Abstract
Background
Protein C (PC) is one of the most important physiological inhibitors of coagulation proteases. Hereditary PC deficiency causes a predisposition to venous thrombosis (VT). The genetic characteristics of PC deficiency in the Chinese population remain unknown.
Methods
Thirty-four unrelated probands diagnosed with hereditary PC deficiency were investigated. PC activity and antigen levels were measured. Mutation analysis was performed by sequencing the PROC gene. In silico analyses, including PolyPhen-2, SIFT, multiple sequence alignment, splicing prediction, and protein molecular modeling were performed to predict the consequences of each variant identified. One recurrent mutation and its relative risk for thrombosis in relatives were analyzed in 11 families. The recurrent mutation was subsequently detected in a case (VT patients)-control study, and the adjusted odds ratio (OR) for VT risk was calculated by logistic regression analysis.
Results
A total of 18 different mutations, including 12 novel variants, were identified. One common mutation, PROC c.565C>T (rs146922325:C>T), was found in 17 of the 34 probands. The family study showed that first-degree relatives bearing this variant had an 8.8-fold (95%CI = 1.1–71.6) increased risk of venous thrombosis. The case-control (1003 vs. 1031) study identified this mutation in 5.88% patients and in 0.87% controls, respectively. The mutant allele conferred a high predisposition to venous thrombosis (adjusted OR = 7.34, 95%CI = 3.61–14.94). The plasma PC activity and antigen levels in heterozygotes were 51.73±6.92 U/dl and 75.17±4.84 U/dl, respectively.
Conclusions
This is the first study on the genetic background of PC deficiency in the Chinese population. The PROC c.565C>T mutation is the most frequent cause of PC deficiency as well as a prevalent risk factor for VT in Chinese individuals. The inclusion of this variant in routine thrombophilic detection may improve the diagnosis and prevention of venous thrombosis.
Introduction
Protein C, the key component of the PC anticoagulant system, is an important vitamin K-dependent protein that regulates the physiological coagulation cascade by inactivating factors Va and VIIIa upon activation by thrombin [1]–[3]. The mature PC molecule is a single- chain, 62-kDa glycoprotein that is synthesized by hepatocytes as a 461-amino acid precursor from which a 42-amino acid signal peptide is cleaved. Protein C is composed of a γ-carboxy-glutamic acid residue (Gla) domain, two epidermal growth factor (EGF)-like domains, a short activation peptide, and a serine protease domain [4], [5]. Thrombin cleaves PC at Arg169, removes the activation peptide and generates activated protein C (APC) [6]. In addition to its anticoagulant properties, APC has anti-inflammatory and cytoprotective functions, which are exerted when APC activating protease activated receptor-1 (PAR-1) [7], [8]. The human protein C gene (PROC) is located on chromosome 2q13-q14 and comprises nine exons spanning 11 kb [9], [10].
The most common genetic risk factors for venous thrombosis in Whites, factor V Leiden (FV R506Q) and prothrombin G20210A polymorphism, are rare in Eastern populations [11]–[13]. Therefore, the three main physiological anticoagulant (antithrombin, protein C, and protein S) deficiencies are important risk factors in Asians, and their diagnosis is of great clinical interest [14], [15]. Hereditary protein C deficiency (OMIM#176860) is usually inherited as an autosomal dominant trait and is associated with an increased risk of venous thrombosis and hereditary thrombophilia. Heterozygous individuals have an approximately 7-fold increased risk of venous thrombosis compared with normal individuals [16]. The homozygous (or compound heterozygous) state of protein C deficiency is much more rare [17]. The overlap in the plasma protein C levels between healthy individuals and PC heterozygotes makes the discrimination between these two groups solely based on a single plasma measurement challenging [18]. Genetic determination is likely to be more powerful.
To date, a number of mutations in the PROC gene associated with PC deficiency have been identified, but only a few of these variants were observed in the Chinese population, and the genetic characteristics of PC deficiency in Chinese patients have not been studied. Here, we report the results of the genetic investigation of protein C deficiency in 34 Chinese families and present a prevalent causative variant for PC deficiency and venous thrombosis.
Methods
Ethics Statement
This study was approved by the ethics committee of Union Hospital, Huazhong University of Science and Technology and complied with the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from all participants.
Protein C-deficient patients and sample collection
A total of 34 unrelated probands diagnosed with hereditary protein C deficiency and registered at the Hubei Clinical and Research Center of Thrombosis and Hemostasis from 2008 to 2010 were investigated. The diagnosis was based on repeated low plasma protein C activity measurements and a personal or family history of symptomatic thromboembolic diseases.
Blood samples were collected from all subjects during a non-acute phase of VT (at least one month after a VT incident), and the patients had not received anticoagulant therapy for at least two weeks before the collection. The blood sample was collected by venipuncture into a vacutainer tube containing 1/10 volume of 0.105 mol/L trisodium citrate and was immediately centrifuged at 2000× g for 15 min. The platelet-poor plasma was stored at −80°C until assayed. High molecular weight genomic DNA was isolated from white blood cells and standardized to 50 ng/µl.
Coagulation assays
Protein C, protein S, and antithrombin activities were assayed on a Sysmex CA 7000 Analyzer (Sysmex, Japan) using commercial reagents obtained from Dade Behring-Siemens Healthcare Diagnostics, Germany. Protein C and antithrombin activities were measured using a chromogenic substrate method. The APC-cofactor activity of protein S was evaluated using a clotting method. Protein C antigen was further tested by an enzyme-linked immunosorbent assay using ZYMUTEST Protein C (Hyphen BioMed, France). The normal ranges of these tests in our lab were established in 78 healthy subjects.
