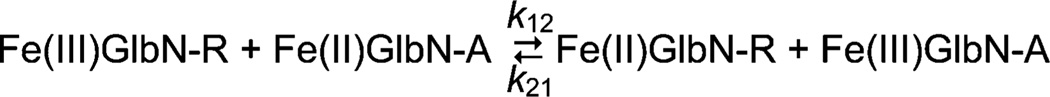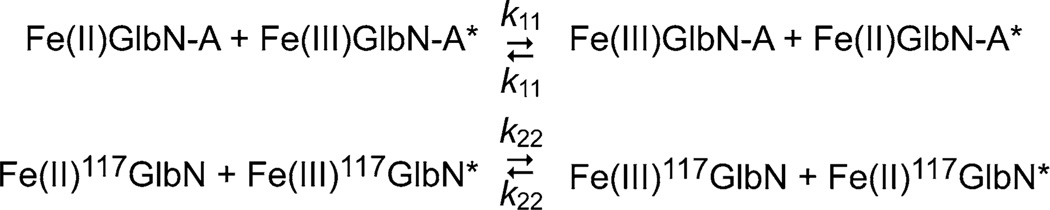Abstract
Many heme proteins undergo covalent attachment of the heme group to a protein side chain. Such posttranslational modifications alter the thermodynamic and chemical properties of the holoprotein. Their importance in biological processes makes them attractive targets for mechanistic studies. We have proposed a reductively driven mechanism for the covalent heme attachment in the monomeric hemoglobins produced by the cyanobacteria Synechococcus sp. PCC 7002 and Synechocystis sp. PCC 6803 (GlbN) (Nothnagel et al. in J Biol Inorg Chem 16:539–552, 2011). These GlbNs coordinate the heme iron with two axial histidines, a feature that distinguishes them from most hemoglobins and conditions their redox properties. Here, we uncovered evidence for an electron exchange chain reaction leading to complete heme modification upon substoichiometric reduction of GlbN prepared in the ferric state. The GlbN electron self-exchange rate constants measured by NMR spectroscopy were on the order of 102–103 M−1 s−1 and were consistent with the proposed autocatalytic process. NMR data on ferrous and ferric Synechococcus GlbN in solution indicated little dependence of the structure on the redox state of the iron or cross-link status of the heme group. This allowed the determination of lower bounds to the cross-exchange rate constants according to Marcus theory. The observations illustrate the ability of bishistidine hemoglobins to undergo facile interprotein electron transfer and the chemical relevance of such transfer for covalent heme attachment.
Keywords: Truncated hemoglobin, 2/2 hemoglobin, Heme posttranslational modification, Hybrid b/c heme
Introduction
Hemoglobins are well known for their ability to bind and release the dioxygen required for vertebrate metabolism. Genomic initiatives, however, have brought to light a rich collection of hemoglobin sequences from all three kingdoms of life. They show that the hemoglobin superfamily has existed for billions of years and that its members are capable of performing a range of physiological tasks in diverse environments [1]. Modern nonvertebrate hemoglobins contain clues for understanding the modulation of heme prosthetic group chemistry and the molecular evolution of hemoglobin function [2].
GlbN is the monomeric 2/2 or “truncated” hemoglobin of the oxygenic cyanobacteria Synechococcus sp. PCC 7002 [3, 4] and Synechocystis sp. PCC 6803 [5, 6]. The two proteins are 59% identical in sequence and belong to a growing group of hemoglobins that ligate the heme iron with two histidines (proximal and distal) in the reduced (ferrous) and oxidized (ferric) states. Synechococcus and Synechocystis GlbNs are nevertheless capable of exogenous ligand binding by displacement of the distal histidine, but they differ from all other hemoglobins in that they undergo covalent heme attachment [7, 8]. The posttranslational modification (PTM), depicted in Fig. 1, occurs rapidly in the reduced state in the absence of exogenous ligand. The function of Synechocystis GlbN is unknown; Synechococcus GlbN protects the cyanobacterial cell against damage from nitrate metabolism [4], likely through enzymatic processes requiring redox cycling of the heme iron. Both forms of Synechococcus GlbN, with and without the PTM, have been extracted from the cell [4].
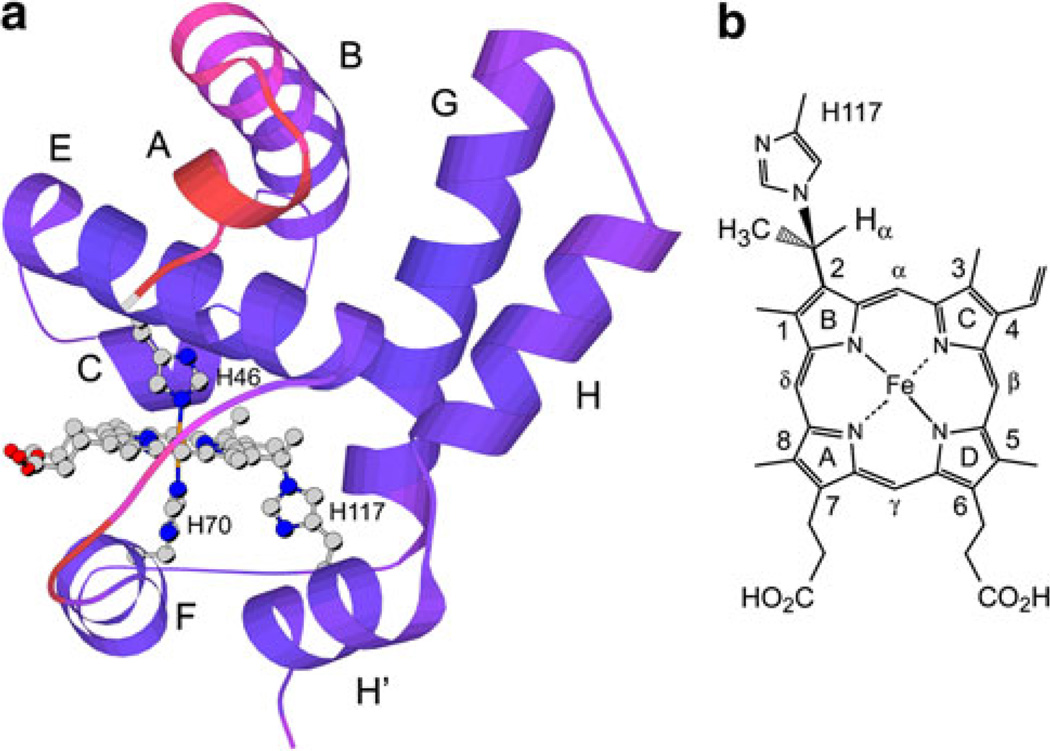
Fig. 1.
a Solution structure of the hemoglobin from Synechococcus sp. PCC 7002 with cross-linked heme (GlbN-A) in the ferric state [4]. A low-energy member of the 2KSC family is shown with redder colors indicating larger ensemble root mean square deviation from the average. Helices, axial histidines (His46, distal, and His70, proximal), and reactive histidine (His117) are labeled. b The structure of the His117–heme covalent linkage [7]
Various heme–protein cross-links have been documented in cytochromes [9, 10] and peroxidases [11]. In these proteins, covalent linkages are proposed to alter holoprotein stability, reactivity, and redox properties. Covalent linkages are expected to contribute to the functional diversification of the hemoglobin superfamily as well; therefore, the mechanism and consequences of the heme PTM in GlbN merit further investigation.
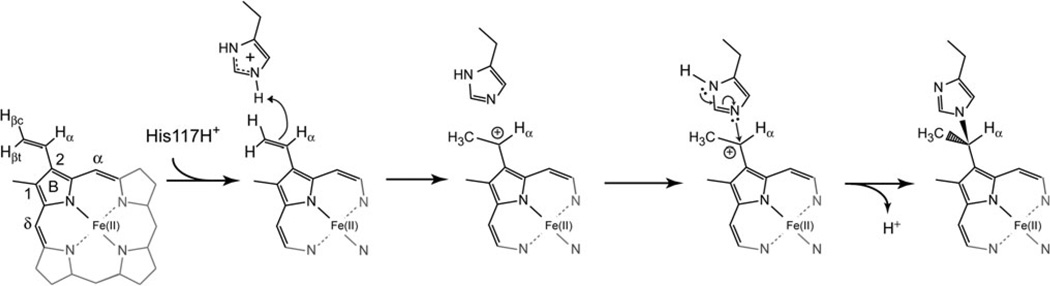
The PTM of Fe(III)GlbN via Fe(II)GlbN does not require an enzyme or dioxygen [7, 12]. Observations thus far are consistent with an electrophilic addition involving the protonation of the 2-vinyl Cβ [12], as outlined in Fig. 2. An intriguing aspect of this particular PTM is that, despite the apparent simplicity of the alkylation reaction, the histidine–heme bond has been found only in the GlbN of the two cyanobacteria studied here. It is not clear whether the rarity hints at subtle molecular determinants or results mainly from the absence of histidines at appropriate locations. In pursuing the description of the reductive mechanism, we investigated the intrinsic ability of Synechococcus and Synechocystis GlbNs to undergo electron transfer (ET) and we characterized the structure of the reduced state of posttranslationally modified Synechococcus GlbN in solution, a form for which no prior data were available. Our results demonstrate that inter-GlbN electron exchange is readily detected by NMR spectroscopy, not only within a redox pair (self-exchange), but also between cross-linked and non-cross-linked GlbN (cross-exchange). This latter process illustrates how a heme modification is amplified and propagated through a GlbN solution.
