Abstract
The role of autophagy in disease pathogenesis following viral infection is beginning to be elucidated. We have previously reported that hepatitis C virus (HCV) infection in hepatocytes induces autophagy. However, the biological significance of HCV induced autophagy has not been clarified. Autophagy has recently been identified as a novel component of innate immune system against viral infection. In the present study, we have shown that knockdown of autophagy related protein Beclin1 or ATG7 in immortalized human hepatocytes (IHH) inhibited HCV growth. Beclin1 or ATG7 knockdown IHH when infected with HCV exhibited an increased expression of IFN-β, OAS-1, IFN-α and IFI27 mRNAs of the interferon signaling pathways as compared to infection of control IHH. Subsequent study demonstrated that HCV infection in autophagy impaired IHH displayed caspase activation, PARP cleavage and apoptotic cell death.
Conclusion
The disruption of autophagy machinery in HCV infected hepatocytes activated IFN signaling pathway, and induced apoptosis. Together, these results suggest that HCV induced autophagy impairs innate immune response.
HCV infection affects nearly 3.3 million people and is the most common cause of cirrhosis and hepatocellular carcinoma (HCC) in the United States (1). The current approved therapy for treatment of HCV is pegylated interferon in combination with ribavirin (2, 3). Although several advances have shown promises in improving the management of HCV infection, nevertheless it remains a major health problem (4–6). HCV is a member of the Flaviviridae family, and its genome contains a positive-strand RNA of approximately 9.6 kb long. HCV genome encodes a polyprotein precursor of about 3,000 amino acids, which is cleaved by both viral and host proteases into structural (core, E1, E2, and p7) and nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins. HCV infected cells accumulate lipid droplets and play an important role in the assembly of virus particles (7–9).
Autophagy is a catabolic process by which the cells remove their own damaged organelles and long lived proteins for the maintenance of cellular homeostasis. During autophagy, the double membrane vesicles engulf the damaged organelles and eventually fuse with the lysosomes for degradation. Autophagy is upregulated in response to extra- or intracellular-stress and signals, such as starvation, growth factor deprivation, endoplasmic reticulum (ER) stress, and pathogen infection (10, 11). Viruses are obligate intracellular parasites and their survival is linked to their ability to subvert cellular antiviral defenses and to regulate cellular processes necessary for their own replication. We have shown that HCV genotype 1a or genotype 2a infection induces autophagy in IHH (12). Autophagy induction was subsequently reported with HCV genotype 1b or genotype 2a subgenomic replicon or infection with HCV genotype 2a (clone JFH1) in Huh7.5 cells (13–16). Although HCV induced autophagy is established, whether autophagy helps in host cell survival or is beneficial for HCV multiplication remains unknown. Some of the viruses, such as cytomegalovirus, Kaposi’s Sarcoma associated herpes virus, and human herpes simplex virus-1 (HSV-1), have evolved strategies to suppress autophagy for their own survival (17). In HSV-1 infection, ICP34.5 protein suppresses autophagy by binding to Beclin1 and blocks initiation of autophagy (18). Certain viruses, such as mouse hepatitis virus, poliovirus, coxsackievirus, and dengue virus, exploit the elements of autophagy system for their own replication (19). In mammalian system, Beclin1 recruits other autophagy proteins to initiate the formation of pre-autophagosomal membrane. ATG7 is required in conjunction with ATG12 and ATG5 to form autophagosomes. We have reported previously that HCV infection induces Beclin1 expression (12). In this study, we have demonstrated that HCV infection in autophagy knockdown cells activates IFN signaling pathway and apoptosis.
Materials and Methods
Cell culture, transfection and HCV infection
Immortalized human hepatocytes (IHH) and human hepatoma (Huh 7.5) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing10% fetal bovine serum, 100 U/ml of penicillin G, and 100 μg/ml of streptomycin at 37°C in a 5% CO2 atmosphere. We have grown HCV genotypes 1a (clone H77) and genotype 2a (clone JFH1) in IHH as previously described (20). HCV genotype 2a was initially grown in Huh 7.5 cells in this study. For infection, IHH (3 × 105 cells) were incubated with HCV genotype 1a (clone H77, 5.3 ×104 IU) and genotype 2a (clone JFH1, 1.2 ×105 IU) in a minimum volume of medium. After 8 h of virus adsorption on hepatocytes, DMEM supplemented with 5% heat-inactivated fetal bovine serum was added. IHH, transfected with control (scrambled) or Beclin1 siRNA or ATG7 siRNA using lipofectamine 2000 (Invitrogen), were similarly infected with cell culture grown HCV. The siRNAs used in this study was mixture of three siRNAs and were purchased from Santa Cruz (the sequences are not disclosed). HCV titer was measured as previously described (20). Briefly, HCV infectivity from 3 day culture supernatant was titrated by an end-point dilution assay in a 96-well plate. Virus inocula were serially diluted and used to infect 6 replicate wells of naïve IHH growing in a microtiter plate. Three days post-infection, the cells were washed, fixed with cold methanol, incubated with mouse monoclonal NS5A-specific antibody (HL1126, kindly provided by Chen Liu) at 4°C overnight. After washing with PBS, cells were incubated with anti-mouse Ig conjugated with Alexa 488 (Invitrogen) for 60 min at room temperature and visualized under fluorescence microscope.
