Abstract
A growing body of information on the biology of miRNAs has revealed new insight into their roles in normal homeostasis and pathology of disease. miRNAs control all steps of the cellular expression machinery acting through a “single miRNA/multiple targets” or “multiple miRNAs/single target” mechanism. They have profound impact on the regulation of signaling pathways, which govern common and specific functions across different cellular phenotypes. There is increasing evidence that various diseases share similar disturbances in gene expression networks. Since miRNAs have both common and varying effects in different cellular contexts, they might also influence overlapping signaling pathways in different organs and disease entities. Here, we review this concept for two miRNAs highly abundant in the brain, miR-124 and miR-126, and their potential role in diseases of the brain.
Keywords: miRNAs, miR-124, miR-126, CNS disorders, brain diseases, signaling pathways, neurodegeneration, cancer, psychiatric, tumor
Introduction
There is increasing evidence for overlapping miRNA functions in different disease entities, such as cancer and neurodegeneration (Du and Pertsemlidis, 2011). Conceptually, this is based on the common belief that a miRNA target in one cell type would also be a target in a different cellular context. However, this is not necessarily the case as cell type and cellular environment (genetic and epigenetic status, sequence, expression levels and stoichiometry of direct and indirect targets, and miRNPs) also seemingly contribute to miRNA functions. There are several levels to assess miRNA regulations across different diseases. On the one hand, one can look at miRNA profiles and determine their expression levels in relation to the pathological condition. Profiling has been done in multiple studies and there is a tremendous effort in developing improved platforms for higher throughput and sensitivity (Krichevsky, 2007, Sonntag, 2010). This also goes together with other technologies, such as laser microdissection, to target specific cell populations. Data from these profiling studies can be used to correlate miRNAs with known molecular networks and signaling pathways to predict functional relations (Sonntag, 2010). On the other hand, one can look at experimentally validated relationships between miRNAs and their targets. While this is more feasible for specific diseases, such as cancer, in which tissue material, cell lines or animal models are available, it is more difficult for neurodegenerative or psychiatric disorders, for which these experimental tools are less accessible.
Conceptually, the potential roles of miRNAs in neurodegeneration have been well established (Barbato, et al., 2009, Bushati and Cohen, 2008, Eacker, et al., 2009, Hebert and De Strooper, 2009, Hebert, et al., 2009, Nelson, et al., 2008, Singh, 2007, Sonntag, 2010), but so far, there is only very little information on the function of these molecules in disease processes. Similarly, there is emerging evidence that miRNAs are involved in psychiatric diseases, including bipolar disorder, schizophrenia, and addiction (Miller and Wahlestedt, 2010, Xu, et al., 2010). However, like in neurodegeneration, their functions remain largely unknown. In contrast, oncogenic and tumor suppressor functions for a number of key miRNAs in cancer are much better understood. In fact, many known miRNA functions and target interactions have been discovered in cancer cells. In addition, there is already an emerging field in cancer research to develop new therapeutic strategies using miRNAs and other regulatory RNAs. Translational research that is based on miRNA profiling, experimentally validated data, and knowledge about the individual biology of disease entities could provide a powerful approach to develop common miRNA-based therapeutic and diagnostic strategies (Du and Pertsemlidis, 2011).
Here we describe converging miRNA functions in the brain exemplified for one key miRNA, miR-124, and discuss a translational approach to predict the roles of a less well characterized miRNA, miR-126, in relation to various brain disorders.
miR-124: the most abundant miRNA in the CNS
miR-124 (also called miR-124a) is the most abundant miRNA in the brain. The miR-124 family was detected in 46 animal species from Caenorhabditis to Homo sapiens (Guo, et al., 2009). Its abundance in embryonic and adult cortical tissues of various mammalian species ranges from 5% to 48% of all miRNAs expressed (Lagos-Quintana, et al., 2002, Landgraf, et al., 2007); Krichevsky, unpublished data), suggesting its key function in the CNS. Due to its extraordinary enrichment in metazoan nervous systems through more than 500 million years of evolution (Peterson, et al., 2009), miR-124 is one of the best-studied miRNAs in various organisms (reviewed by (Gao, 2010), by both basic biologists interested in molecular mechanisms of miRNA regulation and neuroscientists investigating brain development and function. In mammalian brain development, miR-124 expression increases during the prenatal period, reaching its peak level by the end of fetal development and remains highly expressed in the postnatal brain (Krichevsky, et al., 2003). This miRNA is not expressed or is detected only at low levels in neural stem and progenitor cells, but it is induced strikingly with the exit of neuroblasts from the cell cycle during neuronal differentiation and highly expressed in differentiating and mature neurons (Krichevsky, et al., 2006, Maiorano and Mallamaci, 2009, Sempere, et al., 2004). Since astrocytes do not express miR-124 (Smirnova, et al., 2005), this molecule was considered neuron-specific. Indeed, miR-124 is widely expressed in virtually all postmitotic neurons in all CNS regions, with the exception of the pituitary gland in adult humans, mice, and rats, and its expression is relatively low in the ventricular zone in the embryonic brain (Bak, et al., 2008, Deo, et al., 2006, Landgraf, et al., 2007, Olsen, et al., 2009). Similarly, miR-124 is expressed in all differentiating and mature neurons in the chick spinal cord (Cao, et al., 2007, Visvanathan, et al., 2007) as well as in all differentiating cells throughout the larval zebrafish brain and retina (Wienholds, et al., 2005). Interestingly, in Aplysia, miR-124 is abundantly expressed in sensory neurons, but it is almost undetectable in motor neurons (Rajasethupathy, et al., 2009). In C. elegans, it is also mostly confined to sensory neurons (Clark, et al., 2010), suggesting functional divergence of this miRNA during the phylogenetic transition from invertebrates to vertebrates.
miR-124 in differentiation and functions of neurons
The striking induction of miR-124 expression during neuronal differentiation and brain development, its confinement to the CNS, and high expression levels in mature neurons suggest unique and important roles this molecule may play in neurogenesis and neuronal function. Indeed, a number of groups have reported that miR-124, alone or in combination with other factors, can promote the neuronal phenotype during differentiation of uncommitted cells in vitro. For example, transient overexpression of miR-124 and mir-9/9* in murine embryonic stem cell (ESC) derived neural progenitor cells significantly increased the proportion of differentiated βIII-tubulin-positive neurons in culture (Krichevsky, et al., 2006). Stable overexpression of miR-124 induced neuronal markers and promoted neurite extension in cultures of embryonal carcinoma P19, neuroblastoma CAD and N2a, and isolated cortical progenitor cells (Maiorano and Mallamaci, 2009, Makeyev, et al., 2007, Visvanathan, et al., 2007). This previous work has culminated in a recent report showing that a combination of miR-124 with miR-9/9* and several neurogenic transcription factors induces conversion of human fibroblasts to neurons that exhibit functional synapses, action potentials and a gene expression profile characteristic of mature neurons (Yoo, et al., 2011). Therefore, miR-124 has emerged as an integral part of the genetic circuitry that directs neural fate determination.
While the exact functions of miR-124 in the CNS in vivo are still not completely well defined and in part controversial (Cao, et al., 2007, Visvanathan, et al., 2007), several studies point to an instructive role of this miRNA in promoting both embryonic and adult neurogenesis. For example, in the developing spinal cord in the chick neural tube, miR-124 has been implicated in the stimulation of neuronal differentiation (Visvanathan, et al., 2007). In mice, gain- and loss-of-function experiments have also confirmed an involvement of miR-124 in the temporal progression of neurogenesis in the adult subventricular zone (SVZ) (Cheng, et al., 2009). Generation of miR-124 knockout animals would further shed light to this field; however, this is technically-challenging as it would require triple knockouts, because miR-124 is expressed from three independent genetic loci. Interestingly, abrogating the single miR-124 locus in C. elegance did not cause any gross abnormalities in the development of the nervous system (Clark, et al., 2010).