Fig. 2.
Proposed GlbN reductive cross-linking mechanism. Only a portion of the iron(II) protoporphyrin IX molecule is shown
Materials and methods
Protein expression and purification
The procedures used to produce recombinant GlbNs (Synechococcus wild-type GlbN and H117A GlbN, and Synechocystis wild-type GlbN) have been reported previously [3, 6, 12, 13] and were followed with minor adaptations. A description is provided in the electronic supplementary material, along with the reported extinction coefficients used for protein concentration determination. When required for clarity, we refer to GlbN with noncovalently attached heme as GlbN-R and to GlbN with covalently attached heme as GlbN-A [12].
Generation of Fe(II)GlbN from Fe(III)GlbN under microoxic conditions
The glucose oxidase/d-glucose/catalase (GODCAT) enzymatic O2 elimination system [14] was used to generate microoxic conditions in solution. The concentration of the GODCAT components depended on the nature of the experiment. Final reagent concentrations were 0.04–0.7 mg/mL glucose oxidase, 0.04–0.15 mg/mL catalase, and 0.1–0.2% d-glucose (mass/volume). High buffer concentrations (200–250 mM) were used to minimize pH changes caused by GODCAT enzymatic turnover and the oxidation of dithionite. Under these conditions, negligible changes in pH occurred over time.
Dithionite stock solutions were prepared immediately before use. The concentration was estimated by mass or by absorbance measurement with an extinction coefficient of 8 mM−1 cm−1 at 314 nm [15]. Dithionite oxidation eliminates the 314-nm absorbance, which provided a convenient means of assessing the depletion of the reductant.
Fe(II)GlbN UV/vis samples
Fe(III)GlbN and GODCAT solutions were incubated together for approximately 20 min prior to the addition of 200-fold dithionite. During this time, the concentration of the samples was measured and their stability assessed by the constancy of their spectrum in the 250–700-nm region. Upon addition of reductant, the cuvette was stopper-sealed and the content mixed by inversion of the cuvette prior to data acquisition.
Fe(II)GlbN NMR samples
Fe(III)GlbN and GODCAT solutions were incubated for approximately 1 h prior to treatment with dithionite. During this time, the protein concentration was determined by absorbance measurement, and 1H 1D and 1H–15N 2D heteronuclear single quantum coherence (HSQC) NMR data were collected to confirm sample homogeneity. Fe(III) GlbNs were stable in the presence of GODCAT. Fe(III) GlbN (+GODCAT) solutions were placed in a glove box continuously purged with 99.9% argon. GlbN reduction (30–100%) was achieved by adding a small volume of dithionite stock solution also prepared under argon. The protein solution was mixed by pipette and transferred to a Shigemi NMR tube, which was sealed with Parafilm. GlbN redox mixtures prepared in this manner underwent some initial oxidation but eventually reached a stable Fe(II)/Fe(III) ratio, which was maintained from days to weeks. This enabled the acquisition of multidimensional NMR spectra. For kinetic conversion experiments, the dead time between the addition of dithionite and the initiation of NMR data acquisition was approximately 12 min.
Kinetic experiments (optical)
Electronic absorption spectroscopy, manual mixing
Electronic absorption spectra were collected at room temperature using a Varian Cary 50-Bio spectrophotometer. Protein concentrations were 5–11 µM. Kinetic experiments monitoring the conversion of Synechococcus Fe(III)GlbN-R to Fe(II)GlbN-A upon treatment with 200-fold excess dithionite were conducted in 250 mM borate, pH 9.2 in the presence of GODCAT. The dead time was about 15 s. Absorbance data were collected every 1 nm over the range 600–500 nm, using a 0.2-s averaging time. Data collection was performed in triplicate.
Electronic absorption spectroscopy, rapid mixing stopped-flow
Electronic absorption spectra were collected at 24 °C using a Hi-Tech Scientific stopped-flow spectrophotometer equipped with a photodiode-array detector. Syringes contained either Synechocystis GlbN-R (+GODCAT) in buffer or 200-fold excess dithionite. Solutions were mixed by stopped-flow (200 µL reaction volume). Final reaction concentrations were 5 µM GlbN, 1 mM dithionite, and 200 mM buffer. Reaction mixing times (dead times) ranged between 100 ms at low pH and 750 ms at pH 8.9, but in all cases were such that reliable rate constants could be obtained from the data. During the mixing time, complete GlbN-R reduction occurred. Reactions were conducted at pH 5.3 (acetate), pH 5.7 (acetate/phosphate), pH 6.0 (phosphate), p1H or p2H 6.7 (phosphate), pH 7.5 (phosphate), pH 8.3 [tris(hydroxymethyl)aminomethane], pH 8.9 [tris(hydroxymethyl)aminomethane], and pH 9.5 (glycine). A total of 200 spectra were recorded per run in the wavelength range 320–704 nm.
Determination of PTM rate (optical data)
Singular value decomposition and global nonlinear least-squares fitting were performed on optical spectra using the program KinTek Explorer (KinTek) [16, 17] or a combination of SciLab 5.2 for singular value decomposition and Savuka [18] for global fitting. Two or three abstract kinetic vectors were determined to be significant on the basis of their large singular values and high autocorrelation coefficients.
The pH dependence of the Synechocystis GlbN cross-linking reaction was modeled assuming the existence of two Fe(II)GlbN states in rapid equilibrium: a protonated, “reactive” form that can serve as a H+ donor in the Markovnikov addition reaction, and a deprotonated, “unreactive” form that cannot act as a general acid. With this model, the pH dependence data were adequately fit using a single apparent pKa and Hill coefficient in Eq. 1 with the program KaleidaGraph 3.6 (Figure S1):
| (1) |
NMR spectroscopy
Samples and acquisition
Synechococcus and Synechocystis GlbN samples were prepared in 225–250 mM phosphate (pH 7.1) or borate (pH 9.2) buffer in 95:5 1H2O/2H2O. Protein concentration ranged from 0.6 to 4.9 mM. NMR spectra were collected at 298 K with Bruker Avance-600, Bruker Avance II-600, or Varian Inova 800 spectrometers, each equipped with a cryoprobe. 1H chemical shifts were referenced to 2,2-dimethylsilapentane-5-sulfonic acid through the 1H2O line (4.76 ppm). Water-presaturation 1D data and 1H–15N 2D HSQC data were collected as described previously [19]. To confirm backbone HN assignments determined using ZZ exchange methods, triple-resonance HNCACB and CBCA(CO)NH experiments were performed on Synechococcus wild-type Fe(II)GlbN-A prepared by treatment with fivefold excess dithionite as described earlier.
Processing
NMR data were processed using either TopSpin 2.1 (Bruker BioSpin, Rheinstetten, Germany) or NMRPipe 3.0 [20]. Spectra were analyzed using the programs Sparky [21] and TopSpin 2.1. Further information is provided in our prior NMR work on the same proteins [22, 23].
