [PubMed] [Google Scholar]
- Yasui DH, Peddada S, Bieda MC, et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proceedings of the National Academy of Sciences. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Bierer LM. The relevance of epigenetics to PTSD: Implications for the DSM-V. Journal of Traumatic Stress. 2009;22:427–434. doi: 10.1002/jts.20448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, LeDoux J. Response variation following trauma: a translational neuroscience approach to understanding PTSD. Neuron. 2007;56:19–32. doi: 10.1016/j.neuron.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Zhang T-Y, Meaney MJ. Epigenetics and the environmental regulation of the genome and its function. Annual Review of Psychology. 2010;61:439–466. doi: 10.1146/annurev.psych.60.110707.163625. [DOI] [PubMed] [Google Scholar]
- Zhao H, Shiina H, Greene KL, et al. CpG methylation at promoter site −140 inactivates TGF beta2 receptor gene in prostate cancer. Cancer. 2005;104:44–52. doi: 10.1002/cncr.21135. [DOI] [PubMed] [Google Scholar]
- Zoladz PR, Conrad CD, Fleshner M, Diamond DM. Acute episodes of predator exposure in conjunction with chronic social instability as an animal model of posttraumatic stress disorder. Stress. 2008;11:259–281. doi: 10.1080/10253890701768613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoladz PR, Cooper D, Carter G, Apostolatos H, Patel NA, Diamond DM. Effects of corticosterone and predator stress on alternative splicing of acetylcholinesterase: potential relevance to PTSD-related pathology Program No. 841.3. 2009 Neuroscience Meeting Planner; Chicago, IL. Society for Neuroscience; 2009. Online. [Google Scholar]
- Zoladz PR, Woodson JC, Haynes VF, Diamond DM. Activation of a remote (1-year old) emotional memory interferes with the retrieval of a newly formed hippocampus-dependent memory in rats. Stress. 2010;13:36–52. doi: 10.3109/10253890902853123. [DOI] [PubMed] [Google Scholar]