The PROC gene analysis
The PROC gene was analyzed in each proband by PCR and resequencing. All nine exons, including at least 100 bp flanking intron regions, and the 5′- and 3′-untranslated regions of the human protein C gene were amplified. Detailed amplification conditions and the sequences of the oligonucleotide primers used are available upon request. The amplified fragments were sequenced on an ABI PRISM 3730XL automated sequencer (Applied Biosystems). Identified variants were confirmed on a second PCR product, sequencing on both strands. The variants were designated according to current nomenclature and the recommendations of the Human Genome Variation Society (HGVS, http://www.hgvs.org/mutnomen/) and were checked using the Mutalyzer program [19]–[21]. All new data have been deposited in GenBank under accession numbers NM_000312.3 and NP_000303.1. Each novel variant was then detected in 50 normal individuals (100 alleles) using direct sequencing.
In silico analysis of novel amino acid changes
The possible impact of novel coding sequence changes (amino acid substitutions) on the structure and function of PC was assessed using two bioinformatics tools, Sorting Intolerant From Tolerant (SIFT, http://sift.jcvi.org) and Polymorphism Phenotyping-2 (PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/) [22], [23]. The UniProtKB Protein ID “P04070” and Protein Ensembl ENSP ID “ENSP00000234071” were used, respectively.
The conservation of the affected amino acids were further checked by multiple sequence alignment (HomoloGene, http://www.ncbi.nlm.nih.gov/sites/entrez) with sequences from Pantroglodytes, Canis lupus familiaris, Bos taurus, Mus musculus, Rattus norvegicus, and Gallus gallus.
Splicing efficiency prediction
The effect of exon-intron boundary variants on the efficiency of splice processing was predicted by two web server programs: SplicePort (http://spliceport.cs.umd.edu/) and Alternative Splice Site Predictor (ASSP, http://www.es.embnet.org/~mwang/assp.html) [24], [25].
Protein molecular modeling
The molecular structures of the wild-type and mutant proteins were modeled using Swiss model workspace (http://swissmodel.expasy.org/) [26] and noc-3.01 software based on the tridimensional crystal structure of PC (PDB ID: 3F6U) [27].
Recurrent mutation detection in venous thrombosis patients and controls
The PROC c.565C>T mutation, which was common among the probands, was subsequently detected in an independent case-control study to evaluate the OR of this variant for venous thrombosis in the Chinese population.
This secondary study included consecutive unrelated patients (n = 1003) who were diagnosed with venous thrombosis at the Hubei Clinical and Research Center of Thrombosis and Hemostasis from 2008 to 2011. The thrombosis incidents were validated based on clinical manifestations, D-dimer levels, Doppler ultrasound (for deep vein thrombosis), ventilation perfusion lung scan, and/or computed tomography angiography (for pulmonary embolism). Age- and sex-matched controls (n = 1031) without an individual or family history of VT were enrolled from a community screening program in the same regions during the same time window. Demographic data and acquired thrombotic risk factors were recorded. Blood samples were collected from all participants.
All of the participants included in the case-control study were central Chinese. To test whether this PC variant was also common in populations from other regions of China, we recruited a cohort of 492 healthy individuals (43.9% female) who were evaluated for routine physical check-up in Shanghai (Dongfang Hospital, eastern China) in February 2012. The healthy Shanghai participants ranged in age from 38 to 66 years and had no family history of thrombosis. Blood samples were collected, and genomic DNA was extracted and prepared for genotyping.
The c.565C>T mutation was genotyped using the PCR-RFLP method. The mutagenic primers 5′-TCCTTGAACCCTGCACTGTGGCAA-3′ and 5′-TTTCAGGTGACTACGC- TTCTTCTCCATGC-3′ were employed. The underlined and italicized base G in the lower primer was used to introduce a Hin6 I restriction site into the normal allele. In contrast, the underlined and italicized base A was used to eliminate an adjacent restriction site. Therefore, only the PCR product of the common allele has a Hin6 I recognition site. Altogether, 10 µl of PCR product was digested with 5 U of Hin6 I (Fermentas, Canada) for 3 hours at 37°C and then subjected to 2% agarose gel electrophoresis. To verify the genotyping results determined by PCR/RFLP assay, up to 192 DNA samples were randomly selected and subjected to sequencing.
Family analysis of the recurrent mutation
To avoid bias, probands were excluded, and analyses were performed on first-degree relatives only. Included families were composed of at least two family members (one c.565C>T carrier and one non-carrier). Six families were excluded because they did not meet the criteria. In total, 49 family members from 11 families were enrolled to estimate the thrombotic risk associated with the recurrent mutation. The annual incidence of thrombosis was calculated by dividing the number of events in each group (carriers and non-carriers) by the total number of patient-years of follow-up. For each individual, years of follow-up were defined as the time from the date of birth to either the date of the first episode of venous thrombosis, if any, or December 2011. The relative risks were calculated using only the first thrombotic event during follow-up.
Statistical analysis
For continuous variables, differences between groups were analyzed by a Student's t-test or Mann-Whitney U-test, depending on the normality of the data. A chi-squared test was used for categorical variables. Deviations from Hardy-Weinberg expectations were assessed using both Fisher's Exact test and chi-squared test. Multivariate logistic regression analysis was used to calculate the OR of the c.565C>T mutation for venous thrombosis adjusted for selected confounders (age, gender, smoking status, alcohol abuse, malignant tumor, type 2 diabetes, sedentariness/immobilization, and pregnancy/puerperium). A Cox regression analysis was performed to estimate the relative risk (hazard ratio) for thrombosis in first-degree relatives with the c.565C>T mutation, adjusted for age and gender. Statistical power was estimated using the Power and Sample (PS) size calculation program [28]. A two-tailed P<0.05 was considered statistically significant. Analyses were performed using SPSS version 12.0 software (SPSS Inc., Chicago, IL).