The functions of miR-124 in the adult CNS and mature neurons are relatively poorly studied. So far, it is unclear whether miR-124 is required for fine-tuning gene expression in order to maintain the neuronal phenotype or for regulation of principal neuronal functions such as, e.g., synaptic plasticity. With numerous direct targets identified in the adult brain (see below), it is possible that miR-124 is involved in both of these processes. This notion is supported by the high turnover of miR-124 along with other miRNAs in mature neurons (Krol, et al., 2010), as well as by findings of recent studies in which miR-124 was shown to be a part of a complex regulatory gene expression network mediated by miRNAs in the context of cocaine-induced neuronal adaptations (Chandrasekar and Dreyer, 2009, Chandrasekar and Dreyer, 2011). Also, in Aplysia, where miR-124 is present exclusively in sensory but not motor neurons, its expression is modulated by the neurotransmitter serotonin and, as such, it may have a role in long-term synaptic plasticity by directly controlling CREB and CREB signaling (Rajasethupathy, et al., 2009). Whether miR-124 has a similar function in the mature CNS in vertebrates remains to be investigated.
Numerous miR-124 targets: from HeLA cells to brain
As most other miRNAs, miR-124 is predicted to target hundreds of mRNA transcripts. In fact, due to its high conservation and tissue specificity, miR-124 served as a model for understanding miRNA targeting and establishing target prediction rules. For example, a seminal study by Lim and colleges (Lim, et al., 2005) reported that ectopic overexpression of miR-124 in non-neural HeLa cells led to the repression of 174 non-neuronal mRNAs (expressed at low levels in the brain and at high levels in other tissues) and shifted their expression profile towards that of a neural phenotype. Subsequently, many of the regulated genes have been validated as direct miR-124 targets. Further studies on pro-neural CAD cells (Makeyev, et al., 2007) demonstrated that miR-124 down-regulated approximately 300 conserved targets, preferentially those expressed at low levels in the CNS, and conversely, specific inhibition of miR-124 in primary cortical neurons increased levels of numerous non-neuronal transcripts (Conaco, et al., 2006). Further mechanistic insight into miR-124 regulation was provided by Hendrickson and colleagues (Hendrickson, et al., 2009), who identified direct targets in miR-124 transfected non-neural HEK-293T cells by virtue of their association with Argonaute proteins, which are core components of the miRNA effector complex. Among the approximately 600 mRNAs specifically recruited to Argonaute proteins by miR-124, there was a large dynamic range for both the reduction of mRNA stability and their translation rate with changes in mRNA stability accounting for ~75% of the estimated effect on protein production. As for most other miRNAs, miR-124 targets follow the classical rules: they tend to have sequences in the 3'UTR that are recognized by the 5' seed of miR-124 (Chi, et al., 2009, Wang, et al., 2010). Finally, high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation mapped functional Argonaute-miR-124-mRNA interaction sites in the mouse brain and confirmed direct miR-124 binding to hundreds of transcripts (Chi, et al., 2009). Collectively, these studies provide a comprehensive molecular atlas of miR-124- target gene interactions and imply miR-124’s involvement in establishing and maintaining neuronal identity, as well as neuron function.
Among its numerous mRNA targets miR-124 may also modulate key molecules in signaling pathways that are associated with disease pathogenesis. For example, miR-124 down-regulates the phosphatase SCP1, a component of the anti-neural RE1-silencing transcription repressor complex (REST), whose down-regulation is critical for induction of neurogenesis during CNS development (Visvanathan, et al., 2007). Interestingly, in non-neuronal cells, miR-124 itself is transcriptionally repressed by REST, allowing the persistence of non-neuronal transcripts (Conaco, et al., 2006). A double-negative feedback loop between REST/SCP1 and miR-124 ensures stabilization and maintenance of neuronal gene expression in appropriate pro-neural conditions. Another principal target is PTBP1 (PTB/hnRNP I), a global repressor of alternative pre-mRNA splicing in non-neuronal cells (Makeyev, et al., 2007). During developmental shifts, miR-124 controls down-regulation of PTBP1, and thus the alternative splicing machinery towards production of neuronal-specific transcripts. Strikingly, among alternatively spliced mRNAs that are affected by miR-124, those that are involved in “cytoskeleton organization and biogenesis” and “cell projection” bio-terms are overrepresented. Together with miR-9*, miR-124 also regulates BAF53a, a subunit of the neural-progenitor-specific chromatin-remodeling complex (Yoo, et al., 2011). Mutations abolishing this regulation and causing persistent BAF53a expression lead to defective activity-dependent dendritic outgrowth in neurons. In addition to these “non-neuronal” transcripts, mRNAs associated with mitotic cell cycle progression, cell proliferation, and DNA replication are also overrepresented among miR-124 targets (Makeyev, et al., 2007, Wang and Wang, 2006). Collectively, these data substantiate miR-124’s role in arresting cell cycle and promoting mitotic exit during neurodifferentiation, and instructing morphogenesis of neurons. Disturbances in this regulation could result in dysfunction of neural pathways.
miR-124 in brain tumors
miR-124 regulation of numerous brain transcripts strongly suggests that dysregulation or malfunction of this miRNA might contribute to the pathogenesis of human brain disorders. As brain tumors are diseases of dysregulated neuro-glial cell proliferation and differentiation, the involvement of miR-124 in such pathologies would be anticipated. Indeed, expression profiling of various human brain tumors demonstrated that miR-124 is one of the most strongly and uniformly down-regulated miRNAs in brain neoplasia. In anaplastic astrocytoma, oligodendroglioma, and glioblastoma (GBM), which represent the most common and aggressive primary glial brain tumors in adults, miR-124 is down-regulated 30 to 7,800-fold when compared to non-neoplastic brain tissues and gliosis (Fowler, et al., 2010, Nelson, et al., 2006, Silber, et al., 2008); and Krichevsky, unpublished results). Medulloblastoma (MB), the most common malignant pediatric brain tumor of neuronal linage, thought to arise from aberrant cerebellar development, also exhibits reduced miR-124 levels relative to normal cerebellum (Ferretti, et al., 2008, Li, et al., 2009, Pierson, et al., 2008). miR-124 expression is especially low in MBs associated with activated Sonic Hedgehog signaling (Cho, et al., 2011) and in Gli1high MBs (Ferretti, et al., 2008), a subgroup with relatively poor clinical outcome.
Mechanistically, it is unclear whether decreased levels of miR-124 correlate with neoplastic transformation. As described above, miR-124 is expressed at high levels in differentiated neurons, but not in glial cells or neuronal progenitors. In situ hybridization in oligodendroglioma specimens has demonstrated that miR-124 is expressed in neurons only, but not in tumor cells (Nelson, et al., 2006). The low levels of miR-124 detected in tissue derived from highly invasive tumors might, therefore, be due to residual neurons contaminating tumor specimens. If so, does miR-124 reduction in brain tumors merely reflect the absence of neurons and the abundance of undifferentiated highly proliferative cells in the tumor mass? Or does this miRNA have an active tumor suppressive role in the brain? In other words, would reduction of miR-124 in committed/mature neurons lead to de-differentiation and neoplastic transformation? Or would failure to induce miR-124 expression in neural progenitor/stem cells under certain conditions lead to neoplastic transformation? While these important questions await further experimentation using both in vitro and in vivo model systems, support for miR-124 as a tumor suppressor comes from non-neural systems. Previously considered as neuron-specific, miR-124, in fact, is also expressed in peripheral blood samples, pancreatic β-cells, and some other cells, albeit at lower levels (Agirre, et al., 2009, Liang, et al., 2007). A tumor suppressive role for miR-124 has been reported in acute lymphoblastic leukemia (ALL), where its epigenetic silencing by hypermethylation is an independent negative prognostic factor (Agirre, et al., 2009). Furthermore, overexpression of miR-124 in a xenogeneic mouse model of ALL significantly decreases tumor growth. Hypermethylation of miR-124 promoters has also been reported for colon, breast, lung, and hematological malignancies (Furuta, et al., 2010, Lujambio, et al., 2008, Wong, et al., 2011). However, as of today, there are no data supporting aberrations in any of the three human miR-124 genes and/or their epigenetic modifications in brain cancers, and treatment with demethylating agents does not restore miR-124 expression in GBM and MB cell lines (Li, et al., 2009, Silber, et al., 2008).