The microtubule-associated protein 1 light chain 3 (LC3), a homologue of Apg8p essential for autophagy in yeast, is associated to the autophagosome membranes after processing, and is indispensible for the elongation of autophagic vesicles. Two forms of LC3, LC3-I and -II, are produced post-translationally in various cells. LC3-I is cytosolic, whereas LC3-II is membrane bound. Lipidated LC3 (LC3-II) is a useful marker of autophagic membranes, and autophagosomes are visualized as bright GFP-LC3 puncta by fluorescence microscopy. As readout of induction of autophagy, cells were transfected with LC3-GFP as described previously (12). Cells containing ≥ 3 GFP-LC3 dots were defined as autophagy-positive cells. For LysoTracker red staining, the cells were treated with 1 μM LysoTracker Red DND-99 (Invitrogen) at 37°C for 30 min. Control IHH and Beclin1 knockdown IHH (siBCN1-IHH) or ATG7 knockdown IHH (siATG7-IHH) were starved in Hank’s balanced salts solution (HBSS; Lonza) for 120 min in the presence of LysoTracker red dye. Cells then were fixed in 3.7% formaldehyde, and nuclei were stained with DAPI (Invitrogen). Three-channel optical images (DAPI, GFP, and Lysotracker red) were collected using the sequential scanning mode (405-, 488-, and 543-nm excitation; 450-, 522-, and 595-nm emission, respectively) of the Olympus FV1000 confocal system.
RNA quantitation
Control IHH, siBCN1-IHH or siATG7-IHH were infected with HCV and incubated for 72 h. Total RNA was isolated using Qiagen RNeasy kit (Qiagen, CA). cDNA was synthesized using random hexamer and Superscript III reverse transcriptase kit (Invitrogen, CA). Real time-PCR was performed with the cDNA for RNA quantitation using Taqman Gene Expression PCR master Mix (Applied Biosystems) and FAM-MGB probe for IFI-27 (Hs00271467_m1), IFN-β (Hs02621180_sl), 2′–5′ oligoadenylate synthetase 1 (OAS-1) (Hs00973637_m1), and IFNA1 (Hs00353738_s1). FAM-MGB probe for GAPDH (Hs99999905_m1) was used as an endogenous control.
Cell death assay
HCV infected control IHH, siBCN1-IHH or siATG7-IHH were assayed for cell viability/death after 72 h of infection using Live/Dead two-color fluorescence assay according to the manufacturer’s instruction (Molecular Probes, CA). Infected IHH were washed in PBS and exposed to 4 μm calcein-AM and 2 μM ethidium homodimer in PBS for 30 min at room temperature. Dye uptake was detected for fluorescein (for calcein in live cells) and Texas red (for ethidium homodimer in dead cells). Image analysis to quantify Live/Dead cells was performed using Image J software.
Immunoblot analysis
HCV infected IHH, siBCN1-IHH or siATG7-IHH were lysed using sodium dodecyl sulfate sample buffer. Proteins were subjected to electrophoresis on polyacrylamide gel and transferred onto nitrocellulose membrane. The membrane was probed with an antibody for PARP (Poly ADP-Ribose Polymerase), caspase-9 or caspase-3 (Cell Signaling Technology, MA). Proteins were detected using an enhanced chemiluminescent ECL Western blot substrate (Pierce, IL). The membrane was reprobed for tubulin as an internal control protein.
