Abstract
The protein tyrosine phosphatase, SHP-1, is a negative regulator of proinflammatory signaling and autoimmune disease. We have previously reported reduced SHP-1 expression in peripheral blood leukocytes of subjects with multiple sclerosis (MS). Recent evidence indicates that virus-induced DNA methylation of the SHP-1 promoter is responsible for aberrant silencing of SHP-1 expression and function in hematopoietic cells that might relate to inflammatory diseases. In the present study, bisulfite sequencing of the SHP-1 promoter demonstrated that over a third of MS subjects had abnormally high promoter methylation. As SHP-1 is deficient in MS leukocytes and SHP-1-regulated proinflammatory genes are correspondingly upregulated, we propose that increased SHP-1 promoter methylation may relate in part to decreased SHP-1 expression and increased leukocyte-mediated inflammation in MS.
Keywords: Bisulfite sequencing, epigenetics, immune regulation, multiple sclerosis, PBMC, protein tyrosine phosphatase
1. Introduction
Multiple sclerosis is a chronic inflammatory demyelinating disease of the central nervous system (Kornek and Lassmann, 2003). A recent analysis of multiple sclerosis (MS) patient tissue discovered that activated transcription factors, such as NF-κB and STATs, are present in MS lesions and peripheral blood mononuclear cells (PBMC) of MS patients (Gobin et al., 2001, Frisullo et al., 2006, Christophi et al., 2008b, Christophi et al., 2011), suggesting that altered regulation of cytokine signaling that occurs in MS patients may be responsible for promoting expression of proinflammatory genes and inflammatory demyelination in MS lesions.
SHP-1, a protein tyrosine phosphatase, contains two SH2 domains and functions as a negative regulator of cytokine signaling through both NF-κB and STATs (David et al., 1995, Jiao et al., 1996). When motheaten mice (me/me), which genetically lack SHP-1 gene expression, are subjected to either virus-induced or autoantigen-induced demyelinating diseases, an increased level of inflammatory demyelination relative to that seen in wild type mice is observed (Deng et al., 2002, Massa et al., 2002). Abnormally low levels of SHP-1 in mice (hypomorphic mutations) also cause increased susceptibility to autoimmune and innate inflammatory disease (Croker et al., 2008, Croker et al., 2011). Moreover, reduced SHP-1 in lymphocytes has been related to human lymphoproliferative diseases (Oka et al., 2002, Oka et al., 2001, Zhang et al., 2000, Koyama et al., 2003). These results indicate that SHP-1 plays an important role in lymphocyte activation and proliferation, which may be important to inflammatory demyelinating immune reactions in white matter observed in MS.
We and others previously described that the anti-inflammatory gene SHP-1 is reduced in PBMC of MS compared to normal subjects both at the mRNA and protein levels which may relate to increased inflammatory activity of these cells in the CNS. Further analysis indicated that both lymphocytes and myeloid cells of MS patients have lower amounts of SHP-1 protein (Christophi et al., 2008c, Feng et al., 2002). Moreover, decreased SHP-1 correlated with specific repression of promoter 2 (hematopoietic specific) relative to promoter 1 (epithelial specific) (Tsui et al., 2002) transcripts (Christophi et al., 2008a, Christophi et al., 2009a). Therefore, reduced promoter 2 transcriptional activity may be responsible for reduced expression of SHP-1 in PBMC in MS.
Several studies have described the reduction of SHP-1 expression in various inflammatory and lymphoproliferative disorders. In the latter, DNA methylation within promoter 2 of the SHP-1 gene was responsible for a strong reduction of SHP-1 mRNA and protein (Howell et al., 2000, Koyama et al., 2003, Oka et al., 2002, Nakase et al., 2009, Ruchusatsawat et al., 2006, Oka et al., 2001). In the present study, we addressed the question of whether similar methylation patterns may relate to our previous observations of decreased expression of promoter 2 transcripts in MS. To do this, we used bisulfite genomic sequencing to analyze DNA methylation of specific and functionally relevant CpG sites within the human SHP-1 promoter 2 in MS subject peripheral blood leukocytes. We discovered that SHP-1 promoter 2 in more than a third of MS subjects is modified by extensive CpG methylation, and that this DNA methylation occurs in a region responsible for reduction of SHP-1 promoter 2 activity.