Regardless of whether reduction of miR-124 contributes to brain tumorigenesis or not, it could still be a potential therapeutic agent. If overexpression of miR-124 can shift HeLa and HEK cells toward expressing neuronal genes, it is plausible that supplementation of miR-124 might also promote differentiation of both glial and neural tumors. Indeed, it has been demonstrated that ectopic overexpression of miR-124, alone or in combination, can induce cell cycle arrest and appearance of the neuronal marker βIII-tubulin in human glioma cells in vitro (Silber, et al., 2008). Overexpression of miR-124 in the glioma cell line A172 also decreases cell migration and invasion without affecting their proliferation (Fowler, et al., 2010). In addition, in several MB cell lines, significant effects of miR-124 ectopic expression on proliferation and growth have been reported (Li, et al., 2009). Mechanistically, restoration of miR-124 in glioma and other malignant brain tumors may slow tumor progression by repressing various miR-124 targets. Indeed, numerous validated targets are highly expressed in GBM and have demonstrated functions in gliomagenesis. These include factors of the cell cycle machinery that are directly regulated by miR-124, such as CDK6, which is an important regulator of the G1 to S-phase progression and is highly active in brain tumor cells (Lujambio, et al., 2008, Pierson, et al., 2008, Silber, et al., 2008). PTBP1, a principal miR-124 target and regulator of splicing (Makeyev, et al., 2007), and see above), is aberrantly overexpressed in glioma, and its removal inhibits glioma proliferation and migration (Cheung, et al., 2009). GTPase activating protein 1 (IQGAP1), Laminin γ1 (LAMC1), and β1-integrin (ITGB1) are three additional well-established targets (Cao, et al., 2007, Chi, et al., 2009, Fowler, et al., 2010, Lee, et al., 2010, Lim, et al., 2005) with important roles in tumor cell invasion, and frequently elevated in gliomas (Kawataki, et al., 2007, McDonald, et al., 2007, Piao, et al., 2009). miR-124 also targets regulator of sex determining region Y-box 2 (SOX9) (Cheng, et al., 2009), a transcription factor and stem cell marker, which is highly expressed in various malignant brain tumors and is involved in tumor cell proliferation (Sutter, et al., 2010, Swartling, et al., 2009). It should be noted that the reduced levels of miR-124 in GBM might contribute to or may even directly result in the de-repression of these molecules in the tumor cells. In our analysis of The Cancer Genome Atlas, the largest available database of molecular characteristics for 260 GBM patients (2008), we have determined that the expression of PTBP1, LAMC1, IQGAP1, ITGB1, CDK6, and SOX9 is strongly negatively correlated with miR-124 expression, with correlation coefficients ranging from −0.22 to −0.57 and p-values <0.002 (Krichevsky, unpublished results). These data further suggest that restoring miR-124 levels/activity by its ectopic overexpression in cancer cells should perhaps be a focus of future therapy developments for brain tumors.
miR-124 in neurodegeneration
A contribution of miR-124 to neurodegenerative disorders, such as Alzheimer’s disease (AD), Parkinson disease (PD), and Huntington’s disease (HD) is unknown. Several reports indicate a possible reduction of miR-124 levels in human brain regions affected by AD and HD (Lukiw, 2007, Packer, et al., 2008, Smith, et al., 2011), while other screens do not observe this decrease (Cogswell, et al., 2008, Johnson, et al., 2008). It should, however, be noted that reduced levels of miR-124 sometimes observed in the neurodegenerative hetero-cellular tissues could very likely be due to the reduced proportion of neurons, in which the miRNA is expressed. More refined analysis of specific cell populations affected by a disease should be carried out to draw any meaningful conclusion. High levels of neuronal miR-124 were also detected by in situ hybridization in human entorhinal and transentorhinal cortex in both healthy and AD subjects (Nelson and Wang, 2010). In HD, Johnson et al. reported that miR-124 targets in the caudate are statistically enriched and represent 2.2% of all up-regulated genes arguing for miR-124 down-regulation and function in this disease (Johnson and Buckley, 2009, Johnson, et al., 2008). Interestingly, the transcription factor REST, whose function is highly linked to miR-124 (see above), has been suggested as one of the factors involved in neuronal cell death in the striatum of HD patients, further supporting a potential role of this miRNA in HD pathogenesis. Still, no functional studies investigating the role of miR-124 in these neurodegenerative diseases have been reported. It has recently been demonstrated that miR-124 can shift the splicing patterns of Amyloid Precursor protein (APP), a key protein in AD pathology, and this reaction is mediated by PTBP1, which is a known target of miR-124 (Smith, et al., 2011). However, the relevance of this finding to APP biogenesis and amyloid accumulation in AD is unclear.
A new and surprising function of miR-124 has recently been discovered in our studies of microglia, the CNS-resident cells of the macrophage linage (Ponomarev, et al., 2011). Microglia isolated from the mouse brain and spinal cord, but not peripheral macrophages isolated from other tissues, express high levels of miR-124. These normally quiescent cells in the healthy adult CNS become activated in areas of trauma and infection in the aging human brain, as well as in neurodegenerative and neuroinflammatory diseases, such as multiple sclerosis (MS). Microglia activation is the principal pathology in these conditions. Interestingly, miR-124 has emerged as a key regulator of microglia quiescence in the CNS. Activated microglia down-regulate miR-124 expression in vitro and miR-124 levels correlate inversely with the activation state of microglia and macrophages in the CNS. Ectopic expression of miR-124 in peripheral bone marrow derived macrophages deactivates them by reducing the production of multiple cytokines and markers of activated cells, whereas the inhibition of miR-124 in microglia in vitro and intracranially in vivo activated the cells. Furthermore, systemic administration of miR-124 in experimental autoimmune encephalomyelitis, a mouse model of MS, prevented or ameliorated disease symptoms and enhanced recovery. The mechanism underlying systemic function of this miRNA is through the direct uptake by peripheral macrophages and their deactivation, which in turn, leads to deactivation of dendritic cells and autoimmune T cells. Since miR-124 administration suppressed macrophage activation in vivo, theoretically it could be therapeutic not only for MS, but also for other inflammatory diseases such as type 1-diabetes, rheumatoid arthritis and atherosclerosis.
In contrast to neurons, the functions of miR-124 in monocyte/macrophage cells are primarily mediated by its direct target enhancer-binding protein alpha (C/EBPα), a master transcription factor that controls differentiation of myeloid cells, and further downstream through PU.1, another master regulator of monocyte/macrophage differentiation (Feng, et al., 2008, Zhang, et al., 2004). Similar to its function in differentiation of neural cells, miR-124 may restrict proliferation and promote differentiation of monocytes into adult microglia. Thus, miR-124 may be viewed as a master regulator of differentiation and maturation of various cell types in the CNS. Investigation of this miRNA as a principal element of molecular networks that drive distinct cell fates like neurons and microglia, may improve our understanding of diverse brain disorders, from brain tumors to neurodegenerative conditions and autoimmune diseases such MS.
Identifying new miRNA players in brain diseases
Although a role of miR-124 in normal brain functioning and in diseases has been intensively investigated, corresponding data of the involvement of other brain-enriched miRNAs are sparse. Because the living human brain is not easily accessible to experimental manipulations, the identification and characterization of miRNAs in the pathophysiology of brain diseases, including both neurologic and psychiatric disorders, is challenging. As such, studying postmortem brains from subjects with these disorders is often the only viable alternative approach. In fact, there have been many reports of mRNA expression profile abnormalities in various neurologic and psychiatric conditions. The number of studies that examine miRNA expression profiles in these conditions is also rapidly increasing. By associating specific miRNAs that are differentially expressed in disease states with disturbances in signaling pathways revealed by mRNA expression profiling, it is possible to begin to understand the roles of miRNAs in the molecular pathophysiology of brain dysfunction (Sonntag, 2010). However, several biological and technical confounds that are intrinsic to this approach need to be considered. For example, a number of miRNAs have been implicated in the pathophysiology of schizophrenia (SZ), a prevalent psychiatric illness that is characterized by debilitating cognitive and psychotic symptoms. Such findings came entirely from profiling miRNA expression in homogenized gray matter tissues obtained from postmortem brains of subjects with the disorder (Beveridge, et al., 2010, Beveridge, et al., 2008, Guo, et al., 2010, Kim, et al., 2010, Kocerha, et al., 2009, Mellios, et al., 2008, Perkins, et al., 2007, Santarelli, et al., 2011). However, similar to the discussion above for AD, these findings in SZ have not been consistent between studies. In fact, in most cases, even within the same cortical region (e.g. dorsolateral prefrontal cortex), the miRNAs that were differentially expressed in different studies seldom overlap. In instances when they do overlap, the same miRNAs were altered in opposite directions (i.e. up- versus down-regulation) in different studies (e.g. (Beveridge, et al., 2010, Santarelli, et al., 2011)). This inconsistency may in part be due to the different technologies utilized for expression profiling, variable platform specificity, as well as various statistical methods used for data analysis.