Results
Knockdown of Beclin1 reduces HCV growth
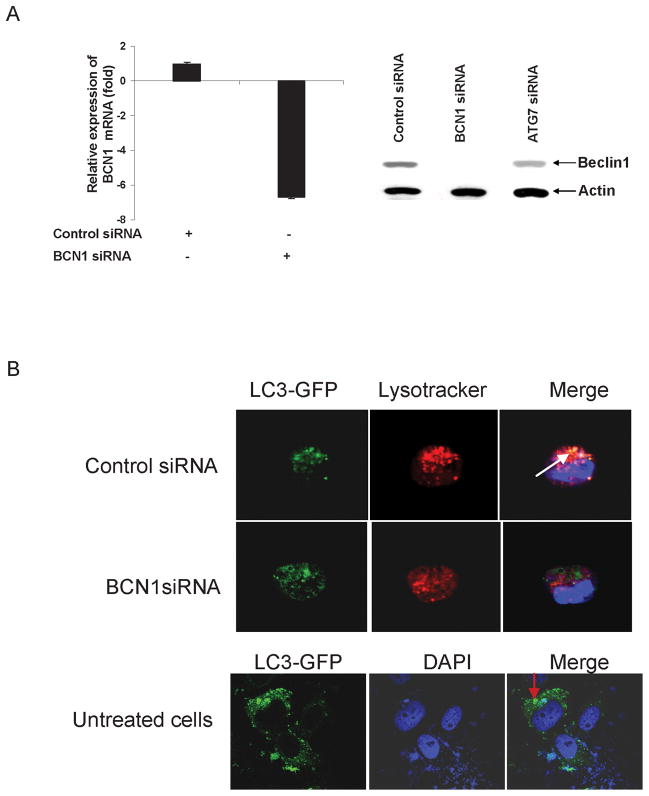
Beclin1 is one of the upstream molecules, which recruits other autophagy proteins to initiate the autophagy signaling pathway (21, 22). Beclin1 forms complex with Vsp34 and PI3K and promotes autophagic vesicles formation (22). We chose to use Beclin1 in examining whether knockdown of autophagy gene alter HCV growth. IHH were transfected with control (scrambled) or Beclin1 siRNA, and expression of Beclin1 was examined at the mRNA and protein levels. A significant inhibition of Beclin1 at RNA (~ 6 fold) and protein (> 90%) levels were observed following siRNA treatment (Fig. 1, panel A). IHH treated with ATG7 siRNA did not display knockdown of Beclin1 expression, suggesting the specificity of Beclin1 siRNA. We did not observe a difference in cell viability in Beclin1 knockdown cells as compared to control IHH. To examine the induction of autophagy in control IHH or siBCN1-IHH, cells were starved with nutrients. The presence of autolysosomes in cells was assessed using GFP-LC3 and LysoTracker red staining, which stains for acidic organelles such as lysosomes. Clearly, in starved IHH, LC3 was found to be colocalized with LysoTracker red, suggesting the formation of the autolysosomes (Fig. 1, panel B). As expected, autolysosome formation was not observed in starved siBCN1-IHH, or uninfected IHH as shown the distribution of LC3.
Figure 1. Knockdown of Beclin1 in IHH does not induce autophagy.
Panel A: IHH was transfected with 10 μM control (scrambled) siRNA, Beclin1 or ATG7 siRNA and incubated for 5 days. Expression of Beclin1 at the mRNA and protein levels was examined by RT-qPCR and Western blot analysis. Beclin1 was downregulated >6 fold at the mRNA levels and >90% at the protein level. However ATG 7 siRNA had no effect on Beclin1 expression. Panel B: Confocal microscopy of autolysosomes. For starvation, IHH was transfected with control siRNA or Beclin1 siRNA were starved for 120 min in HBSS. Lysosomes were stained by LysoTracker red dye (red), autophagosomes were stained by GFP-LC3 (green). Autolysosomes were stained by LysoTracker red and GFP-LC3 showing a yellow signal. The nuclei were stained with DAPI (blue). Distribution of GFP-LC3 in untreated IHH is shown by arrow in bottom panel.
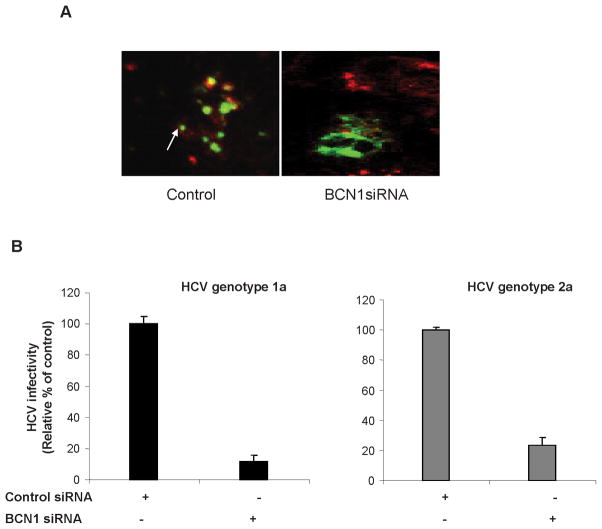
HCV infected control IHH and siBCN1-IHH were transfected with LC3-GFP to examine induction of autophagy. As expected, LC3 redistribution into discrete dots was markedly increased in HCV infected IHH as compared to HCV infected siBCN1-IHH (Fig. 2, panel A). HCV infected IHH displayed LC3 II expression, while a predominant LC3 I band was observed in HCV infected siBCN1-IHH (data not shown). Next, control IHH and siBCN1-IHH were infected with cell culture grown HCV genotype 1a or genotype 2a (H77 or JFH1 clone) for 3 days. We determined the effect of Beclin1 knockdown on the release of infectious virus particles. HCV titer in Beclin1 knockdown infected hepatocytes was reduced as compared to HCV infected control IHH (Fig. 2, panel B). Therefore, the impairment of autophagy machinery reduces HCV production in IHH.