Band-selective 1D and 2D 15N–1H HSQC spectroscopy
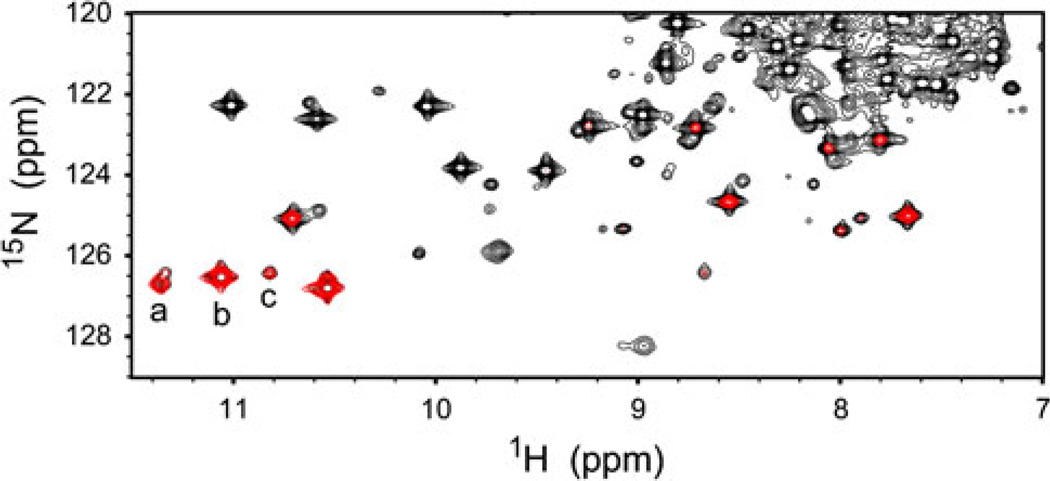
We used an 15N-edited 1D experiment (band-selective 1D 15N–1H HSQC) to collect high signal-to-noise ratio data rapidly. The sequence was derived from a water suppression through gradient-tailored excitation (WATERGATE) HSQC with water flip-back. 15N band selectivity was achieved by addition of a 20-ms RE-BURP [24] centered at 130.5 ppm (Synechococcus GlbN) or 131.5 ppm (Synechocystis GlbN) at the end of the minimal evolution period. Excited signals were retained by phase cycling of the selective 15N pulse. The excitation bandwidth (approximately 900 Hz) was determined using a 2D version of the experiment as shown in Fig. 3 (a complete explanation is provided in the electronic supplementary material). Each 1D data point was collected in 1.5 min (Synechocystis GlbN) or 2.5 min (Synechococcus GlbN).
Fig. 3.
Portion of 1H–15N heteronuclear single quantum coherence spectra collected 5 h after substoichiometric reduction of a Synechococcus Fe(III)GlbN-R sample (GlbN-R is GlbN with noncovalently attached heme). Black conventional flip-back WATERGATE spectrum, red band-selected spectrum. Cross peaks a, b, and c are from Thr80 HN in Fe(II)GlbN-A, Fe(III)GlbN-A, and Fe(III)GlbN-R, respectively
Two-dimensional 15N–1H ZZ exchange NMR spectroscopy
Electron self-exchange (ESE) rates were measured in Synechococcus wild-type GlbN-A and H117A GlbN, and Synechocystis GlbN-A at 600 MHz with an 15N ZZ exchange experiment [25]. To eliminate the effects of heteronuclear cross-correlated relaxation, an even number of 1.3-ms G3 1H-selective inversion pulses [26] were applied every 10 ms during the mixing period (τmix), with the 1H carrier positioned 4 ppm downfield of the H2O resonance (4.76 ppm). The duration and positioning of these pulses ensured minimal perturbation of the water resonance, which was thereby maintained close to thermal equilibrium for all values of τmix.
The mixing time was varied in the following order (ms): 497, 994, 45, 633, 249, 339, 1,288, 791, 136, 497, 1,582 for Synechococcus wild-type GlbN-A; 497, 994, 294, 746, 136, 1,582, 68, 1,288, 610, 384, 203, 497 for Synechococcus H117A GlbN; and 497, 294, 136, 1,288, 633, 1,582, 1,084, 384, 836, 68, 497 for Synechocystis wild-type GlbN-A. Duplicate τmix (497 ms) data points were used to estimate peak volume uncertainty (approximately 10%). The Synechococcus wild-type GlbN-A sample contained approximately 40–45% Fe(II) and 60–55% Fe(III) protein (2.0 mM total GlbN). H117A GlbN ESE kinetics were measured on an approximately 1:3 Fe(II)/Fe(III) protein sample (1.7 mM total GlbN). The Fe(II)/Fe(III) redox mixture of Synechocystis wild-type GlbN-A was prepared at a total protein concentration of approximately 4.9 mM with about 15% Fe(II) protein and 85% Fe(III) protein.
Resolved cross-peaks were integrated to generate evolution curves. These were globally fitted and provided forward and reverse apparent first-order rate constants for each residue. The sum of the two constants was divided by the total protein concentration to obtain the second-order rate constant for the self-exchange reaction. Additional information is provided in the electronic supplementary material.
Three-dimensional 1H–15N–1H sensitivity-enhanced ZZ exchange NMR spectroscopy
A 3D sensitivity-enhanced version of the 15N ZZ exchange experiment [25] was used to transfer the previously determined backbone 15N–1H assignments of Synechococcus wild-type Fe(III)GlbN-A [23] to Fe(II)GlbN-A through longitudinal magnetization exchange. The additional 1H frequency-labeling period considerably increased the number of resolved resonances since many NHs exhibit a degenerate 15N chemical shift in ferrous and ferric GlbN-A. The ZZ mixing time was set to 484 ms to maximize cross-peak intensity. Data were collected at 800 MHz to enhance chemical shift dispersion. The pulse sequence is shown in Figure S2.
Heteronuclear relaxation measurements
15N T1 measurements were performed on Synechococcus wild-type GlbN-A in the Fe(II) and Fe(III) oxidation states using a standard pulse sequence [27] with a WATERGATE modification [28]. The experiments were performed at 600 MHz using the Bruker spectrometers mentioned before. The sample conditions were approximately 1.5 mM GlbN-A at pH 7.2 in 20 mM or 100 mM phosphate buffer containing 10% 2H2O. Fe(II)GlbN-A was generated by the addition of a fivefold molar excess dithionite in the presence of GODCAT. The T1 delays were set randomly to 14, 105*, 203, 301, 448, 595, 742*, 1,183, and 2,408 ms for Fe(III)GlbN-A and to 14, 630*, 1,400, and 1,975* ms for Fe(II)GlbN-A (where the asterisks indicate duplicate time points for error analysis); the delay between transients was 3 s. The data were analyzed using the jackknife method of error analysis with the program Curvefit (A.G. Palmer, Columbia University) through the use of the script Sparky2rate (J.P. Loria, Yale University).
Structural calculations
Backbone 1H, 15N, 13Cα, and 13Cβ chemical shifts (pH 7.1) for Synechococcus wild-type Fe(II)GlbN-A at pH 7.1–7.2 were used as input for the program TALOS+ [29]. In addition, the Fe(II)GlbN-A chemical shifts were subtracted from those of Fe(III)GlbN-A [23] to obtain Δδ for comparison with the pseudocontact contribution, δpc, at 114 HN and 18 CαH sites. The pseudocontact shift is expressed as
| (2) |
in the frame of reference that diagonalizes χ, the magnetic susceptibility tensor. In Eq. 2, ri represents the distance between the 1H of interest (i) and the Fe center, θi and ϕi are the angular coordinates, and Δχax and Δχrh are the axial and rhombic components of χ [30]. The Δδ values were tabulated for use as restraints in refinement with XPLOR-NIH [31] (version 2.23) and the PARA restraint module [32]. All other previously deposited 2KSC restraints (nuclear Overhauser effects, dihedral angles, and hydrogen bonds) [4] were also applied. Model 2 of the Fe(III)GlbN-A ensemble 2KSC was chosen and subjected to a Gaussian/DELPHIC torsional potential [33] as previously reported [4]. The program Numbat [34] was used to obtain starting Δχax and Δχrh values from the experimental shifts and 2KSC model 2.
Results and discussion
Kinetics of the cross-linking reaction as a function of pH
The reactive histidine of Synechocystis GlbN-R, His117, has pKa ~ 6.9 in Fe(III)GlbN-R [8]. Preliminary pH dependence data [12] led us to propose that in addition to serving as the nucleophile, His117 also served as the proton donor to form the 2-vinyl α carbocation (Fig. 2). The change in the optical spectrum on going from Fe(II)GlbN-R to Fe(II)GlbN-A is small but sufficient to follow the cross-linking reaction after complete reduction with dithionite [12]. Above pH 9, the reaction proceeded with an observed rate constant, kxl, below 10−2 s−1 (5 × 10−3 s−1 at pH 9.2) and could be monitored by manual mixing experiments. Between pH 8.9 and pH 5.7, stopped-flow rapid mixing was required as the observed rate constant increased to 0.9 s−1. Below pH 5.7, the ferrous heme dissociated from the apoprotein, as demonstrated by scavenging with horse apomyoglobin (data not shown). This additional equilibrium complicated the conversion mechanism and rendered the fits unreliable. The observed reaction rate constant, plotted as a function of pH, is shown in Figure S1. The curve displayed an inflection at pH ~ 6.8, arguing against water acting as the proton donor and supporting the hypothesis that His117 acted as the general acid in the rate-determining protonation of Fe(II) heme 2-vinyl Cβ as shown in Fig. 2.
