Abstract
Purpose
Myopia, or nearsightedness, is highly prevalent in Asian countries and is considered a serious public health issue globally. High-grade myopia can predispose individuals to myopic maculopathy, premature cataracts, retinal detachment, and glaucoma. A recent study implicated zinc finger protein 644 isoform 1 (ZNF644) variants with non-syndromic high-grade myopia in a Chinese-Asian population. Herein we focused on investigating the role for ZNF644 variants in high-grade myopia in a United States (US) cohort.
Methods
DNA from a case cohort of 131 subject participants diagnosed with high-grade myopia was screened for ZNF644 variants. Spherical refractive error of -≤-6.00 diopters (D) in at least one eye was defined as affected. All coding, intron/exon boundaries were screened using Sanger sequencing. Single nucleotide allele frequencies were determined by screening 672 ethnically matched controls.
Results
Sequencing analysis did not detect previously reported mutations. However, our analysis identified 2 novel single nucleotide variants (c.725C>T, c.821A>T) in 2 high-grade myopia individuals- one Caucasian and one African American, respectively. These variants were not found in normal controls. A rare variant - dbsSNP132 (rs12117237→c.2119A>G) - with a minor allele frequency of 0.2% was present in 6 additional cases, but was also present in 5 controls.
Conclusions
Our study has identified two novel variants in ZNF644 associated with high-grade myopia in a US cohort. Our results suggest that ZNF644 may play a role in myopia development.
Introduction
Myopia is a common ocular disorder resulting from excessive axial elongation of the globe [1,2]. High myopia in particular can predispose one to several ocular complications such as myopic maculopathy, premature cataracts, retinal detachment, and glaucoma, and is considered a public health concern in numerous countries around the world [3]. Myopia prevalence rates vary world-wide. A recent study by Vitale et al. [4] reported that 33.1% of the USA population greater than the age of 20 has some degree of myopia. Studies have shown that prevalence rates in Asia countries are higher. For example, 84% of school children in Taiwan develop myopia by the age of 18 [5,6]. Additionally, more than 85% of Hong Kong Chinese school children aged between 13 and 15 years are myopic [7]. The economic impact of refractive error management can be substantial- vision impairment correction costs account for 3.8 to 7.2 billion dollars annually in the USA alone [8].
Accepted as a common complex disorder, myopia is thought to be influenced by both genetic and environmental factors [9,10]. To date, over 20 genetic loci have been mapped using linkage analysis for both common myopia and high myopia [11-17]. A recent plethora of genome wide association studies have shown positive associations with refractive error [14,18-25]. Of late, large parallel sequencing techniques have been used to identify causal genes for ocular disorders including myopia. Rare ocular diseases such as retinitis pigmentosa and familial exudative vitreoretinopathy were among the first where causative genes were successfully identified using exome sequencing [26-28]. More recently, Mordechai et al. [29] identified a leprecan-like 1 (LEPREL1) mutation as causal for autosomal recessive high-grade axial myopia in a large, consanguinous Bedouin Israeli kindred. The mutation identified in their study demonstrated an autosomal recessive mode of inheritance with variable expressivity. As the lone finding for high myopia and autosomal dominant inheritance, Shi et al. [30] recently used exome sequencing to identify mutations in zinc finger protein 644 isoform 1 (ZNF644) – first in a large pedigree with autosomal dominant high myopia, and then replicated in a Chinese cohort. ZNF644 is a transcription factor gene, and is expressed in the retina and retinal pigment epithelium (RPE) [30]. To our knowledge, the role of ZNF644 has not been studied in a myopic USA cohort composed primarily of Caucasians. We therefore screened for ZNF644 mutations in a high-grade myopia USA population data set. The identification of mutations in ZNF644 in other ethnicities supplements existing knowledge of the etiology and heterogeneity of this debilitating eye disorder.
Methods
Patient information
Informed consent was obtained from all participants before entering the study, with approval by the Institutional Review Board according to the principles of the Declaration of Helsinki. High-grade myopia cases were defined as individuals with a spherical refractive error of greater than or equal to −6.00 diopters (D) in at least one eye. From venous blood samples, genomic DNA was extracted using AutoPure LS® DNA Extractor and PUREGENE™ reagents (Gentra Systems Inc., Minneapolis, MN). DNA for 672 ethnically matched Caucasian healthy control participants was purchased commercially (The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Canada). Although the controls had no documented ocular malformations, refractive error information was not available for these individuals. Additionally, 50 ethnically matched African American participant controls with refractive error data were ascertained, and blood for DNA extraction was collected for this study.