Figure 2. Knockdown of Beclin1 suppresses infectious HCV particle production.
Panel A: Control IHH and siBCN1-IHH were infected HCV genotype1a for 48 h and then transfected with LC3-GFP. Induction of autophagy was examined 48 h after transfection. Confocal microscopy examination for subcellular localization of GFP-LC3 visualized by intrinsic fluorescence (green), and HCV NS5A protein (red) in merged image panels. Punctate localization of LC3 on autophagic vacuoles (shown by arrow) was clearly visible in HCV-infected control IHH, not in siBCN1-IHH. Panel B: HCV (genotype 1a or 2a) infectivity was measured from culture supernatants of virus infected siBCN1-IHH or control IHH at serial dilutions, and expressed as percentage of that in control cells.
HCV infection enhances IFN signaling in autophagy knockdown IHH
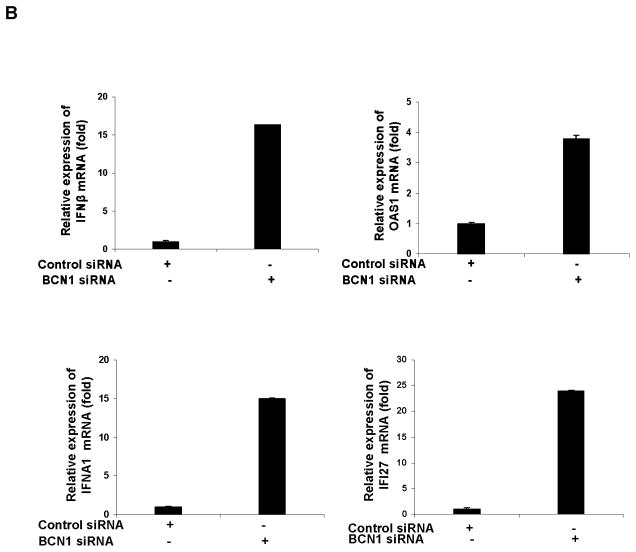
We have seen earlier that HCV infected IHH displayed inhibition of IFN-α and IFI27 expression (23). Next, we asked whether knockdown of autophagy protein following HCV infection modulates the IFN signaling pathway. For this, the expression level of IFN-β, OAS1, IFN-α, and IFI27 from control IHH and siBCN1-IHH was initially examined to verify if knockdown of Beclin1 has an effect on interferon signaling pathway. A significant difference in expression of these genes in control IHH and siBCN1-IHH was not observed (data not shown). We next examined the status of a number of interferon stimulated genes (ISGs) in HCV infected control IHH and siBCN1-IHH. We have shown earlier that HCV infection in IHH upregulates IFN-%beta; and OAS1 (24). Here, the mRNA level of IFN-β and OAS1 was 6–10 fold higher in HCV genotype 1a (panel A) or genotype 2a (panel B) infected siBCN1-IHH as compared to virus infected control IHH (Fig. 3). Since HCV infection of IHH inhibits IFN-α and IFI27 mRNA expression, we wanted to examine whether IFN-α synthesis is altered in HCV infected siBCN1-IHH. The results displayed a significant increase in IFN-α and its downstream molecule IFI27 expression in HCV infected siBCN1-IHH as compared to HCV infected control IHH (Fig. 3). Together, these results indicated that HCV induced autophagy suppresses the expression of interferon stimulated genes.
Figure 3. HCV infection upregulates IFN signaling molecules in Beclin1 knockdown IHH.
Total cellular RNA was extracted from HCV genotype 1a (panel A) or genotype 2a (panel B) infected siBCN1-IHH or control IHH after 3 days of infection. Intracellular mRNA expressions of IFN-β, 2′-5′ OAS1, IFNA1 or IFI27 was measured by real-time RT-PCR. GAPDH was used as an internal control. The fold changes of mRNA are presented after normalizing with mock infected control. The results are presented as mean from four independent experiments with standard errors (P < 0.001).