Results
Mutation profiles
Eighteen different genetic variants (12 novel) were identified in 32 of the 34 probands (94.1%). No candidate mutations were found in the other two subjects, most likely due to gross insertion/deletion of the PROC gene. The mutation profiles consisted of 14 missense mutations, two splice site mutations, one small deletion, one small duplication, and one 3′-untranslated region variant. Each novel variant identified in a proband was shown to cosegregate with a low plasma PC level in the family. All of the novel variants were absent in all 50 healthy individuals, suggesting that they are not common polymorphisms. The mutation profiles are summarized in Table 1.
Table 1. Mutation profiles, laboratory, and clinical data of for patients with PC deficiency.
| Patient No. | Age | Clinical data | PC∶A (U/dl) | PC∶Ag (U/dl) | NT exchange | AA substitution | Ref. |
| PC1 | 15 | DVT | 6.3 | 12.6 | c.889G>C c.1258G>T | p.Asp297His p.Val420Leu | novel novel |
| PC3 | 64 | DVT | 40.4 | 48.3 | c.*73C>T | - | novel |
| PC4 | 33 | DVT | 52.9 | 58.8 | c.524G>A | p.Cys175Tyr | novel |
| PC6 | 32 | DVT/MVT | 36.4 | 49.2 | c.716dupG | p.Ala240GlyfsX17 | novel |
| PC7 | 55 | DVT | 52.1 | 31.5 | c.632G>A | p.Arg211Gln | [32] |
| PC11 | 24 | DVT/PE | 9.9 | 9.2 | c.349_352del c.541T>G | p.Phe118AlafsX16 p.Phe181Val | Novel [33] |
| PC12 | 45 | DVT | 61.7 | 53.4 | c.400+5G>A | - | [29] |
| PC15 | 65 | DVT | 49.3 | 61.7 | not found | - | - |
| PC16 | 46 | DVT | 44.1 | 53.3 | c.316T>G | p.Cys106Gly | novel |
| PC17 | 46 | DVT/PE | 57.7 | 65.2 | c.889G>C c.891C>T | p.Asp297His | novel novel |
| PC19 | 44 | DVT | 30.4 | 76.7 | c.935C>T | p.Ser312Leu | [34] |
| PC20 | 37 | DVT | 40.9 | 38.3 | c.658C>T | p.Arg220Trp | [29] |
| PC22 | 49 | DVT | 48.5 | 53.3 | not found | - | - |
| PC25 | 42 | DVT/AIS | 39.4 | 26.7 | c.541T>G c.980A>T | p.Phe181Val p.Glu327Val | [33] novel |
| PC27 | 46 | DVT | 48.7 | 42.4 | c.208A>G | p.Lys70Glu | novel |
| PC28 | 54 | DVT | 40.0 | 53.0 | c.237+5G>A | - | novel |
| PC29 | 48 | DVT | 60.0 | 88.1 | c.669C>A | p.Ser223Arg | novel |
| PC31 | 64 | DVT | 44.2 | 42.4 | c.541T>G | p.Phe181Val | [31] |
| others | 38–67 | DVT | 40.4–62.3 | 68.7–83.0 | c.565C>T | p.Arg189Trp | [34] |
Mutations were designated according to the HGVS nomenclature for variants, comparing with the NCBI Reference Sequences NM_000312.3 and NP_000303.1. Patients with two nucleotide substitutions indicated compound heterozygotes. NT = nucleotide. AA = amino acid. Ref. = References. DVT = deep vein thrombosis. MVT = mesenteric vein thrombosis. PE = pulmonary embolism. AIS = acute ischemic stroke.
Missense mutations
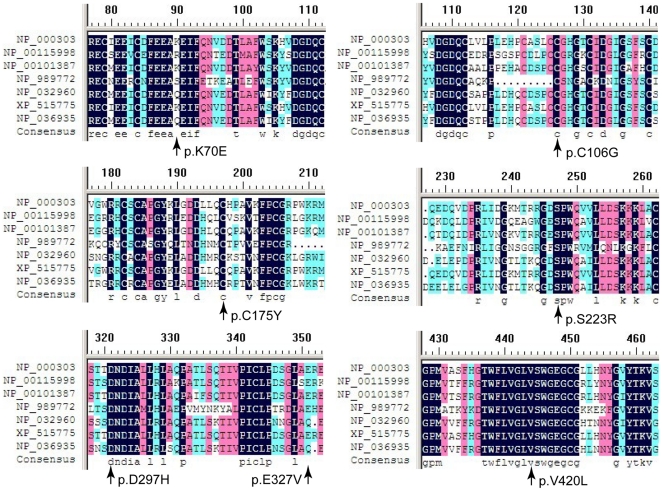
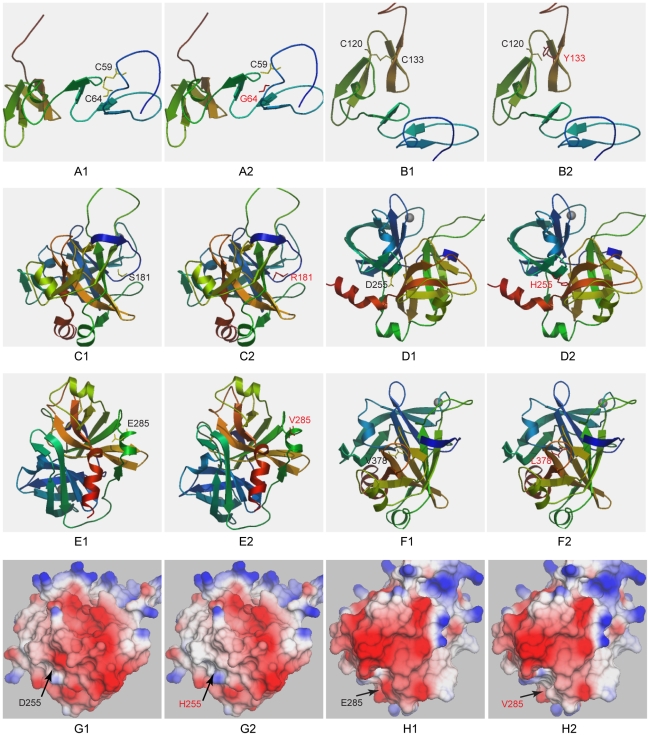
A total of 14 missense mutations resulted in amino acid substitutions. Eight are reported for the first time here. A multiple sequence alignment among selected species showed that seven of the newly identified mutations led to substitutions of highly conserved amino acids (Figure 1). Although Lys70 (basic amino acid) is not conserved, Glu (acidic amino acid) is never present at this site in the evaluated species. Novel missense mutations of p.Cys175Tyr, p.Asp297His, and p.Val420Leu were predicted to be deleterious by both the SIFT and PolyPhen-2 algorithms. The other four mutations (p.Lys70Glu, p.Cys106Gly, p.Ser223Arg, and p.Glu327Val) were predicted to be damaging by one algorithm. The location of each affected amino acid and the correlative structure of mutant PC are depicted in Figure 2.