The pKa of His117 in Synechococcus Fe(III)GlbN-R is approximately 6.1 [4]. Synechococcus GlbN-R is not as stable as Synechocystis GlbN-R at low pH (data not shown), and therefore a full pH dependence of the cross-linking rate was not obtained. Given the similarity between the two proteins [8], the reaction mechanism was assumed to be conserved. Conversion to GlbN-A following reduction was expected to be observable by manual mixing methods at high pH. We chose a pH of 9.2 as a compromise between rate of reaction and alkaline stability and we determined the kinetics as above. Addition of excess dithionite to Fe(III)GlbN-R reduced the entire protein population within the dead time of the experiment (less than 1 s). Conversion to GlbN-A occurred in an apparent first-order process as described by Eq. 3:
| (3) |
Isosbestic points were detected during the first 40 min of reaction (approximately eight half-lives), before heme bleaching caused by extended incubation with excess reducing agent became significant. A typical time course is shown in Figure S3. At pH 9.2, the observed rate constant extracted from the visible spectra was kxl = (2.3 ± 0.1) × 10−3 s−1. This led to the expectation that better than 90% conversion to Fe(II)GlbN-A would be achieved within the time required to prepare an NMR sample and set up the experiment (approximately 12 min).
Kinetics of reaction upon substoichiometric Fe(III)GlbN-R reduction
A simple scenario predicts that if a substoichiometric amount of reducing agent is added to Fe(III)GlbN-R at pH 9.2, the reaction mixture will in a short time contain Fe(III)GlbN-R and Fe(II)GlbN-A. Heme proteins, however, can undergo electron exchange, and the composition and stability of the mixture will depend on the rate of interprotein ET. Thus, it is possible that Fe(III)GlbN-R, Fe(III)GlbN-A, Fe(II)GlbN-R, and Fe(II)GlbN-A coexist at various times in solution. NMR spectroscopy was used to investigate this aspect of the PTM.
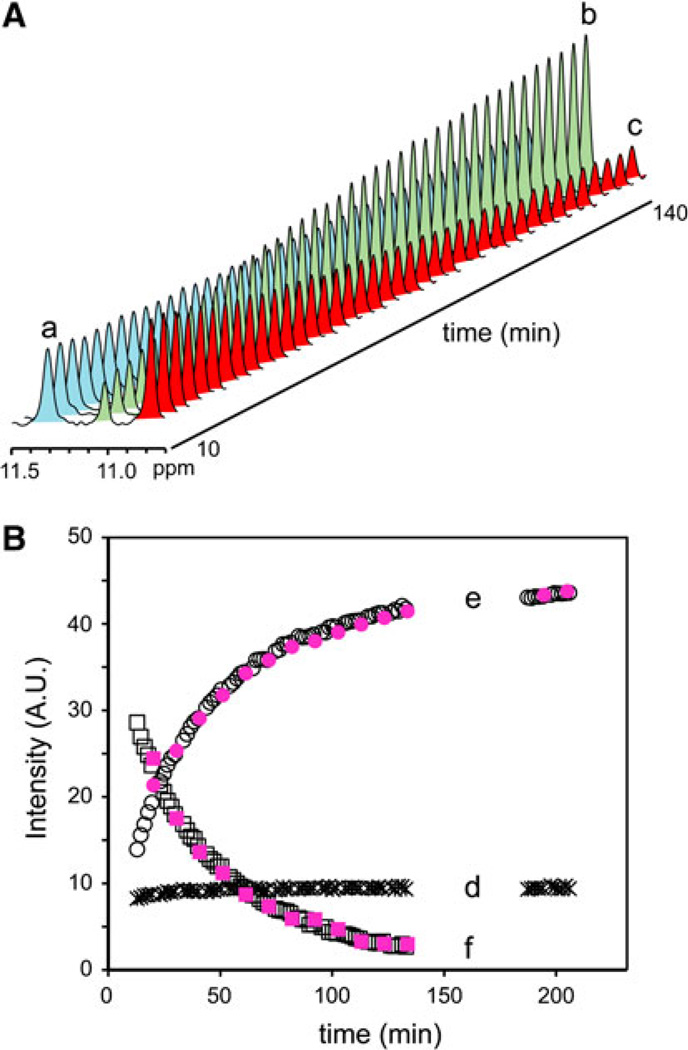
In mixtures of GlbN-R and GlbN-A, several heme signals from Fe(III)GlbN-R and Fe(III)GlbN-A (both paramagnetic, S = 1/2 complexes) are resolved in the downfield region of the 1H spectrum [8]. These signals, which report on relative concentrations, can be followed as a function of reaction progress. In contrast, no signal from Fe(II)GlbN-R or Fe(II)GlbN-A (diamagnetic, S = 0 complexes) could be reliably monitored by 1D observation. Two-dimensional 1H–15N HSQC data offered the desired spectral resolution, but insufficient temporal resolution. An 15N band-selective 1D HSQC experiment was used to circumvent this problem. In both GlbNs, the downfield-shifted amide of residue 80 (Thr in Synechococcus GlbN and Asn in Synechocystis GlbN) was chosen as a probe because the oxidation state and cross-link status perturbed the 1H shift without significantly affecting the 15N shift. This is shown in Fig. 3 for Synechococcus GlbN Thr80. Band selection eliminated overlapping signals and provided 1D 1H spectra suitable for kinetic analysis (Fig. 4A).
Fig. 4.
A Stacked plot of the 1D spectra obtained with 15N band selection after substoichiometric reduction of Synechococcus Fe(III)GlbN-R. The time course of the GlbN-R → GlbN-A conversion is illustrated with the signals from Thr80 HN in Fe(II)GlbN-A (light blue, a), Fe(III)GlbN-A (light green, b), and Fe(III)GlbN-R (red, c). The signal of Fe(II)GlbN-A initially contained intensity from unresolved Thr80 HN in Fe(II)GlbN-R (Figure S4). B The same experiment with Synechocystis GlbN. The signals are from Asn80 HN in Fe(II)GlbN-A (asterisk, d), Fe(III)GlbN-A (open circle, e), and Fe(III)GlbN-R (open square, f), and heme 5-CH3 in Fe(III)GlbN-R (filled square) and Fe(III)GlbN-A (filled circle). The heme signals were scaled arbitrarily to coincide with the Asn80 signals. Simulation and fits of the data are shown in the electronic supplementary material
In the experiment shown in Fig. 4A, the reaction was initiated by addition of dithionite such that approximately 40% of a 2 mM Synechococcus Fe(III)GlbN-R NMR sample was reduced. As predicted by the kinetics of cross-link formation (Eq. 3) determined optically, a burst of Fe(II)GlbN-A production occurred during the dead time. But, this initial production of Fe(II)GlbN-A was followed by slow formation of Fe(III)GlbN-A at the expense of Fe(III)GlbN-R. A slight intensity decrease was observed in Fe(II)GlbN-A, which also occurred with the inert H117A variant (Figure S5). This minor phase was attributed to reoxidation unrelated to cross-link propagation. Data collected after 17 h confirmed the complete disappearance of Fe(III)GlbN-R.
The same experiment was performed with Synechocystis GlbN (Fig. 4B). The sample was reduced with dithionite to obtain approximately 15% Fe(II)GlbN-R at the beginning of the reaction. As observed with Synechococcus GlbN, cross-link propagation continued to completion. Additionally, Synechocystis GlbN appeared to be less susceptible to autoxidation, and the level of reduced protein (Fig. 4B, trace d) was nearly constant throughout the conversion.
A parsimonious model of the results invokes electron exchange between Fe(II)GlbN-A and Fe(III)GlbN-R (Scheme 1) combined with Fe(II)GlbN-R cross-linking (Eq. 3). The bimolecular exchange step effectively recycles electrons to form reactive Fe(II)GlbN-R until all Fe(III)GlbN-R is exhausted. The kinetic data were fit accordingly to yield K12 = k12/k21 and a lower bound for k12 (Figures S6–S9). For Synechococcus GlbN (Fig. 4A), K12 was approximately 0.1 and k12 > 4 M−1 s−1. For Synechocystis GlbN (Fig. 4B), K12 was approximately 0.25 and k12 > 10 M−1 s−1. In both cases, the left-hand side of Scheme 1 was thermodynamically favored.
Scheme 1.
