Abstract
Through epigenetic modifications, specific long-term phenotypic consequences can arise from environmental influence on slowly evolving genomic DNA. Heritable epigenetic information regulates nucleosomal arrangement around DNA and determines patterns of gene silencing or active transcription. One of the greatest challenges in the study of epigenetics as it relates to disease is the enormous diversity of proteins, histone modifications and DNA methylation patterns associated with each unique maladaptive phenotype. This is further complicated by a limitless combination of environmental cues that could alter the epigenome of specific cell types, tissues, organs and systems. In addition, complexities arise from the interpretation of studies describing analogous but not identical processes in flies, plants, worms, yeast, ciliated protozoans, tumor cells and mammals. This review integrates fundamental basic concepts of epigenetics with specific focus on how the epigenetic machinery interacts and operates in continuity to silence or activate gene expression. Topics covered include the connection between DNA methylation, methyl-CpG-binding proteins, transcriptional repression complexes, histone residues, histone modifications that mediate gene repression or relaxation, histone core variant stability, H1 histone linker flexibility, FACT complex, nucleosomal remodeling complexes, HP1 and nuclear lamins.
Key words: DNA methyltransferases, methyl-CpG-binding proteins, H2A.Z, high mobility group proteins, SWI-SNF, nucleosomal remodeling complexes, heterochromatin proteins, epigenetics
Introduction
The human genome is composed of billions of sequence arrangements containing a bioinformatics code that controls how genes are expressed. This code is further dependent upon heritable non-static epigenetic arrangement of histone scaffolding that surrounds the DNA and comprises the “epigenome.” The historical transitional evolution of the human genome is believed to have occurred through a number of processes, one being the altered sequence and re-arrangement of transposable elements located at segments of non-coding DNA. It is believed that the greater the complexity of an organism, the greater amount of non-coding DNA. In humans, protein-coding regions of DNA account for <1.6% of the genome.1 Transposable elements, also referred to as “jumping genes,” have accumulated throughout millions of years as evolutionary ancient DNA in the form of transposons and retrotransposons, which are reverse transcribed long-terminal repeat (LTR) retroviruses.2 Today, active non-LTR retrotransposons (i.e., Alu and LINEs) perpetuate transgenerational genetic diversities through genomic DNA variation among humans. While the evolution of DNA occurs at a slow pace, expedient heritable changes to the epigenome allow dynamic and flexible modification to suit rapid environmental adaptation.3 While the epigenome has more influence on the temporal phenotype, the collective effects of change to the genome and the epigenome contribute to observable physical or biochemical characteristics of an organism.
Throughout the life cycle, dynamic epigenetic control over the phenotype is influenced by a time component responsible for maturation and senescence from conception to adulthood. Environmental epigenetic factors affecting long-term phenotypic change are largely initiated during in utero/perinatal periods, when introduction to the external world is being established.4 It is believed that since epigenetic patterns are inherited through mitosis, the earlier the stage of development, the more critical the environmental impact on the resulting phenotype. During fetal development, environmental cues can induce the modification of a pliable epigenome, which can result in long-term changes in gene expression that occur in a self-sustaining manner in the absence of the original stimulus.5 Adverse gestational conditions that arise from inadequate healthcare, poor nutrition, socioeconomic disadvantage and racial disparities are often associated with long-lasting phenotypic consequences in adults, yielding greater risk of diabetes and heart disease,6,7 as well as low birth weight and congenital defects in progeny.8–10 It is now becoming evident that these effects are inextricably linked to altered epigenetic patterns. Offspring exposed to gestational malnutrition due to extended famine in certain populations also show higher prevalence of adult onset obesity and schizophrenia, tantamount to altered DNA methylation patterns for specific genes such as insulin like growth factor 1/2 and the obesity factor gene leptin.11 Altered epigenetic patterns acquired during early development involve changes in DNA methylation patterns, genomic imprinting,12 histone modifications and the establishment of specific expression profiles of non-coding miRNAs.13,14
Given the enormous impact of early epigenetic programming and the serious nature of related developmental conditions, such as Praeder-Willi, Angelman and Beckwith-Wiedemann syndromes, neural tube defects, adult onset psychiatric disorders, obesity, cancer and schizophrenia,15–19 considerable attention is given to the “nurture of the epigenome” prior to birth. A number of community outreach projects such as the CDC's National Center on Birth Defects and Developmental Disabilities promote awareness about reducing the risk of epigenetic related defects such as spina bifida by suggesting an adequate intake of epigenetic-related nutrients (e.g., choline, vitamin B12, B6 and folate) during pregnancy.20–25 The epigenome appears to remain pliable after birth and during the first years of life. This is evidenced by correlations described in infants exposed to stress or lack of emotional nurturing who show overactive hypothalamic-pituitary-adrenal stress response, glucocorticoid feedback or decreased hypothalamic corticotropin-releasing factor.26 Once established in the offspring, epigenetic marks can become transgenerational, continuing transmittance to future descendants—including the very trait of maternal nurturing in females.27 The longevity of transgenerational epigenomic inheritance pattern is further influenced by the severity and repetition of a similar environmental stimulus among individuals of the same lineage. If the stimuli are discontinued, phenotypic traits could dissipate after the first or second generation.27,28 In other instances, longer lasting epigenetic changes adversely affect the phenotype of the third or fourth generation,29 often initiated by environmental factors adverse to human health that perpetuate aberrant patterns of transgenerational transmission of phenotype.30
The purpose of this review is to simplify the enormous complexity of epigenetic biochemistry that links nuclear DNA to the environment. On one hand, the concept of epigenetics is relatively simple in that it describes a means by which genes are either turned on or off by a heritable epigenome. On the other hand, the environmental and biological controls that mediate these events are extraordinary in number, compounded by instances of similar methylation events that have opposite effects when occurring at different histone amino acids (e.g., H3K36me3 and H3K9me3) and by variation in the interpretation of studies performed in diverse organisms such as flies, plants, worms, yeast, ciliated protozoans, tumor cells and mammals.31
DNA Methylation
More than 30 million base pairs of the human genome are CG dinucleotides (CpGs). CpG sites are target platforms for methylation, which consist of the covalent attachment of a methyl group to the 5′ position of the cytosine (C). CpG methylation patterns correlate with transcriptional silencing patterns observed in 60–90% of the human genes.32 Methylated CpGs reinforce silencing by a number of processes including direct ability to block transcription initiation complexes from binding to DNA promoter regions, inhibition of RNA polymerase (RNAP) and recruitment of transcriptional repressor complexes that bind through Krüppel-like C2H2 zinc fingers, such as Kaiso, and human zinc finger and BTB domain-containing proteins 4 and 38 (ZBTB4 and ZBTB38).33–35 Methylation of CpG sites is performed by DNA methyltransferases DNMT3A and DNMT3B, which carry out de novo methylation, and DNMT1, which ensures heritable epigenome replication through cell division.36 A closer look at DNMT1 shows that this enzyme not only methylates DNA, but also docks directly to methyl-CpG binding proteins (MBPs), such as MeCP2, MBD2 and MBD3. MBPs can equally dock to constrictive histone enzymes, such as histone deacetylases HDAC1 and HDAC2, human H3K9 methyltransferase (Suv39 h1) and heterochromatin protein 1 (HP1), all synergistic components of gene silencing.
DNMT1 ensures that silencing remains heritable during cell division by binding to the replication fork, through its UHRF1 domain, and to methylated CpGs, through its SRA domain.37 UHRF1 and SRA domains function synergistically in the recruitment of histones that bear constrictive epigenetic marks (e.g., H3K9me3) and constrictive enzymes, such as G9a and HDAC1.38,39 DNMT1 is further stabilized by a UHRF1/HDAC1 complex containing USP7 deubiquitinase, also called “herpes virus-associated ubiquitin-specific protease,” which prevents DNMT1 proteosomal degradation.39 In addition, UHRF1 both reinforces silencing at methylated CpG sites and ensures transcription at unmethylated CpG islands. CpG islands are open regions of DNA that are highly accessible to transcription initiation complexes. At transcription start sites, UHRF1 degrades DNMT1 by ubiquitin E3 ligase proteosomal targeting39,40 and ensures gene expression by interacting with histone expansive enzymes such as acetyltransferase Tip60, which promotes nucleosomal expansion.38–40 The bob and weave nature of UHRF1 contributes to the replication of DNA methylation patterns. Coordinated histone scaffold inheritance patterns occur by enabling both DNMT1 deubiquitination (stability) and DNMT1 ubiquitination (proteosomal degradation).41 De novo DNA methylation enzymes, such as DNMT3a and b, work in a similar way in that they recruit constrictive silencing elements, such as transcriptional repressor domains (TRDs), a repression protein of 58 kDa also known as Zfp238 that is critical for embryonic development, HDAC1, Suv39 h1 and HP1.30 All of these enzymes are tightly interwoven within compact nucleosomes surrounding methylated CpG DNA.42 While de novo DNMT3s establish silencing patterns in response to environmental triggers, silencing inheritance patterns maintained throughout cell replication are carried forward by DNMT1.
Once silencing expression patterns are established by DNA methylation, epigenetic inheritance patterns are further solidified by small non-coding RNAs (miRNAs) that ensure regional silencing through degradation of unwanted mRNAs, stunting mRNA maturation or blocking promoter areas associated with mRNA to be silenced.1,17,43 miRNAs can also influence the epigenome itself. This is exemplified by the fact that GpG islands are often occupied by miRNAs that specifically degrade mRNA transcripts that encode for proteins involved in silencing or histone constriction, such as MeCP2, DNMTs, HDACs and EZH244 (all of which are discussed further below). In other words, methylation patterns control transcription of non-coding miRNAs, which, in turn, may assist in silencing by destroying any potential mRNA that may have eluded repression.
