Abstract
Little is known about the regulatory mechanisms that allow Porphyromonas gingivalis to survive in the oral cavity. Here we characterize the sigma factor SigH, one of six extracytoplasmic (ECF) sigma (σ) factors encoded in the P. gingivalis genome. Our results indicate that sigH expression is upregulated by exposure to molecular oxygen, suggesting that sigH plays a role in adaptation of P. gingivalis to oxygen. Furthermore, several genes involved in oxidative stress protection, such as sod, trx, tpx, ftn, feoB2 and the hemin uptake hmu locus, are downregulated in mutant deficient in SigH designated as V2948. ECF σ consensus sequences were identified upstream of the transcriptional start sites of these genes, consistent with the SigH-dependent regulation of these genes. Growth of V2948 was inhibited in the presence of 6% oxygen when compared to the wild-type W83 strain, while in anaerobic conditions both strains were able to grow. In addition, reduced growth of V2948 was observed in the presence of peroxide and thiol-oxidizing reagent, diamide when compared to the W83 strain. The SigH-deficient strain V2948 also exhibited reduced hemin uptake, consistent with the observed reduced expression of genes involved in hemin uptake. Finally, survival of V2948 was reduced in the presence of host cells compared to the wild-type W83 strain. Collectively, our studies demonstrate that SigH is a positive regulator of gene expression required for survival of the bacterium in the presence of oxygen and oxidative stress, hemin uptake, and virulence.
Keywords: ECF sigma factor, regulon, oxidative stress, hemin uptake, host-pathogen, Porphyromonas gingivalis
Introduction
Regulation of gene expression in response to environmental changes is a required adaptive response that allows bacteria to grow and survive. This is especially important for pathogenic bacteria that have to adapt to various host environments. Adaptation to such changes involves differential expression of genes involved in bacterial survival and virulence (Bashyam and Hasnain, 2004;Staron et al., 2009;Lewis et al., 2009).
Bacterial RNA polymerase (RNAP) is a multimeric protein comprised of a core polymerase (E) that contains a beta, beta’, two alpha subunits and a dissociable specificity factor sigma (σ). While there is one core RNAP (E), there are multiple σ factors that guide RNAP to selected promoters and provide some specificity to transcription initiation. All bacteria have one essential (housekeeping) σ factor that is required for basal transcription of most genes and activates the expression of genes required for everyday cell viability. However, many bacterial genomes also encode alternative σ factors that direct RNAP to transcribe genes in response to environmental stimuli (Helmann, 2002b;Campbell et al., 2008). One such factor is σ70, the extracytoplasmic function sigma factor (ECF-σ) (Potvin et al., 2008).
Ninety percent of the 1873 ECF-σ sequences belong to only four bacterial phyla: Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria (Staron et al., 2009). Some members of the Bacteroidetes phylum encode a large number of σ factors (> 30/genome) (Staron et al., 2009), suggesting that regulation by ECF-σ factors is especially important in these bacteria. Porphyromonas gingivalis, a gram-negative anaerobic bacterium of the Bacteroidetes phylum, is a major etiological agent in adult-onset periodontal disease (Slots et al., 1986). It is also an excellent model bacterium due to its similarity to other medically significant organisms such as Bacteroides fragilis, Prevotella intermedia, and Tannerella forsythia which are implicated in oral or intestinal diseases. Some of our previous work has suggested that novel forms of regulation exist in P. gingivalis (He et al., 2006;Anaya-Bergman et al., 2010). Indeed, the P. gingivalis W83 genome encodes six putative ECF-σ factors and recent studies have shown role of these factors in regulating response to oxidative stress, gingipain activity and hemagglutination in P. gingivalis (Dou et al., 2010;Kikuchi et al., 2009;Nelson et al., 2003).
One mechanism that allows P. gingivalis to sustain itself in the oral cavity is high aerotolerance and the ability to protect itself against reactive oxygen species (ROS). ROS, generated by the incomplete reduction of oxygen (Storz et al., 1990), are much more reactive than molecular oxygen and can cause severe damage to nucleic acids, cell membranes, and proteins (Farr and Kogoma, 1991), which can lead to mutagenesis and cell death. Several enzymes involved in oxidative stress protection have been identified in P. gingivalis. For instance, Fe/Mn–containing superoxide dismutase has been shown to play a role in aerotolerance in P. gingivalis (Amano et al., 1990;Nakayama, 1994) and Dps and AphC contribute to peroxide resistance in P. gingivalis (Ueshima et al., 2003;Johnson et al., 2004). Also, rubrerythrin (Rbr) was identified in P. gingivalis and was shown to play a role in protection from hydrogen peroxide and molecular oxygen (Sztukowska et al., 2002). Both, Dps and Rbr are required for P. gingivalis virulence (Ueshima et al., 2003;Mydel et al., 2006). Other proteins potentially involved in oxidative stress protection have been reported in P. gingivalis, including ferritin and several thioredoxins, though the role of these proteins in oxidative stress protection remains to be established (Kikuchi et al., 2005;Ratnayake et al., 2000).
Although expression of most of the genes described above has been shown to be regulated by OxyR (Diaz et al., 2006;Ohara et al., 2006), here we show that OxyR is not the sole regulator of genes involved in oxidative stress protection in P. gingivalis. Oxidative stress response mechanisms have been extensively studied in the related bacterium B. fragilis and have demonstrated the presence of catalase (KatB), ferritin, and thioredoxin systems in this bacterium (Reott et al., 2009;Rocha and Smith, 2004;Rocha and Smith, 1997;Rocha and Smith, 1995;Rocha et al., 2007). Oxygen-dependent transcription of genes of the Trx/Tpx system in B. fragilis was demonstrated to be OxyR-independent, suggesting that other antioxidant homeostasis regulators must be functional in the Bacteroidetes phylum.
We hypothesized that ECF-σ factors might be involved in the maintenance of oxidative stress homeostasis in Bacteroidetes. This hypothesis was supported by our data demonstrating that the SigH ECF-σ factor (PG1827) is upregulated in the presence of oxygen (Lewis et al., 2009). We further showed that a SigH-deficient mutant exhibits reduced growth in the presence of oxygen and a reduced ability to survive in the presence of host cells, which supports our hypothesis that ECF-σ factors play a role in oxidative stress protection in P. gingivalis and suggests a role for these factors in P. gingivalis virulence. Finally, we propose a mechanism for SigH mediated adaptation to oxygen based on results of microarray analysis.
Materials and Methods
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Supplementary Table 1. The W83 strain was cultured in an anaerobic atmosphere composed of 10% H2, 10% CO2, and 80% N2 at 37 °C. Bacteria were maintained on either blood agar plates (TSA II, 5% Sheep Blood) (BBL, Cockeysville, MD) or liquid cultures prepared in brain heart infusion broth (BHI, Difco Laboratories, Detroit, MI) supplemented with hemin (5 µg/ml) (Sigma, St. Louis, MO), yeast extract (5 mg/ml), cysteine (1 mg/ml) (Sigma, St. Louis, MO) and vitamin K3 (1 µg/ml) (Sigma, St. Louis, MO). Growth studies were conducted in BHI media both anaerobically and in the presence of 6% of oxygen[conditions generated as described previously (Lewis et al., 2009)]. To examine growth of the parental and mutant strains overnight cultures were used to inoculate BHI broth to an OD660nm = 0.1. One aliquot was incubated anaerobically while the other was grown in the presence of 6% of oxygen Growth was monitored for 24 h. Cultures to be used for harvesting of cells for subsequent RNA isolation and microarray analysis were inoculated to an OD660nm = 0.2 and grown until they reached logarithmic phase.
Clindamycin (0.5 µg/ml) was used for selection and maintenance of P. gingivalis sigh mutant containing the ermF-ermB cassette (Fletcher et al., 1995). Escherichia coli was grown aerobically at 37 °C in Luria-Bertani (LB) broth or on solid agar. Carbenicillin (50 µg/ml) and erythromycin (300 µg/ml) were added to select for recombinant strains.
Construction of the P. gingivalis sigH mutant strain
The 639 bp sigH gene was amplified using P. gingivalis W83 genomic DNA as a template (primers are listed in Supplementary Table 2) and cloned into a pCR®2.1 vector according to manufacturer’s instructions (Invitrogen, Carlsbad, CA). An ermF-ermAM gene isolated from pVA2198 (Fletcher et al., 1995) was blunt ended using Klenow and ligated into the NruI restriction enzyme site located 158 bp from the 5’ end of the sigH gene. This plasmid was linearized with EcoRI and electroporated into P. gingivalis electrocompetent cells as described previously (Fletcher et al., 1995). Colonies were selected on BHI agar supplemented with clindamycin (0.5 µg/ml) and screened using PCR analysis with primers specific for sigH. Disruption of sigH in predicted mutants was verified by sequencing as well as the absence of sigH transcript following insertion of the erm cassette at 158 bp was confirmed by mRNA sequencing (Supplemental Fig. S1). The mutant strain containing disrupted sigH was designated V2948.
Microarray analysis
RNA was isolated as described previously from mid-logarithmic cultures of P. gingivalis grown under aerobic and anaerobic conditions as described above (Lewis et al., 2009). The concentration of RNA was measured using the NanoDrop spectrophotometer ND-1000. Microarray analysis was conducted using arrays provided by The J Craig Venter Institute (jcvi.org) and previously published protocols were used to prepare probes for cDNA labeling (Lewis et al., 2009). Briefly, cDNA was generated using the Stratagene®FairPlay® III Microarray Labeling Kit according to the manufacturer’s protocol (Stratagene). The cDNA was labeled with Cy-3 or Cy-5 dyes (GE Healthcare) and hybridized to glass microarray slides. An axon 4200A microarray scanner was used to detect hybridized cDNA (Molecular Devices). The images were analyzed and inspected using the GenePix v 6.0 software. Significant statistical differences were determined using the Significance Analysis for Oral Pathogen Microarrays (SAOPMD) tools available at the Bioinformatics Resource for Oral Pathogens (BROP) at The Forsyth Institute (www.brop.org) (Chen et al., 2005). All repeats within and between arrays were combined to generate and analyze the microarray results. Differential gene expression was evaluated based on the change in mRNA expression as represented by the ratio of Cy-5/Cy-3 fluorescence. Microarray results in this study were compared to oxygen-dependent gene regulation in the parental W83 strain published previously (Lewis et al., 2009).
Sensitivity of P. gingivalis to oxidative and thiol stress
BHI media was inoculated with actively growing overnight cultures of wild-type and mutant P. gingivalis strains to an OD660 of 0.1. The cultures were then divided into several aliquots and incubated for 24 hrs with various concentrations of hydrogen peroxide, diamide (thiol oxidizing reagent), and plumbagin (superoxide stress generator) under anaerobic conditions. Culture without oxidative or thiol oxidizing supplements served as controls. Growth was monitored by measuring the optical density of the culture at 660nm. Growth inhibition was assessed by comparing growth rates of bacteria in media that contained oxidative agents and thiol oxidizing supplements to growth of bacteria in control media.
Hemin uptake
Hemin uptake in the W83 and SigH-deficient strain V2948 was measured as described previously (Lewis et al., 2006).