PCR and sequence analysis
ZNF644 (NM_201269.1) encodes for a zinc finger transcription factor which maps to chromosome 1p22.2 (Chromosome1:91,380,860–91,487,671 – GRCh37.p5). The gene comprises 6 exons, five of which are coding. Polymerase chain reaction (PCR) and sequencing primers were designed to cover all coding and untranslated gene regions (UTR) including intron-exon boundaries using the ExonPrimer program (Helmholtz Center, Munich, Germany). Primers were selected to produce amplification products not to exceed 850 base pairs (bp) in size for optimal sequence output and analysis. A total of thirteen primer sets were designed to ensure full coverage of the exons and the flanking intronic regions (Table 1).
Table 1. Primers for PCR and sequencing of ZNF644.
| ZNF644 exon | Forward | Reverse | Product size (base pair) |
|---|---|---|---|
| Exon 1 |
AAAATGCGTCCTTTTGGATG |
GGAGGTGACCTTGTTTGGTT |
492 |
| Exon 2 |
AATGATGGTATTCTGGTTG |
AAGTCAATTATTTGCATTTC |
363 |
| Exon 2* |
ATCAGACCTGGAGAGGCAAA |
TAGTCACATGAAGCCGAGCA |
353 |
| Exon 3.1 |
TCTGTGGTGTAGACAGCTGAA |
TTGTATACATGACGTATTGGACTGTT |
697 |
| Exon 3.2 |
CTTTTTGGGGATCCCAGTTT |
ACGTTGACTCTGCCTGAAGAA |
580 |
| Exon 3.3 |
TGAAAGTAGCAGGTGACTCAGAA |
GTGGATCAGCCAACAACAGA |
778 |
| Exon 3.4 |
CAGGTTCTTCAAGGATGTCATTT |
TGTGGAGAAGAGAGTTCACCTG |
796 |
| Exon 3.5 |
TTCTTTTCAGCAGAATTAAGTTTTTG |
AGCACACGGAGTACTTGCATT |
742 |
| Exon 3.6 |
AAACTGACCACCCTAAAATGAGTT |
TGGAGGGGAAGACTTGGATA |
759 |
| Exon 4 |
GCAGCTTAAACAGGAAGATTGTG |
GAATTAACTCATTTTAGGGTGGTCA |
790 |
| Exon 5 |
TTTTAAGCCTATCTCCAAAAGTTCA |
GAATGCATGCTTCAGGGAAT |
395 |
| Exon 6.1 |
TTAAAAACACATCTTCCACCCTA |
TGAATTGGGAGTTTTGATGTTT |
564 |
| Exon 6.2 |
CGTCTATTCTAAACTGTGTAGTGAGCA |
ACAGTGACATCAGAGCAAATTGA |
829 |
| Exon 6.3 | CATTATATTGACCAATGAGGTGATTC | TGCTTACAGGACAGGTTTGC | 782 |
*Denotes isoform 2 of exon 2, which is an untranslated exon.
Samples were amplified using standard PCR protocol and amplicons were visualized after agarose gel (2%) electrophoresis. Sequencing of the amplicons was then completed on an Applied Biosystems ABI3730 xlrobotics using BigDye™ Terminator 3.1 technology (Applied Biosystems, Inc. [ABI], Foster City, CA). Sequences were analyzed using the Sequencher 5.0™ program (Gene Codes, Ann Arbor, MI), and were compared against the known reference sequence (GRCh37.p5) and analyzed for sequence variation. Single nucleotide variants (SNVs) that were novel and/or coding non-synonymous with a minor allele frequency (MAF) of less than 1% were checked for co-segregation in remaining family member samples.
Genotyping
Allelic discrimination assays were employed to measure the allelic frequencies in 672 Caucasian matched control DNA samples using the TaqMan® SNP Genotyping system (Applied Biosystems). Assays were designed according to Applied Biosystems specifications using a combination of unlabeled primers and minor groove binding (MGB) probes with fluorescently labeled dyes (FAM and VIC) to interrogate the base pair of interest. Reactions were completed and ABI 7900 robotics was used to read the allelic calls for each control sample (Applied Biosystems). SDS v2.4 Software provided by ABI was used to analyze each sample and accurately analyze each genotype call.
Results
Full ophthalmologic exams were performed on all subject participants with one or more individual(s) with high grade myopia. The average cycloplegic spherical refractive error for 131 high myopia cases was −11.22 D for the right eye (OD) and −11.48 for the left eye (OS), and the range was from −6.00 D to −50.00 D (OD) and −5.25 D to −50.00 D (OS) across all cases. Of the 131 case participants screened for ZNF644, 74% (97/131) were Caucasians, 12.2% (16/131) were African Americans, 10% (13/131) were Asians, while 5 individuals (3.8%) were of Hispanic descent or declined to self-identify.
The entire coding and untranslated DNA sequence of ZNF644 was sequenced in 131 high myopia patients. In all, we identified a total of 31 heterozygous SNVs in ZNF644, of which 10 were missense, 7 silent, 9 untranslated, and 5 intronic (Table 2 and Table 3).