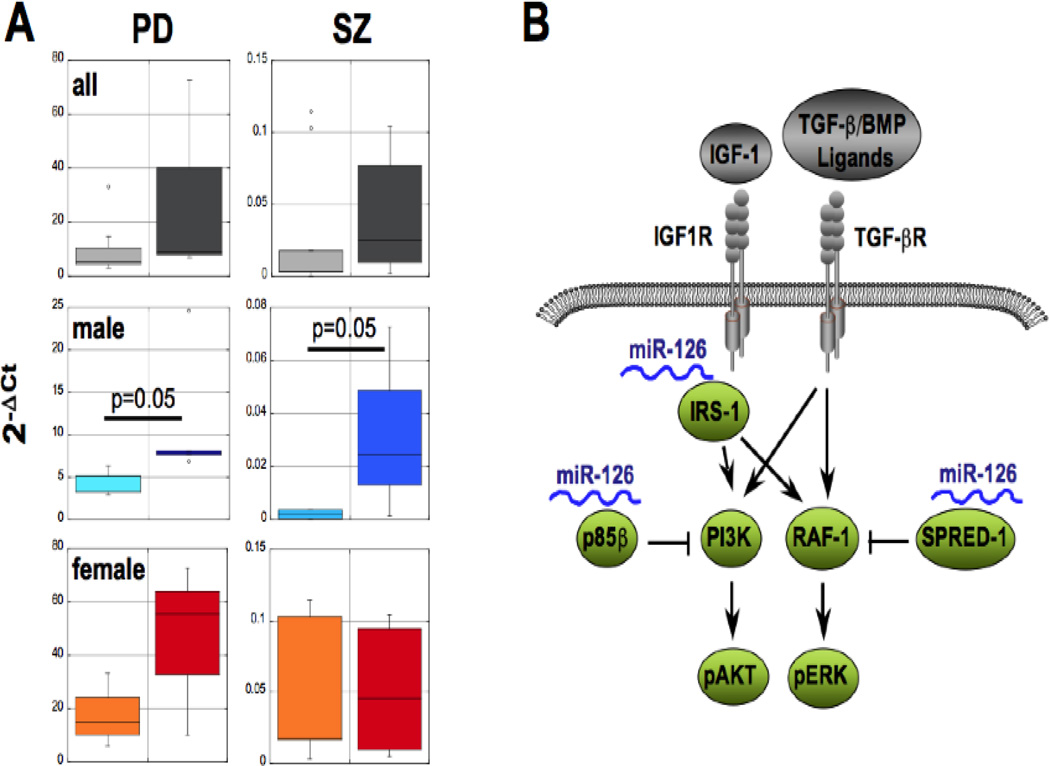
Because of the tremendous heterogeneity in the organization of neural circuits in the cerebral cortex, without knowing which miRNAs are altered in what cell populations, which include many functionally distinct neuronal and glial cell types, it is exceedingly difficult to understand how miRNA dysregulation leads to neural circuit dysfunction and hence the clinical manifestations of various diseases, especially if findings differ between studies. This problem is further compounded by the fact that current understanding of the roles of many of these miRNAs in the regulation of normal brain circuit functions is extremely limited. An informative approach to this problem is the analysis of targeted cell populations using laser-assisted microdissection technology and there is an increasing appreciation of this methodology in studying postmortem brain tissue. For example, we and others have recently determined the complete mRNA expression profiles of laser-microdissected dopamine neurons in PD (Cantuti-Castelvetri, et al., 2007, Elstner, et al., 2011, Simunovic, et al., 2009, Simunovic, et al., 2010) and in the pyramidal neurons in layer 3 of the superior temporal gyrus (STG) of the cerebral cortex in SZ individuals (Woo, unpublished results). These studies delineated a dysregulated expression network of the diseased cells and revealed insight into disturbed signaling pathways, which could be targets of miRNAs (Sonntag, 2010). To test this possibility, we additionally determined the complete miRNA profiles in the same dopamine and pyramidal cell populations using high throughput quantitative RT-PCR technologies. The experiments revealed a set of deregulated miRNAs that was associated with PD- or SZ-relevant signaling pathways, and some of these associations are also seen in other cellular contexts and diseases. Altogether, these data provide the framework for a translational approach of how mRNA/miRNA profiling and data mining could link pathways to miRNA abnormalities in and between different disease entities. In the following, this concept is exemplified by miR-126, which was significantly up-regulated in the male populations in both PD and SZ. This miRNA may be associated with PI3K signaling together with IGF-1, a pathway implicated in the pathogenesis of PD, or with the TGF-β and bone morphogenetic protein (BMP) signaling cascades, which were deregulated in the SZ samples (Fig. 1).
Figure 1.
Expression of miR-126 in PD and schizophrenia (SZ) and its targeting of PI3K signaling pathways. (A) Substantia nigra DA neurons from sporadic PD patients (5 males and 3 females, right bars) or pyramidal neurons in layer 3 of the superior temporal gyrus (STG) of the cerebral cortex from SZ patients (4 males and 5 females, right bars) and aged-matched controls (PD: 5 males and 3 females, SZ: 4 males and 5 females, left bars) were isolated by laser-assisted microdissection from postmortem brain material as previously described (Pietersen, et al., 2011). miRNA expression profiles were performed by Megaplex Human miRNA TaqMan® Arrays (LifeTechnologies) and quantified by the 2−ΔCt method (Livak and Schmittgen, 2001) using RNU44 or MammU6 snoRNA expression for normalization. Data are plotted for all samples combined or males and females separately, and statistical significance determined by 3-way ANOVA and student’s t-test. (B) Schematic of upstream factors in IGF-1/TGF-1β/PI3K signaling and validated targets of miR-126.
miR-126 in different cellular contexts
miR-126 has mainly been studied in endothelial cells and it is implicated in several forms of cancer, angiogenesis, vascularization, erythropoiesis, endothelial/leukocyte interaction, inflammation and immune response (Fish, et al., 2008, Fish and Srivastava, 2009, Grabher, et al., 2010, Harris, et al., 2008, Huang, et al., 2011, Meister and Schmidt, 2011, Oglesby, et al., 2010, Shen, et al., 2008, Zhao, et al., 2011). In endothelial cells, its expression is regulated by the transcription factors Ets-1 and -2, and in tumor cells it can be suppressed by Src tyrosine kinase (Li, et al., 2009). Validated direct targets of miR-126 are listed in Table 1; among them are several factors and pathways that are also important in neural cell function. For example, a major emerging function of miR-126 is its involvement in modulating key molecules in PI3K, MAPK, and AKT signaling as it regulates the expression of both negative regulators, such as Sprouty-related EVH1 domain–containing protein 1 (Spred1) and phosphoinositide kinase 3 regulatory subunit 2 (PIK3R2 or p85β) (Fish, et al., 2008, Guo, et al., 2008, Ishizaki, et al., 2011, Wang, et al., 2008, Zhu, et al., 2011), and pathway activators like insulin receptor substrate 1 (IRS-1) (Ryu, et al., 2011, Zhang, et al., 2008), Vascular Epithelial Growth Factor (VEGF) (Zhu, et al., 2011), and EGF-like domain-containing protein 7 (EGFL7) (Fish, et al., 2008, Kuhnert, et al., 2008, Musiyenko, et al., 2008, Nikolic, et al., 2010, Wang, et al., 2008). Other factors targeted by miR-126 are involved in apoptosis, the transport of ubiquinated proteins in the ubiquitin proteasome/endosome system, and TNF-α signaling (Harris, et al., 2008, Li, et al., 2008, Oglesby, et al., 2010). Recently miR-126 was validated as a regulator of SOX2 in the modulation of cell cycle arrest and apoptosis in gastric cancer (Otsubo, et al.). In the neural context, SOX2 is an important factor in maintaining self-renewal and pluripotency of stem cells and neural progenitors. Taken together, miR-126 appears to be a miRNA that affects cell growth and survival pathways in diverse cellular contexts.