Figure 1. Multiple sequence alignment highlighting the affected amino acids among selected species.
The conservation degree of the affected amino acids was evaluated in sequences from Homo sapiens (NP_000303.1), Bos taurus (NP_001159984.1), Canis lupus familiaris (NP_001013871.1), Gallus gallus (NP_989772.1), Mus musculus (NP_032960.3), Pan troglodytes (XP_515775.3), and Rattus norvegicus (NP_036935.1).
Figure 2. Structure models of the wild-type and mutant PC proteins.
The amino acid number was designated according to the previous nomenclature described in the Human Gene Mutation Database. That is, the first 42 amino acids (signal peptide) is subtracted. The wild-type amino acids are shown in yellow and the mutants are shown in red. The D255H and the E285V mutations are also displayed in a solid surface model (G, H) in which the electrostatic potential is clearly indicated.
Null mutations
Five of the detected variants were suspected to result either in the complete absence of the gene product or in the expression of a non-functional protein; these mutations were defined as null mutations, and four of them are novel. The frameshift mutations c.719dupG and c.349_352del might produce a premature termination codon and lead to the production of a truncated protein. The c.237+5G>A mutation occurs in the donor splice site of intron C. This substitution changed the splice site scores to 0.000 from original scores of 7.137 and 0.180 assigned by the splicing efficiency prediction programs ASSP and SplicePort, respectively. The closest candidate splice site was 46 bp downstream from the normal splice site. No matter whether the cryptic splice site would be used, this should result in an abnormal translation product. The c.*73C>T variant, 73 bp downstream from the stop codon, completely cosegregated in the PC3 family and was absent in 50 control subjects. This mutation might affect the stability of the PC mRNA.
Recurrent mutation detection in patients and controls
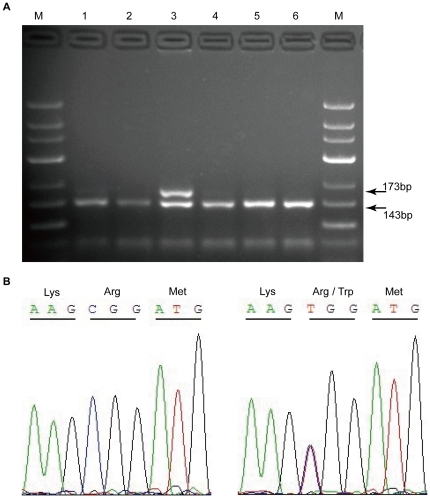
Seventeen of the 34 probands (50%) carried the same mutation, c.565C>T (p.Arg189Trp), in exon 7. The recurrent mutation was then evaluated in an independent case-control study by PCR/RFLP (Figure 3). The genotyping data from the RFLP assays were completely consistent with those obtained by sequencing. The demographic data and acquired thrombotic risk factors of all subjects are presented in Table 2. The heterozygous c.565C>T mutation was identified in 59 (5.88%) venous thrombosis patients and 9 (0.87%) controls. The homozygous genotype was absent in all subjects. As illustrated in Table 3, the observed OR of this mutation was 6.91 (95%CI = 3.42–13.98; P = 4.67×10−10). Multivariate logistical regression analysis revealed that the association between the variant and VT was still significant (OR = 7.34; 95%CI = 3.61–14.94; P = 3.88×10−8) after adjustment for age, gender, smoking status, alcohol abuse, malignant tumor, type 2 diabetes, sedentariness/immobilization, and pregnancy/puerperium in the dominant model. Meanwhile, this mutation was also detected and confirmed in 4 of the 492 (0.81%) healthy participants from Shanghai, indicating that this mutation is not restricted to the central Chinese population and is likely to be prevalent in the general population.
Figure 3. PROC c.565C>T variant detection by PCR-RFLP and direct sequencing.
(A) Electrophoretic patterns following Hin6 I digestion. PCR products were 173 bp. Only amplicons with the wild-type sequences were digested, yielding two bands of 143 bp and 30 bp. The digestion products were separated by 2% agarose gel electrophoresis. M, DNA marker with 50-bp ladder. Lanes 1, 2, and 4–6, normal individuals. Lane 3, heterozygous individual for the variant. (B) Chromatograms obtained by sequencing. Left, wild-type; Right, heterozygote.