Survival of P. gingivalis strains with host cells
Bacterial survival in the presence of eukaryotic cells was determined as described previously (He et al., 2006;Ueshima et al., 2003). The HN4 cell line (Miyazaki et al., 2006) was grown at 37 °C in GIBCO® Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen; Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 U/mL), streptomycin (100 µg/mL), 2.5 µg/mL fungizone, 10 mM HEPES buffer, 1 mM sodium pyruvate, and 2 mM L-glutamine. HN4 cells were incubated in 90% air and 10% CO2. For invasion and adherence assays, HN4 cells were grown in the DMEM media described above without antibiotics. Bacterial infections were performed under anaerobic conditions. Thus, plates containing HN4 cells were transferred to an anaerobic chamber, media was replaced with a de-oxygenated cell media (generated by incubation of the media in anaerobic chamber for 24hr) and the cells were infected with P. gingivalis strains at a multiplicity of infection (MOI) of 100. The plates were incubated anaerobically at 37°C for 30min and subsequently washed. Bacteria were released from the HN4 cells by addition of 1% saponin (Riedel-de Haën 16109). The mixture was then diluted 4:1 with anaerobic BHI media and plated on blood agar plates. Colony forming units (CFUs) were counted following a 7 d incubation under anaerobic conditions. To account for intracellular bacteria the infected HN4s were treated with gentamycin (300 µg/ml) and metronidazole (400 µg/ml) for 60 min to kill extracellular bacteria and surviving intracellular bacteria were released and accounted for as described above.
Transcriptome analysis and determination of transcriptional start sites
RNA was isolated from P. gingivalis W83 and V2948 bacterial cells that were harvested from mid-logarithmic anaerobic cultures using the RNeasy mini kit (Qiagen) according to the manufacturer’s protocol and depleted of ribosomal RNA using the Epicentre Ribo-Zero kit (Epicentre). A cDNA library was constructed using the Illumina cDNA library generation kit (mRNA-seq) as described by the manufacturer (Illumina). The cDNA library was sequenced using the Illumina Genome Analyzer. Sequence reads were aligned to the reference P. gingivalis W83 genome using the CLC Genomic Workbench (CLC Bio). Transcriptional start sites for genes differentially regulated in the SigH-deficient strain V2948 when compared to the parental W83 strain were determined using the P. gingivalis W83 transcriptome data. Differential gene expression was determined by comparing number of reads/gene for W83 and V2948.
Results
Bioinformatic characterization of P. gingivalis SigH
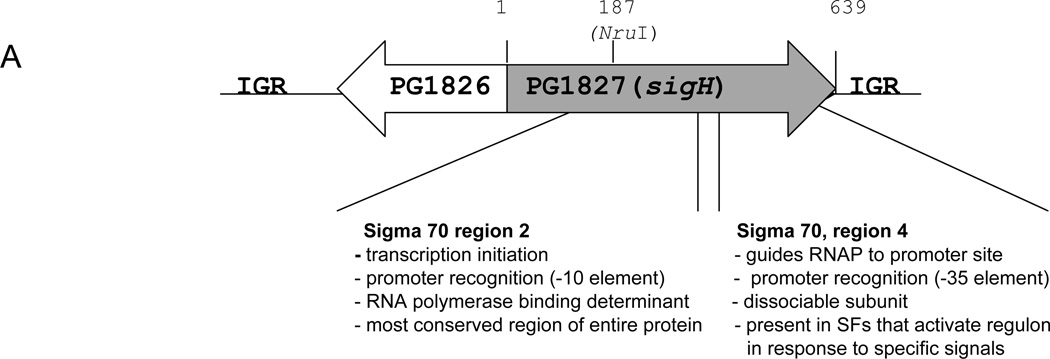
The sigH gene of P. gingivalis (designated as PG1595 on the Oralgen database [oralgen.lanl.gov] and PG1827 on the JCVI database [jcvi.org] codes for a 213 aa protein. Based on sequence similarity determined using the BLAST search (Entrez, NCBI) SigH belongs to the RNA polymerase sigma factor 70 family (Fig. 1A). Regions 56 – 123 are similar to the Sigma – 70 region 2, which binds the −10 promoter region upstream of the initiation start site, while residues 159 – 207 share homology with Sigma – 70 region 4, which binds to a β-1 flap of the RNAP as well as the – 35 promoter region (Fig. 1C) (Murakami and Darst, 2003). The genomic region coding for the SigH protein is unusual, however. In P. gingivalis an open reading frame, PG1826, is encoded immediately upstream of the sigH gene in the opposite direction (Fig. 1A). Furthermore, an anti-σ factor is not encoded after the sigH gene, unlike in other bacteria where the genes encoding ECF-σ factors are flanked by genes encoding anti-sigma factors.
Fig. 1. Characteristics of P. gingivalis SigH.
A. P. gingivalis W83 genomic locus coding for SigH (Oralgen). The grey and open arrows indicate two open reading frames (ORFs), PG1827 (sigH) and PG1826, respectively, and their direction of transcription. Intergenic regions (IRG) flank the two ORFs. The location of start and stop codons as well as the location of the NruI site are expressed in bp. Functional assignments of two regions encoded by sigH based on similarity to Sigma 70 (predicted using Entrez, NCBI) are shown underneath the schematic. B. Alignment of SigH protein sequences from P. gingivalis W83 (P.g. SigH) and Mycobacterium tuberculosis (M.t. SigH). C. Comparison of two putative ECF proteins from P. gingivalis W83, SigH (P.g. SigH) and a protein encoded by PG0162 (ECF1).
Blast analysis showed that residues 34 – 212 share 30% similarity with the sigH gene of Mycobacterium tuberculosis (Manganelli et al., 2002) (Fig. 1B). It also has a paralog, residues 72 – 201 are 25% identical and 49% similar to PG0148 (Oralgen annotation) (PG0162 according to JCVI annotation) (annotated as putative RNA Polymerase ECF σ factor, 70 family and designated here as ECF1) (Fig. 1C).
Further Blast search revealed that P. gingivalis SigH shares similarity with ECF-like sigma proteins from a variety of bacteria but is most closely related to putative σ factors of the Bacteroidetes family including B. fragilis (52% identity and 73% similarity), B. thetaiotamicron (50% identity and 73% similarity), and P. intermedia (27% identity and 50% similarity) (Supplementary Table S3).
The SigH-deficient strain exhibits reduced growth in the presence of oxygen
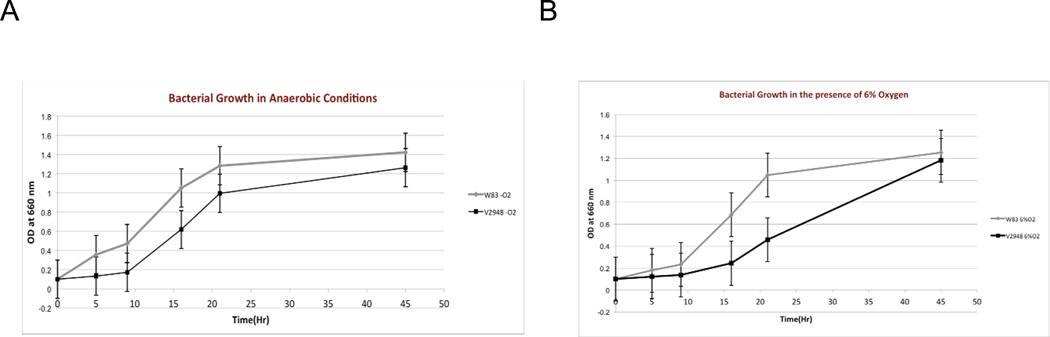
The overexpression of sigH in the presence of oxygen suggests that SigH plays a role in the growth of P. gingivalis in the presence of oxygen (Lewis et al., 2009). To investigate this further we compared the ability of the parental W83 and SigH-deficient V2948 strains to grow under anaerobic and aerobic (6% oxygen) conditions. As shown in Fig. 2A, both strains were able to grow in anaerobic conditions, however, growth of V2948 was slower than the parental strain. V2948 had longer lag phase and was able to grow once it reached OD660nm of 0.2. In the presence of oxygen the wild type W83 strain had longer lag phase when compared to its growth in anaerobic conditions, however, it grew well once it entered logarithmic phase (Fig. 2B). V2948 on the other hand, again had longer lag phase compared to the parental W83 strain, however, it maintained significantly reduced growth thorough logarithmic growth phase when compared to W83. The higher reduction of growth of the SigH-deficient strain in the presence of oxygen indicates it is required for growth and survival of P. gingivalis with oxygen. These results are consistent with a previous report that demonstrated sigH was upregulated in the presence of oxygen in wild-type P. gingivalis (Lewis et al., 2009).
Fig. 2. Effect of oxygen on growth of P. gingivalis strains.
P. gingivalis parental W83 (W83) and SigH-deficient mutant (V2948) were inoculated in BHI media and grown anaerobically (Panel A) as well as in the presence of oxygen (6% of oxygen) (Panel B). Bacterial growth was monitored by measuring optical density of the cultures at 660 nm. Means and standard deviations from two experiments are shown.
Expression of genes involved in oxidative stress protection is reduced in the SigH-deficient strain
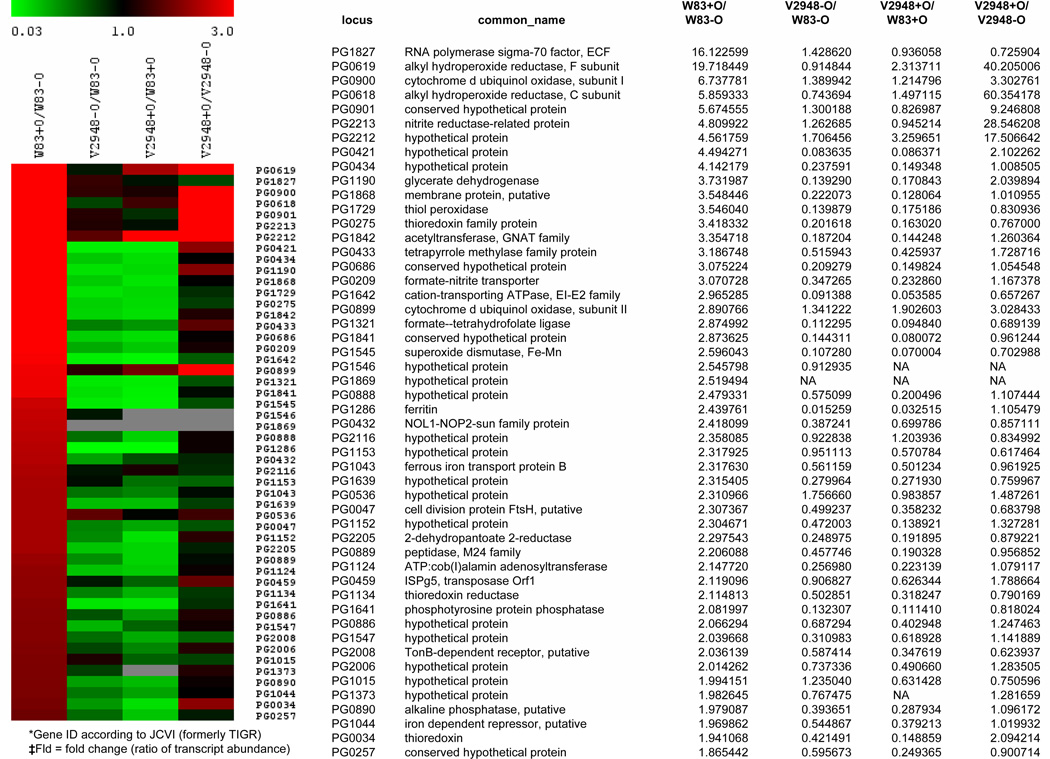
In order to identify genes with altered expression levels in the SigH-deficient strain V2948 we conducted microarray analysis. The analysis was done using RNA derived from cells of bacterial grown both in aerobic and anaerobic conditions. The growth curves of both, the parental W83 and the mutant V2948 strains were similar when higher bacterial inoculums were used (Supplemental Figure S2) thus enabling us to perform such analysis without the additional confounding factor which would be reduced growth rate of bacteria. Such growth dynamics was made possible by using high inoculum to start the cultures (see Materials and Methods section above). Microarray analysis of gene expression in anaerobic conditions showed that two hundred fifty genes exhibited a 1.5 fold reduction in expression in the SigH-deficient strain compared to the wild-type strain (60 most highly regulated genes are shown in Table 1). Some of the genes identified as downregulated were organized in operons (PG0046-47, PG0257-258, PG0421-422, PG0432-435, PG0855-890, PG1042-1044, PG1551-1556, PG1625-26, PG1638-42, PG1866-68, PG2134-35, PG2205-09, PG2216-17). While two of the operons PG1042-1044 and PG1551-1556, were shown previously (Lewis et al., 2006;Dashper et al., 2005) the remaining 11 are yet to be demonstrated and thus our results may also aid in identification of other co-transcribed genes.