Table 2. Summary of affected patients with ZNF644 variants in 131 high-grade myopia cases.
|
Family |
Individual |
Gender |
Ethnicity |
Spherical Refractive Error (D) |
Exon |
Amino Acid Change |
cDNA Position |
Chromosome 1 base pair location* |
Variant Identification† |
Co-segregation |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| OD | OS | ||||||||||
| MYP104 |
IND0603564 |
M |
African American |
−18.00 |
−18.00 |
3 |
T242M |
c.725C>T |
91406186 |
Novel |
N/A |
| MYP163 |
IND0603809 |
F |
Caucasian |
−7.50 |
−6.75 |
3 |
E274V |
c.821A>T |
91406090 |
Novel |
N/A |
| MYP8 |
IND0519772 |
F |
Asian |
−11.00 |
−12.25 |
3 |
H706Y |
c.2116C>T |
91404795 |
Novel |
NO |
| MYP19 |
IND0519791 |
M |
Caucasian |
−19.25 |
−21.00 |
3 |
K707E |
c.2119A>G |
91404792 |
rs12117237 |
YES |
| MYP83 |
IND0603414 |
M |
Caucasian |
−9.75 |
−9.00 |
3 |
K707E |
c.2119A>G |
91404792 |
rs12117237 |
NO |
| MYP89 |
IND0603458 |
M |
Caucasian |
−6.00 |
−6.00 |
3 |
K707E |
c.2119A>G |
91404792 |
rs12117237 |
YES |
| MYP102 |
IND0603552 |
F |
Caucasian |
−16.00 |
−18.00 |
3 |
K707E |
c.2119A>G |
91404792 |
rs12117237 |
N/A |
| MYP113 |
IND0603616 |
F |
Caucasian |
−12.75 |
−13.50 |
3 |
K707E |
c.2119A>G |
91404792 |
rs12117237 |
NO |
| MYP129 |
IND0603676 |
F |
Caucasian |
−5.50 |
−6.25 |
3 |
K707E |
c.2119A>G |
91404792 |
rs12117237 |
N/A |
| MYP2 | IND0519764 | M | Caucasian | −10.75 | −10.50 | 4 | R1100H | c.3299G>A | 91403431 | Novel | NO |
Rare is defined by variants not found in controls or have minor allele frequency of less than 1% . Abbreviations: N/A- not available, OD- right eye, OS- left eye, DS- Diopter Sphere, M- Male, F- Female, *UCSC hg19 browser – GRCh37.p5, †-dbSNP132.
Table 3. Summary of variants identified in ZNF644 in 131 high-grade myopia cases.
| Chromosome 1 base pair location* | Variant type | dbSNP132 | Allele change | Amino acid change | Population |
|---|---|---|---|---|---|
| 91487710 |
UTR |
Novel |
A>G |
N/A |
Caucasian |
| 91487657 |
UTR |
Novel |
C>T |
N/A |
Caucasian |
| 91487013 |
UTR |
Novel |
G>T |
N/A |
Caucasian |
| 91447985 |
Intronic |
rs358691 |
A>G |
N/A |
Caucasian |
| 91406677 |
Synonymous |
rs17131243 |
G>A |
L78L |
African American |
| 91406033 |
Synonymous |
Novel |
G>A |
R293Q |
Caucasian |
| 91405699 |
Synonymous |
rs41286763 |
C>T |
T404T |
Caucasian |
| 91405245 |
Nonsynonymous |
rs17131242 |
A>G |
M556V |
African American¥ |
| 91405215 |
Nonsynonymous |
rs60262072 |
A>T |
T566S |
African American¥ |
| 91404592 |
Synonymous |
Novel |
C>T |
H773H |
Hispanic |
| 91404532 |
Synonymous |
Novel |
C>T |
D793D |
African American |
| 91404530 |
Nonsynonymous |
rs10922938 |
C>T |
A794V |
African American¥ |
| 91404303 |
Nonsynonymous |
rs59922637 |
A>G |
T870A |
African American† |
| 91404256 |
Nonsynonymous |
rs41286761 |
G>T |
E885D |
Caucasian |
| 91383756 |
Intronic |
Novel |
A>G |
N/A |
Caucasian |
| 91383589 |
Intronic |
rs2448020 |
G>T |
N/A |
Multiple |
| 91382635 |
Intronic |
rs17131234 |
G>C |
N/A |
African American |
| 91382406 |
Synonymous |
rs114618312 |
C>T |
A1311A |
African American |
| 91382370 |
Synonymous |
Novel |
C>T |
A1323A |
Asian |
| 91382086 |
UTR |
Novel |
A>T |
N/A |
African American |
| 91381679 |
UTR |
rs1188952 |
C>T |
N/A |
Multiple |
| 91381534 |
UTR |
Novel |
A>C |
N/A |
Caucasian |
| 91381240 |
UTR |
Novel |
C>T |
N/A |
Caucasian |
| 91381181 |
UTR |
Novel |
G>C |
N/A |
African American |
| 91381105 |
UTR |
rs17131232 |
A>T |
N/A |
Multiple |
| 91380797 | Intronic | rs115299241 | C>T | N/A | African American |
Abbreviations: N/A- not applicable, UTR- untranslated region, Multiple - variant found in more than one population, dbSNP132. *GRCh37.p5. ¥-Identified in IND0603564. †-Identified in IND0603416.