2. Materials and Methods
Human subjects
Genomic DNA was isolated from buffy coats of sixty-nine MS subjects from the Multiple Sclerosis Research Center of New York (MSRCNY), New York, NY (archival DNA samples provided by Dr. Bernadette Kalman) and nineteen normal subjects (Tables 1 and 2, respectively). All MS subjects had definite MS (McDonald et al., 2001). Six out of the 42 relapsing-remitting MS (RR) subjects (specimen# 2, 7, 13, 21, 23, and 41) (Table 1) were classified as active at the time of drawing blood (defined as having a moderate to severe relapse within the last 6 months). The remaining 36 RR MS subjects were classified as stable.
Table 1.
MS subjects.
| Specimen No. | Gender | Patient's age |
Disease course |
Current EDSS | No. of years with MS |
|---|---|---|---|---|---|
| 1 | F | 57 | SP | 7.5 | 20 |
| 2 | F | 47 | RR | 1.5 | 12 |
| 3 | M | 58 | RR | 4 | 6 |
| 4 | F | 30 | RR | 3 | 5 |
| 5 | F | 35 | RR | 0.5 | 3 |
| 6 | M | 33 | RR | 1 | 4 |
| 7 | M | 27 | RR | 2 | 7 |
| 8 | F | 52 | RR | 0 | 2 |
| 9 | F | 57 | SP | 1 | 5 |
| 10 | F | 59 | SP | 4 | 26 |
| 11 | F | 54 | PP | 7 | 8 |
| 12 | M | 42 | PP | 6.5 | 10 |
| 13 | F | 27 | RR | 0.5 | 0 |
| 14 | M | 48 | PP | 6 | 8 |
| 15 | F | 47 | RR | 2 | 3 |
| 16 | M | 69 | SP | 4.5 | 17 |
| 17 | F | 49 | SP | 5 | 14 |
| 18 | F | 39 | RR | 1 | 5 |
| 19 | F | 30 | RR | 1 | 11 |
| 20 | M | 60 | PP | 6.5 | 27 |
| 21 | F | 50 | RR | 3.5 | 4 |
| 22 | M | 35 | RR | 0.5 | 3 |
| 23 | M | 37 | RR | 2.5 | 3 |
| 24 | M | 37 | RR | 1 | 6 |
| 25 | F | 33 | RR | 1.5 | 6 |
| 26 | F | 57 | RR | 3.5 | 8 |
| 27 | F | 33 | RR | 0 | 2 |
| 28 | F | 35 | RR | 0 | 2 |
| 29 | M | 39 | RR | 1.5 | 8 |
| 30 | F | 36 | RR | 0 | 8 |
| 31 | F | 41 | RR | 0 | 2 |
| 32 | F | 37 | RR | 5.5 | 9 |
| 33 | F | 53 | RR | 3 | 8 |
| 34 | F | 54 | SP | 6.5 | 19 |
| 35 | F | 34 | SP | 4.5 | 8 |
| 36 | F | 48 | RR | 2 | 7 |
| 37 | F | 57 | SP | 4 | 12 |
| 38 | M | 45 | SP | 7 | 13 |
| 39 | F | 48 | RR | 1.5 | 9 |
| 40 | M | 38 | RR | 0.5 | 4 |
| 41 | F | 36 | RR | 1 | 10 |
| 42 | F | 51 | RR | 0.5 | 8 |
| 43 | M | 32 | RR | 0 | 1 |
| 44 | F | 34 | RR | 0 | 7 |
| 45 | M | 48 | PP | 7 | 15 |
| 46 | F | 31 | RR | 0.5 | 3 |
| 47 | F | 45 | RR | 3 | 7 |
| 48 | F | 38 | RR | 0 | 3 |
| 49 | M | 23 | RR | 1 | 4 |
| 50 | F | 31 | RR | 4.5 | 6 |
| 51 | F | 41 | RR | 3 | 7 |
| 52 | M | 44 | PP | 4 | 7 |
| 53 | F | 37 | RR | 0 | 2 |
| 54 | F | 37 | RR | 2 | 6 |
| 55 | F | 49 | SP | 2.5 | 5 |
| 56 | F | 39 | RR | 1.5 | 7 |
| 57 | F | 57 | SP | 6.5 | 8 |
| 58 | M | 43 | RR | 1 | 8 |
| 59 | F | 27 | RR | 0 | 1 |