Electron transfer between cross-linked (GlbN-A) and non-cross-linked (GlbN-R) hemoglobins
Electron self-exchange in GlbN
To test the plausibility of Scheme 1, we sought to determine the timescale of GlbN ESE. We reasoned that extremely slow rates would not be consistent with the observed progress of the conversion and that extremely fast rates would lead to a different NMR spectral appearance. We used two versions of Synechococcus GlbN: wild-type GlbN-A and H117A GlbN, which is incapable of cross-linking and serves as a GlbN-R surrogate. The ESE reactions are shown in Scheme 2, where the asterisk marks individual molecules and 117GlbN represents the H117A variant. The ESE rate constants (k11 and k22) were determined at pH 9.2 for consistency with the conversion conditions. In both GlbNs, the 1H–15N HSQC spectrum of a mixture of reduced and oxidized states was a superposition of unperturbed parent resonances, indicating that ESE was slow on the chemical shift timescale and that the putative encounter complexes were not populated to an appreciable extent. An 15N ZZ exchange experiment [25] was therefore used for rate measurement.
Scheme 2.
Electron self-exchange in wild-type GlbN-A and H117A GlbN (117GlbN)
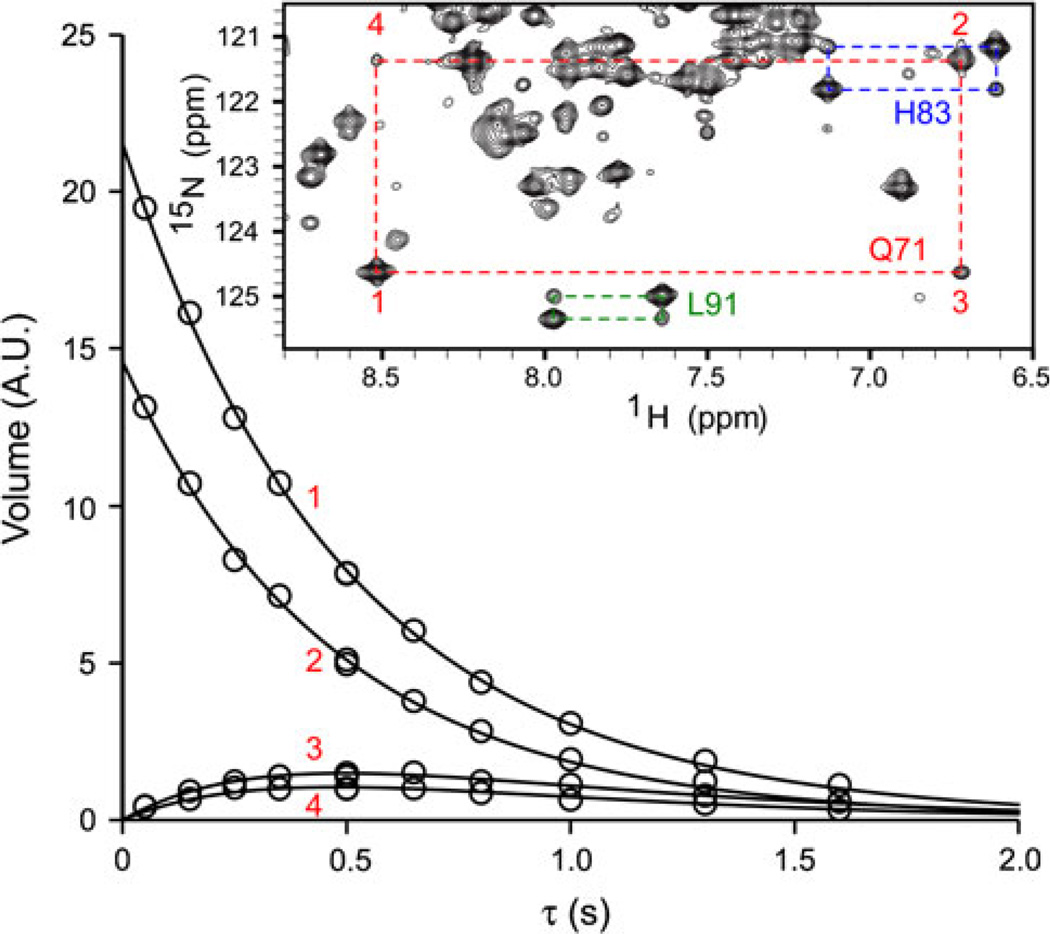
The inset in Fig. 5 shows a portion of a Synechococcus GlbN-A 15N ZZ spectrum (the full spectrum is shown in Figure S10). The pulse sequence generates Nz magnetization, which is allowed to exchange between the two redox partners during the mixing time. Four types of peaks forming a quartet are detected per residue: auto peaks have 15N and 1H chemical shifts corresponding to the oxidized or reduced species as one would observe in the 1H–15N HSQC spectra of the pure species, and exchange peaks have the 15N shift of one species and the 1H shift of the other. As the mixing time is varied, the intensity of the peaks changes according to exchange rates and 15N longitudinal relaxation rates.
Fig. 5.
Electron self-exchange rate determination in Synechococcus GlbN-A using the time dependence of the auto (1, 2) and exchange (3, 4) peaks of Gln71 in the 15N ZZ experiment. The inset shows a small portion of the 15N ZZ exchange spectrum acquired at 600 MHz with τ ~ 0.5 s (pH 9.2, 298 K)
In Fig. 5, the curves represent fits to Gln71 data using established expressions for magnetization exchange in 2D experiments [25, 35]. 15N longitudinal relaxation rates (15N R1) extracted via exchange data fitting were consistent with the values measured independently (Figure S11). Global analysis of several quartets corresponding to residues in various secondary structure elements (e.g., Figure S12) confirmed that Synechococcus GlbN-A and H117A GlbN each registered a single exchange process. The rate constants were k11 = (4.5 ± 0.4) × 102 M−1 s−1 for wild-type GlbN-A and k22 = (3.7 ± 0.5) × 102 M−1 s−1 for H117A GlbN, with confidence intervals determined as described in Figures S13 and S14. The ESE rate constant was also measured for Synechocystis GlbN-A and was found to be (1.4 ± 0.4) × 103 M−1 s−1 (Figure S15), three times faster than for Synechococcus GlbN-A under the same conditions.
Although Synechococcus H117A GlbN differs from Synechococcus GlbN-R in some respects [36], the k22 value (Figure S14) argued for a minimal influence of the heme–protein cross-link on the features controlling ESE. The experimental kESE values were compatible with the proposed inter-GlbN ET-mediated PTM (Scheme 1, Eq. 3) and NMR spectral appearance.
The kinetics of ESE have been determined by NMR spectroscopy in many heme proteins under a broad range of conditions [37]. Measured ESE rate constants range between 102 and 107 M−1 s−1. Values for cytochrome b5 are around 104 M−1 s−1 and those for cytochrome c are around 105 M−1 s−1. Rate constants on the order of 103 M−1 s−1 have been obtained for myoglobin and hemoglobin with bound trimethylphosphine [37]. The GlbN values were on par with these unnatural complexes and were tenfold to 100-fold slower than those for cytochromes b5 and c. We note that the ability to measure ESE in a native form of hemoglobin is directly related to the stable bishistidine coordination of the iron in both the ferric and the ferrous states. The relatively fast GlbN rate constants caution that kinetic analysis of bishistidine globin reactions involving redox processes may have to take into account inter-GlbN electron exchange and its concentration dependence.
The cellular biosynthetic machinery is expected to produce ferrous GlbN-R from apoprotein and heme. Synechococcus GlbN-R, however, can survive the cellular environment without undergoing the PTM and interestingly, when Synechococcus cells are grown under microoxic conditions, the relative levels of GlbN-A are higher than under normal conditions [4]. We have reported that CO, when bound to Fe(II)GlbN-R, retards significantly the conversion to GlbN-A [12]. This behavior is consistent with the proposed mechanism, which calls for stabilization of the trivalent carbocation (carbenium ion) with increased electron density at the heme 2-vinyl (Fig. 2). Similarly, O2 binding is expected to be inhibitory. Whether or not the autocatalytic modification takes place in the cell is unclear as it depends on the rates of a number of processes (e.g., exogenous ligand off rate) and local protein concentration. The ESE rates, however, do suggest that GlbN-R and GlbN-A are capable of efficient ET with each other and other proteins.