From seven synonymous sequence variants detected in our cohort, we identified 4 novel SNVs in four separate individuals, which were either within exon 3 or 6. We also found 9 SNVs in the UTR, where 6 of 7novel variants were unique to an individual. In addition to the exonic variants, 4 intronic variants were previously reported in the dbSNP132 database, while 1 was novel and unique to an individual. All intronic variants observed had high MAFs, and/or were not located near splice-site junctions (Table 2).
Of the 10 missense variants identified, 4 were novel and 6 were reported in the dbSNP132 database (Table 3). One novel missense variant was identified in Caucasian individual IND0603809. This adenine to thymine substitution at position c.821A>T alters glutamic acid to valine (Glu274Val), and was not found in other high myopic case samples. A second novel missense variant was identified on exon 3 in African American individual IND0603564. This cytosine to thymine substitution (c.725C>T) results in a threonine to methionine (Thr242Met) amino acid change was only present in this individual. Due to the lack of additional family members for these individuals, segregation analysis was not possible (Figure 1). Novel variants c.2116C>T (His706Tyr) and c.3299G>A (Arg1100His) were identified in Caucasian individuals IND0519772 and IND0519764, respectively. Neither variant segregated with disease after screening available family members.
Figure 1.
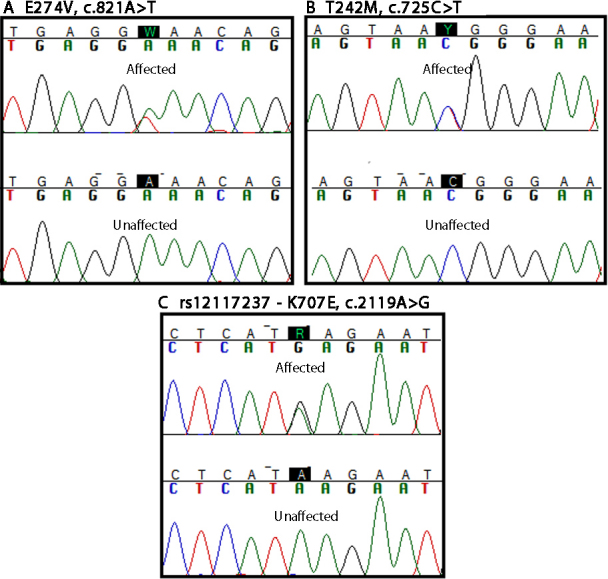
Sequence chromatogram of identified ZNF644 variants. Base pair location in bold depicts the variant change in affected individual compared to an unaffected individual. A: Novel variant identified in individual MYP0603809 with E274V (c.821A>T) change. B: Novel variant identified in individual MYP0603564 with T242M (c.725C>T) change. C: rs12117237 variant (K707E, c.2119A>G) that was present in 5 high myopic cases.
To confirm the rarity of c.821A>T, a TaqMan genotyping assay was employed to screen a DNA sample data set of Caucasian controls. The variant (c.821A>T) was not replicated in 672 Caucasian controls by allelic discrimination genotyping method. The novel variant found in African American individual IND0603564 (c.725C>T) was not replicated in 50 African American control samples via Sanger sequencing (Figure 1).
Four missense variants in dbSNP132 were identified in African American individuals IND0603564 and IND0603416 but were ruled out due to minor allele frequency not meeting criteria below 1% (Table 3). Missense variant rs12117237, the only variant in dbSNP132 with a minor allele frequency of less than 1% in public databases was identified in 6 Caucasian high myopia cases (0.2%). The minor allele was also seen in 0.75% (5/672) of controls. Fisher’s exact two-sided test for rs12117237 demonstrated a p-value of 0.0015 when comparing the prevalence rates of the allele between cases and controls (SAS Institute Inc., Cary, NC). Other family members of the cases were sequenced to determine co-segregation. Two cases had uninformative family members or were simplex cases, and thus were not useful for determining segregation of the variant. Of the 6 myopia cases identified with rs12117237, 4 families (MYP19, MYP83, MYP89, and MYP113) had at least 3 additional family members. These members were sequenced, and MYP19 and MYP89 demonstrated phenotype co-segregation with the SNV (Figure 2).
Figure 2.