Table 2. Data from participants enrolled in the secondary case-control study.
| Variable | Cases | Controls | P | ||
| No. | % | No. | % | ||
| Age (years) | 51.45±14.36 | 50.26±14.52 | 0.063 | ||
| Sex | 0.970 | ||||
| Male | 532 | 53.04% | 546 | 52.96% | |
| Female | 471 | 46.96% | 485 | 47.04% | |
| Smoking | 0.009 | ||||
| Yes | 227 | 22.6% | 185 | 17.9% | |
| No | 776 | 77.4% | 846 | 82.1% | |
| Alcohol drinker | 0.717 | ||||
| Yes | 117 | 11.7% | 115 | 11.2% | |
| No | 886 | 88.3% | 916 | 88.8% | |
| Malignant tumor | 1.207×10−13 | ||||
| Yes | 65 | 6.5% | 5 | 0.5% | |
| No | 938 | 93.5% | 1026 | 99.5% | |
| T2DM | 0.037 | ||||
| Yes | 42 | 4.2% | 26 | 2.5% | |
| No | 961 | 95.8% | 1005 | 97.5% | |
| Sedentariness | 0.031 | ||||
| Yes | 23 | 2.3% | 11 | 1.1% | |
| No | 980 | 97.7% | 1020 | 98.9% | |
| Pregnancy/puerperium | |||||
| Yes | 26 | 2.6% | 14 | 1.4% | 0.045 |
| No | 977 | 97.4% | 1017 | 98.6% | |
Years of age were expressed as mean±standard deviation. Age of cases: age at the first incident. Age of controls: age at enrollment. Age was further divided into three levels: under 40 years, 40 to 60 years, and over 60 years. Sedentariness/immobilization reflects the 4 weeks prior to the venous thrombosis incident. A Chi-squared test was used to compare the differences between groups according to gender, smoking status, alcohol abuse, malignant tumor, type 2 diabetes, sedentariness/immobilization, and pregnancy/puerperium. A Student's t test was used to compare the mean ages of the cases and controls. VT = venous thrombosis. T2DM = type 2 diabetes mellitus.
Table 3. Association between the PROC c.565C>T and venous thrombosis in the Chinese population.
| PROC c.565C>T | Cases | Controls | Without adjustment | After adjustment* | ||||||
| No. | % | No. | % | OR | 95%CI | P | OR | 95%CI | P | |
| C/C | 944 | 94.12% | 1022 | 99.13% | 1 | - | - | 1 | - | - |
| C/T | 59 | 5.88% | 9 | 0.87% | 7.10 | 3.50–14.39 | 3.31×10−10 | 7.34 | 3.61–14.94 | 3.88×10−8 |
| MAF | - | 2.94% | - | 0.44% | 6.91 | 3.42–13.98 | 4.67×10−10 | - | - | - |
Test for H-W equilibrium in controls: P = 0.89. CI = confidence interval. MAF = minor allele frequency.
Data were analyzed by logistic regression adjusted for age, gender, smoking status, alcohol abuse, malignant tumor, type 2 diabetes, sedentariness/immobilization, and pregnancy/puerperium.
Thrombotic risk in first-degree relatives with the c.565C>T mutation
As shown in Table 4, the incidence of thrombosis was greater in first-degree relatives with the c.565C>T mutation (7.9 per 1000 person-years) compared with those without this mutation (0.8 per 1000 person-years), with a relative risk of 8.8 (95%CI = 1.1–71.6) for thrombosis.
Table 4. Family analysis of thrombotic risk in c.565C>T carriers and non-carriers.
| Carriers | Non-carriers | |
| Number | 20 | 29 |
| Male/Female | 12/8 | 16/13 |
| Age at first thrombosis, median (range) | 51.5 (49–61) | 51 |
| Years of follow up | 890 | 1264 |
| Number of VT events | 7 | 1 |
| Events per 1000 person-years (95%CI) | 7.9 (7.6–8.1) | 0.8 |
| Relative risk for thrombosis (95% CI)* | 8.8 (1.1–71.6) | 1 |
Data were analyzed using a Cox regression model adjusted for age and gender.
Discussion
Protein C deficiency is known to be associated with an increased risk of VT and its genetic background has been analyzed in several populations [29]–[31]. In the present study, we systematically investigated the genetic characteristics of PC deficiency in the Chinese population.
A total of 18 different mutations, including one recurrent mutation, were identified in 34 probands. Six of the detected variants (c.400+5G>A, p.Phe181Val, p.Arg189Trp, p.Arg211Gln, p.Arg220Trp, and p.Ser312Leu) have been reported previously as disease-causing mutations [29], [32]–[34].
Eight novel coding sequence variants contributed to seven amino acid exchanges. The variant p.Lys70Glu is located in the Gla domain, which is involved in the interaction between PC and anionic phospholipid surfaces [35]. This alteration creates a new Glu residue and changes the charge nature of this site, which might reduce the efficiency of the interaction with phospholipid membranes. The p.Cys106Gly and p.Cys175Tyr variants eliminate Cys residues that are important for disulfide bond formation in the EGF-1 and EGF-2 domains (Figure 2A–2B), respectively. These mutations might result in the production of an aberrant multimer or high molecular weight complex [36]. Variants in the enzymatic serine protease domain (p.Ser223Arg, p.Asp297His, p.Glu327Val, and p.Val420Leu) are located adjacent to either the serine protease active site triad or the substrate binding region (Figure 2C–2H). These substitutions may impair the structural integrity and stability of the heavy chain and attenuate the catalytic activity of the protein [37].