Table 1.
Functionally validated targets for miR-126.
| Target | Species | Cell System | References |
|---|---|---|---|
| c-Myb | Zebrafish | Hematopoiesis | (Grabher, et al., 2010) |
| CRK | Human | Lung cancer | (Crawford, et al., 2008) |
| HOXA9 | Human | Hematopoiesis | (Shen, et al., 2008) |
| IRS-1 | Human | Breast cancer, hepatocytes | (Ryu, et al., 2011, Zhang, et al., 2008) |
| Pak1 | Zebrafish | Endothelium | (Zou, et al., 2011) |
| PIK3R2 (p85β) | Human | Colon, breast cancer | (Guo, et al., 2008, Zhu, et al., 2011) |
| PLK2 | Human | Leukemia | (Li, et al., 2008) |
| PTNP9 | Human | Erythropoiesis | (Huang, et al., 2011) |
| SLC7A5 | Human | Lung cancer | (Miko, et al., 2011) |
| SOX2 | Human | Gastric cancer | (Otsubo, et al.) |
| Spred1 | Zebrafish, Mouse | Endothelium | (Fish, et al., 2008, Wang, et al., 2008) |
| TOM1 | Human | Epithelium | (Oglesby, et al., 2010) |
| VCAM | Human | Endothelium | (Harris, et al., 2008) |
| VEGFA | Human | Lung, breast cancer | (Liu, et al., 2009, Zhu, et al., 2011) |
A potential role of miR-126 in brain disorders
High expression of miR-126 has been reported in the human and rodent brain (Landgraf, et al., 2007) and in cultured rat motoneurons (Wei, et al., 2010) indicating that it could play a role in neuronal cell function. Nevertheless, there are very little functional data of this miRNA in the CNS. One interesting aspect of miR-126 function in the nervous system could be through its genomic and regulatory link with EGFL7. miR-126 is encoded in intron 7 of the EGFL7 gene and originates from the EGFL7 pre-mRNA (Wang, et al., 2008). Although the EGFL7 mRNA has not been described as a direct target of miR-126, it can regulate EGFL7 expression in different cellular contexts, such as endothelial cells and cancer, probably through indirect mechanisms (Kuhnert, et al., 2008, Meister and Schmidt, 2011, Musiyenko, et al., 2008, Nikolic, et al., 2010). EGFL7 is expressed in the mammalian brain where it regulates Notch signaling in neural stem cells altering their self-renewal and multipotency as well as their differentiation potential towards neuro- and gliogenesis (Schmidt, et al., 2009). Thus, EGFL7 could have implication for boosting adult neurogenesis as a repair mechanism in neurodegenerative disorders and brain injury (Bicker and Schmidt, 2010), and might also contribute to brain neoplasia (Huang, et al., 2010). Although it remains to be determined whether miR-126 is involved in these processes, given its impact on EGFL7 regulation in a magnitude of cell systems, it is likely that similar mechanisms also occur in neurons.
Another potential role of miR-126 in the neuronal context could be derived from its profound impact on regulating factors in key signaling pathways that are also active in neurons and are associated with disease processes. As discussed above, miR-126 is involved in the PI3K pathway, and in particular IGF-1, VEGF or EGFL7 signaling, from which it targets IRS-1, PI3KR2 (p85β), SPRED1, and CRK. Insulin/IGF-1 signaling promotes neurite sprouting, synapse formation, and neuronal survival, and it has been implicated in memory, aging, and neurodegeneration (Bishop, et al., 2010, Nelson and Alkon, 2005). For example in AD and PD, there is evidence that neurons are resistant to IGF-1 and there is a large body of data linking changes in growth factor/IGF-1 signaling to disease pathogenesis and altered neuronal cell function (Craft and Watson, 2004, Freude, et al., 2009, Moloney, et al., 2010, von Bohlen und Halbach and Unsicker, 2009). In experimental animal models of neurotoxicity related to PD pathogenesis, activation of IGF-1 signaling protects against toxic insult to 6-OHDA (Guan, et al., 2000, Quesada, et al., 2008, Quesada and Micevych, 2004, Xu, et al., 2009), whereas disruption of IGF-1 signaling produces enhanced neurotoxicity (Giovannone, et al., 2009). Similar protective effects were also observed in multiple other studies on inflammation and brain injury suggesting a central role of the IGF-1 pathway in cell survival in vivo. In addition to growth factor/IGF-1 signaling, the PI3K pathway is also involved in other aspects of neuronal cell function including mechanisms of dysfunction related to, e.g., oxidative stress, apoptosis, and others. In SZ, TGF-β and BMP signaling in pyramidal neurons in layer 3 of the cortex is among the most significantly dysregulated pathways and bioinformatics analysis indicates that these disturbances may be linked to miRNA dysregulation (Woo, unpublished results). TGF-β/BMP signaling also engages the PI3K pathway and regulates a variety of other signaling cascades, including those that are associated with apoptosis and anoikis (Vachon, 2011), which may play a role in the pathophysiology of decreased dendritic spine density on pyramidal neurons in SZ (Glantz, et al., 2006).
Altogether, the findings discussed above raise the possibility that disturbances in the expression of the genes related to PI3K signaling pathways in PD and SZ may be associated with miR-126 deregulation in neuronal cell populations as well. However, this might not be as straightforward as one would predict from confirmed miRNA/target relationships; regulation of one or multiple targets by a miRNA and its consecutive effects on signaling pathways in one cell type does not necessarily predict the same outcome in a different cellular context (Gabriely, et al., 2011). For example, miR-126 is not associated with the down-regulation of PIK3R2 in small lung cancer (SCLC), TOM1 in breast cancer (MCF7), or SPRED1 in acute myeloid leukemia (AML) cell lines, while it regulates these sane targets in other cell types (Miko, et al., 2011). Another example suggestive of cell type-dependent miRNA functions comes from a recent study on miR-34b and miR-34c in PD (Minones-Moyano, et al., 2011). Both of these miRNAs are members of the miR-34a/b/c family, which is down-regulated in several forms of cancer and targets factors that are involved in cell cycle arrest, senescence, and apoptosis (Hermeking, 2010), and has also been proposed as a potential plasma marker for HD (Gaughwin, et al., 2011, Parekh, 2011). In the Miñones-Moyano et al. study, miRNA profiling in different brain regions revealed down-regulation of miR-34b and miR-34c in tissue from PD patients. Subsequent functional analysis in SH-SY5Y neuroblastoma cells showed that inhibition of miR-34b/c compromised cell viability by impairing mitochondrial function including the production of reactive oxygen species. Although the study did not provide information on specific miR-34b/c targets, it indicated a reduction of Parkin and DJ1, two proteins that are linked to mitochondrial dysfunction in PD, in both miRNA-depleted SH-SY5Y cells and those PD brain regions with decreased miR-34b/c levels. The authors suggest that miR-34b/c deficiency might negatively affect mitochondrial function through an indirect mechanism that includes Parkin and DJ1. Interestingly, in cancer, expression of the miR-34 family members is activated by p53, while their silencing is associated with promoter methylation (Corney, et al., 2007, Kumamoto, et al., 2008, Toyota, et al., 2008). However, it is unclear whether these mechanisms are responsible for the observed down-regulation in PD. A caveat in the Miñones-Moyano study is that miR-34 expression was analyzed in heterogeneous cell populations from brain tissue and its function was not directly linked to the neuronal phenotype. This is an important parameter as the apoptosis-inductive properties of miR-34 in cancer presumably depend on the cellular and genetic context, its expression levels and feed-back loops involving p53 (Hermeking, 2010). At present, it remains to be established whether similar mRNA targets and downstream signaling pathways are also affected by miR-34b/c in neurons. Some evidence comes from a recent study showing that miR-34c is up-regulated in the hippocampus of AD patients and mice, and its identification as a target to rescue learning impairment in a mouse model of AD (Zovoilis, et al., 2011). Interestingly, this function was associated with regulating its target Sirt1, which regulates p53 activity in cancer cells (Yamakuchi and Lowenstein, 2009), and this mechanism has also been implicated in neurite elongation during mouse neural stem cell differentiation (Aranha, et al., 2011). Finally, it needs to be established whether aberrant miRNAs are cause or consequence of disease pathogenesis and whether they confer (neuro)protection or vulnerability. In the case of miR-34, similar to its function in cancer, cognitive disturbances in AD, and neural cell differentiation, down-regulation in Parkinsonian DA neurons that undergo apoptosis (Simunovic, et al., 2009) could be a compensatory anti-apoptotic effect as well.