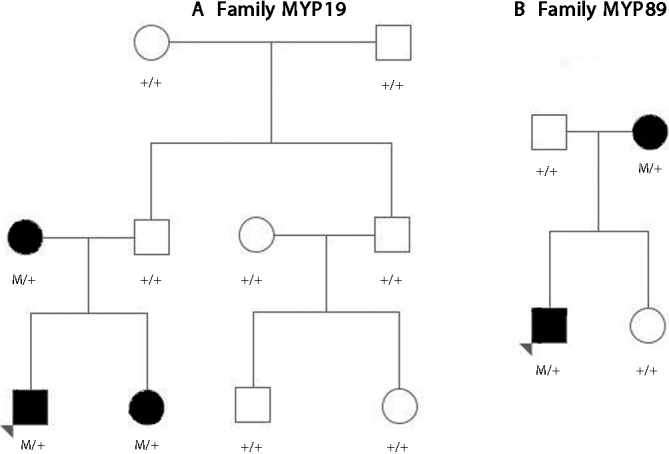
Pedigree and segregation of rs12117237 on MYP19 and MYP89 family. A: Kindred structure and segregation of ZNF644 rs12117237 in MYP19. B: Kindred structure and segregation of ZNF644 rs12117237 in MYP89. Affected individuals are identified by solid squares (male) or circles (females). Normal individuals are identified by open symbols. Colored triangle depicts index case patient. M: 707E recessive allele of ZNF644; +: K707 normal allele of ZNF644.
Discussion
Mutation screening of ZNF644 has successfully identified 2 novel missense variants (c.725C>T, c.821A>T) in two ethnicities within our US population, thus supporting the original report of a causal candidate gene for high-grade myopia (Table 2). The two novel variants from our study (c.725C>T, c.821A>T) localize to a conserved region on the third exon, where Shi et al. [30] previously identified several mutations. The variants were not seen in 1344 and 100 ethnically matched chromosomes, respectively, confirming their rarity. PolyPhen-2 software predicted c.725C>T to be possibly damaging, while c.821A>T was tolerated. With the additional variant information from our study, exon 3 may be a hotspot for susceptibility for high-grade myopia in multiple ethnicities. The clustering of variants in exon 3 may depict the importance in protein domain structures and gene regulatory functions of ZNF644.
The single nucleotide variant rs12117237 was present in 6 Caucasian case samples, but also present in 5 ethnically matched control DNA samples. Public database dbSNP132 reports this variant’s MAF to be 0.2% in a European population, and our p-value of 0.0015 by Fisher’s exact test suggests presence of the alleles likely did not occur by chance. As a result of sequencing additional family members in three high myopia cases for the variant, we discovered 100% co-segregation of the mutation with the disease phenotype in two of four families. PolyPhen-2 software predicts the amino acid change to be tolerated. The variant could be a common rare variant, and proper functional validation would be important to determine whether it is a risk or a causal allele [31]. However, the MAF is in line with prevalence rates for high-grade myopia in a general population and a founder mutation effect may be plausible. Clinical information was missing refractive error from the 5 ethnically matched controls who also presented with the variant in the heterozygous state. The prevalence rate of high myopia is estimated to be 4.5% in the US population [32,33]. Moreover, a recent study suggests that the rarer a variant, the higher the likelihood that the variant functionally alters the protein – thus rs12117237 may be associated with high-grade myopia in a Caucasian population [34]. A larger case-control population data set should be tested to understand the true significance of the association of this variant to high-grade myopia in Caucasians.
ZNF644 is a transcription factor that may play a role in protein domain structures or regulatory functions [30]. Expressed in all tissue types, it follows the trend of ubiquitously expressed genes pathogenic to ocular diseases [35]. To date, it is widely accepted that transcription factor genes in both humans and mouse can play an important role in mammalian eye growth and development. For example, paired-like homeodomain transcription factors 2 and 3 (PITX2, PITX3) have been implicated in Axenfeld-Rieger syndrome and cataracts, respectively [36-38]. Microphthalmia transcription factor (Mitf) is associated with ocular albinism, and paired box 6 (PAX6) has been associated with retinal degeneration, extreme myopia, and corneal innervation [20,39-46]. However, the true function of ZNF644 remains unclear, and molecular characterization of ZNF644 is necessary.
To the best of our knowledge, this is the first successful study confirming a gene implicated with non-syndromic high-grade myopia determined by exome sequencing. We identified two novel variants in ZNF644 in our cohort, in addition to a known variant that demonstrated association. The discovery of previously unidentified variants is not expected due to genetic heterogeneity present in rare complex disease across multiple populations [47]. Ethnic group specific alleles due to founder effect may explain why the previously reported variants were only present in the Asian population whereas the variants discovered in this report appear to be specific to the Caucasian and African American ethnicities. Determining pathogenic rare missense variants remains a challenge for complex diseases. Identification of novel variants in separate ethnicities emphasizes the importance and demonstrates the power of current research approaches.
Acknowledgments
The authors thank all subjects who participated in this study. This research effort was supported by the National Institutes of Health Grant EY014685, The Lew Wasserman Award from Research To Prevent Blindness Inc., and the Duke- National University of Singapore core grant. (T.L.Y.)