| 60 | M | 31 | RR | 1 | 4 |
| 61 | F | 40 | RR | 0 | 6 |
| 62 | F | 52 | RR | 0 | 1 |
| 63 | F | 30 | RR | 0 | 1 |
| 64 | F | 57 | PP | 6 | 4 |
| 65 | F | 27 | RR | 0 | 0 |
| 66 | F | 35 | RR | 1 | 6 |
| 67 | F | 31 | RR | 0 | 3 |
| 68 | M | 28 | RR | 4.5 | 6 |
| 69 | F | 71 | SP | 5 | 8 |
| Summary of MS patients information | |||||
| Number of Males | 20 | ||||
| Number of Females | 49 | ||||
| Average age (total) (years) | 42.2 | ||||
| Average EDSS (total) | 2.44 | ||||
| Average years with MS | 7.07 | ||||
| Number of Relapsing/Remitting (RR) MS | 50 | ||||
| Number of Primary Progressive (PP) MS | 7 | ||||
| Number of Secondary progressive (SP) MS | 12 | ||||
DNA was isolated from buffy coat leukocytes. F = female. M = male. SP = Secondary Progressive type. RR = Relapsing/Remitting. PP = Primary Progressive. No. of years of MS was calculated by subtracting year of diagnosis of MS from the year of DNA collection.
Table 2.
Normal Subjects
| Specimen Number |
Normal subject code |
Gender | Age |
|---|---|---|---|
| 1 | C1 | M | 26 |
| 2 | C2 | F | 34 |
| 3 | C3 | F | 42 |
| 4 | C4 | M | 23 |
| 5 | C5 | M | 25 |
| 6 | C6 | F | 25 |
| 7 | C7 | M | 29 |
| 8 | C8 | F | 26 |
| 9 | C11 | M | 56 |
| 10 | C12 | F | 25 |
| 11 | C13 | M | 56 |
| 12 | C14 | M | 71 |
| 13 | C15 | M | 25 |
| 14 | C16 | F | 25 |
| 15 | C17 | M | 31 |
| 16 | C18 | M | 25 |
| 17 | C19 | F | 27 |
| 18 | C23 | F | 25 |
| 19 | C24 | F | 39 |
| Summary of normal subject | |||
| Number of Male Number of Female Average Age (total) |
10 9 33.4 |
||
DNA from all of the samples was isolated from buffy coat. M = male. F = female
Cell lines and PCR primers
Genomic DNA from 293t cells was used as a positive control for SHP-1 promoter 2 methylation (Nakase et al., 2009). The cells were cultured in DMEM (HyClone, Cat. No. SH30243.01) supplemented with 10% FBS and 2mM glutamine. Genomic DNA was isolated using QIAamp DNA mini kit (Qiagen, Cat. No.51304). A 1,738 bp synthetic PCR product that covers the relevant portion of SHP-1 promoter 2 under investigation was used as a negative control of SHP-1 promoter 2 methylation. The latter was amplified using primer pairs SHP1-F (5’-CAC AGT GGC ATG GAC AAA CTG CAT-3’) and SHP1-R (5’-AGC AGG GAG GAG GGA CAT TGA GA-3’). Platinum Taq DNA polymerase, High Fidelity (Invitrogen, Cat. No.11304-011), was used for the PCR. The cycle conditions were 5’ at 94C° for the initial denaturation, 35 cycles of 30 seconds at 94 C°; 30seconds at 65 C°; 30 seconds at 68 C°, followed by 5’ at 68 C° of the final extension.