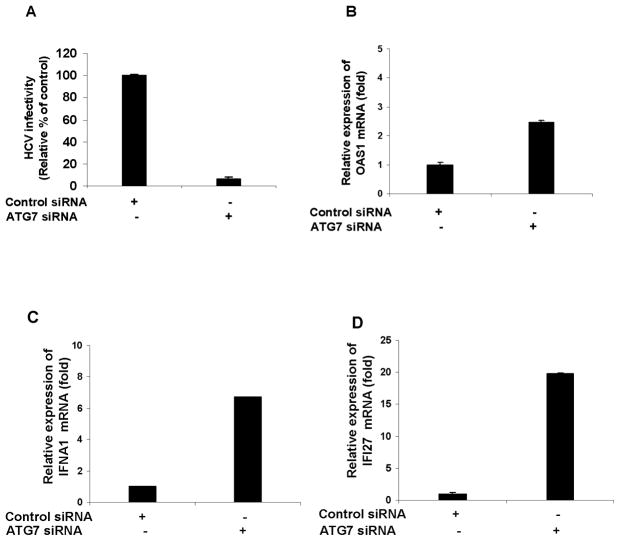
To further confirm the modulation of ISGs in HCV infected autophagy knockdown cells, we used another autophagy protein, ATG7. Knockdown of ATG7 in IHH (siATG7-IHH) does not alter the cell viability or display any off-target effect (data not shown). Control IHH or siATG7-IHH was infected with HCV (genotype 1a) for 72h. Virus release was measured and a decrease in HCV growth in siATG7-IHH as compared to HCV infected control IHH was observed (Fig. 4, panel A). OAS-1 (panel B), IFNA1 (panel C) and IFI27 (panel D) mRNA expression were upregulated in HCV infected siATG7-IHH as compared to HCV infected control IHH (Fig. 4). Similar results were obtained when cells infected with HCV genotype 2a. Together these results suggested that disruption of autophagy machinery induces IFN-signaling pathway in HCV infected hepatocytes.
Figure 4. HCV infection in ATG7 knockdown IHH upregulates IFN signaling molecules.
Virus growth was measured from culture supernatants of HCV (genotype 1a) infected siATG7-IHH or control IHH (panel A). Cellular RNA was extracted from virus infected siATG7-IHH or control IHH after 3 days of infection. Intracellular gene expression of 2′-5′ OAS1 (panel B), IFNA1 (panel C) or IFI27 (panel D) were measured as described in Fig. 3. The results are presented as mean from three independent experiments with standard errors (P < 0.001).
HCV infection in autophagy knockdown IHH induces apoptosis
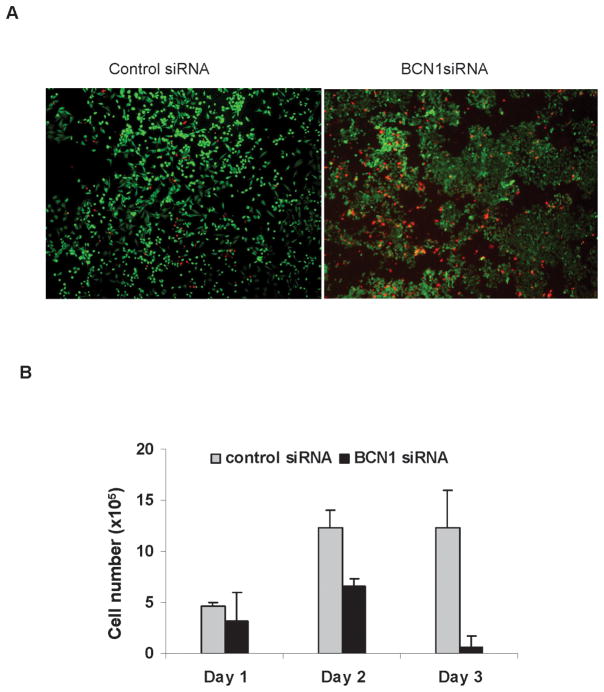
We hypothesized that autophagy promotes survival of HCV infected cells for virus persistence. Therefore, viability of HCV infected control IHH and siBCN1-IHH was examined. For this, cells were initially stained with Calcein AM and ethidium homodimer-1 dye to quantitate live and dead cells (Fig. 5, panel A). The percentage of dead cells (red color) was significantly higher in HCV infected siBCN1-IHH (~70 %) as compared to HCV infected control IHH (~20 %). A time course experiments involving cell viability following HCV infection was performed. A significant inhibition of cell viability was noted in HCV infected siBCN1-IHH as compared to that of control IHH (Fig. 5, panel B).
Figure 5. HCV infection in autophagy knockdown IHH induces apoptotic cell death.