References
- 1.Curtin BJ, Karlin DB. Axial length measurements and fundus changes of the myopic eye. Am J Ophthalmol. 1971;71:42–53. doi: 10.1016/0002-9394(71)91092-0. [DOI] [PubMed] [Google Scholar]
- 2.Curtin BJ. The Myopias: Basic Science and Clinical Management. Philadelphia: Harper & Row; 1985. [Google Scholar]
- 3.Pararajasegaram R. VISION 2020-the right to sight: from strategies to action. Am J Ophthalmol. 1999;128:359–60. doi: 10.1016/s0002-9394(99)00251-2. [DOI] [PubMed] [Google Scholar]
- 4.Vitale S, Ellwein L, Cotch MF, Ferris FL, III, Sperduto R. Prevalence of refractive error in the United States, 1999–2004. Arch Ophthalmol. 2008;126:1111–9. doi: 10.1001/archopht.126.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin LL, Shih YF, Tsai CB, Chen CJ, Lee LA, Hung PT, Hou PK. Epidemiologic study of ocular refraction among schoolchildren in Taiwan in 1995. Optom Vis Sci. 1999;76:275–81. doi: 10.1097/00006324-199905000-00013. [DOI] [PubMed] [Google Scholar]
- 6.Lin LL, Shih YF, Hsiao CK, Chen CJ, Lee LA, Hung PT. Epidemiologic study of the prevalence and severity of myopia among schoolchildren in Taiwan in 2000. J Formos Med Assoc. 2001;100:684–91. [PubMed] [Google Scholar]
- 7.Lam CS, Goldschmidt E, Edwards MH. Prevalence of myopia in local and international schools in Hong Kong. Optom Vis Sci. 2004;81:317–22. doi: 10.1097/01.opx.0000134905.98403.18. [DOI] [PubMed] [Google Scholar]
- 8.Vitale S, Cotch MF, Sperduto R, Ellwein L. Costs of refractive correction of distance vision impairment in the United States, 1999–2002. Ophthalmology. 2006;113:2163–70. doi: 10.1016/j.ophtha.2006.06.033. [DOI] [PubMed] [Google Scholar]
- 9.Cordain L, Eaton SB, Brand MJ, Lindeberg S, Jensen C. An evolutionary analysis of the aetiology and pathogenesis of juvenile-onset myopia. Acta Ophthalmol Scand. 2002;80:125–35. doi: 10.1034/j.1600-0420.2002.800203.x. [DOI] [PubMed] [Google Scholar]
- 10.Hammond CJ, Snieder H, Gilbert CE, Spector TD. Genes and environment in refractive error: the twin eye study. Invest Ophthalmol Vis Sci. 2001;42:1232–6. [PubMed] [Google Scholar]
- 11.Ciner E, Wojciechowski R, Ibay G, Bailey-Wilson JE, Stambolian D. Genomewide scan of ocular refraction in African-American families shows significant linkage to chromosome 7p15. Genet Epidemiol. 2008;32:454–63. doi: 10.1002/gepi.20318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paluru PC, Nallasamy S, Devoto M, Rappaport EF, Young TL. Identification of a novel locus on 2q for autosomal dominant high-grade myopia. Invest Ophthalmol Vis Sci. 2005;46:2300–7. doi: 10.1167/iovs.04-1423. [DOI] [PubMed] [Google Scholar]
- 13.Klein AP, Duggal P, Lee KE, Klein R, Bailey-Wilson JE, Klein BE. Confirmation of linkage to ocular refraction on chromosome 22q and identification of a novel linkage region on 1q. Arch Ophthalmol. 2007;125:80–5. doi: 10.1001/archopht.125.1.80. [DOI] [PubMed] [Google Scholar]
- 14.Lam CY, Tam PO, Fan DS, Fan BJ, Wang DY, Lee CW, Pang CP, Lam DS. A genome-wide scan maps a novel high myopia locus to 5p15. Invest Ophthalmol Vis Sci. 2008;49:3768–78. doi: 10.1167/iovs.07-1126. [DOI] [PubMed] [Google Scholar]
- 15.Nishizaki R, Ota M, Inoko H, Meguro A, Shiota T, Okada E, Mok J, Oka A, Ohno S, Mizuki N. New susceptibility locus for high myopia is linked to the uromodulin-like 1 (UMODL1) gene region on chromosome 21q22.3. Eye. 2009;23:222–9. doi: 10.1038/eye.2008.152. [DOI] [PubMed] [Google Scholar]
- 16.Young TL, Metlapally R, Shay AE. Complex trait genetics of refractive error. Arch Ophthalmol. 2007;125:38–48. doi: 10.1001/archopht.125.1.38. [DOI] [PubMed] [Google Scholar]
- 17.Yu ZQ, Li YB, Huang CX, Chu RY, Hu DN, Shen ZH, Huang W. A genome-wide screening for pathological myopia suggests a novel locus on chromosome 15q12 - 13. Zhonghua Yan Ke Za Zhi. 2007;43:233–8. [PubMed] [Google Scholar]
- 18.Han W, Yap MK, Wang J, Yip SP. Family-based association analysis of hepatocyte growth factor (HGF) gene polymorphisms in high myopia. Invest Ophthalmol Vis Sci. 2006;47:2291–9. doi: 10.1167/iovs.05-1344. [DOI] [PubMed] [Google Scholar]