The three novel null mutations (c.237+5G>A, c.349_352del, c.719dupG) are evidently detrimental because they either impair mRNA splicing by disrupting the consensus sequences of the exon-intron boundary or produce a premature stop codon. Abnormal translation products may not be stable in vivo due to nonsense-mediated mRNA decay [38], [39]. The c.*73C>T mutation is located in the untranslated region of the last exon. The 3′-untranslated region is essential for the posttranscriptional regulation of mRNA expression. Mutations in this region almost always reduce the stability and half-life of transcripts [40], [41].
The recurrent c.565C>T variant (p.Arg189Trp, R147W, or rs146922325:C>T) was identified in 17 probands, accounting for half of the cases of PC deficiency. The retrospective family study showed that first-degree relatives bearing this variant had an 8.8-fold increased risk of venous thrombosis. The statistical power of this analysis is approximately 0.989, with a type I error probability of 0.01. However, the confidence intervals was wide due to the small number of family members enrolled and the low incidence of thrombosis in the non-carrier group. A larger prospective study will be required to confirm our observations.
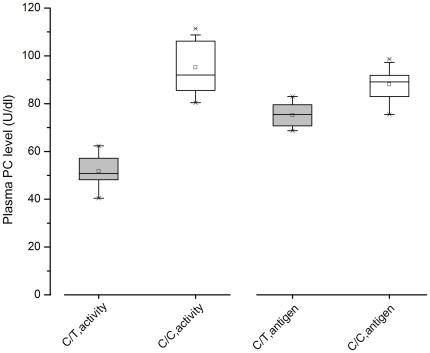
The R147W substitution is adjacent to the EGF-2 domain at the C-terminal of the light chain and may impair the interaction of PC with other molecular such as thrombin-thrombomodulin complex, substrate, or phospholipid [42]. Further functional studies are required to define the deleterious effect of this mutation on the activation of PC, the inactivation of FVa by APC, and the cytoprotective role of APC. This missense mutation was first described in a family with PC deficiency as well as an asymptomatic individual in 1995 [34], [43]. Consistent with previous studies, our analysis showed that the heterozygous state is associated with decreased functional activity (40.4–62.3 U/dl) and a relatively normal antigen level (68.7–83.0 U/dl, Figure 4), indicating type II PC deficiency. This variant is believed to be a rare mutation and the corresponding minor allele frequency in dbSNP is 0.000. Because its prevalence, a case-control study was further conducted. This variant was also present in 9 out of the 1031 control subjects (0.87%) and was significantly associated with VT risk. The statistical power of our analysis is approximately 0.999, with a type I error probability of 0.01. In combination with the genotyping data from the 492 DNA samples from Shanghai, the c.565C>T variant was present in approximately 0.85% (95%CI = 0.38%–1.31%) of the general population. The carrier rate of this variant allele is relatively lower than that of factor V Leiden (5%–10%) and prothrombin G20210A (2%–4%), two polymorphisms that confer moderate predisposition to VT in Whites [44]. However, it is more common than the antithrombin Cambridge II variant (approximately 0.2%) [45] and other rare but severe anticoagulant protein (AT, PC, and PS) mutations. One previous study [46] showed that the estimated prevalence of protein C deficiency in the general Chinese population is approximately 0.29%. In their study, only subjects with PC activity below the 1st percentile were selected for DNA mutation analysis and considered to have “PC deficiency”. Subjects with a PC level between the 1st and 2.5th percentiles might also have a genetic defect. Therefore, these genetic variants might have been missed. The PROC c.565C>T was one such of the missed mutations. When this moderate mutation is taken into account, however, it is clear that PC deficiency has been underestimated and may be more frequent in the Chinese population than in Japanese or European populations [47], [48].
Figure 4. Plasma protein C level of the PROC c.565C>T heterozygous subjects and normal individuals.
The mean, median, interquartile range, and range of protein C levels (activity and antigen) are shown. Gray box, C/T genotype (n = 17). White box, C/C wild-type (n = 20).
In conclusion, we investigated the genetic background of PC deficiency in the Chinese population for the first time and identified one recurrent variant as well as 17 other mutations. The NM_000312.3:c.565C>T mutation is not only the most frequent variant for PC deficiency but also a significant risk factor for venous thrombosis in Chinese individuals. Although this variant was identified about 16 years ago as a rare mutation in Western populations, its high prevalence and potential clinical significance in the Chinese population was not appreciated until now. The prevalence of and the thrombotic risk associated with this mutation in other populations (especially other Asian populations) should be further evaluated. This genetic detection may be included in the routine thrombophilia screening, and carriers of this variant may benefit from early diagnosis and better prevention [49].