Estimation of the ET rate constants
The ET reaction (Scheme 1), by nature of the partners, resembles a self-exchange reaction (Scheme 2). The parallel offers a favorable situation for further analysis of rate constants using Marcus theory. We therefore inspected the prediction of the cross-relation [38]
| (4) |
which estimates the ET rate constant for the cross-reaction (k12, Scheme 1) from the ESE rate constants of individual redox couples (k11 and k22, Scheme 2). In Eq. 4, f12 is an efficiency factor assumed to be unity and K12 is typically calculated with the known midpoint reduction potentials of individual couples [37]. The reactivity of bishistidine Fe(II)GlbN-R, however, complicates direct potential measurement, and instead we used K12 derived from Scheme 1 and Eq. 3. For Synechococcus GlbN, the ET rate constant obtained from Eq. 4 with K12 = 0.1, k11 = 450 M−1 s−1, and k22 = 370 M−1 s−1 was k12 = 130 M−1 s−1, well within the range allowed by kinetic data fitting (k12 > 4 M−1 s−1) and in agreement with slow exchange on the chemical shift timescale. Likewise, using K12 = 0.25 and k11 = k22 = 1,400 M−1 s−1 for Synechocystis GlbN returned k12 = 700 M−1 s−1, also allowed by the conversion data (k12 > 10 M−1 s−1) and consistent with slow exchange. The K12 values corresponded to −60 mV (Synechococcus) and −35 mV (Synechocystis) differences in potential between the GlbN-R and GlbN-A couples (GlbN-R having the lower E°) and suggested a small effect from heme–protein cross-linking.
Structural comparison of the four complexes involved in the conversion
The cross-relation given by Eq. 4 assumes identical work terms for GlbN-R and GlbN-A self-exchange reactions [38]. These terms capture the energy required to bring the reactants into an ET-competent configuration and depend on the structure of the proteins involved. As a first level of verification for the use of Eq. 4, we gathered structural data on all participating species with the expectation that large differences would cast doubt on the validity of the approach and that highly similar partners would transfer electrons via the same interface and mechanism.
Structural similarity has been documented for Synechocystis wild-type GlbN-A and H117A GlbN in the crystalline state, leading to the conclusion that neither reduction nor cross-linking has a dramatic effect on this particular protein [39]. Little difference has been noted between Synechococcus wild-type Fe(III)GlbN-R and Fe(III)GlbN-A in solution [4], or between H117A Fe(III)GlbN and wild-type Fe(III)GlbN-R [36], also in solution. However, no information is available on reduced synechococcal GlbN. We therefore initiated the characterization of Fe(II)GlbN-A. This complex is diamagnetic and amenable to standard NMR experiments.
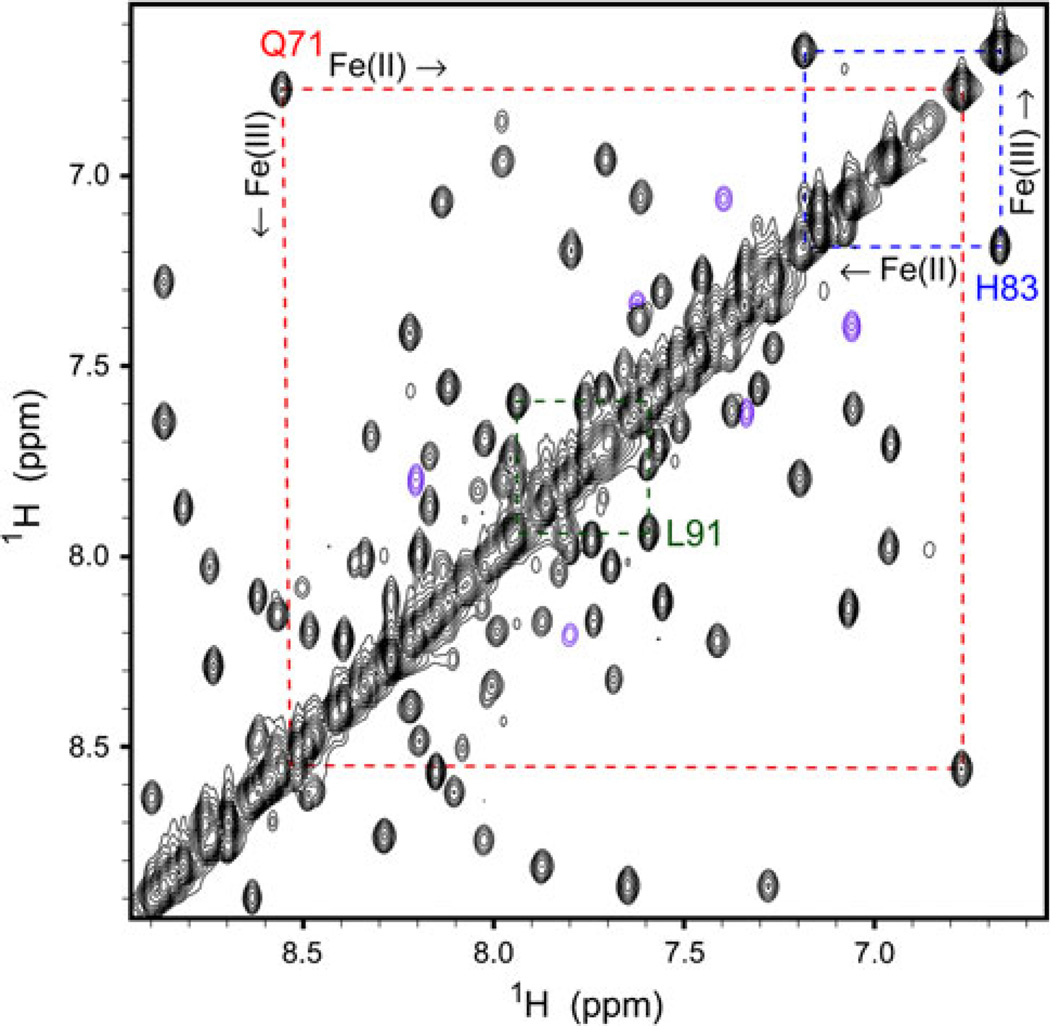
We have assigned the backbone and side chain resonances of Synechococcus Fe(III)GlbN-A [23]. Because exchange between the ferric and ferrous states is slow on the chemical shift timescale, the ZZ experiment can, in principle, be used to transfer assignments from Fe(III)GlbN-A to Fe(II)GlbN-A. The spectral congestion displayed by the redox mixture (Figs. 5, S10) made it clear that a 2D approach would yield little information. The same data revealed that the difference in chemical shift (in hertz) in the 1H dimension was generally much larger than that in the 15N dimension. A 3D H–Nz–N–H exchange experiment was therefore designed to take advantage of the 1H dispersion. Figure 6 illustrates the quality of the data with the 1H–1H projection of the 3D set. Each cross peak connects a known Fe(III)GlbN-A resonance to its counterpart in Fe(II)GlbN-A. A total of 60% of Fe(II)GlbN-A HN assignments were immediately obtained with this approach. Triple resonance backbone-based experiments resolved remaining ambiguities.
Fig. 6.
Portion of the 1H–1H projection of the H–Nz–N–H 3D experiment. Data were collected at 800 MHz (pH 9.2, 298 K) on an approximately 4:6 mixture of Synechococcus Fe(II) and Fe(III)GlbN-A. The mixing time was approximately 0.5 s. Peaks in magenta were folded in the 15N dimension
The backbone chemical shifts of Synechococcus Fe(II)GlbN-A were used to map the secondary structure with the program TALOS+ [29]. The helices of GlbN did not appear to be affected by reduction of the iron (Figure S17). Tertiary structure was assessed indirectly with the pseudocontact shift experienced by backbone protons in Fe(III)GlbN-A. If Fe(III)GlbN-A and Fe(II)GlbN-A are isostructural, Δδ of individual 1Hs is equal to the hyperfine shift, composed of a contact contribution extending over a few bonds from the paramagnetic center and a pseudo-contact contribution (δpc) effective through space [30]. Because δpc is sensitive to the position of the nucleus relative to the paramagnetic center (Eq. 2 [30]), a comparison of experimental Δδ with δpc calculated from the coordinates of one of the structures tests the extent of structural similarity.