- 19.Inamori Y, Ota M, Inoko H, Okada E, Nishizaki R, Shiota T, Mok J, Oka A, Ohno S, Mizuki N. The COL1A1 gene and high myopia susceptibility in Japanese. Hum Genet. 2007;122:151–7. doi: 10.1007/s00439-007-0388-1. [DOI] [PubMed] [Google Scholar]
- 20.Liang CL, Hsi E, Chen KC, Pan YR, Wang YS, Juo SH. A functional polymorphism at 3′UTR of the PAX6 gene may confer risk for extreme myopia in the Chinese. Invest Ophthalmol Vis Sci. 2011;52:3500–5. doi: 10.1167/iovs.10-5859. [DOI] [PubMed] [Google Scholar]
- 21.Metlapally R, Li YJ, Tran-Viet KN, Abbott D, Czaja GR, Malecaze F, Calvas P, Mackey D, Rosenberg T, Paget S, Zayats T, Owen MJ, Guggenheim JA, Young TL. COL1A1, COL2A1 genes and myopia susceptibility: Evidence of association and suggestive linkage to the COL2A1 locus. Invest Ophthalmol Vis Sci. 2009;50:4080–6. doi: 10.1167/iovs.08-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metlapally R, Li YJ, Tran-Viet KN, Bulusu A, White TR, Ellis J, Kao D, Young TL. Common MFRP sequence variants are not associated with moderate to high hyperopia, isolated microphthalmia, and high myopia. Mol Vis. 2008;14:387–93. [PMC free article] [PubMed] [Google Scholar]
- 23.Tang W, Yip S, Yap M, Lo K, Ng P, Lee S, Choi P. Testing for association between COL2A1 and myopia susceptibility in Hong Kong Chinese population. ARVO Annual Meeting; 2004 April 25-29; Fort Lauderdale (FL). [Google Scholar]
- 24.Tang WC, Yip SP, Lo KK, Ng PW, Choi PS, Lee SY, Yap MK. Linkage and association of myocilin (MYOC) polymorphisms with high myopia in a Chinese population. Mol Vis. 2007;13:534–44. [PMC free article] [PubMed] [Google Scholar]
- 25.Wang IJ, Chiang TH, Shih YF, Hsiao CK, Lu SC, Hou YC, Lin LL. The association of single nucleotide polymorphisms in the 5′-regulatory region of the lumican gene with susceptibility to high myopia in Taiwan. Mol Vis. 2006;12:852–7. [PubMed] [Google Scholar]
- 26.Nikopoulos K, Gilissen C, Hoischen A, van Nouhuys CE, Boonstra FN, Blokland EA, Arts P, Wieskamp N, Strom TM, Ayuso C, Tilanus MA, Bouwhuis S, Mukhopadhyay A, Scheffer H, Hoefsloot LH, Veltman JA, Cremers FP, Collin RW. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86:240–7. doi: 10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poulter JA, Ali M, Gilmour DF, Rice A, Kondo H, Hayashi K, Mackey DA, Kearns LS, Ruddle JB, Craig JE, Pierce EA, Downey LM, Mohamed MD, Markham AF, Inglehearn CF, Toomes C. Mutations in TSPAN12 cause autosomal-dominant familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86:248–53. doi: 10.1016/j.ajhg.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Züchner S, Dallman J, Wen R, Beecham G, Naj A, Farooq A, Kohli MA, Whitehead PL, Hulme W, Konidari I, Edwards YJ, Cai G, Peter I, Seo D, Buxbaum JD, Haines JL, Blanton S, Young J, Alfonso E, Vance JM, Lam BL, Pericak-Vance MA. Whole-exome sequencing links a variant in DHDDS to retinitis pigmentosa. Am J Hum Genet. 2011;88:201–6. doi: 10.1016/j.ajhg.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mordechai S, Gradstein L, Pasanen A, Ofir R, El Amour K, Levy J, Belfair N, Lifshitz T, Joshua S, Narkis G, Elbedour K, Myllyharju J, Birk OS. High myopia caused by a mutation in LEPREL1, encoding prolyl 3-hydroxylase 2. Am J Hum Genet. 2011;89:438–45. doi: 10.1016/j.ajhg.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Y, Li Y, Zhang D, Zhang H, Lu F, Liu X, He F, Gong B, Cai L, Li R, Liao S, Ma S, Lin H, Cheng J, Zheng H, Shan Y, Chen B, Hu J, Jin X, Zhao P, Chen Y, Zhang Y, Lin Y, Li X, Fan Y, Yang H, Wang J, Yang Z. Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet. 2011;7:e1002084. doi: 10.1371/journal.pgen.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raychaudhuri S. Mapping rare and common causal alleles for complex human diseases. Cell. 2011;147:57–69. doi: 10.1016/j.cell.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kempen JH, Mitchell P, Lee KE, Tielsch JM, Broman AT, Taylor HR, Ikram MK, Congdon NG, O'Colmain B, Friedman DS. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol. 2004;122:495–505. doi: 10.1001/archopht.122.4.495. [DOI] [PubMed] [Google Scholar]