Cloning and bisulfite sequencing of genomic promoter 2 DNA
One microgram of genomic DNA from MS subjects, normal control subjects, 293t cells, or twenty nanograms of the synthetic 1,738 bp PCR product that covers the SHP-1 promoter 2 was treated with bisulfite using EpiTect Bisulfite Kit (Invitrogen, Cat. No.59104). To analyze the DNA methylation pattern at the SHP-1 promoter 2, three pairs of PCR primers were used as indicated in the text to amplify promoter 2 sequences: Bis-F (CG2)/Bis-R (CG23) pair covers CpG 2 to 23 of SHP-1 promoter 2, Bis-F (CG13)/Bis-R (CG25) pair (Nakase et al., 2009) covers CpG 13 to 25, and Bis-F (CG2)/Bis-R (CG25) pair covers CpG 2 to 25. The sequence and the position of each primer pair are listed in Table 3 and Figure 1, respectively. The PCR mixture consisted of 3 microliters of bisulfite-modified DNA, 1x PCR buffer, 200nM of dNTP mix, 2mM MgSO4, 500nM of each forward and reverse primer, and 1 unit of Platinum Taq DNA polymerase, High Fidelity (Invitrogen, Cat. No.11304-011) in a final volume of 75µl. To avoid possible PCR bias, the final volume of 75µl was divided into 4 tubes. The PCR conditions used were 3 minutes at 94°C for the initial denaturation, 45 cycles of 30 seconds at 94°C; 30seconds at 55°C for Bis-F (CG2)/Bis-R (CG23) pair, 57°C for Bis-F (CG13)/Bis-R (CG25) pair, and 60°C for Bis-F (CG2)/Bis-R (CG25) pair; 1’ at 68°C, was followed by 5’ at 68°C for the final extension. PCR products were gel purified using QIAEX II Gel Extraction kit (Qiagen, Cat. No.20021), cloned into pGEM-T vector (Promega, Cat. No.A3600), transformed into NEB 5-alpha competent E. coli (High Efficiency) (NEB, Cat. No.C2987). Approximately twenty bacteria colonies from multiple sclerosis and normal control subjects, or ten colonies from 293t cells (positive control) and the synthetic 1,738 bp PCR product that covers SHP-1 promoter 2 (negative control), were picked for DNA bisulfite sequencing. Plasmid DNA was isolated with Fast Plasmid mini Kit (5 prime, Cat. No.2300010) and was commercially sequenced (Genewiz, South Plainfield, NJ). BiQ analyzer software (http://biq-analyzer.bioinf.mpiinf.mpg.de/index.php) was used for methylation analysis of each clone. Up to 20 clones for each MS and control subject were produced for analysis in multiple rounds of bisulfite sequencing of sixty-nine MS and nineteen normal control subjects. To assess reliability of each bisulfite sequencing, we included 293t cell genomic DNA (positive methylation control) and a 1,738 bp synthetic PCR-generated sequence (negative methylation control) for each round.
Table 3.
Primers used for Bisulfite sequencing.
| Primers | Primer sequence |
|---|---|
| Bis-F (CG2) | 5’-GGT GAG AAA TTA ATT AGA TAA GG-3’ |
| Bis-F (CG13) | 5’-ATG TTT TTG AGT TTT TGA TTG TAG A-3’ |
| Bis-R (CG23) | 5’-CAA ACA AAA AAC TCA AAA AC-3’ |
| Bis-R (CG25) | 5’-AAT AAC AAA CCC TTA CCT CAC CAT-3’ |
Three combinations of primer pairs used for Bisulfite genomic sequencing experiments are 1) Bis-F (CG13)/Bis-R (CG25), 2) Bis-F (CG2)/Bis-R (CG23) and 3) Bis-F (CG2)/Bis-R (CG25). CG of CG2, CG13, CG23 and CG25 indicates the position of CpG sites on promoter 2 studied for the assay (see Figure 1 for primer orientation and targets).