Table 1.
60 most highly downregulated genes in V2948 when compared to the parental W83 strain.
| locus* | common_name | M† | Fld‡ | t | P | repeat§ |
|---|---|---|---|---|---|---|
| PG1286 | Ferritin | −6.034202 | 0.015259 | −68.479645 | 0.000000 (7.9E-16) | 12 |
| PG0421 | hypothetical protein | −3.579756 | 0.083635 | −43.136924 | 0.000000 (1.3E-13) | 12 |
| PG1642 | cation-transporting ATPase, EI-E2 family, authentic frameshift | −3.451853 | 0.091388 | −34.343927 | 0.000000 (1.5E-12) | 12 |
| PG1545 | superoxide dismutase, Fe-Mn | −3.220545 | 0.107280 | −61.428030 | 0.000000 (2.6E-15) | 12 |
| PG1321 | Formate—tetrahydrofolate ligase | −3.154633 | 0.112295 | −19.001963 | 0.000000 (9.2E-10) | 12 |
| PG1641 | phosphotyrosine protein phosphatase | −2.918043 | 0.132307 | −110.35678 | 0.000000 (4.2E-18) | 12 |
| PG1190 | Glycerate dehydrogenase | −2.843835 | 0.139290 | −26.540702 | 0.000000 (2.5E-11) | 12 |
| PG1729 | thiol peroxidase | −2.837745 | 0.139879 | −34.986562 | 0.000000 (1.2E-12) | 12 |
| PG1841 | conserved hypothetical protein | −2.792744 | 0.144311 | −14.702032 | 0.000000 (1.4E-08) | 12 |
| PG1842 | acetyltransferase, GNAT family | −2.417317 | 0.187204 | −19.516994 | 0.000000 (6.9E-10) | 12 |
| PG0275 | thioredoxin family protein | −2.310300 | 0.201618 | −33.715855 | 0.000000 (1.9E-12) | 12 |
| PG0686 | conserved hypothetical protein | −2.256500 | 0.209279 | −10.910157 | 0.000000 (3.1E-07) | 12 |
| PG1640 | DNA-damage-inducible protein F | −2.223689 | 0.214093 | −15.750918 | 0.000000 (7.4E-08) | 10 |
| PG1868 | membrane protein, putative | −2.170893 | 0.222073 | −22.985489 | 0.000000 (1.2E-10) | 12 |
| PG0434 | hypothetical protein | −2.073447 | 0.237591 | −37.571574 | 0.000000 (5.7E-13) | 12 |
| PG2205 | 2-dehydropantoate 2-reductase | −2.005926 | 0.248975 | −25.635413 | 0.000000 (3.7E-11) | 12 |
| PG1124 | ATP:cob(I)alamin adenosyltransferase, putative | −1.960271 | 0.256980 | −63.692982 | 0.000000 (7.8E-15) | 12 |
| PG1639 | hypothetical protein | −1.836684 | 0.279964 | −25.210544 | 0.000000 (4.4E-11) | 12 |
| PG1547 | hypothetical protein | −1.685091 | 0.310983 | −10.151634 | 0.000001 (6.3E-07) | 12 |
| PG0209 | formate-nitrite transporter | −1.525891 | 0.347265 | −34.171754 | 0.000000 (1.6E-12) | 12 |
| PG0432 | NOL1-NOP2-sun family protein | −1.368698 | 0.387241 | −10.399818 | 0.000000 (4.9E-07) | 12 |
| PG1551 | hmuY protein | −1.355295 | 0.390855 | −29.589327 | 0.000000 (7.7E-12) | 12 |
| PG0890 | alkaline phosphatase, putative | −1.345013 | 0.393651 | −44.945949 | 0.000000 (8.1E-14) | 12 |
| PG0617 | hypothetical protein | −1.329643 | 0.397867 | −11.092075 | 0.000000 (2.6E-07) | 12 |
| PG0034 | Thioredoxin | −1.246425 | 0.421491 | −39.745567 | 0.000000 (3.1E-13) | 12 |
| PG1553 | CobN-magnesium chelatase family protein | −1.235170 | 0.424792 | −6.217059 | 0.000066 | 12 |
| PG1042 | Glycogen synthase, putative | −1.228527 | 0.426753 | −14.040858 | 0.000000 (2.3E-08) | 12 |
| PG0080 | hypothetical protein | −1.210114 | 0.432234 | −3.013580 | 0.014631 | 11 |
| PG2209 | conserved hypothetical protein | −1.174035 | 0.443180 | −16.606236 | 0.000000 (3.9E-09) | 12 |
| PG0889 | peptidase, M24 family | −1.127380 | 0.457746 | −31.351640 | 0.000000 (4.1E-12) | 12 |
| PG1638 | thioredoxin family protein | −1.124267 | 0.458735 | −12.596331 | 0.000000 (7.1E-08) | 12 |
| PG0025 | fumarylacetoacetate hydrolase family protein | −1.110962 | 0.462985 | −14.075698 | 0.000000 (2.2E-08) | 12 |
| PG1152 | hypothetical protein | −1.083132 | 0.472003 | −10.025723 | 0.000001 (7.2E-07) | 12 |
| PG1423 | hypothetical protein | −1.062350 | 0.478851 | −7.263607 | 0.000047 | 10 |
| PG1625 | hypothetical protein | −1.031446 | 0.489220 | −38.348483 | 0.000000 (4.6E-13) | 12 |
| PG0259 | conserved hypothetical protein | −1.017716 | 0.493898 | −17.708272 | 0.000000 (1.9E-09) | 12 |
| PG1556 | conserved hypothetical protein | −1.010324 | 0.496435 | −5.421035 | 0.000421 | 10 |
| PG0047 | Cell division protein FtsH, putative | −1.002205 | 0.499237 | −12.419474 | 0.000000 (8.2E-08) | 12 |
| PG1134 | thioredoxin reductase | −0.991798 | 0.502851 | −59.004766 | 0.000000 (4.1E-15) | 12 |
| PG0278 | hypothetical protein | −0.957113 | 0.515087 | −18.863828 | 0.000000 (9.9E-10) | 12 |
| PG1129 | Ribonucleotide reductase | −0.955183 | 0.515776 | −25.569153 | 0.000000 (3.8E-11) | 12 |
| PG0433 | tetrapyrrole methylase family protein | −0.954717 | 0.515943 | −9.905916 | 0.000004 (3.8E-06) | 10 |
| PG0491 | conserved hypothetical protein | −0.941769 | 0.520594 | −22.970842 | 0.000000 (1.2E-10) | 12 |
| PG0046 | phosphatidate cytidylyltransferase | −0.941199 | 0.520800 | −41.745913 | 0.000000 (1.8E-13) | 12 |
| PG1671 | hypothetical protein | −0.924388 | 0.526904 | −19.534158 | 0.000000 (6.8E-10) | 12 |
| PG0707 | TonB-dependent receptor, putative | −0.923790 | 0.527122 | −15.874653 | 0.000000 (6.3E-09) | 12 |
| PG0340 | hypothetical protein | −0.913792 | 0.530788 | −16.025064 | 0.000000 (5.7E-09) | 12 |
| PG0644 | TonB-linked receptor Tlr, authentic frameshift | −0.889595 | 0.539765 | −10.442068 | 0.000000 (4.8E-07) | 12 |
| PG0783 | hydrolase, putative | −0.882396 | 0.542466 | −21.345972 | 0.000000 (2.6E-10) | 12 |
| PG1044 | Iron dependent repressor, putative | −0.876025 | 0.544867 | −13.317626 | 0.000000 (3.9E-08) | 12 |
| PG2040 | DNA-binding protein, histone-like family | −0.863655 | 0.549559 | −9.787825 | 0.000001 (9.1E-07) | 12 |
| PG1972 | Hemagglutinin protein HagB | −0.844263 | 0.556995 | −11.942377 | 0.000000 (1.2E-07) | 12 |
| PG2115 | Protease PrtT, degenerate | −0.837659 | 0.559551 | −4.432481 | 0.001007 | 12 |
| PG1043 | ferrous iron transport protein B | −0.833517 | 0.561159 | −21.171608 | 0.000000 (2.9E-10) | 12 |
| PG1674 | Hemagglutinin protein HagB, degenerate | −0.824790 | 0.564564 | −2.680892 | 0.021374 | 12 |
| PG2008 | TonB-dependent receptor, putative | −0.767551 | 0.587414 | −4.479731 | 0.000932 | 12 |
| PG0435 | Capsular polysaccharide biosythesis protein, putative | −0.727857 | 0.603800 | −12.252655 | 0.000000 (9.4E-08) | 12 |
| PG0010 | ATP-dependent Clp protease, ATP-binding subunit ClpC | −0.720886 | 0.606725 | −16.979237 | 0.000000 (3.1E-09) | 12 |
| PG2216 | hypothetical protein | −0.720043 | 0.607079 | −4.741918 | 0.000608 | 12 |
| PG1555 | conserved domain protein | −0.719785 | 0.607188 | −8.109344 | 0.000006 (5.7E-06) | 12 |
Gene ID according to JCVI (formerly TIGR)
M = log (aerobic conditions / anaerobic conditions)
Fld = fold change (ratio of transcript abundance in V2948/ W83)
Repeats = number of spots used for the analysis
Genes involved in oxidative stress protection such as sod, tpx, ftn, trx, and feoB2 are noticeably downregulated in V2948. Many of the genes previously reported to be upregulated in the presence of oxygen (Lewis et al., 2009) are downregulated in V2948 (Fig. 3). However, we also noted that a number of genes involved in oxidative stress protection and oxygen metabolism, such as ahpCF (PG0618-0619), cydAB (PG0899-0901), and the reductase-encoding oxygen-induced operon PG2212 −2213, were not affected by the SigH mutation in the V2948 strain, indicating that other regulatory mechanisms of oxidative stress protection are present in P. gingivalis (Fig. 3).