Acknowledgments
We are indebted to all of the participants in this study and medical assistants of our hospital. Our special thanks go to the Department of Vascular Surgery (Union Hospital), the Ministry of Education Key Lab for Environment and Health (Tongji Medical College), and the co-workers at Shanghai Dongfang Hospital.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Basic Scientific Research Program of China (973 Program, No. 2007 CB935803), the National Natural Sciences Foundation of China (No. 30825018), and the State Ministry of Health Key Clinical Construction Project (2010 No. 58). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Esmon CT. The protein C pathway. Chest. 2003;124:26S–32S. doi: 10.1378/chest.124.3_suppl.26s. [DOI] [PubMed] [Google Scholar]
- 2.Dahlbäck B, Villoutreix BO. Molecular recognition in the protein C anticoagulant pathway. J Thromb Haemost. 2003;1:1525–34. doi: 10.1046/j.1538-7836.2003.00299.x. [DOI] [PubMed] [Google Scholar]
- 3.Marlar RA, Kleiss AJ, Griffin JH. Mechanism of action of human activated protein C, a thrombin-dependent anticoagulant enzyme. Blood. 1982;59:1067–72. [PubMed] [Google Scholar]
- 4.Fisher CL, Greengard JS, Griffin JH. Models of the serine protease domain of the human antithrombotic plasma factor activated protein C and its zymogen. Protein Sci. 1994;3:588–99. doi: 10.1002/pro.5560030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perera L, Foley C, Darden TA, Stafford D, Mather T, et al. Modeling zymogen protein C. Biophys J. 2000;79:2925–43. doi: 10.1016/S0006-3495(00)76530-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kisiel W. Human plasma protein C: isolation, characterization, and mechanism of activation by alpha-thrombin. J Clin Invest. 1979;64:761–9. doi: 10.1172/JCI109521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahlbäck B, Villoutreix BO. Regulation of blood coagulation by the protein C anticoagulant pathway. Novel insights into structure-function relationships andmolecular recognition. Arterioscler Thromb Vasc Biol. 2005;25:1311–20. doi: 10.1161/01.ATV.0000168421.13467.82. [DOI] [PubMed] [Google Scholar]
- 8.Mosnier LO, Zlokovic BV, Griffin JH. The cytoprotective protein C pathway. Blood. 2007;109:3161–72. doi: 10.1182/blood-2006-09-003004. [DOI] [PubMed] [Google Scholar]
- 9.Foster DC, Yoshitake S, Davie EW. The nucleotide sequence of the gene for human protein C. Proc Natl Acad Sci U S A. 1985;82:4673–7. doi: 10.1073/pnas.82.14.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plutzky J, Hoskins JA, Long GL, Crabtree GR. Evolution and organization of the human protein C gene. Proc Natl Acad Sci U S A. 1986;83:546–50. doi: 10.1073/pnas.83.3.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertina RM, Koeleman BP, Koster T, Rosendaal FR, Dirven RJ, et al. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature. 1994;369:64–7. doi: 10.1038/369064a0. [DOI] [PubMed] [Google Scholar]
- 12.Poort SR, Rosendaal FR, Reitsma PH, Bertina RM. A common genetic variation in the 3′- untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood. 1996;88:3698–703. [PubMed] [Google Scholar]
- 13.Bounameaux H, Rosendaal FR. Venous thromboembolism: why does ethnicity matter? Circulation. 2011;123:2189–91. doi: 10.1161/CIRCULATIONAHA.111.031690. [DOI] [PubMed] [Google Scholar]
- 14.Roberts LN, Patel RK, Arya R. Venous thromboembolism and ethnicity. Br J Haematol. 2009;146:369–83. doi: 10.1111/j.1365-2141.2009.07786.x. [DOI] [PubMed] [Google Scholar]
- 15.Suehisa E, Nomura T, Kawasaki T, Kanakura Y. Frequency of natural coagulation inhibitor (antithrombin III, protein C and protein S) deficiencies in Japanese patients with spontaneous deep vein thrombosis. Blood Coagul Fibrinolysis. 2001;12:95–9. doi: 10.1097/00001721-200103000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Koster T, Rosendaal FR, Briët E, van der Meer FJ, Colly LP, et al. Protein C deficiency in a controlled series of unselected outpatients: an infrequent but clear risk factor for venous thrombosis (Leiden Thrombophilia Study). Blood. 1995;85:2756–61. [PubMed] [Google Scholar]
- 17.Millar DS, Johansen B, Berntorp E, Minford A, Bolton-Maggs P, et al. Molecular genetic analysis of severe protein C deficiency. Hum Genet. 2000;106:646–53. doi: 10.1007/s004390000315. [DOI] [PubMed] [Google Scholar]
- 18.Allaart CF, Poort SR, Rosendaal FR, Reitsma PH, Bertina RM, et al. Increased risk of venous thrombosis in carriers of hereditary protein C deficiency defect. Lancet. 1993;341:134–8. doi: 10.1016/0140-6736(93)90003-y. [DOI] [PubMed] [Google Scholar]
- 19.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 20.Taschner PE, den Dunnen JT. Describing structural changes by extending HGVS sequence variation nomenclature. Hum Mutat. 2011;32:507–11. doi: 10.1002/humu.21427. [DOI] [PubMed] [Google Scholar]
- 21.Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 22.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 23.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, Marín A. Characterization and prediction of alternative splice sites. Gene. 2006;366:219–27. doi: 10.1016/j.gene.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 25.Dogan RI, Getoor L, Wilbur WJ, Mount SM. SplicePort–an interactive splice-site analysis tool. Nucleic Acids Res. 2007;35:W285–91. doi: 10.1093/nar/gkm407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–92. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt AE, Padmanabhan K, Underwood MC, Bode W, Mather T, et al. Thermodynamic linkage between the S1 site, the Na+ site, and the Ca2+ site in the protease domain of human activated protein C (APC). Sodium ion in the APC crystal structure is coordinated to four carbonyl groups from two separate loops. J Biol Chem. 2002;277:28987–95. doi: 10.1074/jbc.M201892200. [DOI] [PubMed] [Google Scholar]