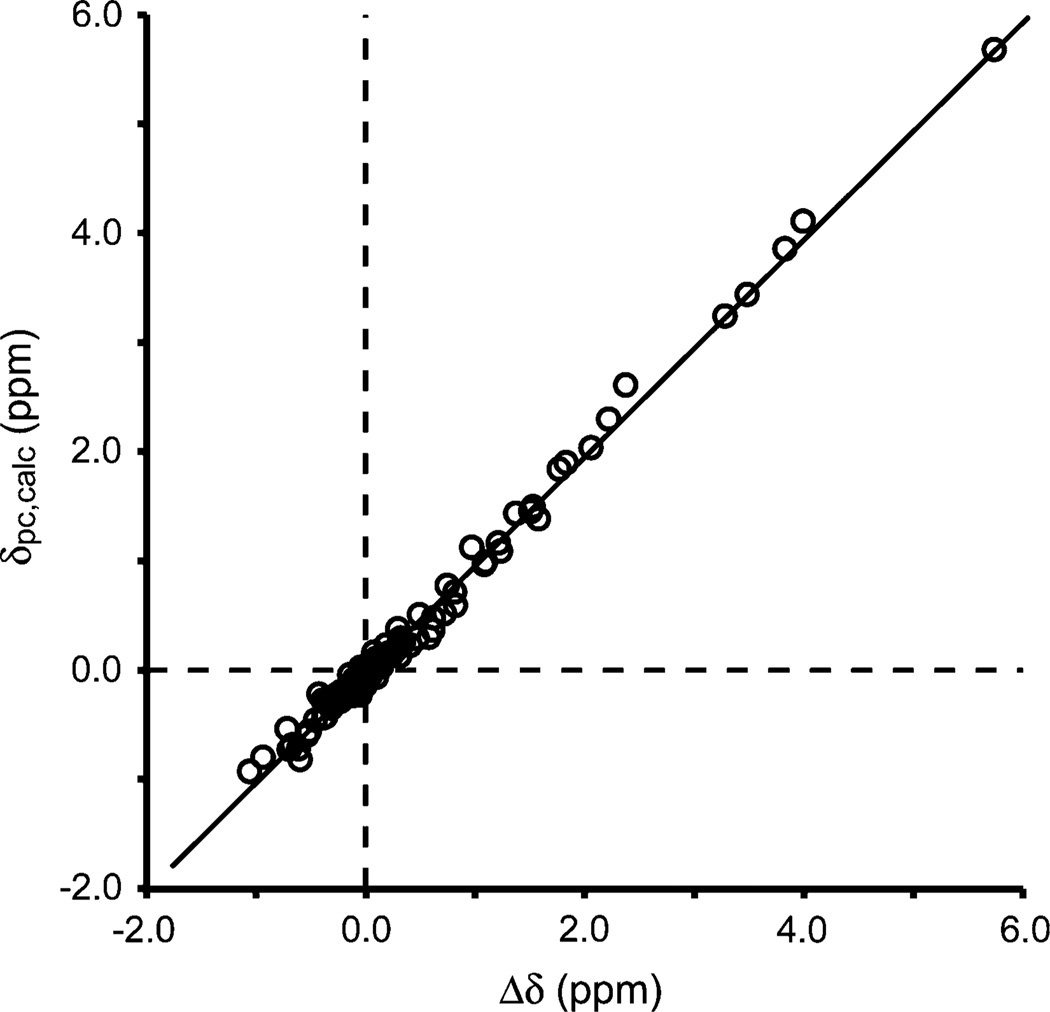
A total of 114 backbone amide 1H and 18 1Hα Δδ values were used as restraints for XPLOR-NIH refinement of a low-energy member of the 2KSC family and determination of the magnetic susceptibility tensor [32, 33]. The refined Synechococcus Fe(III)GlbN-A coordinates were within the bundle of the original ensemble (Figure S18), and calculated δpc values corresponded well to the experimental Δδ values (Fig. 7). For example, Gln71 HN, which is adjacent to the proximal histidine and was used in the evaluation of the self-exchange rate (Fig. 5), experienced a δpc of 1.76 ppm, calculated to be 1.84 ppm. The generally good correlation shown in Fig. 7 supported that the average structures of Fe(II)GlbN-A and Fe(III)GlbN-A were similar.
Fig. 7.
Correlation between observed and calculated pseudocontact shifts for Synechococcus Fe(III)GlbN-A after structural refinement using Δδ as restraints in addition to nuclear Overhauser effects, hydrogen bonds, and dihedral angles (δpc,calc = 0.997Δδ − 0.019, R2 = 0.9924). χ tensor information is provided in the Figures S19–S21
Exchange NMR data collected at pH 9.2 on partially reduced H117A GlbN samples (not shown) indicated that the variant maintained its fold in both oxidation states. We therefore concluded that, as for Synechocystis GlbN, neither reduction nor cross-linking perturbed the structure significantly. In that respect, Scheme 1 (ET) and Scheme 2 (ESE) were considered identical.
The aggregated observations support our application of the cross-relation (Eq. 4) to test the consistency of the kinetic measurements. GlbN emerges from our studies as an excellent system for ET investigations under a complementary array of conditions (e.g., temperature and ionic strength). Thus far, we have found no NMR evidence for a chemically gated mechanism. Characterization of the encounter complex may offer insight into how similar the ET mechanism and intrinsic ET capabilities of GlbN are to those of established ET proteins. The midpoint potential difference and k12 values will remain tentative until additional evidence confirms that the observed rates are governed by ET [40].
Globin function, globin properties, and bishistidine coordination
The best-known globin function is dioxygen transport and storage. Nitric oxide dioxygenation is a second, well-documented role performed by flavohemoglobins [41], certain single domain canonical (3/3) globins [42, 43], and 2/2 globins [44]. It is now recognized that a large number of hemoglobins exist as bishistidine complexes. Because distal histidine coordination can interfere with exogenous ligand binding and identical coordination in the ferric and ferrous states facilitates ET, attention has recently turned to this type of process as yet another function represented in the hemoglobin superfamily [45, 46]. Among bishistidine hemoglobins, the strength of the distal histidine ligation as measured by the equilibrium constant between a “pentacoordinate” His–Fe species and a hexacoordinate His–Fe–His species [47] varies greatly from protein to protein. Two bishistidine hemoglobins in Caenorhabditis elegans (GLB-6 and GLB-26) constitute extreme examples of stable ligation as they do not bind O2 or CO [46]. These hemoglobins have been proposed to serve as ET proteins.
The two GlbNs of interest here bind exogenous ligands. The full complement of distal histidine coordination effects has yet to be explored. Thus far, we have shown that His46 ligation protects the heme from damage by H2O2 [12], which is likely an advantage as GlbNs function under cellular conditions generating peroxides. We also have shown that cyanide binding and the H46A or H46L replacement alter the regiospecificity of the PTM [48]. Synechococcus GlbN has been implicated in the detoxification of reactive nitrogen species [4] and if, as suspected, it is directly involved in nitric oxide or peroxynitrite processing, the reaction mechanism is anticipated to require redox cycling of the iron. Further studies will reveal whether access to a relatively stable bishistidine state in both ferric and ferrous states is an important feature of GlbN related to ET in the context of enzymatic turnover. In any case, comparative studies of hexacoordination stability and ET in other hemoglobins will provide molecular insight into the determinants of ligand binding and ET functions.
Conclusion
We have shown that reducing any amount of bishistidine GlbN in a pool of oxidized bishistidine GlbN will trigger a chain reaction converting the entire sample to GlbN-A. To our knowledge, this is the first example of a redox chain reaction leading to a physiologically pertinent heme modification. We have related this amplification of the modification to the ease with which GlbN exchanges electrons. Our observations highlight the ability of bishistidine hemoglobins to participate in redox reactions and more generally illustrate the ability of a heme protein to catalyze its own PTM.
Supplementary Material
Acknowledgments
This study was supported by National Science Foundation grant MCB-0349409. NMR facilities and resources at Johns Hopkins University were provided by the Biomolecular NMR Center. The authors thank Selena Rice for assistance with the optical measurements, Richard Himes, Ryan Peterson, and Kenneth Karlin for the use of their stopped-flow equipment, and Christopher Falzone for useful discussions and careful reading of the manuscript. Henry Nothnagel’s insight into heme chemistry was essential in the initial phases of the work. Figure 1a was prepared with Molscript [49].
Abbreviations
- ESE
Electron self-exchange
- ET
Electron transfer
- GlbN
Hemoglobin produced by Synechococcus sp. PCC 7002 or Synechocystis sp. PCC 6803
- GlbN-A
GlbN with covalently attached heme
- GlbN-R
GlbN with noncovalently attached heme
- GODCAT
Glucose oxidase/d-glucose/catalase
- HSQC
Heteronuclear single quantum coherence
- PTM
Posttranslational modification
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00775-012-0880-5) contains supplementary material, which is available to authorized users.
Contributor Information
Matthew R. Preimesberger, T.C. Jenkins Department of Biophysics, Johns Hopkins University, Baltimore, MD 21218, USA
Matthew P. Pond, T.C. Jenkins Department of Biophysics, Johns Hopkins University, Baltimore, MD 21218, USA
Ananya Majumdar, Biomolecular NMR Center, Johns Hopkins University, Baltimore, MD 21218, USA.