Fig. 3. Microarray analysis of oxygen and SigH-dependent gene expression in P. gingivalis.
Expression of the fifty genes most upregulated in the parental strain (P. gingivalis W83) in the presence of oxygen (W83+O/W83-O) was compared to that of: a SigH-deficient mutant (V2948 strain) grown anaerobically (V2948 − O/W83-O), V2948 grown erobically (V2948 + O/W83 + O), and V2948 grown in the presence and absence of oxygen (V2948+O/V2948−O). Ratios of gene expression are shown in graphical form on the left and in numerical form on the right. A value greater than one indicates an increase in mRNA expression for the strain labeled with Cy-5 (red color) and conversely, a value less than 1 indicates a decrease in expression for the Cy-5 labeled strain. mRNA probe labeled with Cy3 is designated in green.
We also compared the gene expression profile of W83 and V2948 strains grown in aerobic conditions. Interestingly, many genes affected by SigH mutation in anaerobic conditions were also affected by the mutation in the presence of oxygen (Fig. 3, V2948 + O/W83 + O).
Finally, we compared the gene expression profile of V2948 grown in aerobic conditions to that grown without oxygen. Expression levels of many of the genes downregulated in V2948 were not significantly affected by the presence of oxygen, including genes coding for thiol peroxidase (PG1729), thioredoxin (PG0275), superoxide dismutase (PG1545), ferritin (PG1286), FeoB2 (PG1043), and the formate - nitrite transporter (PG0209) (Fig. 3). These data confirms that the oxygen-dependent expression of those genes is dependent on the presence of SigH. However, a number of oxygen-regulated genes, such as genes coding for alkyl hydroperoxide reductase (PG0618-9), thioredoxin (PG0034), and the nitrite reductase operon (PG2212-13), did exhibit changes in expression levels upon exposure to oxygen in the V2948 strain, suggesting that regulation of these genes is SigH independent (Fig. 3).
We also observed upregulation of gene expression in the absence of SigH (Table 2). Most of the upregulated genes code for transposases. Among other significantly regulated genes are ones coding for putative regulatory proteins (PG1497, PG1535, PG1007, PG1432, PG0928, and PG0121). Finally, genes encoding stress response mechanisms such chaperones (PG0520-21) and ribosomal proteins (PG1960, PG0656, PG1959) were upregulated in V2948.
Table 2.
60 most highly upregulated genes in V2948 when compared to the parental W83 strain.
| locus* | common_name | M† | Fld‡ | t | P | repeat§ |
|---|---|---|---|---|---|---|
| PG1484 | hypothetical protein | 2.099982 | 4.287042 | 26.382838 | 0.000000 (2.7E-11) | 12 |
| PG1482 | conjugative transposon protein TraF | 2.093370 | 4.267439 | 22.848107 | 0.000000 (1.3E-10) | 12 |
| PG1483 | conjugative transposon protein TraE | 1.947643 | 3.857439 | 32.154368 | 0.000000 (3.1E-12) | 12 |
| PG1474 | conjugative transposon protein TraO | 1.937859 | 3.831366 | 40.145591 | 0.000000 (2.7E-13) | 12 |
| PG0606 | hypothetical protein | 1.930285 | 3.811305 | 8.643301 | 0.000003 | 12 |
| PG1480 | conjugative transposon protein TraI | 1.743379 | 3.348184 | 14.201320 | 0.000000 (2.0E-08) | 12 |
| PG1476 | conjugative transposon protein TraM | 1.678199 | 3.200283 | 38.684244 | 0.000000 (4.2E-13) | 12 |
| PG1475 | conjugative transposon protein TraN | 1.672028 | 3.186622 | 8.500293 | 0.000004 | 12 |
| PG1534 | conserved domain protein | 1.609411 | 3.051272 | 10.727336 | 0.000039 | 11 |
| PG1974 | hypothetical protein | 1.583181 | 2.996297 | 49.457197 | 0.000000 (2.8E-14) | 12 |
| PG1477 | hypothetical protein | 1.573150 | 2.975536 | 9.623705 | 0.000001 | 12 |
| PG1479 | conjugative transposon protein TraJ | 1.555254 | 2.938855 | 23.738661 | 0.000000 (8.4E-11) | 12 |
| PG1481 | conjugative transposon protein TraG | 1.540755 | 2.909468 | 34.855289 | 0.000000 (1.3E-12) | 12 |
| PG1478 | conjugative transposon protein TraK | 1.482257 | 2.793854 | 23.184716 | 0.000000 (1.1E-10) | 12 |
| PG1683 | conserved hypothetical protein | 1.433670 | 2.701330 | 39.828657 | 0.000000 (3.0E-13) | 12 |
| PG1010 | ABC transporter, ATP-binding protein | 1.374896 | 2.593492 | 52.412469 | 0.000000 (1.5E-14) | 12 |
| PG1745 | phosphoribulokinase family protein | 1.370986 | 2.586472 | 22.364335 | 0.000000 (1.6E-10) | 12 |
| PG1473 | conjugative transposon protein TraQ | 1.352018 | 2.552689 | 23.740666 | 0.000000 (3.9E-10) | 11 |
| PG1494 | conserved hypothetical protein | 1.291271 | 2.447436 | 18.081931 | 0.000000 (1.6E-09) | 12 |
| PG1684 | Hypothetical protein | 1.218886 | 2.327670 | 126.990104 | 0.000000 (9.0E-19) | 12 |
| PG1497 | DNA-binding protein, histone-like family | 1.166672 | 2.244932 | 12.700840 | 0.000000 (6.5E-08) | 12 |
| PG1535 | transcriptional regulator, putative | 1.154254 | 2.225693 | 34.191452 | 0.000000 (1.6E-12) | 12 |
| PG0906 | lipoprotein, putative | 1.145365 | 2.212022 | 27.472915 | 0.000000 (1.7E-11) | 12 |
| PG1007 | transcriptional regulator, GntR family | 1.101514 | 2.145798 | 28.705630 | 0.000000 (1.1E-11) | 12 |
| PG1663 | ABC transporter, ATP-binding protein | 1.085281 | 2.121788 | 31.306825 | 0.000000 (4.2E-12) | 12 |
| PG1960 | ribosomal protein L28 | 1.074119 | 2.105436 | 24.181677 | 0.000000 (6.9E-11) | 12 |
| PG1119 | flavodoxin, putative | 1.041524 | 2.058401 | 54.008493 | 0.000000 (1.1E-14) | 12 |
| PG1496 | Hypothetical protein | 1.039165 | 2.055038 | 10.057285 | 0.000001 | 12 |
| PG0656 | ribosomal protein L34 | 0.949020 | 1.930561 | 22.713565 | 0.000000 (1.4E-10) | 12 |
| PG1664 | ABC transporter, permease protein, putative | 0.942746 | 1.922183 | 24.905838 | 0.000000 (5.0E-11) | 12 |
| PG0607 | Hypothetical protein | 0.934248 | 1.910894 | 26.892089 | 0.000000 (2.2E-11) | 12 |
| PG1826 | conserved domain protein | 0.901095 | 1.867483 | 104.112734 | 0.000000 (1.0E-18) | 12 |
| PG1008 | Hypothetical protein | 0.895692 | 1.860502 | 29.357205 | 0.000000 (8.4E-12) | 12 |
| PG1009 | Hypothetical protein | 0.885176 | 1.846990 | 42.007912 | 0.000000 (1.7E-13) | 12 |
| PG1959 | ribosomal protein L33 | 0.875783 | 1.835004 | 27.945881 | 0.000000 (1.4E-11) | 12 |
| PG0121 | DNA-binding protein HU | 0.872345 | 1.830637 | 12.626815 | 0.000000 (6.9E-08) | 12 |
| PG1890 | lipoprotein, putative | 0.860410 | 1.815555 | 12.624885 | 0.000000 (6.9E-08) | 12 |
| PG1435 | Integrase | 0.858677 | 1.813375 | 6.251841 | 0.000062 | 12 |
| PG1662 | Hypothetical protein | 0.852168 | 1.805212 | 6.034404 | 0.000085 | 12 |
| PG1432 | Sensor histidine kinase | 0.839072 | 1.788899 | 8.275893 | 0.000009 | 12 |