- 33.Sperduto RD, Seigel D, Roberts J, Rowland M. Prevalence of myopia in the United States. Arch Ophthalmol. 1983;101:405–7. doi: 10.1001/archopht.1983.01040010405011. [DOI] [PubMed] [Google Scholar]
- 34.Zhu Q, Ge D, Maia JM, Zhu M, Petrovski S, Dickson SP, Heinzen EL, Shianna KV, Goldstein DB. A genome-wide comparison of the functional properties of rare and common genetic variants in humans. Am J Hum Genet. 2011;88:458–68. doi: 10.1016/j.ajhg.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKie AB, McHale JC, Keen TJ, Tarttelin EE, Goliath R, van Lith-Verhoeven JJ, Greenberg J, Ramesar RS, Hoyng CB, Cremers FP, Mackey DA, Bhattacharya SS, Bird AC, Markham AF, Inglehearn CF. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet. 2001;10:1555–62. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- 36.Vieira V, David G, Roche O, de la Houssaye G, Boutboul S, Arbogast L, Kobetz A, Orssaud C, Camand O, Schorderet DF, Munier F, Rossi A, Delezoide AL, Marsac C, Ricquier D, Dufier JL, Menasche M, Abitbol M. Identification of four new PITX2 gene mutations in patients with Axenfeld-Rieger syndrome. Mol Vis. 2006;12:1448–60. [PubMed] [Google Scholar]
- 37.Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, Funkhauser C, Daack-Hirsch S, Murray JC. A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet. 1998;19:167–70. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- 38.Idrees F, Bloch-Zupan A, Free SL, Vaideanu D, Thompson PJ, Ashley P, Brice G, Rutland P, Bitner-Glindzicz M, Khaw PT, Fraser S, Sisodiya SM, Sowden JC. A novel homeobox mutation in the PITX2 gene in a family with Axenfeld-Rieger syndrome associated with brain, ocular, and dental phenotypes. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:184–91. doi: 10.1002/ajmg.b.30237. [DOI] [PubMed] [Google Scholar]
- 39.Vetrini F, Auricchio A, Du J, Angeletti B, Fisher DE, Ballabio A, Marigo V. The microphthalmia transcription factor (Mitf) controls expression of the ocular albinism type 1 gene: link between melanin synthesis and melanosome biogenesis. Mol Cell Biol. 2004;24:6550–9. doi: 10.1128/MCB.24.15.6550-6559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leiper LJ, Ou J, Walczysko P, Kucerova R, Lavery DN, West JD, Collinson JM. Control of patterns of corneal innervation by Pax6. Invest Ophthalmol Vis Sci. 2009;50:1122–8. doi: 10.1167/iovs.08-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y, Kondo S, Yokoyama M. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development. 1999;126:5749–58. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- 42.Jones SE, Jomary C, Grist J, Thomas MR, Neal MJ. Expression of Pax-6 mRNA in the retinal degeneration (rd) mouse. Biochem Biophys Res Commun. 1998;252:236–40. doi: 10.1006/bbrc.1998.9631. [DOI] [PubMed] [Google Scholar]
- 43.Hammond CJ, Andrew T, Mak YT, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004;75:294–304. doi: 10.1086/423148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chauhan BK, Yang Y, Cveklova K, Cvekl A. Functional properties of natural human PAX6 and PAX6(5a) mutants. Invest Ophthalmol Vis Sci. 2004;45:385–92. doi: 10.1167/iovs.03-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bitzer M, Schaeffel F. ZENK expression of retinal glucagon amacrine cells in chicks: the effect of defocus presented in vivo, in vitro and under anesthesia. Vision Res. 2006;46:848–59. doi: 10.1016/j.visres.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez-Rodriguez J, Pelcastre EL, Tovilla-Canales JL, Garcia-Ortiz JE, Amato-Almanza M, Villanueva-Mendoza C, Espinosa-Mattar Z, Zenteno JC. Mutational screening of CHX10, GDF6, OTX2, RAX and SOX2 genes in 50 unrelated microphthalmia-anophthalmia-coloboma (MAC) spectrum cases. Br J Ophthalmol. 2010;94:1100–4. doi: 10.1136/bjo.2009.173500. [DOI] [PubMed] [Google Scholar]
- 47.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–7. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]