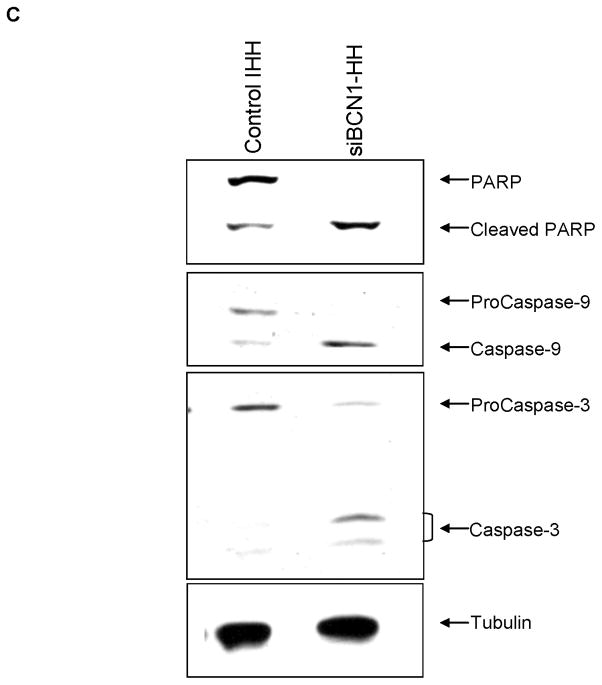
Panel A: Control IHH or siBCN1-IHH infected with HCV were stained with Calcein AM (green color for live cells) and ethidium homodimer-1 (red color for dead cells) dye to quantitate the live and dead cells by fluorescence microscopy. Panel B: Cell viability was examined in HCV infected control IHH or siBCN1-IHH. Number of viable cells was counted at 1, 2 and 3 days by trypan blue exclusion. Results are presented as the mean of three separate experiments with standard errors (P < 0.001). Panel C: HCV infected control IHH or siBCN1-IHH were subjected to Western blot analysis using specific antibody for detection of PARP, caspase-9 or caspase-3. PARP was significantly cleaved to an 86 kD signature peptide in HCV infected siBCN1-IHH as compared to HCV infected control IHH. Caspase 9 and caspase 3 activation were also observed in HCV infected siBCN1-IHH. The blot was reprobed with an antibody to tubulin for comparison of equal protein load.
Subsequently, apoptosis as a possible mechanism of cell death was examined. HCV infected control IHH and siBCN1-IHH were incubated for 72 h. Cell lysates were examined for induction of apoptosis. PARP was significantly cleaved to an 86 kD signature peptide in HCV infected siBCN1-IHH as compared to HCV infected control IHH (Fig. 5, panel C). Our results also demonstrated that Beclin1 knockdown IHH infected with HCV induces caspase-9 and caspase-3 activation. Both procaspase-9 and procaspase-3 were cleaved to 37 kD and 17 kD protein band, respectively (Fig. 5, panel C). Similar results were obtained in HCV infected siATG7-IHH (data not shown). Therefore it is conceivable that autophagy machinery is needed for HCV infected cell survival, and impairment of this pathway induces apoptosis.
Discussion
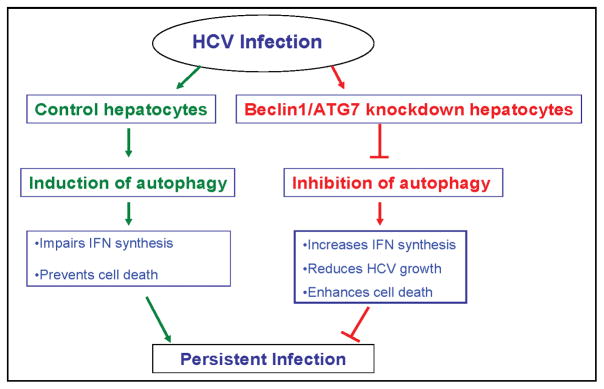
Autophagy is recently identified as a novel component of innate immune system against viral infection. In this study, we have observed that HCV infected siBCN1-IHH does not induce autophagy and virus growth is reduced. We have further demonstrated that HCV infected siBCN1-IHH induces IFN-β, OAS-1, IFN-α and IFI27 mRNA expression, and apoptotic cell death. Similar results were also obtained from ATG7 knockdown HCV infected IHH. We propose that HCV induces autophagy in favor of its own survival, and inhibition of autophagic proteins enhances cell death, and as a result virus growth is reduced (Fig. 6), and may have a potential for future therapeutic modalities.
Figure 6.
Role of autophagy as a proposed mechanism for establishment of persistent HCV infection.
Autophagy plays a key role in recognizing signatures of viral infection, and represents a critical effector mechanism to restrict virus production (25). Upon invasion by a pathogen, the host may initiate autophagosome formation as a cellular defense. Autophagy is proposed to serve as a scaffold for intracellular membrane associated replication for RNA viruses. In rotavirus infected cells, NSP4 protein is involved in virus replication, colocalized with LC3 in double layered vesicular compartment, a site for nascent viral RNA replication (26). In dengue virus infected cells, LC3 colocalizes with double stranded RNA and with NS1 protein suggesting the presence of replication complexes in autophagic vesicles (27). We and others have shown that HCV induces autophagy by accumulation of autophagosomes in infected hepatocytes without colocalization of HCV and autophagy related proteins (12, 13, 16, 28). Knockdown of autophagy proteins in HCV infected cells or replicon bearing hepatocytes does not impair the expression levels of mRNA or proteins (13, 16). Insufficient packaging of viral RNA or blockage in virus release may be a mechanism for suppression of HCV production in autophagy impaired cells. Indeed, further work is necessary in understanding the in-depth mechanism for suppression of infectious virus particle production. The cell type specificity is associated with autophagy machinery. For example, in lung epithelial A549 cells, autophagy machinery favors viral protein accumulation and infectious viral yield (29), while autophagy has no effect on influenza A virus replication and viral titers in mouse embryo fibroblasts (30). Consistent with the previous reports using HCV genotype 2a system in Huh 7 cell line or its derivatives (13, 16), we also observed a reduced infectious HCV particle release in autophagy deficient IHH.