| PG1609 | methylmalonyl-CoA decarboxylase, gamma s. | 0.834169 | 1.782830 | 9.434488 | 0.000001 | 12 |
| PG0536 | Hypothetical protein | 0.812835 | 1.756660 | 18.157552 | 0.000000 (1.5E-09) | 12 |
| PG0928 | response regulator | 0.812297 | 1.756005 | 15.231637 | 0.000000 (9.7E-09) | 12 |
| PG0520 | chaperonin, 60 kDa | 0.810891 | 1.754295 | 11.133896 | 0.000000 (2.5E-07) | 12 |
| PG0609 | Hypothetical protein | 0.797196 | 1.737720 | 4.029668 | 0.002401 | 11 |
| PG1004 | prolyl oligopeptidase family protein | 0.796070 | 1.736364 | 30.484226 | 0.000000 (5.6E-12) | 12 |
| PG1676 | phosphoenolpyruvate carboxykinase (ATP) | 0.787653 | 1.726264 | 38.083595 | 0.000000 (4.9E-13) | 12 |
| PG1634 | Hypothetical protein | 0.787164 | 1.725679 | 10.500075 | 0.000000 (4.5E-07) | 12 |
| PG1786 | Hypothetical protein | 0.786857 | 1.725312 | 13.041755 | 0.000000 (4.9E-08) | 12 |
| PG1586 | batE protein | 0.784310 | 1.722269 | 35.544134 | 0.000000 (1.0E-12) | 12 |
| PG1267 | Hypothetical protein | 0.777617 | 1.714297 | 12.905965 | 0.000000 (5.5E-08) | 12 |
| PG0192 | Cationic outer membrane protein OmpH | 0.775456 | 1.711731 | 15.072637 | 0.000000 (1.1E-08) | 12 |
| PG0292 | chromate transport protein, putative | 0.774012 | 1.710018 | 38.544254 | 0.000000 (4.3E-13) | 12 |
| PG0521 | chaperonin, 10 kDa | 0.771302 | 1.706810 | 12.126001 | 0.000000 (1.0E-07) | 12 |
| PG2212 | Hypothetical protein | 0.771003 | 1.706456 | 7.927241 | 0.000007 | 12 |
| PG0350 | internalin-related protein | 0.770678 | 1.706072 | 33.152570 | 0.000000 (2.2E-12) | 12 |
| PG0681 | Hypothetical protein | 0.769667 | 1.704876 | 12.227690 | 0.000000 (9.6E-08) | 12 |
| PG0293 | secretion activator protein, putative | 0.763501 | 1.697605 | 23.413056 | 0.000000 (9.8E-11) | 12 |
| PG1005 | lipoprotein, putative | 0.760033 | 1.693529 | 25.405906 | 0.000000 (4.0E-11) | 12 |
| PG0138 | malonyl CoA-acyl carrier protein transacylase | 0.746919 | 1.678205 | 17.931087 | 0.000000 (1.7E-09) | 12 |
Gene ID according to JCVI (formerly TIGR)
M = log (aerobic conditions/ anaerobic conditions)
Fld = fold change (ratio of transcript abundance in V2948/W83)
Repeats = number of spots used for the analysis
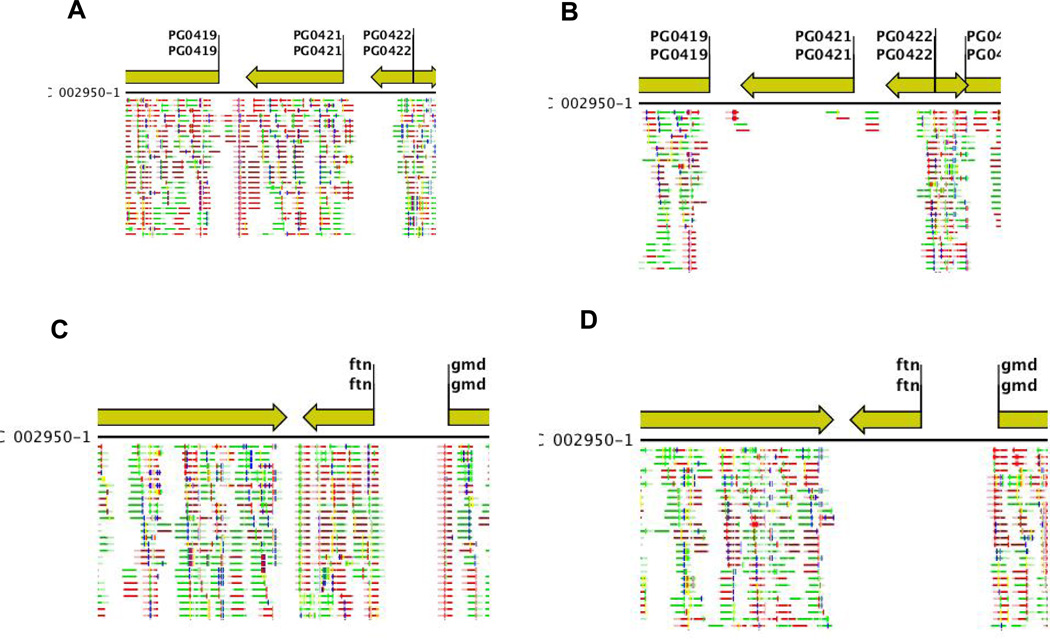
The microarray data was validated by RNAseq analysis. As shown in Supplemental Table S4 most genes detected as regulated in our microarray analysis were also regulated using the RNAseq comparison. Again, 252 genes were upregulated in the W83 strain compared to the V2948 at 2 fold. Such number of regulated genes is very similar to that observed in our microarray analysis. The only difference was the larger fold change indicating that RNAseq is more sensitive method compared to the microarray analysis. Images of the gene-specific reads for the most highly-regulated genes, ftn (PG1286) and PG0421 (Table 1, Supplemental Table S4) as determined using both microarray analysis and RNAseq (Table 1 and Supplemental Table S4, respectively) are shown in Figure 4. The number of reads is drastically reduced in the V2948 strain when compared to the parental W83 strain for both genes. Collectively, these results not only validate our microarray analysis data but also indicate that SigH is absolutely required for transcription of ftn and PG0421.
Fig. 4. Verification of SigH – dependent expression of ftn and PG0421.
Gene expression in P. gingivalis wild-type strain (W83) (Panels A and C) and SigH-deficient mutant (V2948) (Panels B and D) was examined using RNAseq. The green arrows indicate open reading frames (ORFs) and their direction of transcription. Reads derived using RNAseq are shown below each ORF. Expression of PG0421 is shown in Panels A and B. Expression of ftn (PG1286) is shown in Panels C and D.
The SigH-deficient strain exhibits reduced survival in the presence of oxidative and thiol stress
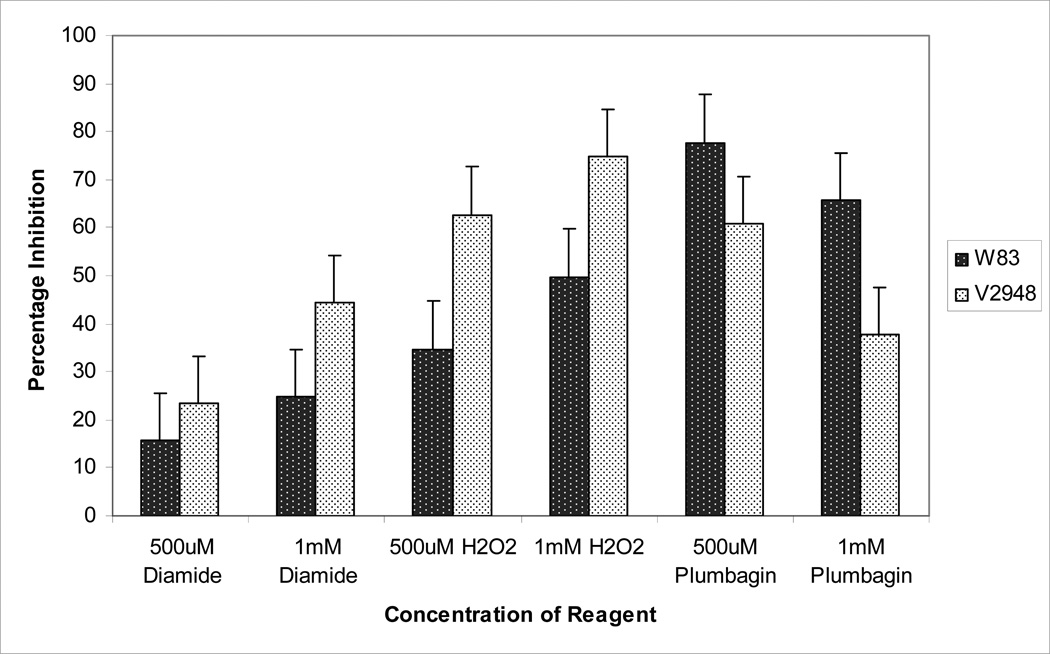
We further examined the ability of the W83 and V2948 strains to grow in the presence of thiol oxidizing agent - diamide, peroxide, and superoxide-generating agent plumbagin. As shown in Fig. 5, significant growth inhibition is observed in both strains in growth media supplemented with diamide in a dose-dependent manner. However, the inhibition in the presence of 1mM of diamide was 2-fold higher in the SigH-deficient V2948 strain when compared to the wild-type strain (Fig. 5), demonstrating that V2948 is more susceptible to thiol stress. The presence of hydrogen peroxide also inhibited the growth of both strains in a dose-dependent manner, though growth inhibition in the V2948 strain was approximately two-fold higher in the presence of 500 µM of peroxide (Fig. 5). Thus, SigH appears to play a role in the upregulation of mechanisms required for growth in the presence of peroxide and thiol oxidizing stress. Plumbagin inhibited growth of both strains; interestingly, the inhibition of V2948 strain was lower than that of the W83 strain (Fig. 5).
Fig. 5. Sensitivity of P. gingivalis strains to thiol and oxidative stress.
P. gingivalis wild-type strain (W83) and SigH-deficient mutant (V2948) were inoculated in BHI media and divided into aliquots that were then supplemented with various concentrations of diamide, hydrogen peroxide, or plumbagin. Unsupplemented BHI cultures served as controls. The ability of the various compounds to inhibit microbial growth was determined following a 12 hr anaerobic incubation by comparing growth of the bacteria in the presence and absence of the compound. Mean and error bars indicating standard deviations using triplicate samples are shown. Experiments were conducted three times with similar results.
Hemin uptake is reduced in the SigH-deficient mutant
Our microarray results indicate that expression of the major hemin uptake locus, hmu, as well as other genes potentially involved in hemin uptake (PG0707, PG0644, PG2008), is reduced in the absence of SigH in the V2948 strain. We examined hemin uptake in the parental and mutant strains and found that hemin uptake was in fact significantly reduced in the SigH-deficient V2948 strain (Table 3). These results are in agreement with our microarray findings and demonstrate that the V2948 strain has a reduced ability to take up hemin.
Table 3.
Hemin uptake in P. gingivalis strains
| Strain | Hemin uptake* at 10 min | Hemin uptake* at 30 min |
|---|---|---|
| W83 | 1010 ± 55.9 | 1365 ± 48.0 |
| V2498 | 281 ± 52.6 | 506 ± 53.4 |
the numbers show hemin uptake that was calculated by subtracting passive hemin binding/uptake (done by performing the assay on ice) from total hemin uptake (done by performing the assay at 37°C).