Juliette T. J. Lecomte, Email: lecomte_jtj@jhu.edu, T.C. Jenkins Department of Biophysics, Johns Hopkins University, Baltimore, MD 21218, USA.
References
- 1.Vinogradov SN, Moens L. J Biol Chem. 2008;283:8773–8777. doi: 10.1074/jbc.R700029200. [DOI] [PubMed] [Google Scholar]
- 2.Kakar S, Sturms R, Tiffany A, Nix JC, DiSpirito AA, Hargrove MS. Biochemistry. 2011;50:4273–4280. doi: 10.1021/bi2002423. [DOI] [PubMed] [Google Scholar]
- 3.Scott NL, Falzone CJ, Vuletich DA, Zhao J, Bryant DA, Lecomte JTJ. Biochemistry. 2002;41:6902–6910. doi: 10.1021/bi025609m. [DOI] [PubMed] [Google Scholar]
- 4.Scott NL, Xu Y, Shen G, Vuletich DA, Falzone CJ, Li Z, Ludwig M, Pond MP, Preimesberger MR, Bryant DA, Lecomte JTJ. Biochemistry. 2010;49:7000–7011. doi: 10.1021/bi100463d. [DOI] [PubMed] [Google Scholar]
- 5.Couture M, Das TK, Savard PY, Ouellet Y, Wittenberg JB, Wittenberg BA, Rousseau DL, Guertin M. Eur J Biochem. 2000;267:4770–4780. doi: 10.1046/j.1432-1327.2000.01531.x. [DOI] [PubMed] [Google Scholar]
- 6.Scott NL, Lecomte JTJ. Protein Sci. 2000;9:587–597. doi: 10.1110/ps.9.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vu BC, Jones AD, Lecomte JTJ. J Am Chem Soc. 2002;124:8544–8545. doi: 10.1021/ja026569c. [DOI] [PubMed] [Google Scholar]
- 8.Vu BC, Vuletich DA, Kuriakose SA, Falzone CJ, Lecomte JTJ. J Biol Inorg Chem. 2004;9:183–194. doi: 10.1007/s00775-003-0512-1. [DOI] [PubMed] [Google Scholar]
- 9.Bowman SE, Bren KL. Nat Prod Rep. 2008;25:1118–1130. doi: 10.1039/b717196j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson AR, Elmore BO, Yang C, Ferrara JD, Hooper AB, Wilmot CM. Biochemistry. 2007;46:8340–8349. doi: 10.1021/bi700086r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L, Wojciechowski G, Ortiz de Montellano PR. J Biol Chem. 2006;281:18983–18988. doi: 10.1074/jbc.M602307200. [DOI] [PubMed] [Google Scholar]
- 12.Nothnagel HJ, Preimesberger MR, Pond MP, Winer BY, Adney EM, Lecomte JTJ. J Biol Inorg Chem. 2011;16:539–552. doi: 10.1007/s00775-011-0754-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vuletich DA, Falzone CJ, Lecomte JTJ. Biochemistry. 2006;45:14075–14084. doi: 10.1021/bi061532g. [DOI] [PubMed] [Google Scholar]
- 14.Englander SW, Calhoun DB, Englander JJ. Anal Biochem. 1987;161:300–306. doi: 10.1016/0003-2697(87)90454-4. [DOI] [PubMed] [Google Scholar]
- 15.Di Iorio EE. Methods Enzymol. 1981;76:57–72. doi: 10.1016/0076-6879(81)76114-7. [DOI] [PubMed] [Google Scholar]
- 16.Johnson KA, Simpson ZB, Blom T. Anal Biochem. 2009;387:30–41. doi: 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 17.Johnson KA. Methods Enzymol. 2009;467:601–626. doi: 10.1016/S0076-6879(09)67023-3. [DOI] [PubMed] [Google Scholar]
- 18.Bilsel O, Zitzewitz JA, Bowers KE, Matthews CR. Biochemistry. 1999;38:1018–1029. doi: 10.1021/bi982365q. [DOI] [PubMed] [Google Scholar]
- 19.Falzone CJ, Vu BC, Scott NL, Lecomte JTJ. J Mol Biol. 2002;324:1015–1029. doi: 10.1016/s0022-2836(02)01093-8. [DOI] [PubMed] [Google Scholar]
- 20.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 21.Goddard TD, Kneller DG. Sparky. Vol. 3. San Francisco: University of California; 2006. [Google Scholar]
- 22.Falzone CJ, Lecomte JTJ. J Biomol NMR. 2002;23:71–72. doi: 10.1023/a:1015337100759. [DOI] [PubMed] [Google Scholar]
- 23.Pond MP, Vuletich DA, Falzone CJ, Majumdar A, Lecomte JTJ. Biomol NMR Assign. 2009;3:211–214. doi: 10.1007/s12104-009-9177-1. [DOI] [PubMed] [Google Scholar]
- 24.Geen H, Freeman R. J Magn Reson. 1991;93:93–141. [Google Scholar]
- 25.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 26.Emsley L, Bodenhausen G. Chem Phys Lett. 1990;165:469–476. [Google Scholar]
- 27.Kay LE, Torchia DA, Bax A. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 28.Piotto M, Saudek V, Sklenár V. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 29.Shen Y, Delaglio F, Cornilescu G, Bax A. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emerson SD, La Mar GN. Biochemistry. 1990;29:1556–1566. doi: 10.1021/bi00458a029. [DOI] [PubMed] [Google Scholar]
- 31.Brünger AT. X-PLOR, version 3.1. A system for X-ray crystallography and NMR. New Haven: Yale University Press; 1992. [Google Scholar]
- 32.Banci L, Bertini I, Cavallaro G, Giachetti A, Luchinat C, Parigi G. J Biomol NMR. 2004;28:249–261. doi: 10.1023/B:JNMR.0000013703.30623.f7. [DOI] [PubMed] [Google Scholar]
- 33.Schwieters CD, Kuszewski JJ, Clore GM. Prog NMR Spectrosc. 2006;619(48):47–62. [Google Scholar]
- 34.Schmitz C, Stanton-Cook MJ, Su XC, Otting G, Huber T. J Biomol NMR. 2008;621(41):179–189. doi: 10.1007/s10858-008-9249-z. [DOI] [PubMed] [Google Scholar]
- 35.Jeener J, Meier BH, Bachmann P, Ernst RR. J Chem Phys. 1979;71:4546–4553. [Google Scholar]
- 36.Vu BC, Nothnagel HJ, Vuletich DA, Falzone CJ, Lecomte JTJ. Biochemistry. 2004;43:12622–12633. doi: 10.1021/bi048726l. [DOI] [PubMed] [Google Scholar]
- 37.Simonneaux G, Bondon A. Chem Rev. 2005;105:2627–2646. doi: 10.1021/cr030731s. [DOI] [PubMed] [Google Scholar]
- 38.Marcus RA, Sutin N. Biochim Biophys Acta. 1985;811:265–322. [Google Scholar]
- 39.Hoy JA, Smagghe BJ, Halder P, Hargrove MS. Protein Sci. 2007;16:250–260. doi: 10.1110/ps.062572607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davidson VL. Acc Chem Res. 2000;33:87–93. doi: 10.1021/ar9900616. [DOI] [PubMed] [Google Scholar]
- 41.Bonamore A, Boffi A. IUBMB Life. 2008;60:19–28. doi: 10.1002/iub.9. [DOI] [PubMed] [Google Scholar]
- 42.Gardner PR. J Inorg Biochem. 2005;99:247–266. doi: 10.1016/j.jinorgbio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Gardner PR, Gardner AM, Brashear WT, Suzuki T, Hvitved AN, Setchell KD, Olson JS. J Inorg Biochem. 2006;100:542–550. doi: 10.1016/j.jinorgbio.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Ouellet H, Ouellet Y, Richard C, Labarre M, Wittenberg B, Wittenberg J, Guertin M. Proc Natl Acad Sci USA. 2002;99:5902–5907. doi: 10.1073/pnas.092017799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fago A, Mathews AJ, Moens L, Dewilde S, Brittain T. FEBS Lett. 2006;580:4884–4888. doi: 10.1016/j.febslet.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 46.Kiger L, Tilleman L, Geuens E, Hoogewijs D, Lechauve C, Moens L, Dewilde S, Marden MC. PLoS ONE. 2011;6:e20478. doi: 10.1371/journal.pone.0020478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kakar S, Hoffman FG, Storz JF, Fabian M, Hargrove MS. Biophys Chem. 2010;152:1–14. doi: 10.1016/j.bpc.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nothnagel HJ, Love N, Lecomte JT. J Inorg Biochem. 2009;103:107–116. doi: 10.1016/j.jinorgbio.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Kraulis P. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.