ATG5 has been shown to be essential for the production of type I interferon in plasmacytoid dendritic cells infected with vesicular stomatitis virus by a mechanism presumed to involve autophagy mediated delivery of viral genetic material to endosomal TLRs (31). On the other hand, several studies have shown that the absence or knockdown of autophagy genes in certain cell types can result in enhanced production of type I interferon or other cytokines, including proinflammatory molecules (11, 32–34). In agreement with the later, we have seen that HCV infection in Beclin1 or ATG7 knockdown IHH increases IFN-β, OAS-1, IFN-α synthesis and enhances IFI27 mRNA. The Atg5-Atg12 conjugate interacts with cytoplasmic viral sensors, the retinoic acid inducible helicases RIG-I/Mda5 and their adaptor protein IPS-1/MAVS, to suppress the activity of such helicases in stimulating the production of type I interferon (32). HCV infection also cleaves these helicases and interferes in IFN signaling pathway (35, 36). Knockdown of Beclin1 in IHH does not induce IFN related gene expression, and Beclin1 knockdown cells infected with HCV does not induce autophagy. Therefore it is possible that the autophagic machinery as well as HCV infection may suppress innate immune signaling by directly inhibiting the interactions with these helicases and their adaptor proteins. Thus, the autophagic machinery may serve a dual function in innate immune signaling, acting not only to modulate antiviral type I interferon responses in host cells, but also to ensure homeostatic balance by preventing excess innate immune activation in other cell types.
Autophagy is also involved in biological pathways and possesses a dual role in mediating cell survival and cell death. Autophagy acts as a cell survival mechanism in tobacco mosaic virus where it restricts the virus to spread from infected to healthy tissue and regulates the programmed cell death in neighboring uninfected cells (37). In autophagy protein deficient cells, influenza A virus induces apoptosis, suggesting that autophagy favors survival of infected cells (30). We have also documented an increase of apoptotic cell death in HCV infected autophagy impaired IHH, reflecting that autophagy promotes cell survival. It is possible that HCV mediated autophagy may sustain cell viability during virus induced stress in order to prevent apoptosis. IFN-α can induce cell death by modulating Jak-STAT or PI3-K/Akt signaling pathway, STAT3 activation or cytokine induction (38). Therefore, IFN induction in autophagy impaired HCV infected cells may mediate apoptotic cell death. Autophagy has been suggested to extend the survival time of human parvovirus B19-infected erythroid cells during viral expansion (39). The hepatitis B virus (HBV)-encoded transcriptional transactivator protein X (HBx) upregulates Beclin1 expression and stimulate autophagy (40). We have shown previously that HCV infection enhances Beclin1 (12). Therefore, it is plausible that HCV induces autophagy to prolong cell survival. This process may help to initiate the development of liver disease progression, including HCC, through an as yet uncharacterized mechanism.
In conclusion, the results from this study revealed: (i) HCV infection in autophagy knockdown cells induces IFN signaling pathway, and (ii) enhances hepatocyte death. Therefore, the autophagy serves as a pivotal entity which may help HCV in establishing persistent infection, reducing antiviral innate immunity, and promoting cell survival. The interaction of virus with the autophagy machinery involves multiple pathways which have only just begun to be characterized. However, other mechanisms, either simultaneously or subsequently, may also be involved in the establishment of chronic HCV infection in humans.
Acknowledgments
This work was supported by research grants DK081817 and AI065535 (R.B.R.), and 5U54AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (R.R.) from the National Institutes of Health.
We thank Charlie Rice, Takaji Wakita, Chen Liu for providing HCV clones and HCV NS5A antibody,and Leonard Grosso for helping to measure the genome copy number of HCV (IU/ml).