The SigH-deficient strain exhibits reduced survival in the presence of host cells
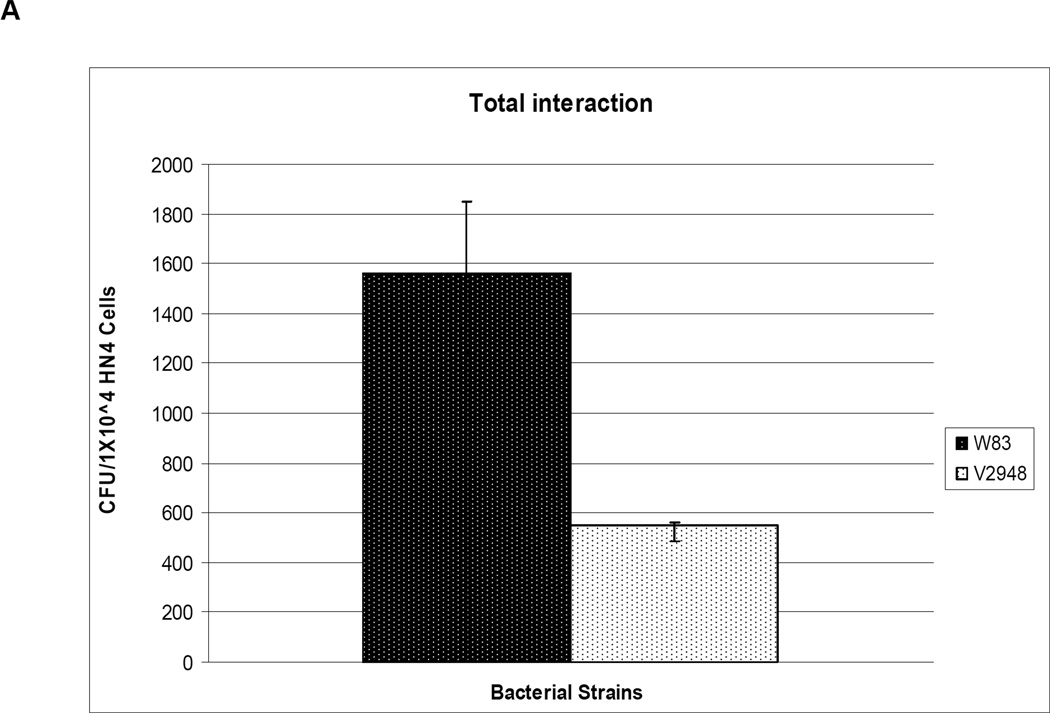
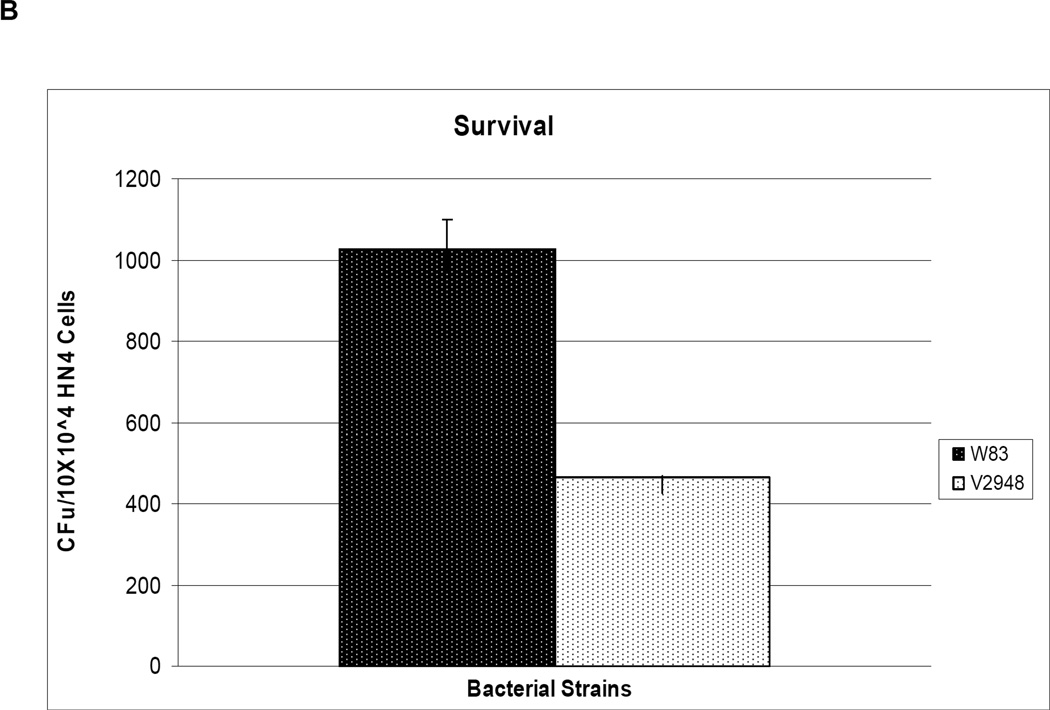
One mechanism by which a host organism defends against bacterial infections is by releasing reactive oxygen species. Since our results indicate that the SigH-deficient mutant V2948 strain had reduced expression levels of genes involved in protection from oxidative stress, we reasoned that this strain may have a decreased ability to survive in the presence of host cells. We incubated host cells with both the wild-type W83 and SigH-deficient mutant V2948 strains and observed that 75% fewer colonies were recovered on plates inoculated with bacterial samples from the incubations conducted with the V2948 strain (Fig. 6A). Similarly, 50% fewer colonies were recovered with V2948 compared to the W83 strain when only internalized bacteria were accounted for (Fig. 6B). To determine whether the ability to invade HN4s was the same for both strains we performed flow cytometry analysis using FITC-labeled bacteria. As shown in Supplemental Table S4 the invasion efficiencies were similar for both W83 and V2948 strains. Thus, the reduced recovery of live cells from HN4s demonstrates that the SigH-deficient mutant strain V2948 exhibits a reduced ability to survive in the presence of host cells.
Fig. 6. Role of SigH in survival of P. gingivalis with host cells.
P. gingivalis wild type (W83) and SigH-deficient mutant (V2948) strains were incubated for 30 min with HN4 cells. Total bacteria (extracellular and intracellular) (Panel A) or intracellular bacteria (Panel B) recovered from host cells were plated on blood agar plates and incubated for 7 days anaerobically. The number of colony forming units/ml (number of colonies on blood plates) from the host-bacteria mixture is shown. Mean and error bars indicating standard deviations from triplicate samples are shown.
Determination of SigH regulon
We determined the transcriptome of P. gingivalis W83 grown in anaerobic conditions. By aligning the sequence reads to the reference P. gingivalis W83 genome, we were able to determine the transcriptional start sites of genes (Supplemental Figure S3). To determine the SigH regulon we examined the transcriptional start sites of genes that were downregulated in the SigH mutant when compared to the parental W83 strain and located the promoter sequences of these genes.
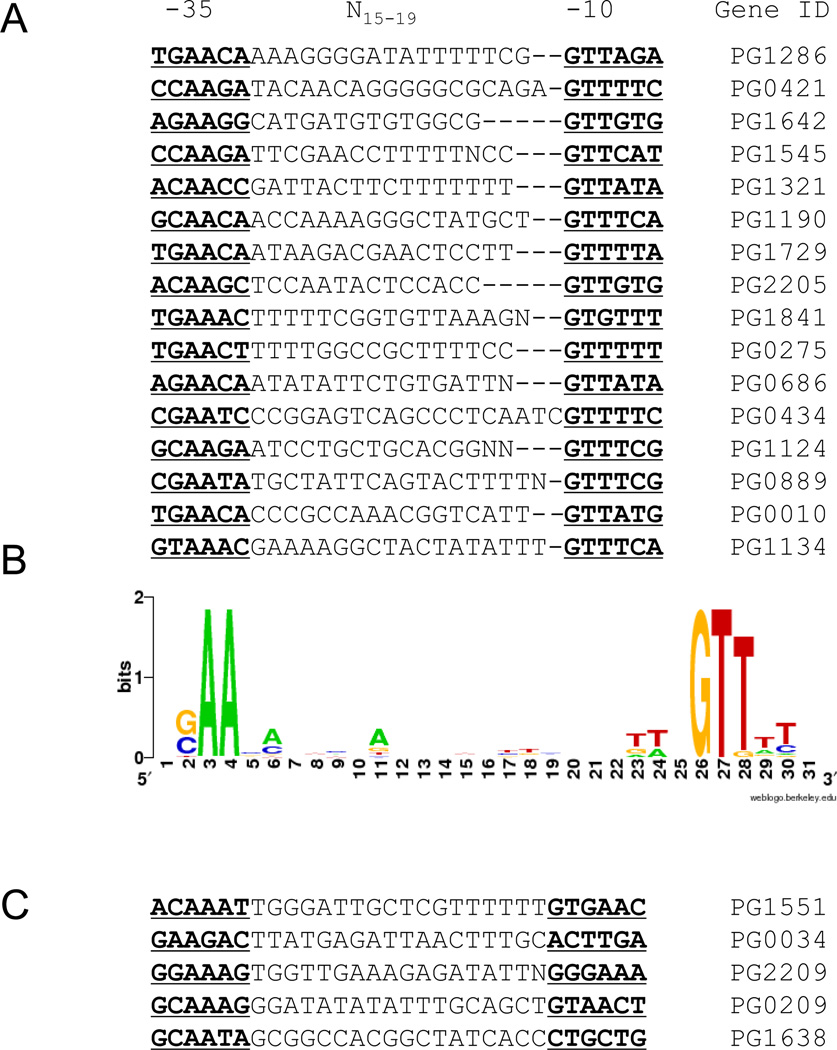
Examination of the promoter sequences of 15 genes regulated by exposure to oxygen revealed that the putative SigH promoter sequences differ from promoters recognized by typical primary sigma factors that bind −35 TTGACA and −10 TATAAT sequences (Helmann, 1995) (Fig. 7). Our study shows the presence of a “C/GAAG” motif in the −35 promoter region and “TGG” sequences in the −10 promoter region (sequences in bold and underlined in Fig. 7A). The sequence similarity among the various promoters is illustrated in Fig. 7B. The presence of SigH recognition sequences (Raman et al., 2001;Song et al., 2008) upstream of genes regulated by SigH is consistent with these genes being part of SigH regulon. However, we did not detect the consensus SigH recognition sequence upstream of some of the genes (Fig. 7C) that had altered expression in the absence of SigH (Table 1, Fig. 3) indicating that they may not be directly regulated by SigH.
Fig. 7. SigH target promoters in P. gingivalis W83.
A. 16 genes downregulated in the SigH mutant V2948 were used to determine transcriptional start sites using transcriptome analysis data. Regions upstream of start sites were examined for the presence of promoter sequences. Putative −35 and −10 sequences are in bold and underlined.
B. Consensus sequence of P. gingivalis SigH-dependent promoters. Sequence logo was generated using WebLogo (http://weblogo.berkley.edu). The height of the letters corresponds to their conservation within promoter sequences. C. Alignment of promoter sequences of 5 genes downregulated in V2948 that do not have the consensus sequence as shown in panel B.
Discussion
Gene regulation mechanisms of anaerobic Bacteroidetes are not well understood. Genomic analysis has revealed that numerous ECF σ factors are encoded in Bacteroidetes species, suggesting a significant role for these proteins in gene regulation (Staron et al., 2009). Our previous study has shown that P. gingivalis sigH (PG1827) coding for a putative ECF σ factor is drastically upregulated upon exposure to oxygen (Lewis et al., 2009). Bioinformatics analysis revealed that SigH has characteristics typical of other ECF σ factors (Staron et al., 2009). Here we show that SigH plays an important role in adaptation of the bacterium to oxygen, oxidative stress protection, metal homeostasis, and survival with host cells. Such results indicate that SigH plays important role in P. gingivalis ability to survive in oral cavity.
Although such results are consistent with the role of the SigH protein in protection against oxidative stress in other bacteria such as Mycobacterium tuberculosis and Salmonella enterica (Bang et al., 2005;Manganelli et al., 2002), the P. gingivalis SigH shares a relatively low degree of similarity with the mycobacterial SigH. Indeed, this σ factor belongs to the group of “unclassified” ECF σ factors described by Staron et al (Staron et al., 2009). A low degree of similarity was also observed between SigH (PG1827) and FecI (Braun et al., 2003). FecI plays a role in metal homeostasis, suggesting that SigH may also have similar role. Our observation that expression of two genes coding for metal/hemin transport feoB2 (PG1443) and the hmu operon is reduced supports such involvement. The finding that P. gingivalis SigH is most similar to the SigH of the Bacteroidetes family suggests that our results may be informative about the regulatory mechanisms of Bacteroidetes.
Typically, ECF σ factors are regulated by anti-σ factors that are encoded upstream or downstream of the σ factor genes (Staron et al., 2009). The genomic organization of the sigH locus is unconventional compared to loci of other ECF-σ factors (Staron et al., 2009). Scrutinizing microarray analysis results we noted that SigH in P. gingivalis is significantly upregulated upon exposure to oxygen (Lewis et al., 2009) and this oxygen-dependent regulation is still present in the SigH- and OxyR-deficient strains (Lewis et al, unpublished), indicating that regulators other than SigH or OxyR play a role in modulating expression of this protein. Although regulation at the transcriptional level has been observed for other ECF sigma proteins, this regulation primarily involved an autoregulatory mechanism whereby the σ factor regulated its own promoter (Staron et al., 2009). The observation that oxygen-dependent regulation is still present in the SigH-deficient mutant suggests that SigH is not autoregulated. Thus, the mechanism by which SigH is regulated needs to be further investigated.