Methyl-CpG-binding proteins.
Once CpG methylation patterns are established, silencing is continually reinforced by a series of methyl-CpG-binding proteins that directly dock to CpGs through their methyl-CpG binding domains (MBD).45,46 In humans, MBD proteins include MBD1, MBD2, MBD3, MBD4 and MeCP2. The basic function of MBD proteins is to secure a primary attachment to methylated CpG and a secondary attachment to the surrounding histone scaffolding in order to enable constriction by further docking of DNMTs, histone methyltransferases, HDACs and ATPase chromatin remodeling complexes. These components work together to compress chromatin into heterochromatin at transcription start sites throughout the genome.47
MBD1. MBD1 forms a primary attachment to methylated CpG sites, a secondary attachment to histone constriction enzymes such as SETDB1/Suv39 h1, which methylates H3K9, a tertiary attachment to DNMT1, and a quaternary attachment to a TRD protein.48 As a complex, MBD1-TRD further recruits HP1, HDACs and MCAF1. MCAF1 houses a homeobox-containing zinc finger protein that ensures that dominant transcriptional silencing occurs through the powerful MBD1-SETDB1-TRD-MCAF1 multi repression complex.49–51 Many studies corroborate a very important role for MBD1 in establishing DNA areas to be silenced; its capabilities involve bringing the DNMT1 and the histone methylation enzymes within the MBD1-SETDB1-TRD-MCAF1 complex in close proximity.29,52 Absence of MBD1 results in loss of heterochromatin formation.50
MBD2-MBD4. MBD2 forms a primary attachment to methylated CpG and a secondary attachment to the Mi2-NuRD complex. The Mi2-NuRD complex houses the histone constriction enzymes HDAC1 and HDAC2, the H4 chaperones RbAp48/p46, the chromatin remodeling factor Mi-2 and metastasis-associated MTA1-like, MTA2, Tpase and p66α/p66β, which bind directly to histone N-terminal tails.48,53 The Mi2-NuRD repression complex is a formidable deacetylation powerhouse32 and, when docked to MBD2, is capable of adjoining to DNMT1, MDB1 (via RbAp48) and MBD3.54 MBD4 is unique in that it functions as a DNA repair enzyme that maintains methylated CpG motifs via a DNA N-glycosylase.46,48
MeCP2. MeCP2 is a prominent silencing mark that is heavily embedded in heterochromatin located throughout the lamina circumscribing the nuclear envelope.55 Similar to MBD1, MeCP2 also contains a TRD unit that, in this case, binds to the pre-initiation transcription machinery (TFIIB) that prevents transcription by RNAP II and docks to the paired amphipathic helix protein Sin3A.56,57 Yeast Sin3 (Sin3A and Sin3B) serve as co-repressors that bring histone deacetylase activity in very close proximity to genes targeted for silencing.58 Sin3 is a deacetylase powerhouse that houses HDAC1, HDAC2, the histone-binding proteins RbAp46/RbAp48, which anchor the Sin3 complex onto nucleosomes, the polypeptides SAP30/SAP18, which stabilize the Sin3A-HDAC interaction to DNA-bound transcription factors to enable repression, and transcriptional repressors such as Mad/Max proteins, which recruit mSin3A-HDAC directly to gene areas to be silenced.59–64 Paired amphipathic helix domains within Sin3 serve as a protein-protein glue to ensure association of HDAC1 to HDAC2, and of Sin3 to N-CoR and Mad/Max.65 In addition, mSin3A and Sin3B also interact with histone methyltransferase ESET via a tudor domain that aids in the establishment of the H3K9me3 constrictive histone modifications.66 As a unit, the MeCP2-Sin3-HDAC complex further serves as platform for CDK2AP1, which enforces nucleosomes to remain in compact formation.66 MeCP2-Sin3-HDAC complexes also interact with the Mi2-NuRD repression complex and with N-CoR1. N-CoR1 is a member of the ISWI class of ATPase/Snf2 h nucleosomal remodeling proteins (discussed below) and contains TIP5, which is capable of interacting with DNMTs and Sin3 and of recruiting constrictive HDACs and methyltransferases to silence promoter regions of DNA.32,46,48,67–69 N-CoR is also reported to bind to un-liganded nuclear hormone receptors, which block the hormonal activated dissociation of the Sin3 complex (inducing nucleosomal expansion) and inhibit the recruitment of HATs (inducing histone relaxation) involved with gene activation.70
Other human MBD proteins. Additional human proteins that contain MBD domains have been identified, suggesting a role in collaborative silencing. These include: (1) BAZ2A/TIP5 and BAZ2B; (2) KMT1F/CLLD8, KMT1E/SETDB1 and (3) KIAA1461/MBD5 and KIAA1887/MBD6.45,71 Little is known about the function of BAZ2B. There is evidence to suggest that it can dock to methylated DNA through its primary MBD domain, also having a secondary DNA-binding homeobox and different transcription factor domain, a tertiary tandem C4HC3 zinc-fingerlike domain and a quaternary AT hook domain, which is capable of binding to DNA, also being part of the N-CoR complex. BAZ proteins are likely to be involved in the recruitment of HDAC1, DNMTs and ISWI-ATPase nucleosomal constriction machinery close to DNA promoter regions targeted for silencing.72,73
In the case of KMT1F/CLLD8, co-existence of a MBD domain on pre-SET/SET brings together constrictive methylase SETDB1 in conjunction with Suv39 h1.45,74–76 MBD5 is unique in that it harbors a secondary proline-tryptophan-tryptophan-proline domain, which can allow direct docking to chromatin, and is generally concentrated in areas with DNMT3a/3b, suggesting a role in newly established patterned heterochromatin.71 All of these studies clearly show the importance of MBD proteins in bringing together methylated CpGs and reinforcements that ultimately model and shape the histone scaffold to constrict around methylated areas of DNA targeted for silencing.
Mutations or defects in any of the above mentioned processes, including DNMTs or MBPs, can lead to developmental disorders such as Rett syndrome (MeCP2),77 Angelman syndrome, defects in brain development, autism (MBD1),78,79 or immunodeficiency-centromeric instability and facial anomalies syndrome (DNMT3b).80,81 Similarly, a deficiency in MBD5 is associated with developmental disorders, specifically within the central nervous system, that manifest themselves as speech difficulties, seizures, microcephaly or behavioral disorders.82
Histone Modifications
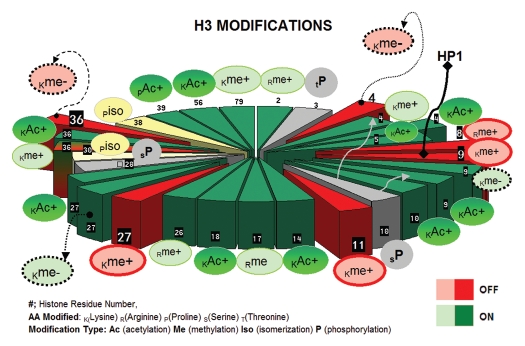
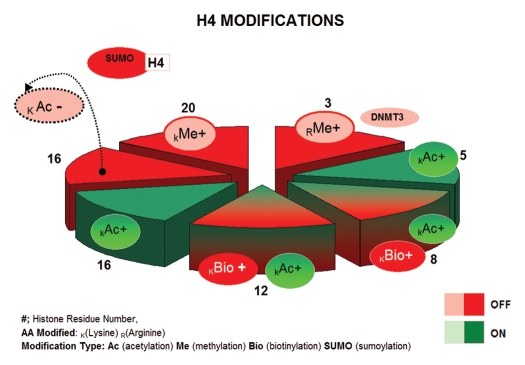
Once CpG sites are methylated and associate with MPD-transcription repression complexes, the next order of macromolecular control is given by the distribution and stability of the histone units. Each individual histone octamer is comprised of two copies of H2A/H2B dimer cores and H3/H4 tetramers, which wrap around 146 base pairs of DNA.83,84 Repeating histone units make up the composition of nucleosomes and nucleosomes make up higher order chromatin. Histone octamer components contain a structured domain and an unstructured N-terminal tail of varying length that protrudes outward from the nucleosome, being readily subject to modifications known as “histone marks.” Histone marks are established by covalent interactions that alter the electrostatic charge and, therefore, histone shape and DNA-histone affinity. The compilation of histone marks make up what is termed “the histone code.” Given that there are at least four amino acid residues that are subject to modification (i.e., lysine, serine, tyrosine and arginine), and more than six types of modifications (i.e., methylation, acetylation, phosphorylation, ubiquitination, biotinylation, sumoylation and proline isomerization), the number of possible combinations comprising the histone code is extraordinarily high (Figs. 1 and 2 summarize described modifications in histone H3 and histone H4, respectively). The histone code is further reinforced by diverse proteins that contain a series of domains, such as CHDs, PHDs, tutors or bromodomains, many found in ATP-dependent nucleosomal remodelers that either (1) disrupt the association between DNA and histones in a ubiquitin-independent manner (inducing expression) or (2) constrict the nucleosomes close to methylated CpG in DNA (inducing silencing).
Figure 1.
H3 histone modifications. Slice numbers represent residue positions. Green denotes histone marks typically associated with gene activation; red represents histone marks generally associated with transcriptional repression. Modifications are denoted at specific histone residues as Iso, isomerated; Ac+, acetylated; Me+, methylated (variable methylation can include me1, me2, me3); P, phosphorylated; me-, de-methylated and Ac-, de-acetylated. Residues are K, lysine; R, Arginine; S, Serrine; T, Threonine; P, Proline.
Figure 2.