- 28.Dupont WD, Plummer WD., Jr Power and sample size calculations. A review and computer program. Control Clin Trials. 1990;11:116–28. doi: 10.1016/0197-2456(90)90005-m. [DOI] [PubMed] [Google Scholar]
- 29.Reitsma PH, Poort SR, Allaart CF, Briët E, Bertina RM. The spectrum of genetic defects in a panel of 40 Dutch families with symptomatic protein C deficiency type I: heterogeneity and founder effects. Blood. 1991;78:890–4. [PubMed] [Google Scholar]
- 30.Gandrille S, Aiach M. Identification of mutations in 90 of 121 consecutive symptomatic French patients with a type I protein C deficiency. The French INSERM Network on Molecular Abnormalities Responsible for Protein C and Protein S deficiencies. Blood. 1995;86:2598–605. [PubMed] [Google Scholar]
- 31.Grundy CB, Schulman S, Tengborn L, Kakkar VV, Cooper DN. Two different missense mutations at Arg 178 of the protein C (PROC) gene causing recurrent venous thrombosis. Hum Genet. 1992;89:685–6. doi: 10.1007/BF00221964. [DOI] [PubMed] [Google Scholar]
- 32.Poort SR, Pabinger-Fasching I, Mannhalter C, Reitsma PH, Bertina RM. Twelve novel and two recurrent mutations in 14 Austrian families with hereditary protein C deficiency. Blood Coagul Fibrinolysis. 1993;4:273–80. doi: 10.1097/00001721-199304000-00009. [DOI] [PubMed] [Google Scholar]
- 33.Miyata T, Sakata T, Zheng YZ, Tsukamoto H, Umeyama H, et al. Genetic characterization of protein C deficiency in Japanese subjects using a rapid and nonradioactive method for single-stand conformational polymorphism analysis and a model building. Thromb Haemost. 1996;76:302–11. [PubMed] [Google Scholar]
- 34.Reitsma PH, Bernardi F, Doig RG, Gandrille S, Greengard JS, et al. Protein C deficiency: a database of mutations, 1995 update. On behalf of the Subcommittee on Plasma Coagulation Inhibitors of the Scientific and Standardization Committee of the ISTH. Thromb Haemost. 1995;73:876–89. [PubMed] [Google Scholar]
- 35.Foster DC, Rudinski MS, Schach BG, Berkner KL, Kumar AA, et al. Propeptide of human protein C is necessary for gamma-carboxylation. Biochemistry. 1987;26:7003–11. doi: 10.1021/bi00396a022. [DOI] [PubMed] [Google Scholar]
- 36.Beckmann RJ, Schmidt RJ, Santerre RF, Plutzky J, Crabtree GR, et al. The structure and evolution of a 461 amino acid human protein C precursor and its messenger RNA, based upon the DNA sequence of cloned human liver cDNAs. Nucleic Acids Res. 1985;13:5233–47. doi: 10.1093/nar/13.14.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greengard JS, Fisher CL, Villoutreix B, Griffin JH. Structural basis for type I and type II deficiencies of antithrombotic plasma protein C: patterns revealed by three-dimensional molecular modelling of mutations of the protease domain. Proteins. 1994;18:367–80. doi: 10.1002/prot.340180407. [DOI] [PubMed] [Google Scholar]
- 38.Singh G, Lykke-Andersen J. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem Sci. 2003;28:464–6. doi: 10.1016/S0968-0004(03)00176-2. [DOI] [PubMed] [Google Scholar]
- 39.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, et al. Deciphering the splicing code. Nature. 2010;465:53–9. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 40.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nat Med. 2000;6:637–41. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 41.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2001;3:REVIEWS0004. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wacey AI, Pemberton S, Cooper DN, Kakkar VV, Tuddenham EG. A molecular model of the serine protease domain of activated protein C: application to the study of missense mutations causing protein C deficiency. Br J Haematol. 1993;84:290–300. doi: 10.1111/j.1365-2141.1993.tb03067.x. [DOI] [PubMed] [Google Scholar]
- 43.Tait RC, Walker ID, Reitsma PH, Islam SI, McCall F, et al. Prevalence of protein C deficiency in the healthy population. Thromb Haemost. 1995;73:87–93. [PubMed] [Google Scholar]
- 44.Dahlbäck B. Advances in understanding pathogenic mechanisms of thrombophilic disorders. Blood. 2008;112:19–27. doi: 10.1182/blood-2008-01-077909. [DOI] [PubMed] [Google Scholar]
- 45.Corral J, Hernandez-Espinosa D, Soria JM, Gonzalez-Conejero R, Ordonez A, et al. Antithrombin Cambridge II (A384S): an underestimated genetic risk factor for venous thrombosis. Blood. 2007;109:4258–63. doi: 10.1182/blood-2006-08-040774. [DOI] [PubMed] [Google Scholar]
- 46.Zhu T, Ding Q, Bai X, Wang X, Kaguelidou F, et al. Normal ranges and genetic variants of antithrombin, protein C and protein S in the general Chinese population. Results of the Chinese Hemostasis Investigation on Natural Anticoagulants Study I Group. Haematologica. 2011;96:1033–40. doi: 10.3324/haematol.2010.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miletich J, Sherman L, Broze G., Jr Absence of thrombosis in subjects with heterozygous protein C deficiency. N Engl J Med. 1987;317:991–6. doi: 10.1056/NEJM198710153171604. [DOI] [PubMed] [Google Scholar]
- 48.Sakata T, Okamoto A, Mannami T, Matsuo H, Miyata T. Protein C and antithrombin deficiency are important risk factors for deep vein thrombosis in Japanese. J Thromb Haemost. 2004;2:528–30. doi: 10.1111/j.1538-7836.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y, Liang L, Zhai Z, He H, Xie W, et al. Pulmonary Embolism Incidence and Fatality Trends in Chinese Hospitals from 1997 to 2008: A Multicenter Registration Study. PLoS One. 2011;6:e26861. doi: 10.1371/journal.pone.0026861. [DOI] [PMC free article] [PubMed] [Google Scholar]