To determine the role of SigH in P. gingivalis we characterize a mutant V2948 in which the gene encoding SigH was disrupted by an erm cassette. We observe that the SigH-deficient V2948 strain is significantly impaired in growth in the presence of oxygen as well as is more sensitive to peroxide and thiol oxidizing stress. The reduced growth of V2948 with peroxide reinforces the results of Dou et al (Dou et al., 2010). Importantly, such reduced growth of the mutant strains is consistent with the observed reduction in expression levels of genes involved in oxidative stress protection. The majority of genes involved in protection from oxidative stress that are upregulated in the presence of oxygen in the wild-type strain, such as superoxide dismutase (PG1545), glycerate dehydrogenase (PG1190), thioredoxins (PG0034, PG0275, PG1134, and PG1638), are significantly downregulated in the V2948 strain. Superoxide dismutase is required for protection of P. gingivalis from atmospheric oxygen (Nakayama, 1994). Glycerate dehydrogenase may also have an antioxidative role as hydroxypyruvate is known to interact with peroxide (Perera et al., 1997). However, the reduced sensitivity of V2948 to superoxide-generating reagent, plumbagin, indicates that other mechanisms involved in protection from superoxide stress are enhanced in V2948.
The increased sensitivity of V2948 to thiol oxidizing reagent, diamide, may be explained by the observation that all four genes encoding the thioredoxin (Trx/Tpx) system as well as a gene PG1729 coding for thiol peroxidase are downregulated in the V2948 strain, suggesting that these genes are regulated by SigH. The thioredoxin system is the major player in regulation of the redox homeostasis and thiol peroxidase was shown to have an antioxidant role in other bacteria (Wan et al., 1997;Zhou et al., 1997). The induction of the Trx/Tpx system in the presence of oxygen was also observed in B. fragilis (Reott et al., 2009;Sund et al., 2008) and was OxyR-independent, similar to our observation that expression levels were significantly altered by the absence of regulator other than OxyR, the ECF sigma factor.
Besides genes coding for oxidative stress protection mechanisms genes encoding proteins mediating metal homeostasis were also downregulated in V2948. We observed that ferritin-encoding gene, PG1286, was the most downregulated gene in the SigH-deficient mutant V2948 strain. The absence of ftn-specific transcript in V2948 indicates that SigH is absolutely required for transcription of the gene. Ferritin is an iron-binding protein protein was shown to play a role in provision of iron in P. gingivalis grown under low-iron concentrations (Ratnayake et al., 2000). It is likely that iron may be required for the function of some oxidative-stress enzymes. Other downregulated genes included the hmu operon (Lewis et al., 2006) and the feoB2 (PG1043) locus coding for manganese transport protein FeoB2 (He et al., 2006). While FeoB2 and manganese are required for growth of P. gingivalis in the presence of oxygen, elevated binding of hemin to the surface of P. gingivalis may also have anti-oxidant capacity (Smalley et al., 2000). However, hemin uptake studies showed that hemin transport was significantly reduced in the V2948 strain, suggesting that the intracellular concentration of iron/hemin is also affected. These results suggest that SigH may also play a role in metal homeostasis in P. gingivalis. Since metal homeostasis plays a significant role in oxidative stress protection in P. gingivalis and other bacteria, it’s not surprising that these two mechanisms might be connected by a common factor.
Our results also show that growth of V2948 is impaired under anaerobic conditions, possibly due to a reduced ability to acquire nutrients such as hemin. This interpretation is guided by our observation of significantly longer lag phase in V2948. The reduction of expression of genes coding for thioredoxins in V2948 could also lead to alteration of the intracellular redox status thus affecting the structure and function of many proteins containing cysteines. Furthermore, there were other genes downregulated in V2948 that code for virulence mechanisms such as the two loci (PG0890 and PG1641) encoding phosphatases. While the role of PepP encoded by PG890 is unknown, the phosphotyrosine protein phosphatase encoded by PG1641 plays a role in the regulation of numerous processes in P. gingivalis (Maeda et al., 2008).
Though many of the genes involved in oxidative stress protection exhibit reduced expression in the SigH mutant, the antioxidative alkyl hydroperoxide reductase, shown to play a major role in oxidative stress protection in P. gingivalis (Johnson et al., 2004), had unaltered expression in the absence of SigH, indicating other regulatory mechanisms play a role in modulating the oxygen-dependent expression of those genes. One of the other ECF σ factors encoded in the P. gingivalis genome could be involved in regulating this gene (Nelson et al., 2003), possibly the ECFs encoded by PG0162 and PG1660, known to play a significant role in growth of P. gingivalis in the presence of peroxide (Dou et al., 2010). Our results show that SigH is similar to ECF1 protein encoded by PG0162 indicating that the protein may also plays a role in regulating genes coding for proteins mediating oxidative stress protection.
Determining the DNA binding site of a transcription factor helps in defining the regulon of that factor. We identified genes regulated by SigH using microarray analysis and combined this with transcriptome data to identify the transcriptional start sites for these genes. In many cases, upstream of the start sites we detected typical ECF sigma factor binding sites (Raman et al., 2001;Song et al., 2008), supporting our hypothesis that the genes identified in the microarray analysis are directly regulated by SigH. We identified a consensus binding site for SigH by aligning putative promoter sequences of genes that exhibited reduced expression in the absence of SigH. The SigH consensus sequence of P. gingivalis is similar to that of other sigma factors (Staron et al., 2009; Helmann, 2002b) and contains the typical “C/GAAG” motif in the −35 region as well as “GTT” rich sequences in the −10 region. We continued to observe expression of genes regulated by SigH, although at low levels, in the SigH-deficient strain. This low level expression may be due to activation by other σ factors. Indeed, overlapping activation by multiple σ factors has been described in other bacteria (Wade et al., 2006).
It is known that P. gingivalis RNA polymerase differs from that of E. coli (Klimpel and Clark, 1990). Also, the primary σ differs in the Bacteroidetes phylum when compared to other bacterial species (Vingadassalom et al., 2005). This, combined with the fact that multiple σ factors are involved in gene regulation, complicates the identification of promoter sites in the phylum. Promoter sites in P. gingivalis have been predicted using consensus promoter sequences of the primary σ factor (−35 “TTGACA” and −10 “TATAAT”) (Helmann, 1995). However, as the σ factor dictates promoter specificity, such predictions based on primary σ factor from other species may not be the best way to identify promoters in P. gingivalis. Indeed, the significant difference of the SigH consensus promoter sequence and the consensus P. gingivalis promoter sequence as identified by Jackson et al (Jackson et al., 2000) highlights the limitations of predicting consensus sequences when using the known consensus sequence of only one σ factor. Elucidating the role that the numerous ECF σ factors of P. gingivalis play in gene regulation and defining their regulons will be a significant advancement in our understanding of the regulatory networks in this bacterium.
Understanding how P. gingivalis adapts to the presence of oxygen is an important biological question as the oral environments inhabited by P. gingivalis are not completely anaerobic (Mettraux et al., 1984;Hanioka et al., 2000;Tanaka et al., 1998). Indeed, higher oxygen levels would be expected in supragingival environments which would inhibit growth of the bacterium. Also, reactive oxygen and nitrogen species are secreted by host cells and other oral bacteria. We show that the SigH-deficient V2948 strain has a reduced ability to survive in the presence of eukaryotic cells. This suggests that SigH plays a particularly important role when the bacterium is present in the periodontal pocket in contact with host cells mounting an ROS response to fight the invading bacteria. Such response would be expected to include mechanisms directly removing the oxidizing reagents as well as repairing oxidized molecules (thioredoxin system). Taken together, our results demonstrate that SigH plays an important role in protecting P. gingivalis from stresses encountered in the oral environment and that inhibition of this factor could lead to reduction of P. gingivalis growth and survival in both supragingival and subgingival locations.
Supplementary Material
Acknowledgements
This research was supported by USPHS grants 5R01DE016124, R01DE018039 and 5R21DE019005 from the National Institute of Dental and Craniofacial Research awarded to Dr. Janina Lewis. The P. gingivalis W83 genomic sequence was obtained from (TIGR) (http://www.tigr.org) and Los Alamos Oral Pathogen Sequence Database (http://www.oralgen.lanl.gov).
We thank Drs. Todd Kitten and Paul Fawcett for their help with the microaerophilic system and with the Axon microarray imager and image processing using GenePix software, respectively.
Footnotes
Portion of the work reported in this paper is included in Sara Sarrafee’s Master Thesis submitted in May, 2009.