Thus, multiple factors depending on cell type, activation, and environment can influence miRNA expression and signaling making their functions not necessarily universal. Currently, much of the mechanisms that govern cell type-specific target regulation by miRNAs are unclear underscoring the complexity of expression networks across different cell systems. The challenge will be to delineate how a certain (or multiple) miRNA(s) is/are regulated by and regulate(s) a particular cellular phenotype and how this regulation contributes to normal or abnormal cell function and consequently to disease processes.
Conclusions
Although there has been significant progress in delineating the basic biology of miRNAs and their roles in cancer, current knowledge of the regulation of brain functions by miRNAs in both normal and disease states is comparatively limited. This review highlights what we believe is an effective strategy to study the roles of miRNA in brain circuit dysfunction in neurologic and neuropsychiatric disorders. The strategy involves characterization of miRNA expression changes in specific cell types within neural circuits known to be dysfunctional in the disorder of interest, and then correlative analysis of these changes in relation to mRNA expression. This approach should facilitate the development of an overarching biological framework within which specific pathophysiological hypotheses can be conceptualized and subsequently tested in in vitro and/or in vivo experimental systems.
Acknowledgments
The authors want to acknowledge the following funding sources and support: Massachusetts’ Alzheimer's Disease Research Center and the Harvard NeuroDiscovery Center, and NIH/NINDS NS067335 for Dr. Kai-C. Sonntag. The miRNA profiling was performed in collaboration with Dr. Yulei Wang (Life Technologies Corporation™); Grants R01MH076060 and Boston CIDAR, "Vulnerability to Progression in Schizophrenia”, and P50MH080272 from NIMH, as well as Life Technologies Corporation™ for providing some of the necessary reagents for Dr. Tsung-Ung W. Woo; Dr. Anna Krichevsky is a recipient of The Sontag Foundation Distinguished Scientist Award, and her research is supported by grants from The Sontag Foundation, the Alzheimer's Association (NIRG-09-132844), and NIH/NCI R01CA138734.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agirre X, Vilas-Zornoza A, Jimenez-Velasco A, Martin-Subero JI, Cordeu L, Garate L, San Jose-Eneriz E, Abizanda G, Rodriguez-Otero P, Fortes P, Rifon J, Bandres E, Calasanz MJ, Martin V, Heiniger A, Torres A, Siebert R, Roman-Gomez J, Prosper F. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69:4443–4453. doi: 10.1158/0008-5472.CAN-08-4025. [DOI] [PubMed] [Google Scholar]
- 3.Aranha MM, Santos DM, Sola S, Steer CJ, Rodrigues CM. miR-34a Regulates Mouse Neural Stem Cell Differentiation. PLoS One. 2011;6:e21396. doi: 10.1371/journal.pone.0021396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbato C, Ruberti F, Cogoni C. Searching for MIND: microRNAs in neurodegenerative diseases. J Biomed Biotechnol. 2009;2009 doi: 10.1155/2009/871313. 871313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15:1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- 8.Bicker F, Schmidt MH. EGFL7: a new player in homeostasis of the nervous system. Cell Cycle. 2010;9:1263–1269. doi: 10.4161/cc.9.7.11091. [DOI] [PubMed] [Google Scholar]
- 9.Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–535. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bushati N, Cohen SM. MicroRNAs in neurodegeneration. Curr Opin Neurobiol. 2008;18:292–296. doi: 10.1016/j.conb.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Cantuti-Castelvetri I, Keller-McGandy C, Bouzou B, Asteris G, Clark TW, Frosch MP, Standaert DG. Effects of gender on nigral gene expression and parkinson disease. Neurobiol Dis. 2007;26:606–614. doi: 10.1016/j.nbd.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandrasekar V, Dreyer JL. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol Cell Neurosci. 2009;42:350–362. doi: 10.1016/j.mcn.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Chandrasekar V, Dreyer JL. Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology. 2011;36:1149–1164. doi: 10.1038/npp.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung HC, Hai T, Zhu W, Baggerly KA, Tsavachidis S, Krahe R, Cote GJ. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain. 2009;132:2277–2288. doi: 10.1093/brain/awp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L, Eberhart CG, Lau CC, Olson JM, Gilbertson RJ, Gajjar A, Delattre O, Kool M, Ligon K, Meyerson M, Mesirov JP, Pomeroy SL. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark AM, Goldstein LD, Tevlin M, Tavare S, Shaham S, Miska EA. The microRNA miR-124 controls gene expression in the sensory nervous system of Caenorhabditis elegans. Nucleic Acids Res. 2010;38:3780–3793. doi: 10.1093/nar/gkq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 21.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 23.Craft S, Watson GS. Insulin and neurodegenerative disease: shared and specific mechanisms. Lancet Neurol. 2004;3:169–178. doi: 10.1016/S1474-4422(04)00681-7. [DOI] [PubMed] [Google Scholar]
- 24.Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Otterson GA, Nuovo G, Marsh CB, Nana-Sinkam SP. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373:607–612. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 25.Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- 26.Du L, Pertsemlidis A. Cancer and neurodegenerative disorders: pathogenic convergence through microRNA regulation. J Mol Cell Biol. 2011;3:176–180. doi: 10.1093/jmcb/mjq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elstner M, Morris CM, Heim K, Bender A, Mehta D, Jaros E, Klopstock T, Meitinger T, Turnbull DM, Prokisch H. Expression analysis of dopaminergic neurons in Parkinson's disease and aging links transcriptional dysregulation of energy metabolism to cell death. Acta Neuropathol. 2011;122:75–86. doi: 10.1007/s00401-011-0828-9. [DOI] [PubMed] [Google Scholar]
- 29.Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E, Screpanti I, Bozzoni I, Gulino A. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27:2616–2627. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fish JE, Srivastava D. MicroRNAs: opening a new vein in angiogenesis research. Sci Signal. 2009;2 doi: 10.1126/scisignal.252pe1. pe1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fowler A, Thomson D, Giles K, Maleki S, Mreich E, Wheeler H, Leedman P, Biggs M, Cook R, Little N, Robinson B, McDonald K. miR-124a is frequently down-regulated in glioblastoma and is involved in migration and invasion. Eur J Cancer. 2010;47:953–963. doi: 10.1016/j.ejca.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 34.Freude S, Schilbach K, Schubert M. The role of IGF-1 receptor and insulin receptor signaling for the pathogenesis of Alzheimer's disease: from model organisms to human disease. Curr Alzheimer Res. 2009;6:213–223. doi: 10.2174/156720509788486527. [DOI] [PubMed] [Google Scholar]
- 35.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 36.Gabriely G, Teplyuk NM, Krichevsky AM. Context effect: microRNA-10b in cancer cell proliferation, spread and death. Autophagy. 2011;7 doi: 10.4161/auto.7.11.17371. [DOI] [PubMed] [Google Scholar]
- 37.Gao FB. Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 2010;5:25. doi: 10.1186/1749-8104-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaughwin PM, Ciesla M, Lahiri N, Tabrizi SJ, Brundin P, Bjorkqvist M. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington's disease. Hum Mol Genet. 2011;20:2225–2237. doi: 10.1093/hmg/ddr111. [DOI] [PubMed] [Google Scholar]