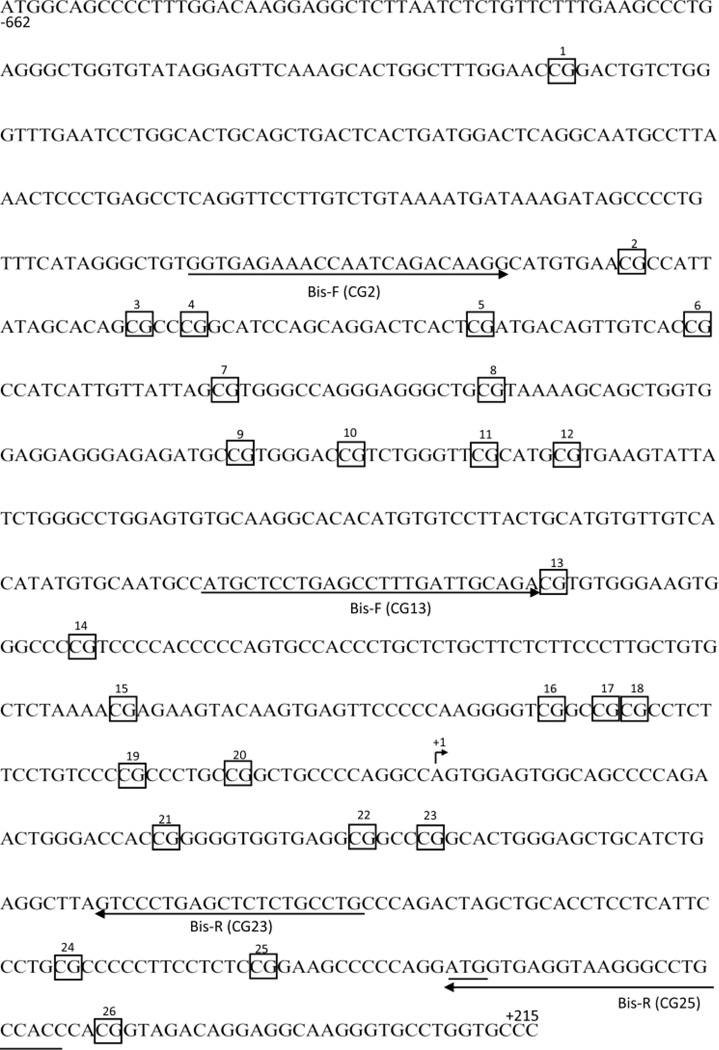
Figure 1.
Assay designs for analysis of SHP-1 gene promoter 2 methylation by Bisulfite genomic sequencing. Arrows indicate placement and direction of PCR primers. Three primer pairs were used to analyze 26 potential methylation sites in the human SHP-1 gene promoter 2 (boxed CpGs are numbered 1–26). Primers, Bis-F (CG13) and Bis-R (CG25) are from Nakase, K et al. 2009. Bis-F (CG2) and Bis-R (CG23) are designed using BiSearch online software. Both the transcriptional and translational start sites are labeled +1 and by underlining, respectively. The nucleotide numbering at the beginning and end of the promoter region displayed is relative to the transcriptional start site (+1).