H4 histone modifications. Slice numbers represent residue positions. Green denotes histone marks typically associated with gene activation; red represents histone marks generally associated with transcriptional repression. Modifications are denoted at specific histone residues as Bio+, biotinylated; Ac+, acetylated; Me+, methylated (variable methylation can include me1, me2, me3); Ac-, de-acetylated and SUMO, sumoylated.
The most extensively studied modifications to H3/H4 tetramers include (1) hyperacetylation, mediated by histone acetylases (HATs) such as GNAT/PCAF and (2) deacetylation by HDACs. HDACs are docked to transcriptional co-repressor silencing complexes such as Sin3, N-CoR and Mi2-NuRD, whereas HATs are in close proximity to CREB protein/p300 co-activators and transcription initiation complexes, such as TAFII250,64,65 around actively transcribed genes.65 A second set of highly investigated modifications to H3/H4 tails involve (1) the transfer of methyl groups by histone methyltransferases (HMTs), which are enzymes that have a highly conserved SET domain and (2) removal of methyl groups by histone demethylases (HKDMs), such as jmjC domain-containing histone demethylase LSD1. The methylation of histones can either provoke gene repression (as in the case of H3K9me3) or gene expression (as in the case of H3K4me3). A complete examination of known histone modifications is outside the scope of this review but, in brief, histone modifications create histone marks that are recognized by diverse supporting elements to perpetuate nucleosomal constriction or relaxation. A good example of this would be the case of DNMTs, which play a role in silencing by binding to unmethylated H3K4, and H3K9 methyltransferase enzyme SETDB1. DNMTs and SETDB1 interact with MBD1, which, in turn, dock to methylated CpGs.85 In contrast, gene expression involves the phosphorylation of H3S10, which leads to the displacement of constrictive HP1 from H3K9me3-marked chromatin, methylation of H3K4, acetylation of H3K9 and dissociation of DNMT3a, all events that facilitate chromatin decondensation in stable euchromatin.25 It is also reported that, in some cases, histone residues can be modified in ways that promote transcription or silencing, as in the case of H3K9, which can be trimethylated (to promote gene repression) or acetylated (to promote transcriptionally active genes).86 Additionally, a number of other modifications such as sumoylation of H4 and biotinylation of H4K8 and H4K12 are generally believed to be associated with transcriptional repression, although this may not always be the case and needs to be investigated further.87,88
Nucleosomal Positioning
The nucleosomal structure is predicted by the characteristics of the collective assembly of individual histone octamers that circumscribe 146 base pairs of DNA, which are further adjoined by a piece of 15 base pairs of DNA woven through the connective linker histone H1. Constricted nucleosomes are packaged tightly around DNA, serving as a multi-composite mechanical barricade by which transcription initiation complexes are denied access to DNA.57 In the opposite manner, expansive, nucleosome-free, non-methylated CpG islands are associated with active gene expression. In these regions, biochemical forces eject nucleosomes (by sliding and twisting) away from DNA to ensure an open unmethylated loop of DNA is highly accessible by transcriptional complexes and RNAP II at 5′ promoter gene regions.48 The three most recognized events associated with how nucleosomes twist and slide involve (1) the stability of the histone cores H2A/H2B; (2) the integrity of the H1 linker and (3) the supporting tortional movement enabled by the ATPase-driven chromatin remodeling machinery.
Histone core stability.
Nucleosomal ejection is initiated by a destabilized H2A histone core that occurs through an exchange process with an unstable dimer. The unstable dimer is collapsible, highly acetylated and enables chromatin expansion and nucleosome ejection at areas of unmethylated DNA targeted for active expression.89 Unstable histone variants such as H2A.Z most often coincide with transcription start sites90,91 and are highly concentrated in euchromatin marked with acetylated H3K9, H3K18, H3K27 and H4 tails circumscribing open 5′ promoter regions of genes.92,93 While methylation of histone tails is primarily associated with silencing, triple methylation at H2AR at serine residues within the H3 tail by enzymes such as PRMT5 are also associated with H2A.Z core exchange, unit collapse, nucleosomal ejection and active gene expression.94,95
While there are many questions as to the sequence of events that initiate core exchange, it has been suggested that H2A cores are subject to modification both before and after core exchange. Mammalian H2A contains two lysine residues (K5 and K9) that are subject to acetylation by Tip60 and CBP/300, respectively, and initiate instability.89 Moreover, a serine residue (S1) that can be phosphorylated by MSK1 kinase and an arginine residue (R3) that can be methylated by PMT5 are also thought to play a role in triggering core exchange. Modifications to the core variants, such as phosphorylation of H2Av in flies, can lead to active recruitment of HATs and proteins containing bromodomains (BRDs), such as Tip60/ATPase/helicases (in eukaryotes) and the NuA4 acetyltransferase/Swr1 complex (in yeast).96–99 Collectively, these forces serve to relax histones through core exchange. Core variant acetylation and any subsequent recognition by BRDs located on reposition machinery assist to move nucleosomes away from DNA to allow transcription. A number of studies support this model in diverse organisms. For example, BRDs on SWI/SNF ATPase nucleosomal repositioning complexes, such as Swr1/Bdf1 in yeast and BRG/SNF5/BAF47 in mammals, recognize acetylated unstable H2A cores and assist in driving the nucleosomes away from unmethylated areas of DNA.97 Moreover, energy driven ATPase chromatin-remodeling complexes can aid in core dimer exchange, such as in the case of SNF2-related CBP activator protein, the human ortholog of Swr1, TIP60 TIP48 and TIP49 HAT complex, which enhances acetylation to facilitate H2A.Z-H2B dimer exchange into the nucleosome and its subsequent ejection.98 SNF2-related CBP activator protein seems to provide the energy to displace the core, as knockdown via siRNA decreases deposition of H2A.Z and acetylated H2A.Z at gene promoter regions.100
A number of complementing molecular events contribute to core exchange and instability. It has been reported that acetylation of the nucleosomal ATPase subunit within chromatin remodeling proteins may create stability for the entire unit, such as that observed for acetylated BAF (stabilized by BAF155), which blocks TRIP12/E3 ubiquitin proteosomal degradation.101 Histone chaperones, such as FACT, nucleoplasmin and NAP-1, also assist in this process.102 FACT subunits facilitate phosphorylation of H2A, which is required for core exchange, as well as dimer ejection, nucleosomal collapse103,104 and activation of RNAPs.105 The function of FACT can be likened to a zipper effect that ejects nucleosomes along transcription start sites occupying unmethylated areas of DNA.
In contrast, core variant exchange with an H2A of greater stability is often associated with gene silencing,106 such as in the case of H2ABbd or macroH2A-H2AB, which evoke inhibition of p300-dependent histone acetylation, attenuation of RNAP II and gene silencing.107,108 Modifications to the H2B core are also involved in transcription control and include sumoylation, acetylation and ubiquitination.89
H1-the histone linker.
Disruption of the H1 linker is evident along transcription start sites that are aligned in proximity to unstable histone core variants such as H2A.Z109 and hyperacetylated H3 and H4 tails. In mammals, these modifications occur alongside high mobility group (HMG) proteins, which serve to loosen tension of the H1 linker, enabling nucleosomal expansion.110,111 HMGB1–4 proteins are believed to dock to the H1 linker via nucleosomal binding domains causing a break of integrity in association with nucleosomal ejection around promoter gene regions.112,113 HMGN1 is associated not only with disruption of the H1 linker, but also with increased acetylation of H3K14 and with increased HAT/PCAF within the vicinity of where it docks to nucleosomes, events all associated with transcription.114 HMGs have dynamic capability as they can also bind DNA through an HMG-box, a feature that is likely to stabilize and facilitate ejection of DNA from nucleosomes.111,115 Other identified HMG proteins include HMGN3a/3b and HMGN5,115 which are present in euchromatin.116 There is very little information at this point about the regulatory roles of post-translational modifications to the H1 linker, such as ADP-ribosylation or methylation,117 but there is some indication that phosphorylation of H1 increases the dissociation rate of unstable cores within nucleosomes.118 However, most studies allude to the importance of histone H1 linker elasticity in regulating gene expression through loss of nucleosomal integrity.
Nucleosomal remodeling complexes.
SWI-SNF. ATP-dependent chromatin remodeling complexes were first discovered in yeast and named SWI (after yeast mating-type switching)-SNF (after sucrose non fermenting) nucleosome remodeling complexes. The SWI-SNF complexes in yeast contain a Swi2/Snf2 ATPase subunit. A similar complex in yeast, the RSC, contains an Sth1 ATPase subunit; the mammalian SWI-SNF complex contains a hBrm ATPase subunit (also known as SNF2α/SMARCA2), a Brg1 ATPase subunit (also known as SNF2β/SMARCA4) or a collective unit referred to as mammalian BAF.32 SWI/SNF BRG1 and hBRM ATPases are typically associated with gene expression and reportedly present in the vicinity of unmethylated CpG islands and heavily acetylated histone tails.