- 39.Giovannone B, Tsiaras WG, de la Monte S, Klysik J, Lautier C, Karashchuk G, Goldwurm S, Smith RJ. GIGYF2 gene disruption in mice results in neurodegeneration and altered insulin-like growth factor signaling. Hum Mol Genet. 2009;18:4629–4639. doi: 10.1093/hmg/ddp430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glantz LA, Gilmore JH, Lieberman JA, Jarskog LF. Apoptotic mechanisms and the synaptic pathology of schizophrenia. Schizophr Res. 2006;81:47–63. doi: 10.1016/j.schres.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 41.Grabher C, Payne EM, Johnston AB, Bolli N, Lechman E, Dick JE, Kanki JP, Look AT. Zebrafish microRNA-126 determines hematopoietic cell fate through c-Myb. Leukemia. 2010;25:506–514. doi: 10.1038/leu.2010.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guan J, Krishnamurthi R, Waldvogel HJ, Faull RL, Clark R, Gluckman P. N-terminal tripeptide of IGF-1 (GPE) prevents the loss of TH positive neurons after 6-OHDA induced nigral lesion in rats. Brain Res. 2000;859 doi: 10.1016/s0006-8993(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 43.Guo AY, Sun J, Jia P, Zhao Z. A novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC Syst Biol. 2010;4:10. doi: 10.1186/1752-0509-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo L, Sun B, Sang F, Wang W, Lu Z. Haplotype distribution and evolutionary pattern of miR-17 and miR-124 families based on population analysis. PLoS One. 2009;4:e7944. doi: 10.1371/journal.pone.0007944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 48.Hebert SS, Horre K, Nicolai L, Bergmans B, Papadopoulou AS, Delacourte A, De Strooper B. MicroRNA regulation of Alzheimer's Amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 49.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 51.Huang CH, Li XJ, Zhou YZ, Luo Y, Li C, Yuan XR. Expression and clinical significance of EGFL7 in malignant glioma. J Cancer Res Clin Oncol. 2010;136:1737–1743. doi: 10.1007/s00432-010-0832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang X, Gschweng E, Van Handel B, Cheng D, Mikkola HK, Witte ON. Regulated expression of microRNAs-126/126* inhibits erythropoiesis from human embryonic stem cells. Blood. 2011;117:2157–2165. doi: 10.1182/blood-2010-08-302711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishizaki T, Tamiya T, Taniguchi K, Morita R, Kato R, Okamoto F, Saeki K, Nomura M, Nojima Y, Yoshimura A. miR126 positively regulates mast cell proliferation and cytokine production through suppressing Spred1. Genes Cells. 2011;16:803–814. doi: 10.1111/j.1365-2443.2011.01529.x. [DOI] [PubMed] [Google Scholar]
- 54.Johnson R, Buckley NJ. Gene dysregulation in Huntington's disease: REST, microRNAs and beyond. Neuromolecular Med. 2009;11:183–199. doi: 10.1007/s12017-009-8063-4. [DOI] [PubMed] [Google Scholar]
- 55.Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol Dis. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 56.Kawataki T, Yamane T, Naganuma H, Rousselle P, Anduren I, Tryggvason K, Patarroyo M. Laminin isoforms and their integrin receptors in glioma cell migration and invasiveness: Evidence for a role of alpha5-laminin(s) and alpha3beta1 integrin. Exp Cell Res. 2007;313:3819–3831. doi: 10.1016/j.yexcr.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 57.Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, van den Oord EJ, Riley BP, Kendler KS, Vladimirov VI. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124:183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, Sales N, Willoughby D, Elmen J, Hansen HF, Orum H, Kauppinen S, Kenny PJ, Wahlestedt C. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci U S A. 2009;106:3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krichevsky AM. MicroRNA profiling: from dark matter to white matter, or identifying new players in neurobiology. Scientific World Journal. 2007;7:155–166. doi: 10.1100/tsw.2007.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, Oertner TG, Schratt G, Bibel M, Roska B, Filipowicz W. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 63.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 64.Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S, Yokota J, Harris CC. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68:3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 66.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee MR, Kim JS, Kim KS. miR-124a is important for migratory cell fate transition during gastrulation of human embryonic stem cells. Stem Cells. 2010;28:1550–1559. doi: 10.1002/stem.490. [DOI] [PubMed] [Google Scholar]
- 68.Li KK, Pang JC, Ching AK, Wong CK, Kong X, Wang Y, Zhou L, Chen Z, Ng HK. miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Hum Pathol. 2009;40:1234–1243. doi: 10.1016/j.humpath.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 69.Li X, Shen Y, Ichikawa H, Antes T, Goldberg GS. Regulation of miRNA expression by Src and contact normalization: effects on nonanchored cell growth and migration. Oncogene. 2009;28:4272–4283. doi: 10.1038/onc.2009.278. [DOI] [PubMed] [Google Scholar]
- 70.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, Bohlander SK, Zhang DE, Larson RA, Le Beau MM, Thirman MJ, Golub TR, Rowley JD, Chen J. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 73.Liu B, Peng XC, Zheng XL, Wang J, Qin YW. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66:169–175. doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 74.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 75.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 77.Maiorano NA, Mallamaci A. Promotion of embryonic corticocerebral neuronogenesis by miR-124. Neural Dev. 2009;4:40. doi: 10.1186/1749-8104-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McDonald KL, O'Sullivan MG, Parkinson JF, Shaw JM, Payne CA, Brewer JM, Young L, Reader DJ, Wheeler HT, Cook RJ, Biggs MT, Little NS, Teo C, Stone G, Robinson BG. IQGAP1 and IGFBP2: valuable biomarkers for determining prognosis in glioma patients. J Neuropathol Exp Neurol. 2007;66:405–417. doi: 10.1097/nen.0b013e31804567d7. [DOI] [PubMed] [Google Scholar]
- 80.Meister J, Schmidt MH. miR-126 and miR-126*: new players in cancer. Scientific World Journal. 2011;10:2090–2100. doi: 10.1100/tsw.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miko E, Margitai Z, Czimmerer Z, Varkonyi I, Dezso B, Lanyi A, Bacso Z, Scholtz B. miR-126 inhibits proliferation of small cell lung cancer cells by targeting SLC7A5. FEBS Lett. 2011;585:1191–1196. doi: 10.1016/j.febslet.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 83.Miller BH, Wahlestedt C. MicroRNA dysregulation in psychiatric disease. Brain Res. 2010;1338:89–99. doi: 10.1016/j.brainres.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Minones-Moyano E, Porta S, Escaramis G, Rabionet R, Iraola S, Kagerbauer B, Espinosa-Parrilla Y, Ferrer I, Estivill X, Marti E. MicroRNA profiling of Parkinson's disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 85.Moloney AM, Griffin RJ, Timmons S, O'Connor R, Ravid R, O'Neill C. Defects in IGF-1 receptor, insulin receptor and IRS-1/2 in Alzheimer's disease indicate possible resistance to IGF-1 and insulin signalling. Neurobiol Aging. 2010;31:224–243. doi: 10.1016/j.neurobiolaging.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 86.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med. 2008;86:313–322. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nelson PT, Wang WX. MiR-107 is reduced in Alzheimer's disease brain neocortex: validation study. J Alzheimers Dis. 2010;21:75–79. doi: 10.3233/JAD-2010-091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nelson TJ, Alkon DL. Insulin and cholesterol pathways in neuronal function, memory and neurodegeneration. Biochem Soc Trans. 2005;33:1033–1036. doi: 10.1042/BST20051033. [DOI] [PubMed] [Google Scholar]
- 91.Nikolic I, Plate KH, Schmidt MH. EGFL7 meets miRNA-126: an angiogenesis alliance. J Angiogenes Res. 2010;2:9. doi: 10.1186/2040-2384-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oglesby IK, Bray IM, Chotirmall SH, Stallings RL, O'Neill SJ, McElvaney NG, Greene CM. miR-126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. J Immunol. 2010;184:1702–1709. doi: 10.4049/jimmunol.0902669. [DOI] [PubMed] [Google Scholar]