Footnotes
Potential conflict of interest: Nothing to report
References
- 1.Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, Alter MJ. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med. 2006;144:705–714. doi: 10.7326/0003-4819-144-10-200605160-00004. [DOI] [PubMed] [Google Scholar]
- 2.Lee WM, Dienstag JL, Lindsay KL, Lok AS, Bonkovsky HL, Shiffman ML, et al. Evaluation of the HALT-C Trial: pegylated interferon as maintenance therapy for chronic hepatitis C in previous interferon nonresponders. Control Clin Trials. 2004;25:472–492. doi: 10.1016/j.cct.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Schiff ER. Emerging strategies for pegylated interferon combination therapy. Nat Clin Pract Gastroenterol Hepatol. 2007;4:S17–S21. doi: 10.1038/ncpgasthep0691. [DOI] [PubMed] [Google Scholar]
- 4.Andersson K, Chung RT. Hepatitis C virus in the HIV-infected patient. Clin Liver Dis. 2006;10:303–320. doi: 10.1016/j.cld.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Bortolotti F, Verucchi G, Cammà C, Cabibbo G, Zancan L, Indolfi G, et al. Long-term course of chronic hepatitis C in children: from viral clearance to end-stage liver disease. Gastroenterology. 2008;134:1900–1907. doi: 10.1053/j.gastro.2008.02.082. [DOI] [PubMed] [Google Scholar]
- 6.Williams R. Global challenges in liver disease. Hepatology. 2006;44:521–526. doi: 10.1002/hep.21347. [DOI] [PubMed] [Google Scholar]
- 7.McLauchlan J. Hepatitis C virus: viral proteins on the move. Biochem Soc Trans. 2009;37:986–990. doi: 10.1042/BST0370986. [DOI] [PubMed] [Google Scholar]
- 8.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 9.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 10.Dreux M, Chisari FV. Viruses and the autophagy machinery. Cell Cycle. 2010;9:1295–1307. doi: 10.4161/cc.9.7.11109. [DOI] [PubMed] [Google Scholar]
- 11.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizui T, Yamashina S, Tanida I, Takei Y, Ueno T, Sakamoto N, et al. Inhibition of hepatitis C virus replication by chloroquine targeting virus-associated autophagy. J Gastroenterol. 2010;45:195–203. doi: 10.1007/s00535-009-0132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy. 2009;5:937–945. doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- 17.Lin LT, Dawson PW, Richardson CD. Viral interactions with macroautophagy: a double-edgedsword. Virology. 2010;402:1–10. doi: 10.1016/j.virol.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Sir D, Ou JH. Autophagy in viral replication and pathogenesis. Mol Cells. 2010;29:1–7. doi: 10.1007/s10059-010-0014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanda T, Basu A, Steele R, Wakita T, Ryerse JS, Ray R, et al. Generation of infectious hepatitis C virus in immortalized human hepatocytes. J Virol. 2006;80:4633–4639. doi: 10.1128/JVI.80.9.4633-4639.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 2007;17:839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 23.Raychoudhuri A, Shrivastava S, Steele R, Dash S, Kanda T, Ray R, et al. Hepatitis C virus infection impairs IRF-7 translocation and interferon-{alpha} synthesis in immortalized human hepatocytes. J Virol. 2010 Sep 1; doi: 10.1128/JVI.00900-10. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanda T, Steele R, Ray R, Ray RB. Hepatitis C virus infection induces the beta interferon signaling pathway in immortalized human hepatocytes. J Virol. 2007;81:12375–12381. doi: 10.1128/JVI.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HK, Iwasaki A. Autophagy and antiviral immunity. Curr Opin Immunol. 2008;20:23–29. doi: 10.1016/j.coi.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkova Z, Crawford SE, Trugnan G, Yoshimori T, Morris AP, Estes MK. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J Virol. 2006;80:6061–6071. doi: 10.1128/JVI.02167-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panyasrivanit M, Khakpoor A, Wikan N, Smith DR. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. J Gen Virol. 2009;90:448–456. doi: 10.1099/vir.0.005355-0. [DOI] [PubMed] [Google Scholar]
- 28.Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010;91:2230–2237. doi: 10.1099/vir.0.022186-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Z, Jiang X, Liu D, Fan Z, Hu X, Yan J, et al. Autophagy is involved in influenza A virus replication. Autophagy. 2009;5:321–328. doi: 10.4161/auto.5.3.7406. [DOI] [PubMed] [Google Scholar]
- 30.Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Rämer PC, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009;6:367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 32.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 35.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Herzer K, Hofmann TG, Teufel A, Schimanski CC, Moehler M, Kanzler S, et al. IFN-alpha-induced apoptosis in hepatocellular carcinoma involves promyelocytic leukemia protein and TRAIL independently of p53. Cancer Res. 2009;69:855–862. doi: 10.1158/0008-5472.CAN-08-2831. [DOI] [PubMed] [Google Scholar]
- 39.Nakashima A, Tanaka N, Tamai K, Kyuuma M, Ishikawa Y, Sato H, et al. Survival of parvovirus B19-infected cells by cellular autophagy. Virology. 2006;349:254–263. doi: 10.1016/j.virol.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 40.Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]