BAF type remodeling proteins contain BRDs, which bind to acetylated lysines with high specificity.119 It is now becoming more apparent that the human genome encodes for dozens of BRD-containing proteins, many of which play a role in promoting transcription. These include the HAT enzymes PCAF and GCN5, the Tip60 acetyltransferase, the co-activator CBP, the transcriptional protein TAF1, BRD2, BPTF, SNF2L4, BAZ2A, BAZ2B, and the nuclear proteins Sp100, Sp110 and Sp140.119–121 Moreover, many proteins that contain BRDs also have other domains including PHD, PWWP, B-box type zinc finger, Ring finger, SAND, FY Rich, SET, TAZ zinc finger, helicase, ATPase, BAH, WD40 repeat and MBDs.119 The complexity of BRD-containing proteins role in the regulation of gene expression can be exemplified by the BRDs in human SWI/SNF BRG1 and hBRM ATPases, which have dynamic capacity to bind H3K14ac, H4K8ac, H4K5ac and H4K122ac, as well as DNA, via AT-hook or zinc finger binding motifs.122–125 This is also exemplified by the Brg-1 component of remodeling complex BAF, which contains two bromodomains (BD1 and BD2) with capacity to trigger transcription through association with transcription factor E2F, transcription mediators such as CDK8 and TRAP220, RNAP II,119 and with ability to dock to acetylated histone tails.126 Other human BRD proteins, such as Brd4, are also recruited to acetylated H3/H4 tails or phosphorylated H3S10 and, in turn, recruit enzymes that phosphorylate RNAP II and recruit P-TEFb to transcriptional start sites.127 The nucleosomal reposition machinery, therefore, not only enforces ejection of histones, but also recruits transcriptional machinery and provides the torsional stress on chromatin required to open a DNA loop out of the helix, which is further mediated in part by RAD54 in Swi2/Snf2 (yeast) in co-operation with Rad51, which destabilizes nucleosomes along stretches of DNA associated with eviction.128
SWI/SNF Swi2/Snf2 complexes are also known to contain actin-related proteins Arp7p and Arp9, which are the human equivalent of β-actin/BAF53α/BAF53β. These subunits assist in the ability of euchromatin to build scaffolding between neighboring chromatin fibers or proteins, possibly involved in maintaining the open and distant location of nucleosomes away from DNA to allow for gene expression. For example, BAF53 is usually associated with active transcription and has the capability to form direct complexes with TIP49, TIP48 (which is necessary for the assembly and functional activity of the TIP60 acetyltransferase complex), TRRAP and several HATs.129 Mutations in nucleosomal reposition complexes are associated with serious developmental problems including mental retardation, facial dimorphism, urogenital abnormalities and α-thalassemia.130
ACF. Other remodeling complexes are involved in gene silencing, such as human ATP-dependent chromatin assembly and remodeling factor, hACF, or its yeast counterpart, ISW2, which arrange nucleosomes by a sliding motion toward formation of organized heterochromatin.131,132 Human ACF and yeast ISW2 bring nucleosomes toward a state of compaction in close proximity to methylated CpGs, HDACs, TRD-Sin3A/Rpd3 and MeCP2. ACF docks solidly to the nucleosome via PHD fingers in the core histones.133 Deletion of PHD modules abolishes ACF subunit Acf1-directed nucleosome repressive mobilization.133 ACF is a member of the SF2 family of ISWI-class of ATPases, which includes DExx-box proteins such as helicases and nucleic acid translocases, SNF2 h and CHRAC. Acf1 is involved in the strategic deposition of histones in periodic nucleosome arrangement around methylated CpG DNA targeted for silencing and is assisted by the chaperones NAP-1 and CAF-1.69 The CHRAC complex serves primarily as a lubricant to enable nucleosomes to slide with little friction along DNA; the Acf1 subunit provides a slipping sliding motion by which nucleosomal silencing arrays are fashioned.134,135 CHRAC subunits also include NURF, topoisomerase136,137 and HMGB1, which are believed to lubricate areas of distorted DNA to allow smooth entry along a compact nucleosome,138 working collectively with Acf1, which assists in DNA repair.139
CHD/Mi2-NuRD. Repressive remodeling complexes of the SNF2-like DNA helicase/ATPase category also include NuRD, which contains Mi-2 subunits and histone deacetylases HDAC1 and HDAC2. Mi-2 subunits of the NuRD complex belong to the CHD family of proteins that includes CHD1, CHD2, CHD3/Mi-2α and CHD4/Mi-2β, the latter of which comprise the catalytic ATP hydrolyzing subunits in the complex. CHD3 and CHD4 contain PHD-zinc finger domains, two chromodomains and a SWI2/SNF2-type ATPase/helicase domain. While the PHD fingers and chromodomains within this complex have the ability to bind repressive histone marks H3K9me3 and H3K4me3, respectively,140,141 each of these subunits can act as an autonomous repressor complex when separated from HDAC/NuRD in Drosophila melanogaster.142 Current research is now suggesting a dual co-repressor/co-activator nature of Mi2-NuRD, given the ability of its Mi-2 PHD-zinc finger domain to affiliate with constrictive histone marks (e.g., H3K9me) and expansive histone marks (e.g., H3K9ac); nevertheless, the latter has not been fully explored58,143–145 In general, this complex was found associated with nucleosomal constriction and transcriptional repression in the majority of the research literature.
Heterochromatin Proteins
Heterochromatin proteins HP1α and HP1β are the major tethering elements that bring together most, if not all, silencing elements by adjoining Suv39 h1, H3K9me3 by a HP1 N-terminal chromodomain, MeCP2, methylated CpGs, MBD1/SETDB1/CAF-1 and human Swi/Snf ATPases, via their chromoshadow PXVXL motif.25,146–148 HP1 proteins are involved in nucleosomal constriction and counteract expansive chromatin remodeling complexes such as Brg1 and Brm.148 Moreover, HP1 can also interact with BAHD1, which interacts directly with MBD1 and HDAC5, allowing for organized heterochromatin that lacks acetylated H4 and is enriched in H3K27me3.149 The histone mark H3K9me3 seems to be of utmost importance in the stability of constrictive HP1, which, if lost by phosphorylation of H3S10, can lead to nucleosomal expansion and formation of euchromatin.25 As previously mentioned, H3S10 is a general histone expansive mark and signals recruitment of bromodomain proteins (which recognize acetylated H3/H4) and RNA initiation and elongation factors toward transcriptional start sites.127 HP1 is dynamic, having ability to interact with constrictive HDACs, a number of repression complexes and DNMT1, bringing together a plethora of silencing agents into position.150 While HP1α and HP1β are associated with silencing, other forms of HP1, such as HP1-type C, are found in euchromatin interacting with FACT subunit SSRP1, which is responsible for guiding FACT toward active genes to promote nucleosomal instability by H2AX exchange and RNAP activation.103–105,151,152
Macromolecular Chromatin
Euchromatin.
At the macromolecular level, transcriptionally active euchromatin is housed at the inner nucleoplasm/center of chromosome territories, while silenced heterochromatin is arranged at the outer circular perimeter of the nuclear membrane. Euchromatin contains a combination of unstable core variants, HATs, G9a, HMG proteins and a low occupancy of linker H1.116,146 While euchromatin may contain both activation marks (e.g., H3K4me3 and H3K36me3) and some silencing marks (e.g., H3K9me2 or HP1α), a hallmark component of active genes is the loss of H3K9 methylation at transcription start sites.153 In euchromatin, abundant ribosomal unit transcription factories called nucleoli are easily visualized, which synthesize ribosomal subunit proteins that will be assembled later into the small and large subunits of ribosomes.67,154
Heterochromatin and lamins.
Silenced heterochromatin is comprised of two categories. Constitutive heterochromatin is considered to be static and irreversible through the lifetime of the organism, whereas facultative heterochromatin can change patterns through epigenetic alteration.151 At the nuclear periphery, silenced heterochromatin contains a combination of MBD proteins, repression complexes, H3K9me2, H3K9me3, HP1α, Suv39 h1 and methylated DNA.152 Within heterochromatin, Suv39 h1 can become a target of the SIRT enzyme, which maintains deacetylation at H4K16, contributing toward the reinforced silencing within heterochromatin.155 DNA methylation within heterochromatin must be sustained, as its loss could initiate the dismantlement of chromatin, acetylation of histone H4, and H3K4 di- and trimethylation, all events associated with relaxation of nucleosomes.156 The nuclear lamina is required to lock the peripheral position of heterochromatin within the nuclear envelope to maintain silencing.32 The nuclear lamina itself is a type of glue that secures heterochromatin organization into position and in organized fashion to sustain gene silencing. Lamins directly interact with H3K9me3 and MeCP2/Sin3A, which bind to lamin B, HP1γ and HP1α.55,157 Lamins also adhere to lamin-associated polypeptides and, at the inner membrane, bind with emerin, Sun1 and Sun2, which anchor nesprin-3 on the outer side, where it binds to cytoskeletal actin via a Klarsicht/ANC-1/Syne homology domain. Moreover, on the outside membrane, nesprin-2 forms complexes with kinesin-1 motor protein apparatus by associating with and recruiting KLC1 to the outer nuclear membrane, which requires lamin A and C. Ultimately, the kinesin-1/nesprin-2/SUN complex spans the entire nuclear envelope in order to anchor cytoskeletal structures to nuclear lamina.158
Given the important role of lamins in these processes, it is not surprising that lamin A and lamin C mutations are associated with severe disorganization of chromatin and degenerative diseases, collectively termed as laminopathies. One example is the Hutchinson-Gilford progeria syndrome, which corresponds to a loss of coordinated transcriptional control of the progerin protein.157,159 In addition, more than 200 pathogenic mutations are associated with the LMNA gene, most of which result in developmental disorders of skeletal, heart, muscle and neurological systems, such as Emery-Dreifuss muscular dystrophies, AD-limb girdle muscular dystrophy, lipodystrophy, neuropathy, cardiomyopathy, dermopathy, Atypical Werner Syndrome, hepatic stenosis, hyperpigmentation, fertility problems and accelerated aging syndromes.160,161
Summary
While there are few absolutes in the field of epigenetics, a general line of thought to simplify a very complex matter could include the following. Epigenetics may be conceptualized as control over how genomic DNA is expressed. This control is initiated by DNMTs that methylate CpGs, which are then tagged by MBD proteins attached to potent repression complexes. Repression complexes, in turn, control modifications to histones H3 and H4 tails, which perpetuate constriction and make stable modifications to the histone cores H2A and H2B to prevent histone unit ejection/nucleosomal displacement. These collective events are associated with stabilized tension of the histone H1 linker. HP1α and HP1β proteins tether silencing elements—from methylated CpGs to ATPase remodeling machinery—together, in order to tightly crowd methylated DNA close to the nucleosomes, thereby blocking transcription elements. The positioning of silenced heterochromatin along the nuclear envelope is carried on by lamins; centrally positioned euchromatin remaining transcriptionally active. Within euchromatin, unmethylated CpG islands remain open to transcription initiation complexes by opposing processes such as hyperacetylation of histone tails, H2A core variant exchange, HMG proteins binding, which disrupt the H1 linker, and the FACT complex and ATPase reposition machinery, which promotes continual nucleosomal expansion. Silencing and transcription patterns are altered by environmental cues and the transmittance of epigenetic information is heritable through mitosis and meiosis. While this describes a very basic model, the complexities of epigenetic control in regulating the phenotype seem infinite.