- 93.Olsen L, Klausen M, Helboe L, Nielsen FC, Werge T. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. PLoS One. 2009;4:e7225. doi: 10.1371/journal.pone.0007225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Otsubo T, Akiyama Y, Hashimoto Y, Shimada S, Goto K, Yuasa Y. MicroRNA-126 inhibits SOX2 expression and contributes to gastric carcinogenesis. PLoS One. 2011;6:e16617. doi: 10.1371/journal.pone.0016617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parekh V. Neurodegenerative disease: miR-34b-a novel plasma marker for Huntington disease? Nat Rev Neurol. 2011;7:304. doi: 10.1038/nrneurol.2011.68. [DOI] [PubMed] [Google Scholar]
- 97.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays. 2009;31:736–747. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- 99.Piao Y, Lu L, de Groot J. AMPA receptors promote perivascular glioma invasion via beta1 integrin-dependent adhesion to the extracellular matrix. Neuro Oncol. 2009;11:260–273. doi: 10.1215/15228517-2008-094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pierson J, Hostager B, Fan R, Vibhakar R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol. 2008;90:1–7. doi: 10.1007/s11060-008-9624-3. [DOI] [PubMed] [Google Scholar]
- 101.Pietersen CY, Lim MP, Macey L, Woo TU, Sonntag KC. Neuronal type-specific gene expression profiling and laser-capture microdissection. Methods Mol Biol. 2011;755:327–343. doi: 10.1007/978-1-61779-163-5_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Quesada A, Lee BY, Micevych PE. PI3 kinase/Akt activation mediates estrogen and IGF-1 nigral DA neuronal neuroprotection against a unilateral rat model of Parkinson's disease. Dev Neurobiol. 2008;68:632–644. doi: 10.1002/dneu.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Quesada A, Micevych PE. Estrogen interacts with the IGF-1 system to protect nigrostriatal dopamine and maintain motoric behavior after 6-hydroxdopamine lesions. J Neurosci Res. 2004;75:107–116. doi: 10.1002/jnr.10833. [DOI] [PubMed] [Google Scholar]
- 105.Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ryu HS, Park SY, Ma D, Zhang J, Lee W. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS One. 2011;6:e17343. doi: 10.1371/journal.pone.0017343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry. 2011;69:180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 108.Schmidt MH, Bicker F, Nikolic I, Meister J, Babuke T, Picuric S, Muller-Esterl W, Plate KH, Dikic I. Epidermal growth factor-like domain 7 (EGFL7) modulates Notch signalling and affects neural stem cell renewal. Nat Cell Biol. 2009;11:873–880. doi: 10.1038/ncb1896. [DOI] [PubMed] [Google Scholar]
- 109.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, Bergers G, Weiss WA, Alvarez-Buylla A, Hodgson JG. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Simunovic F, Yi M, Wang Y, Macey L, Brown LT, Krichevsky AM, Andersen SL, Stephens RM, Benes FM, Sonntag KC. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson's disease pathology. Brain. 2009;132:1795–1809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Simunovic F, Yi M, Wang Y, Stephens R, Sonntag KC. Evidence for gender-specific transcriptional profiles of nigral dopamine neurons in Parkinson disease. PLoS One. 2010;5:e8856. doi: 10.1371/journal.pone.0008856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Singh SK. miRNAs: from neurogeneration to neurodegeneration. Pharmacogenomics. 2007;8:971–978. doi: 10.2217/14622416.8.8.971. [DOI] [PubMed] [Google Scholar]
- 115.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 116.Smith P, Al Hashimi A, Girard J, Delay C, Hebert SS. In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J Neurochem. 2011;116:240–247. doi: 10.1111/j.1471-4159.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- 117.Sonntag KC. MicroRNAs and deregulated gene expression networks in neurodegeneration. Brain Res. 2010;1338:48–57. doi: 10.1016/j.brainres.2010.03.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sutter R, Shakhova O, Bhagat H, Behesti H, Sutter C, Penkar S, Santuccione A, Bernays R, Heppner FL, Schuller U, Grotzer M, Moch H, Schraml P, Marino S. Cerebellar stem cells act as medulloblastomainitiating cells in a mouse model and a neural stem cell signature characterizes a subset of human medulloblastomas. Oncogene. 2010;29:1845–1856. doi: 10.1038/onc.2009.472. [DOI] [PubMed] [Google Scholar]
- 119.Swartling FJ, Ferletta M, Kastemar M, Weiss WA, Westermark B. Cyclic GMP-dependent protein kinase II inhibits cell proliferation, Sox9 expression and Akt phosphorylation in human glioma cell lines. Oncogene. 2009;28:3121–3131. doi: 10.1038/onc.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 121.Vachon PH. Integrin signaling, cell survival, and anoikis: distinctions, differences, and differentiation. J Signal Transduct. 2011;2011 doi: 10.1155/2011/738137. 738137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.von Bohlen und Halbach O, Unsicker K. Neurotrophic support of midbrain dopaminergic neurons. Adv Exp Med Biol. 2009;651:73–80. doi: 10.1007/978-1-4419-0322-8_7. [DOI] [PubMed] [Google Scholar]
- 124.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang WX, Wilfred BR, Xie K, Jennings MH, Hu YH, Stromberg AJ, Nelson PT. Individual microRNAs (miRNAs) display distinct mRNA targeting "rules". RNA Biol. 2010;7:373–380. doi: 10.4161/rna.7.3.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang X, Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wei H, Wang C, Zhang C, Li P, Wang F, Zhang Z. Comparative profiling of microRNA expression between neural stem cells and motor neurons in embryonic spinal cord in rat. Int J Dev Neurosci. 2010;28:545–551. doi: 10.1016/j.ijdevneu.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 128.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 129.Wong KY, So CC, Loong F, Chung LP, Lam WW, Liang R, Li GK, Jin DY, Chim CS. Epigenetic inactivation of the miR-124-1 in haematological malignancies. PLoS One. 2011;6:e19027. doi: 10.1371/journal.pone.0019027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xu B, Karayiorgou M, Gogos JA. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res. 2010;1338:78–88. doi: 10.1016/j.brainres.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xu L, Chen WF, Wong MS. Ginsenoside Rg1 protects dopaminergic neurons in a rat model of Parkinson's disease through the IGF-I receptor signalling pathway. Br J Pharmacol. 2009;158:738–748. doi: 10.1111/j.1476-5381.2009.00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 133.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ, Ma D. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008;377:136–140. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- 135.Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, Shigematsu H, Levantini E, Huettner CS, Lekstrom-Himes JA, Akashi K, Tenen DG. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 136.Zhao S, Wang Y, Liang Y, Zhao M, Long H, Ding S, Yin H, Lu Q. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63:1376–1386. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]
- 137.Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, Bai Y, Shen Y, Yuan W, Jing Q, Qin Y. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351:157–164. doi: 10.1007/s11010-011-0723-7. [DOI] [PubMed] [Google Scholar]
- 138.Zou J, Li WQ, Li Q, Li XQ, Zhang JT, Liu GQ, Chen J, Qiu XX, Tian FJ, Wang ZZ, Zhu N, Qin YW, Shen B, Liu TX, Jing Q. Two functional microRNA-126s repress a novel target gene p21-activated kinase 1 to regulate vascular integrity in zebrafish. Circ Res. 2011;108:201–209. doi: 10.1161/CIRCRESAHA.110.225045. [DOI] [PubMed] [Google Scholar]
- 139.Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, Farinelli L, Delalle I, Schmitt A, Falkai P, Bahari-Javan S, Burkhardt S, Sananbenesi F, Fischer A. microRNA-34c is a novel target to treat dementias. EMBO J. 2011;30:4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]