Reference List
- Amano A, Shizukuishi S, Tamagawa H, Iwakura K, Tsunasawa S, Tsunemitsu A. Characterization of superoxide dismutases purified from either anaerobically maintained or aerated Bacteroides gingivalis. J Bacteriol. 1990;172:1457–1463. doi: 10.1128/jb.172.3.1457-1463.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaya-Bergman C, He J, Jones K, Miyazaki H, Yeudall A, Lewis JP. Porphyromonas gingivalis ferrous iron transporter FeoB1 influences sensitivity to oxidative stress. Infect Immun. 2010;78:688–696. doi: 10.1128/IAI.00108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang IS, Frye JG, McClelland M, Velayudhan J, Fang FC. Alternative sigma factor interactions in Salmonella: sigma and sigma promote antioxidant defences by enhancing sigma levels. Mol Microbiol. 2005;56:811–823. doi: 10.1111/j.1365-2958.2005.04580.x. [DOI] [PubMed] [Google Scholar]
- Bashyam MD, Hasnain SE. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect Genet Evol. 2004;4:301–308. doi: 10.1016/j.meegid.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Braun V, Mahren S, Ogierman M. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr Opin Microbiol. 2003;6:173–180. doi: 10.1016/s1369-5274(03)00022-5. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase sigma factor activity: a structural perspective. Curr Opin Microbiol. 2008;11:121–127. doi: 10.1016/j.mib.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Abbey K, Deng WJ, Cheng MC. The bioinformatics resource for oral pathogens. Nucleic Acids Res. 2005;33:W734–W740. doi: 10.1093/nar/gki361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashper SG, Butler CA, Lissel JP, Paolini RA, Hoffmann B, Veith PD, et al. A novel Porphyromonas gingivalis FeoB plays a role in manganese accumulation. J Biol Chem. 2005;280:28095–28102. doi: 10.1074/jbc.M503896200. [DOI] [PubMed] [Google Scholar]
- Diaz PI, Slakeski N, Reynolds EC, Morona R, Rogers AH, Kolenbrander PE. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J Bacteriol. 2006;188:2454–2462. doi: 10.1128/JB.188.7.2454-2462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Osbourne D, McKenzie R, Fletcher HM. Involvement of extracytoplasmic function sigma factors in virulence regulation in Porphyromonas gingivalis W83. FEMS Microbiol Lett. 2010;312(1):24–32. doi: 10.1111/j.1574-6968.2010.02093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr SB, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher HM, Schenkein HA, Morgan RM, Bailey KA, Berry CR, Macrina FL. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanioka T, Tanaka M, Takaya K, Matsumori Y, Shizukuishi S. Pocket oxygen tension in smokers and non-smokers with periodontal disease. J Periodontol. 2000;71:550–554. doi: 10.1902/jop.2000.71.4.550. [DOI] [PubMed] [Google Scholar]
- He J, Miyazaki H, Anaya C, Yu F, Yeudall WA, Lewis JP. Role of Porphyromonas gingivalis FeoB2 in metal uptake and oxidative stress protection. Infect Immun. 2006;74:4214–4223. doi: 10.1128/IAI.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD. OxyR: a molecular code for redox sensing? Sci STKE. 2002a;2002:e46. doi: 10.1126/stke.2002.157.pe46. [DOI] [PubMed] [Google Scholar]
- Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol. 2002b;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- Jackson CA, Hoffmann B, Slakeski N, Cleal S, Hendtlass AJ, Reynolds EC. Porphyromonas gingivalis promoter sequence. FEMS Microbiol Lett. 2000;186:133–138. doi: 10.1111/j.1574-6968.2000.tb09094.x. [DOI] [PubMed] [Google Scholar]
- Johnson NA, Liu Y, Fletcher HM. Alkyl hydroperoxide peroxidase subunit C (ahpC) protects against organic peroxides but does not affect the virulence of Porphyromonas gingivalis W83. Oral Microbiol Immunol. 2004;19:233–239. doi: 10.1111/j.1399-302X.2004.00145.x. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Ohara N, Sato K, Yoshimura M, Yukitake H, Sakai E, et al. Novel stationary-phase-upregulated protein of Porphyromonas gingivalis influences production of superoxide dismutase, thiol peroxidase and thioredoxin. Microbiology. 2005;151:841–853. doi: 10.1099/mic.0.27589-0. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Ohara N, Ueda O, Hirai K, Shibata Y, Nakayama K, Fujimura S. Porphyromonas gingivalis mutant defective in a putative extracytoplasmic function sigma factor shows a mutator phenotype. Oral Microbiol Immunol. 2009;24:377–383. doi: 10.1111/j.1399-302X.2009.00526.x. [DOI] [PubMed] [Google Scholar]
- Klimpel KW, Clark VL. The RNA polymerases of Porphyromonas gingivalis and Fusobacterium nucleatum are unrelated to the RNA polymerase of Escherichia coli. J Dent Res. 1990;69:1567–1572. doi: 10.1177/00220345900690090601. [DOI] [PubMed] [Google Scholar]
- Lewis JP, Iyer D, Anaya-Bergman C. Adaptation of Porphyromonas gingivalis to microaerophilic conditions involves increased consumption of formate and reduced utilization of lactate. Porphyromonas gingivalis. Microbiology. 2009;155:3758–3774. doi: 10.1099/mic.0.027953-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JP, Plata K, Yu F, Rosato A, Anaya C. Transcriptional organization, regulation and role of the Porphyromonas gingivalis W83 hmu haemin-uptake locus. Microbiology. 2006;152:3367–3382. doi: 10.1099/mic.0.29011-0. [DOI] [PubMed] [Google Scholar]
- Maeda K, Tribble GD, Tucker CM, Anaya C, Shizukuishi S, Lewis JP, et al. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol. 2008;69:1153–1164. doi: 10.1111/j.1365-2958.2008.06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol Microbiol. 2002;45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- Mettraux GR, Gusberti FA, Graf H. Oxygen tension (pO2) in untreated human periodontal pockets. J Periodontol. 1984;55:516–521. doi: 10.1902/jop.1984.55.9.516. [DOI] [PubMed] [Google Scholar]
- Meuric V, Gracieux P, Tamanai-Shacoori Z, Perez-Chaparro J, Bonnaure-Mallet M. Expression patterns of genes induced by oxidative stress in Porphyromonas gingivalis. Oral Microbiol Immunol. 2008;23:308–314. doi: 10.1111/j.1399-302X.2007.00429.x. [DOI] [PubMed] [Google Scholar]
- Miyazaki H, Patel V, Wang H, Ensley JF, Gutkind JS, Yeudall WA. Growth factor-sensitive molecular targets identified in primary and metastatic head and neck squamous cell carcinoma using microarray analysis. Oral Oncol. 2006;42:240–256. doi: 10.1016/j.oraloncology.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- Mydel P, Takahashi Y, Yumoto H, Sztukowska M, Kubica M, Gibson FC, III, et al. Roles of the host oxidative immune response and bacterial antioxidant rubrerythrin during Porphyromonas gingivalis infection. PLoS Pathog. 2006;2:e76. doi: 10.1371/journal.ppat.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K. Rapid viability loss on exposure to air in a superoxide dismutase-deficient mutant of Porphyromonas gingivalis. J Bacteriol. 1994;176:1939–1943. doi: 10.1128/jb.176.7.1939-1943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara N, Kikuchi Y, Shoji M, Naito M, Nakayama K. Superoxide dismutase-encoding gene of the obligate anaerobe Porphyromonas gingivalis is regulated by the redox-sensing transcription activator OxyR. Microbiology. 2006;152:955–966. doi: 10.1099/mic.0.28537-0. [DOI] [PubMed] [Google Scholar]
- Perera A, Parkes HG, Herz H, Haycock P, Blake DR, Grootveld MC. High resolution 1H NMR investigations of the reactivities of alpha-keto acid anions with hydrogen peroxide. Free Radic Res. 1997;26:145–157. doi: 10.3109/10715769709097793. [DOI] [PubMed] [Google Scholar]
- Potvin E, Sanschagrin F, Levesque RC. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2008;32:38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- Raman S, Song T, Puyang X, Bardarov S, Jacobs WR, Jr, Husson RN. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J Bacteriol. 2001;183:6119–6125. doi: 10.1128/JB.183.20.6119-6125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnayake DB, Wai SN, Shi Y, Amako K, Nakayama H, Nakayama K. Ferritin from the obligate anaerobe Porphyromonas gingivalis purification, gene cloning and mutant studies. Microbiology. 2000;146(Pt 5):1119–1127. doi: 10.1099/00221287-146-5-1119. [DOI] [PubMed] [Google Scholar]
- Reott MA, Parker AC, Rocha ER, Smith CJ. Thioredoxins in redox maintenance and survival during oxidative stress of Bacteroides fragilis. J Bacteriol. 2009;191:3384–3391. doi: 10.1128/JB.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha ER, Smith CJ. Biochemical and genetic analyses of a catalase from the anaerobic bacterium Bacteroides fragilis. J Bacteriol. 1995;177:3111–3119. doi: 10.1128/jb.177.11.3111-3119.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha ER, Smith CJ. Regulation of Bacteriodes fragilis katB mRNA by oxidative stress and carbon limitation. J Bacteriol. 1997;179:7033–7039. doi: 10.1128/jb.179.22.7033-7039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha ER, Smith CJ. Transcriptional regulation of the Bacteroides fragilis ferritin gene (ftnA) by redox stress. Microbiology. 2004;150:2125–2134. doi: 10.1099/mic.0.26948-0. [DOI] [PubMed] [Google Scholar]
- Rocha ER, Tzianabos AO, Smith CJ. Thioredoxin reductase is essential for thiol/disulfide redox control and oxidative stress survival of the anaerobe Bacteroides fragilis. J Bacteriol. 2007;189:8015–8023. doi: 10.1128/JB.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slots J, Bragd L, Wikstrom M, Dahlen G. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J Clin Periodontol. 1986;13:570–577. doi: 10.1111/j.1600-051x.1986.tb00849.x. [DOI] [PubMed] [Google Scholar]
- Smalley JW, Birss AJ, Silver J. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the mu-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide. FEMS Microbiol Lett. 2000;183:159–164. doi: 10.1111/j.1574-6968.2000.tb08951.x. [DOI] [PubMed] [Google Scholar]
- Song T, Song SE, Raman S, Anaya M, Husson RN. Critical role of a single position in the -35 element for promoter recognition by Mycobacterium tuberculosis SigE and SigH. J Bacteriol. 2008;190:2227–2230. doi: 10.1128/JB.01642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staron A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol. 2009;74:557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- Storz G, Tartaglia LA. OxyR: a regulator of antioxidant genes. J Nutr. 1992;122:627–630. doi: 10.1093/jn/122.suppl_3.627. [DOI] [PubMed] [Google Scholar]
- Storz G, Tartaglia LA, Farr SB, Ames BN. Bacterial defenses against oxidative stress. Trends Genet. 1990;6:363–368. doi: 10.1016/0168-9525(90)90278-e. [DOI] [PubMed] [Google Scholar]
- Sund CJ, Rocha ER, Tzianabos AO, Wells WG, Gee JM, Reott MA, et al. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol Microbiol. 2008;67:129–142. doi: 10.1111/j.1365-2958.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- Sztukowska M, Bugno M, Potempa J, Travis J, Kurtz DM., Jr Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol Microbiol. 2002;44:479–488. doi: 10.1046/j.1365-2958.2002.02892.x. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Hanioka T, Takaya K, Shizukuishi S. Association of oxygen tension in human periodontal pockets with gingival inflammation. J Periodontol. 1998;69:1127–1130. doi: 10.1902/jop.1998.69.10.1127. [DOI] [PubMed] [Google Scholar]
- Ueshima J, Shoji M, Ratnayake DB, Abe K, Yoshida S, Yamamoto K, Nakayama K. Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect Immun. 2003;71:1170–1178. doi: 10.1128/IAI.71.3.1170-1178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingadassalom D, Kolb A, Mayer C, Rybkine T, Collatz E, Podglajen I. An unusual primary sigma factor in the Bacteroidetes phylum. Mol Microbiol. 2005;56:888–902. doi: 10.1111/j.1365-2958.2005.04590.x. [DOI] [PubMed] [Google Scholar]
- Wade JT, Roa DC, Grainger DC, Hurd D, Busby SJ, Struhl K, Nudler E. Extensive functional overlap between sigma factors in Escherichia coli. Nat Struct Mol Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- Wan XY, Zhou Y, Yan ZY, Wang HL, Hou YD, Jin DY. Scavengase p20: a novel family of bacterial antioxidant enzymes. FEBS Lett. 1997;407:32–36. doi: 10.1016/s0014-5793(97)00302-5. [DOI] [PubMed] [Google Scholar]
- Wang B, Shi Q, Ouyang Y, Chen Y. [Progress in oxyR regulon- the bacterial antioxidant defense system--a review] Wei Sheng Wu Xue Bao. 2008;48:1556–1561. [PubMed] [Google Scholar]
- Wu J, Lin X, Xie H. OxyR is involved in coordinate regulation of expression of fimA and sod genes in Porphyromonas gingivalis. FEMS Microbiol Lett. 2008;282:188–195. doi: 10.1111/j.1574-6968.2008.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wan XY, Wang HL, Yan ZY, Hou YD, Jin DY. Bacterial scavengase p20 is structurally and functionally related to peroxiredoxins. Biochem Biophys Res Commun. 1997;233:848–852. doi: 10.1006/bbrc.1997.6564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.