Acknowledgments
This research was supported by a grant from NIH NCRR RCMI program (G12RR 03020).
Abbreviations
- ACF
ATP-dependent chromatin assembly and remodeling factor
- Arp7p
actin-related protein 7p
- BAF53
actin-related protein Nβ
- BAH
bromo adjacent homolog
- BAHD1
bromo adjacent homology domain-containing protein 1
- BAZ2A
bromodomain adjacent to zinc finger domain protein 2A
- BAZ2B
bromodomain adjacent to zinc finger domain protein 2B
- BPTF
BRD and plant homeodomain [PHD] finger-containing transcription factor
- BRD
bromodomain
- Brg-1
Brahma-related Gene 1
- CAF-1
chromatin assembly factor-1
- CDC
Centers for Disease Control and Prevention
- CDK2AP1
cyclin-dependent kinase 2 associated protein 1
- CDK8
cyclin-dependent protein kinase 8
- CHD
chromo-helicase/ATPase-DNA binding
- CHD3
chromodomain helicase DNA binding protein 3
- CHRAC
chromatin accessibility complex
- CREB
cAMP responsive element binding protein
- DDT domain
DNA binding homeobox and different transcription factor
- DNMT1
DNA methyltransferase 1
- DNMT3A
DNA methyltransferase 3a
- DNMT3B
DNA methyltransferase 3b
- EZH2
histone-lysine N-methyltransferase
- FACT
facilitates chromatin transcription
- G9a
H3-K9 methyltransferase
- GNAT
Gcn5-related N-acetyltransferase
- H1
histone linker
- H2A
histone H2A
- H2A.x
H2A histone family member x
- H2A.Z
H2A histone family member Z
- H2Av
H2A histone family member V
- H2B
histone H2b
- H3/H4
histone H3/histone H4
- H3K27(me3)
histone H3 lysine 27 triple methylated
- H3K9(me3)
histone H3 lysine 9 triple methylated
- H4K20(me3)
histone H4 lysine 20 triple methylated
- H4K5(-ac)
histone H4 lysine 5 deacetylated
- hACF
human ATP-dependent chromatin assembly and remodeling factor
- HATs
histone acetyltransferases
- hBrm
human brahma
- HDAC1
histone deacetylase 1
- HDAC2
histone deacetylase 2
- HGPS
hutchinson-gilford progeria syndrome
- HKDM
histone demethylase
- HMG
high mobility group
- HMGN
high-mobility group nucleosome binding domain
- HMTs
histone methyltransferase
- HP1
heterochromatin protein 1
- HP1α
heterochromatin protein 1α
- HP1β
heterochromatin protein 1β
- ISWI
member of the SWI2/SNF2 family of ATPases, is the catalytic subunit of NURF, CHRAC and ACF
- JMJD
(JmjC)-domain-containing histone demethylases
- KASH
klarsicht/ANC-1/Syne homology
- KLC1
kinesin light chain 1
- KMT1E SETDB1
histone methyltransferase
- KMT1F/CLLD8
lysine methyltransferase
- LMNA
lamin A/C gene
- LSD1
histone demethylase
- LTR
long terminal repeats
- MBD
methyl binding domains
- MBD1
methyl-CpG-binding domain (MBD) protein 1
- MBD2
methyl-CpG-binding domain (MBD) protein 2
- MBD3
methyl-CpG-binding domain (MBD) protein 3
- MBD4
methyl-CpG-binding domain (MBD) protein 4
- MBPs
methyl-CpG binding proteins
- MCAF1
MBD1-containing chromatin associated factor
- MeCP2
methyl-CpG-binding protein
- Mi2-NuRD
Mi-2/nucleosome remodeling and histone de-acetylation chromatin remodeling complex miRNAs, microRNA
- mRNA
messenger RNA
- MSK1
mitogen- and stress-activated protein kinase 1
- MTA1-like
metastasis associated 1-like
- MTA2
metastasis associated 2
- NAP-1
nucleosome assembly protein-1
- NBDs
nucleosomal binding domains
- N-CoR1
nuclear receptor co-repressor 1
- NE
nuclear envelope
- NuRD
nucleosome remodeling and deacetylation
- NURF
nucleosome remodeling factor
- ORC2
origin recognition complex protein
- PAH
paired amphipathic helix
- PCAF
p300 cAhlP response element binding protein CREBI-binding protein associated factor
- PHD
plant homeo domain-zinc finger domains
- RNP II
RNA polymerase II
- pre-SET/SET
histone lysine methyltransferases
- PRMT5
histone-arginine N-methyltransferase 5
- P-TEFb
positive transcription elongation factor b
- PWWP
proline-tryptophan-tryptophan-proline domain
- RbAp48/p46
Rb-associated histone deacetylase complex
- RP58
repression protein of 58 kDa
- SAP18
Sin3A-associated protein, 18 kDa
- SAP30
Sin3A-associated protein, 30 kDa
- SET
su(var)39, enhancer of zeste, trithorax domains
- SETDB1
SET domain bifurcated 1
- SETDB1/Suv39 h1
histone methyltransferase
- Sin3
transcriptional co-repressor Sin3
- Sin3A
SIN3 homolog A, transcription regulator (yeast)
- Sin3B
SIN3 homolog B, transcription regulator (yeast)
- SIRT
silent information regulator 2
- SMARC
SWI/SNF related, matrix associated, actin dependent regulator of chromatin
- SMARCA4
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
- SNF
sucrose non fermenting
- Sp100
SP100 nuclear antigen
- Sp110
SP110 nuclear body protein
- Sp140
SP140 nuclear body protein
- SRA
SET and RING associated
- SRCAP
SNF2-related CBP activator protein
- SUN
Sad1 and UNC84 domain containing 1
- Suv39 h1
suppressor called variegation 3-9 homologue 1
- SWI/SNF
SWItch/sucrose non fermentable yeast nucleosome remodeling complex
- TAF1
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor
- TFIIB
general transcription factor IIB
- TIP48
components of the histone acetylase/chromatin remodeling complex
- TIP5
termination factor TTFI-interacting protein 5
- Tip60
K(lysine) acetyltransferase 5
- Tpase
thymidine phosphorylase
- TRAP220
thyroid receptor-associated protein complex 220
- TRDs
transcriptional repressor domains
- TRIP12
thyroid hormone receptor interactor 12
- TRRAP
transformation/transcription domain-associated protein
- UHRF1
ubiquitin-like with PHD and ring finger domains 1
- USP7
ubiquitin specific peptidase 7 (herpes virus-associated)
- WD40
beta-transducin repeat
- ZBTB38
zinc finger and BTB domain containing 38
- ZBTB4
zinc finger and BTB domain containing 4
References
- 1.Costa FF. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wade PA, Archer TK. Epigenetics: environmental instructions for the genome. Environ Health Perspect. 2006;114:140–141. doi: 10.1289/ehp.114-a140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martínez-Frías ML. Can our understanding of epigenetics assist with primary prevention of congenital defects? J Med Genet. 2010;47:73–80. doi: 10.1136/jmg.2009.070466. [DOI] [PubMed] [Google Scholar]
- 5.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuzawa CW, Sweet E. Epigenetics and the embodiment of race: developmental origins of US racial disparities in cardiovascular health. Am J Hum Biol. 2009;21:2–15. doi: 10.1002/ajhb.20822. [DOI] [PubMed] [Google Scholar]
- 7.Thaye ZM, Kuzawa CW. Biological memories of past environments: epigenetic pathways to health disparities. Epigenetics. 2011;6:798–803. doi: 10.4161/epi.6.7.16222. [DOI] [PubMed] [Google Scholar]
- 8.Lu MC, Kotelchuck M, Hogan V, Jones L, Wright K, Halfon N. Closing the Black-White gap in birth outcomes: a life-course approach. Ethn Dis. 2010;20:62–76. [PMC free article] [PubMed] [Google Scholar]
- 9.Dailey DE. Social stressors and strengths as predictors of infant birth weight in low-income African American women. Nurs Res. 2009;58:340–347. doi: 10.1097/NNR.0b013e3181ac1599. [DOI] [PubMed] [Google Scholar]
- 10.Brooks PE. Ethnographic evaluation of a research partnership between two African American communities and a university. Ethn Dis. 2010;20:21–29. [PubMed] [Google Scholar]
- 11.Heijmans BT, Tobi EW, Lumey LH, Slagboom PE. The epigenome: archive of the prenatal environment. Epigenetics. 2009;4:526–531. doi: 10.4161/epi.4.8.10265. [DOI] [PubMed] [Google Scholar]
- 12.Gong L, Pan YX, Chen H. Gestational low protein diet in the rat mediates Igf2 gene expression in male offspring via altered hepatic DNA methylation. Epigenetics. 2010;5:619–626. doi: 10.4161/epi.5.7.12882. [DOI] [PubMed] [Google Scholar]
- 13.Fryer AA, Nafee TM, Ismail KM, Carroll WD, Emes RD, Farrell WE. LINE-1 DNA methylation is inversely correlated with cord plasma homocysteine in man: a preliminary study. Epigenetics. 2009;4:394–398. doi: 10.4161/epi.4.6.9766. [DOI] [PubMed] [Google Scholar]
- 14.Fu Q, Yu X, Callaway CW, Lane RH, McKnight RA. Epigenetics: intrauterine growth retardation (IUGR) modifies the histone code along the rat hepatic IGF-1 gene. FASEB J. 2009;23:2438–2449. doi: 10.1096/fj.08-124768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maccani MA, Marsit CJ. Epigenetics in the placenta. Am J Reprod Immunol. 2009;62:78–89. doi: 10.1111/j.16000897.2009.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan D, Cushnie DW, Neaga OR, Lawrance AK, Rozen R, Trasler JM. Strain-specific defects in testicular development and sperm epigenetic patterns in 5,10-methylenetetrahydrofolate reductase-deficient mice. Endocrinology. 2010;151:3363–3373. doi: 10.1210/en.20091340. [DOI] [PubMed] [Google Scholar]
- 17.Coppedè F. One-carbon metabolism and Alzheimer's disease: focus on epigenetics. Curr Genomics. 2010;11:246–260. doi: 10.2174/138920210791233090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duthie SJ. Epigenetic modifications and human pathologies: cancer and CVD. Proc Nutr Soc. 2011;70:47–56. doi: 10.1017/S0029665110003952. [DOI] [PubMed] [Google Scholar]
- 19.de Vogel S, Wouters KA, Gottschalk RW, van Schooten FJ, de Goeij AF, de Bruïne AP, et al. Dietary methyl donors, methyl metabolizing enzymes and epigenetic regulators: diet-gene interactions and promoter CpG island hypermethylation in colorectal cancer. Cancer Causes Control. 2011;22:1–12. doi: 10.1007/s10552-010-9659-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 21.Zeisel SH. Epigenetic mechanisms for nutrition determinants of later health outcomes. Am J Clin Nutr. 2009;89:1488–1493. doi: 10.3945/ajcn.2009.27113B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehedint MG, Niculescu MD, Craciunescu CN, Zeisel SH. Choline deficiency alters global histone methylation and epigenetic marking at the Re1 site of the calbindin 1 gene. FASEB J. 2010;24:184–195. doi: 10.1096/fj.09140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davison JM, Mellott TJ, Kovacheva VP, Blusztajn JK. Gestational choline supply regulates methylation of histone H3, expression of histone methyltransferases G9a (Kmt1c) and Suv39 h1 (Kmt1a), and DNA methylation of their genes in rat fetal liver and brain. J Biol Chem. 2009;284:1982–1989. doi: 10.1074/jbc.M807651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James SJ, Melnyk S, Jernigan S, Pavliv O, Trusty T, Lehman S, et al. A functional polymorphism in the reduced folate carrier gene and DNA hypomethylation in mothers of children with autism. Am J Med Genet B Neuropsychiatr Genet. 2010;153:1209–1220. doi: 10.1002/ajmg.b.31094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals. Cell Mol Life Sci. 2009;66:596–612. doi: 10.1007/s00018-008-8432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meaney MJ, Diorio J, Francis D, Widdowson J, LaPlante P, Caldji C, et al. Early environmental regulation of forebrain glucocorticoid receptor gene expression: implications for adrenocortical responses to stress. Dev Neurosci. 1996;18:49–72. doi: 10.1159/000111395. [DOI] [PubMed] [Google Scholar]
- 27.Skinner MK, Guerrero-Bosagna C. Environmental signals and transgenerational epigenetics. Epigenomics. 2009;1:111–117. doi: 10.2217/epi.09.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Titus-Ernstoff L, Troisi R, Hatch EE, Hyer M, Wise LA, Palmer JR, et al. Offspring of women exposed in utero to diethylstilbestrol (DES): a preliminary report of benign and malignant pathology in the third generation. Epidemiology. 2008;19:251–257. doi: 10.1097/EDE.0b013e318163152a. [DOI] [PubMed] [Google Scholar]
- 29.Weinhold B. Epigenetics: the science of change. Environ Health Perspect. 2006;114:160–167. doi: 10.1289/ehp.114-a160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tal O, Kisdi E, Jablonka E. Epigenetic contribution to covariance between relatives. Genetics. 2010;184:1037–1050. doi: 10.1534/genetics.109.112466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim SJ, Tan TW, Tong JC. Computational Epigenetics: the new scientific paradigm. Bioinformation. 2010;4:331–337. doi: 10.6026/97320630004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakao M. Epigenetics: interaction of DNA methylation and chromatin. Gene. 2001;278:25–31. doi: 10.1016/S03781119(01)00721-1. [DOI] [PubMed] [Google Scholar]
- 33.Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26:169–181. doi: 10.1128/MCB.26.1.169-81.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasai N, Nakao M, Defossez PA. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res. 2010;38:5015–5022. doi: 10.1093/nar/gkq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet. 2010;70:101–141. doi: 10.1016/B978-012-380866-0.60005-8. [DOI] [PubMed] [Google Scholar]
- 36.Xu F, Mao C, Ding Y, Rui C, Wu L, Shi A, et al. Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs. Curr Med Chem. 2010;17:4052–4071. doi: 10.2174/092986710793205372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tittle RK, Sze R, Ng A, Nuckels RJ, Swartz ME, Anderson RM, et al. Uhrf1 and Dnmt1 are required for development and maintenance of the zebrafish lens. Dev Biol. 2011;350:50–63. doi: 10.1016/j.ydbio.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unoki M. Current and potential anticancer drugs targeting members of the UHRF1 complex including epigenetic modifiers. Recent Pat Anticancer Drug Discov. 2011;6:116–130. doi: 10.2174/157489211793980024. [DOI] [PubMed] [Google Scholar]
- 39.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bronner C. Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation and the histone code. Sci Signal. 2011;4:3. doi: 10.1126/scisignal.2001764. [DOI] [PubMed] [Google Scholar]
- 41.Kessler BM, Fortunati E, Melis M, Pals CE, Clevers H, Maurice MM. Proteome changes induced by knockdown of the deubiquitylating enzyme HAUSP/USP7. J Proteome Res. 2007;6:4163–4172. doi: 10.1021/pr0702161. [DOI] [PubMed] [Google Scholar]
- 42.Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, et al. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siomi MC, Tsukumo H, Ishizuka A, Nagami T, Siomi H. A potential link between transgene silencing and poly(A) tails. RNA. 2005;11:1004–1011. doi: 10.1261/rna.2280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handel AE, Ebers GC, Ramagopalan SV. Epigenetics: molecular mechanisms and implications for disease. Trends Mol Med. 2010;16:7–16. doi: 10.1016/j.molmed.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Roloff TC, Ropers HH, Nuber UA. Comparative study of methyl-CpG-binding domain proteins. BMC Genomics. 2003;4:1. doi: 10.1186/1471-2164-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fatemi M, Wade PA. MBD family proteins: reading the epigenetic code. J Cell Sci. 2006;119:3033–3037. doi: 10.1242/jcs.03099. [DOI] [PubMed] [Google Scholar]
- 47.Katryniok C, Schnur N, Gillis A, von Knethen A, Sorg BL, Looijenga L, et al. Role of DNA methylation and methyl-DNA binding proteins in the repression of 5-lipoxygenase promoter activity. Biochim Biophys Acta. 2010;1801:49–57. doi: 10.1016/j.bbalip.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 49.Uchimura Y, Ichimura T, Uwada J, Tachibana T, Sugahara S, Nakao M, et al. Involvement of SUMO modification in MBD1- and MCAF1-mediated heterochromatin formation. J Biol Chem. 2006;281:23180–23190. doi: 10.1074/jbc.M602280200. [DOI] [PubMed] [Google Scholar]
- 50.Ichimura T, Watanabe S, Sakamoto Y, Aoto T, Fujita N, Nakao M. Transcriptional repression and heterochromatin formation by MBD1 and MCAF/AM family proteins. J Biol Chem. 2005;280:13928–13935. doi: 10.1074/jbc.M413654200. [DOI] [PubMed] [Google Scholar]
- 51.Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, et al. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39 h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem. 2003;278:24132–24138. doi: 10.1074/jbc.M302283200. [DOI] [PubMed] [Google Scholar]
- 52.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 53.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 54.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guarda A, Bolognese F, Bonapace IM, Badaracco G. Interaction between the inner nuclear membrane lamin B receptor and the heterochromatic methyl binding protein, MeCP2. Exp Cell Res. 2009;315:1895–1903. doi: 10.1016/j.yexcr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 56.Kaludov NK, Wolffe AP. MeCP2 driven transcriptional repression in vitro: selectivity for methylated DNA, action at a distance and contacts with the basal transcription machinery. Nucleic Acids Res. 2000;28:1921–1928. doi: 10.1093/nar/28.9.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martins RP, Krawetz SA. Towards understanding the epigenetics of transcription by chromatin structure and the nuclear matrix. Gene Ther Mol Biol. 2005;9:229–246. [PMC free article] [PubMed] [Google Scholar]
- 58.McDonel P, Costello I, Hendrich B. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int J Biochem Cell Biol. 2009;41:108–116. doi: 10.1016/j.biocel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rottmann S, Speckgens S, Lüscher-Firzlaff J, Lüscher B. Inhibition of apoptosis by MAD1 is mediated by repression of the PTEN tumor suppressor gene. FASEB J. 2008;22:1124–1134. doi: 10.1096/fj.07-9627com. [DOI] [PubMed] [Google Scholar]
- 60.Wahlström T, Henriksson M. Mnt takes control as key regulator of the myc/max/mxd network. Adv Cancer Res. 2007;97:61–80. doi: 10.1016/S0065-230X(06)97003-1. [DOI] [PubMed] [Google Scholar]
- 61.Rottmann S, Menkel AR, Bouchard C, Mertsching J, Loidl P, Kremmer E, et al. Mad1 function in cell proliferation and transcriptional repression is antagonized by cyclin E/CDK2. J Biol Chem. 2005;280:15489–15492. doi: 10.1074/jbc.C400611200. [DOI] [PubMed] [Google Scholar]
- 62.Anderson DM, Arredondo J, Hahn K, Valente G, Martin JF, Wilson-Rawls J, et al. Mohawk is a novel homeobox gene expressed in the developing mouse embryo. Dev Dyn. 2006;235:792–801. doi: 10.1002/dvdy.20671. [DOI] [PubMed] [Google Scholar]
- 63.McCallum SA, Bazan JF, Merchant M, Yin J, Pan B, de Sauvage FJ, et al. Structure of SAP18: a ubiquitin fold in histone deacetylase complex assembly. Biochemistry. 2006;45:11974–11982. doi: 10.1021/bi060687l. [DOI] [PubMed] [Google Scholar]
- 64.Hassig CA, Schreiber SL. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/S13675931(97)80066-X. [DOI] [PubMed] [Google Scholar]
- 65.Davie JR. Covalent modifications of histones: expression from chromatin templates. Curr Opin Genet Dev. 1998;8:173–178. doi: 10.1016/S0959-437X(98)80138-X. [DOI] [PubMed] [Google Scholar]
- 66.Yang L, Mei Q, Zielinska-Kwiatkowska A, Matsui Y, Blackburn ML, Benedetti D, et al. An ERG (ets-related gene)-associated histone methyltransferase interacts with histone deacetylases 1/2 and transcription corepressors mSin3A/B. Biochem J. 2003;369:651–657. doi: 10.1042/BJ20020854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bártová E, Horáková AH, Uhlírová R, Raska I, Galiová G, Orlova D, et al. Structure and epigenetics of nucleoli in comparison with non-nucleolar compartments. J Histochem Cytochem. 2010;58:391–403. doi: 10.1369/jhc.2009.955435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 69.Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pile LA, Schlag EM, Wassarman DA. The SIN3/RPD3 deacetylase complex is essential for G(2) phase cell cycle progression and regulation of SMRTER corepressor levels. Mol Cell Biol. 2002;22:4965–4976. doi: 10.1128/MCB.22.14.4965-76.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laget S, Joulie M, Le Masson F, Sasai N, Christians E, Pradhan S, et al. The human proteins MBD5 and MBD6 associate with heterochromatin but they do not bind methylated DNA. PLoS One. 2010;5:11982. doi: 10.1371/journal.pone.0011982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doerks T, Copley R, Bork P. DDT—a novel domain in different transcription and chromosome remodeling factors. Trends Biochem Sci. 2001;26:145–146. doi: 10.1016/S09680004(00)01769-2. [DOI] [PubMed] [Google Scholar]
- 73.Zhou Y, Grummt I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol. 2005;15:1434–1438. doi: 10.1016/j.cub.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 74.Hung MS, Shen CK. Eukaryotic methyl-CpG-binding domain proteins and chromatin modification. Eukaryot Cell. 2003;2:841–846. doi: 10.1128/EC.2.5.841-6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Falandry C, Fourel G, Galy V, Ristriani T, Horard B, Bensimon E, et al. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J Biol Chem. 2010;285:20234–20241. doi: 10.1074/jbc.M109.052399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mabuchi H, Fujii H, Calin G, Alder H, Negrini M, Rassenti L, et al. Cloning and characterization of CLLD6, CLLD7 and CLLD8, novel candidate genes for leukemogenesis at chromosome 13q14, a region commonly deleted in B-cell chronic lymphocytic leukemia. Cancer Res. 2001;61:2870–2877. [PubMed] [Google Scholar]
- 77.Rastegar M, Hotta A, Pasceri P, Makarem M, Cheung AY, Elliott S, et al. MECP2 isoform-specific vectors with regulated expression for Rett syndrome gene therapy. PLoS One. 2009;4:6810. doi: 10.1371/journal.pone.0006810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Allan AM, Liang X, Luo Y, Pak C, Li X, Szulwach KE, et al. The loss of methyl-CpG binding protein 1 leads to autism-like behavioral deficits. Hum Mol Genet. 2008;17:2047–2057. doi: 10.1093/hmg/ddn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li X, Barkho BZ, Luo Y, Smrt RD, Santistevan NJ, Liu C, et al. Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. J Biol Chem. 2008;283:27644–27652. doi: 10.1074/jbc.M804899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- 81.Lu Q, Qiu X, Hu N, Wen H, Su Y, Richardson BC. Epigenetics, disease and therapeutic interventions. Ageing Res Rev. 2006;5:449–467. doi: 10.1016/j.arr.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 82.Williams SR, Mullegama SV, Rosenfeld JA, Dagli AI, Hatchwell E, Allen WP, et al. Haploinsufficiency of MBD5 associated with a syndrome involving microcephaly, intellectual disabilities, severe speech impairment and seizures. Eur J Hum Genet. 2010;18:436–441. doi: 10.1038/ejhg.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 84.Bentley GA, Lewit-Bentley A, Finch JT, Podjarny AD, Roth M. Crystal structure of the nucleosome core particle at 16 A resolution. J Mol Biol. 1984;176:55–75. doi: 10.1016/00222836(84)90382-6. [DOI] [PubMed] [Google Scholar]
- 85.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2:657–669. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Villar-Garea A, Israel L, Imhof A. Analysis of histone modifications by mass spectrometry. Current protocols in protein science/editorial board, Coligan JE, et al. 2008;14:14. doi: 10.1002/0471140864.ps1410s51. [DOI] [PubMed] [Google Scholar]
- 87.Filenko NA, Kolar C, West JT, Smith SA, Hassan YI, Borgstahl GE, et al. The role of histone H4 biotinylation in the structure of nucleosomes. PLoS One. 2011;6:16299. doi: 10.1371/journal.pone.0016299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Metzler-Guillemain C, Depetris D, Luciani JJ, Mignon-Ravix C, Mitchell MJ, Mattei MG. In human pachytene spermatocytes, SUMO protein is restricted to the constitutive heterochromatin. Chromosome Res. 2008;16:761–782. doi: 10.1007/s10577-008-1225-7. [DOI] [PubMed] [Google Scholar]
- 89.Wyrick JJ, Parra MA. The role of histone H2A and H2B post-translational modifications in transcription: a genomic perspective. Biochim Biophys Acta. 2009;1789:37–44. doi: 10.1016/j.bbagrm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 90.Conerly ML, Teves SS, Diolaiti D, Ulrich M, Eisenman RN, Henikoff S. Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome Res. 2010;20:1383–1390. doi: 10.1101/gr.106542.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim JH, Saraf A, Florens L, Washburn M, Workman JL. Gcn5 regulates the dissociation of SWI/SNF from chromatin by acetylation of Swi2/Snf2. Genes Dev. 2010;24:2766–2771. doi: 10.1101/gad.1979710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tolstorukov MY, Kharchenko PV, Goldman JA, Kingston RE, Park PJ. Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res. 2009;19:967–977. doi: 10.1101/gr.084830.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sarcinella E, Zuzarte PC, Lau PN, Draker R, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol Cell Biol. 2007;27:6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Subtil-Rodríguez A, Reyes JC. BRG1 helps RNA polymerase II to overcome a nucleosomal barrier during elongation, in vivo. EMBO Rep. 2010;11:751–757. doi: 10.1038/embor.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M, et al. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem. 2010;285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 99.Gévry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wong MM, Cox LK, Chrivia JC. The chromatin remodeling protein, SRCAP, is critical for deposition of the histone variant H2A.Z at promoters. J Biol Chem. 2007;282:26132–26139. doi: 10.1074/jbc.M703418200. [DOI] [PubMed] [Google Scholar]
- 101.Keppler BR, Archer TK. Ubiquitin-dependent and ubiquitin-independent control of subunit stoichiometry in the SWI/SNF complex. J Biol Chem. 2010;285:35665–35674. doi: 10.1074/jbc.M110.173997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gill J, Yogavel M, Kumar A, Belrhali H, Jain SK, Rug M, et al. Crystal structure of malaria parasite nucleosome assembly protein: distinct modes of protein localization and histone recognition. J Biol Chem. 2009;284:10076–10087. doi: 10.1074/jbc.M808633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, et al. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 104.Van Lijsebettens M, Grasser KD. The role of the transcript elongation factors FACT and HUB1 in leaf growth and the induction of flowering. Plant Signal Behav. 2010;5:715–717. doi: 10.4161/psb.5.6.11646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Birch JL, Tan BC, Panov KI, Panova TB, Andersen JS, Owen-Hughes TA, et al. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 2009;28:854–865. doi: 10.1038/emboj.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development. 2010;137:3785–3794. doi: 10.1242/dev.051805. [DOI] [PubMed] [Google Scholar]
- 107.Doyen CM, An W, Angelov D, Bondarenko V, Mietton F, Studitsky VM, et al. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol Cell Biol. 2006;26:1156–1164. doi: 10.1128/MCB.26.3.1156-64.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wan Y, Chiang JH, Lin CH, Arens CE, Saleem RA, Smith JJ, et al. Histone chaperone Chz1p regulates H2B ubiquitination and subtelomeric anti-silencing. Nucleic Acids Res. 2010;38:1431–1440. doi: 10.1093/nar/gkp1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Braunschweig U, Hogan GJ, Pagie L, van Steensel B. Histone H1 binding is inhibited by histone variant H3.3. EMBO J. 2009;28:3635–3645. doi: 10.1038/emboj.2009.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cuddapah S, Schones DE, Cui K, Roh TY, Barski A, Wei G, et al. Genomic profiling of HMGN1 reveals an association with chromatin at regulatory regions. Mol Cell Biol. 2011;31:700–709. doi: 10.1128/MCB.00740-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 112.Thakar A, Gupta P, Ishibashi T, Finn R, Silva-Moreno B, Uchiyama S, et al. H2A.Z and H3.3 histone variants affect nucleosome structure: biochemical and biophysical studies. Biochemistry. 2009;48:10852–10857. doi: 10.1021/bi901129e. [DOI] [PubMed] [Google Scholar]
- 113.Postnikov Y, Bustin M. Regulation of chromatin structure and function by HMGN proteins. Biochim Biophys Acta. 2010;1799:62–68. doi: 10.1016/j.bbagrm.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lim JH, West KL, Rubinstein Y, Bergel M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J. 2005;24:3038–3048. doi: 10.1038/sj.emboj.7600768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rochman M, Malicet C, Bustin M. HMGN5/NSBP1: a new member of the HMGN protein family that affects chromatin structure and function. Biochim Biophys Acta. 2010;1799:86–92. doi: 10.1016/j.bbagrm.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, et al. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol Cell. 2009;35:642–656. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Weiss T, Hergeth S, Zeissler U, Izzo A, Tropberger P, Zee BM, et al. Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenetics Chromatin. 2010;3:7. doi: 10.1186/1756-8935-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iizuka M, Smith MM. Functional consequences of histone modifications. Curr Opin Genet Dev. 2003;13:154–160. doi: 10.1016/S0959-437X(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 119.Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- 120.Denis GV. Duality in bromodomain-containing protein complexes. Front Biosci. 2001;6:849–852. doi: 10.2741/Denis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tang L, Nogales E, Ciferri C. Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog Biophys Mol Biol. 2010;102:122–128. doi: 10.1016/j.pbiomolbio.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang B, Chambers KJ, Faller DV, Wang S. Reprogramming of the SWI/SNF complex for co-activation or co-repression in prohibitin-mediated estrogen receptor regulation. Oncogene. 2007;26:7153–7157. doi: 10.1038/sj.onc.1210509. [DOI] [PubMed] [Google Scholar]
- 123.Shen W, Xu C, Huang W, Zhang J, Carlson JE, Tu X, et al. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46:2100–2110. doi: 10.1021/bi0611208. [DOI] [PubMed] [Google Scholar]
- 124.Umehara T, Nakamura Y, Wakamori M, Ozato K, Yokoyama S, Padmanabhan B. Structural implications for K5/K12-di-acetylated histone H4 recognition by the second bromodomain of BRD2. FEBS Lett. 2010;584:3901–3908. doi: 10.1016/j.febslet.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J Proteome Res. 2006;5:502–511. doi: 10.1021/pr050430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nakamura Y, Umehara T, Nakano K, Jang MK, Shirouzu M, Morita S, et al. Crystal structure of the human BRD2 bromodomain: insights into dimerization and recognition of acetylated histone H4. J Biol Chem. 2007;282:4193–4201. doi: 10.1074/jbc.M605971200. [DOI] [PubMed] [Google Scholar]
- 127.Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000 Biol Rep. 2009;1:98. doi: 10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dupaigne P, Lavelle C, Justome A, Lafosse S, Mirambeau G, Lipinski M, et al. Rad51 polymerization reveals a new chromatin remodeling mechanism. PLoS One. 2008;3:3643. doi: 10.1371/journal.pone.0003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee K, Kang MJ, Kwon SJ, Kwon YK, Kim KW, Lim JH, et al. Expansion of chromosome territories with chromatin decompaction in BAF53-depleted interphase cells. Mol Biol Cell. 2007;18:4013–4023. doi: 10.1091/mbc.E07-05-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barresi V, Ragusa A, Fichera M, Musso N, Castiglia L, Rappazzo G, et al. Decreased expression of GRAF1/OPHN-1-L in the X-linked alpha thalassemia mental retardation syndrome. BMC Med Genomics. 2010;3:28. doi: 10.1186/1755-8794-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chioda M, Vengadasalam S, Kremmer E, Eberharter A, Becker PB. Developmental role for ACF1-containing nucleosome remodellers in chromatin organisation. Development. 2010;137:3513–3522. doi: 10.1242/dev.048405. [DOI] [PubMed] [Google Scholar]
- 132.Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Eberharter A, Vetter I, Ferreira R, Becker PB. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J. 2004;23:4029–4039. doi: 10.1038/sj.emboj.7600382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Eberharter A, Ferrari S, Längst G, Straub T, Imhof A, Varga-Weisz P, et al. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J. 2001;20:3781–3788. doi: 10.1093/emboj/20.14.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Guschin D, Geiman TM, Kikyo N, Tremethick DJ, Wolffe AP, Wade PA. Multiple ISWI ATPase complexes from Xenopus laevis. Functional conservation of an ACF/CHRAC homolog. J Biol Chem. 2000;275:35248–35255. doi: 10.1074/jbc.M006041200. [DOI] [PubMed] [Google Scholar]
- 136.Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 137.Längst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/S0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 138.Bonaldi T, Längst G, Strohner R, Becker PB, Bianchi ME. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 2002;21:6865–6873. doi: 10.1093/emboj/cdf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell. 2010;40:976–987. doi: 10.1016/j.molcel.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 140.Musselman CA, Mansfield RE, Garske AL, Davrazou F, Kwan AH, Oliver SS, et al. Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem J. 2009;423:179–187. doi: 10.1042/BJ20090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, et al. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J Biol Chem. 2011;286:11779–11791. doi: 10.1074/jbc.M110.208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kunert N, Wagner E, Murawska M, Klinker H, Kremmer E, Brehm A. dMec: a novel Mi-2 chromatin remodelling complex involved in transcriptional repression. EMBO J. 2009;28:533–544. doi: 10.1038/emboj.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ramírez J, Hagman J. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics. 2009;4:532–536. doi: 10.4161/epi.4.8.10108. [DOI] [PubMed] [Google Scholar]
- 144.Snow JW, Orkin SH. Translational isoforms of FOG1 regulate GATA1-interacting complexes. J Biol Chem. 2009;284:29310–29319. doi: 10.1074/jbc.M109.043497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zegerman P, Canas B, Pappin D, Kouzarides T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]
- 146.Lomberk G, Mathison AJ, Grzenda A, Urrutia R. The sunset of somatic genetics and the dawn of epigenetics: a new frontier in pancreatic cancer research. Curr Opin Gastroenterol. 2008;24:597–602. doi: 10.1097/MOG.0b013e32830b111d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 148.Lavigne M, Eskeland R, Azebi S, Saint-André V, Jang SM, Batsché E, et al. Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression. PLoS Genet. 2009;5:1000769. doi: 10.1371/journal.pgen.1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bierne H, Tham TN, Batsche E, Dumay A, Leguillou M, Kernéis-Golsteyn S, et al. Human BAHD1 promotes heterochromatic gene silencing. Proc Natl Acad Sci USA. 2009;106:13826–13831. doi: 10.1073/pnas.0901259106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Munshi A, Shafi G, Aliya N, Jyothy A. Histone modifications dictate specific biological readouts. J Genet Genomics. 2009;36:75–88. doi: 10.1016/S1673-8527(08)60094-6. [DOI] [PubMed] [Google Scholar]
- 151.Kwon SH, Workman JL. HP1c casts light on dark matter. Cell Cycle. 2011;10:625–630. doi: 10.4161/cc.10.4.14796. [DOI] [PubMed] [Google Scholar]
- 152.Sedivy JM, Banumathy G, Adams PD. Aging by epigenetics—a consequence of chromatin damage? Exp Cell Res. 2008;314:1909–1917. doi: 10.1016/j.yexcr.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Riddle NC, Minoda A, Kharchenko PV, Alekseyenko AA, Schwartz YB, Tolstorukov MY, et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2011;21:147–163. doi: 10.1101/gr.110098.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Montanaro L, Govoni M, Orrico C, Treré D, Derenzini M. Location of rRNA transcription to the nucleolar components: disappearance of the fibrillar centers in nucleoli of regenerating rat hepatocytes. Cell Struct Funct. 2011;36:49–56. doi: 10.1247/csf.10017. [DOI] [PubMed] [Google Scholar]
- 155.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 156.Sugimura K, Fukushima Y, Ishida M, Ito S, Nakamura M, Mori Y, et al. Cell cycle-dependent accumulation of histone H3.3 and euchromatic histone modifications in pericentromeric heterochromatin in response to a decrease in DNA methylation levels. Exp Cell Res. 2010;316:2731–2746. doi: 10.1016/j.yexcr.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 157.Kubben N, Voncken JW, Demmers J, Calis C, van Almen G, Pinto Y, et al. Identification of differential protein interactors of lamin A and progerin. Nucleus. 2010;1:513–525. doi: 10.4161/nucl.1.6.13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schneider M, Lu W, Neumann S, Brachner A, Gotzmann J, Noegel AA, et al. Molecular mechanisms of centrosome and cytoskeleton anchorage at the nuclear envelope. Cell Mol Life Sci. 2011;68:1593–1610. doi: 10.1007/s00018-010-0535-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Raška I. Importance of molecular cell biology investigations in human medicine in the story of the Hutchinson-Gilford progeria syndrome. Interdiscip Toxicol. 2010;3:89–93. doi: 10.2478/v10102-010-0018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schirmer EC. The epigenetics of nuclear envelope organization and disease. Mutat Res. 2008;647:112–121. doi: 10.1016/j.mrfmmm.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 161.Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]