Statistical analysis of SHP-1 promoter 2 methylation
After bisulfite sequencing of DNA clones, the percent methylation (# methylated sites per total number of CpG sites) of each clone was scored. For each subject, the frequency of clones that were methylated at 30%, 40%, 50%, and 60% of the total CpG residues was determined, in the region spanning CpG 2–23. We did not determine methylation status beyond the 60% level because previous reports have reported that this is a stringent functional criterion (Nakase et al., 2009). Statistical comparisons of the relative frequencies of subjects that met these criteria in the MS and control groups were performed using a Fisher's Exact Test. We also calculated an odds ratio to determine if there was an increased probability of MS subjects achieving different levels of methylation compared to controls using the Haldane modification when necessary to account for zero values in the ratios. These odds ratios were determined to be significant at the p < 0.05 level when the lower limit of the 95% confidence interval exceeded 1.0 (Table 4A). For comparison, we also determined the relative frequencies of positive and negative control DNA clones that met the same criteria (Table 4B). In addition to the subject-level analyses, we also compared the MS and control groups using the Fisher’s Exact Test and Haldane odds ratios based on the results of individual clones, in the region spanning CpG 13–25 (Table 5A), and listed the results from the positive and negative control DNA clones separately (Table 5B).
Table 4.
| A Percentage of subjects with various levels of promoter 2 methylation | ||||
|---|---|---|---|---|
| Subjects (Ss) | ||||
| Normal (N=19) |
MS (N=58) |
Fisher's Exact Test P value |
Haldane Odds Ratio (95th CI) |
|
| Total # clones sequenced | 568 | 1428 | ||
| Ss w/clones > 30% meth | 10.5% (2/19) | 39.7% (23/58) | 0.01522 | 4.6 (1.1–19.2) |
| Ss w/clones > 40% meth | 0.0% (0/19) | 37.9% (22/58) | 0.00055 | 24.0 (1.4–418.1) |
| Ss w/clones > 50% meth | 0.0% (0/19) | 36.2% (21/58) | 0.00084 | 22.4 (1.3–389.2) |
| Ss w/clones > 60% meth | 0.0% (0/19) | 36.2% (21/58) | 0.00084 | 22.4 (1.3–389.2) |
| B Percentage of clones with various levels of promoter 2 methylation in NegCtrl (1,738) and PosCtrl (293t) DNA | ||
|---|---|---|
| % NegCtrl (1738) clones |
% PosCtrl (293) clones |
|
| Total # clones sequenced | 178 | 216 |
| clones > 30% meth | 0.0% (0/178) | 77.3% (167/216) |
| clones > 40% meth | 0.0% (0/178) | 76.9% (166/216) |
| clones > 50% meth | 0.0% (0/178) | 75.9% (164/216) |
| clones > 60% meth | 0.0% (0/178) | 72.2% (156/216) |
Bisulfite sequencing results from primer pairs that span CpG 2–23 and 2–25 were combined for this analysis. Note the increased percentage of MS subjects with clones displaying elevated promoter 2 methylation compared to normal subjects. PosCtrl=Positive Control. NegCtrl=1738bp synthetic DNA negative control. Meth=methylation. 95th CI=95th percentile confidence interval. Ss=subjects.
Table 5.
| A Incidence of highly-methylated promoter 2 clones in MS and Normal Ss | ||||
|---|---|---|---|---|
| Subjects (Ss) | ||||
| Normal (N=14) |
MS (N=29) |
Fisher's Exact Test P value |
Haldane Odds Ratio (95th CI) |
|
| Total # clones sequenced | 568 | 870 | ||
| clones > 50% meth | 0.97% (5/518) | 5.29% (46/870) | 0.00001 | 5.3 (2.2–12.8) |
| clones > 60% meth | 0.97% (5/518) | 5.06% (44/870) | 0.00002 | 5.0 (2.1–12.3) |
| B Percentage of clones with various levels of promoter 2 methylation in NegCtrl (1,738) and PosCtrl (293t) DNA | ||
|---|---|---|
| % NegCtrl (1738) clones |
% PosCtrl (293) clones |
|
| Total # clones sequenced | 147 | 204 |
| clones > 50% meth | 0.0% (0/147) | 83.3% (170/204) |
| clones > 60% meth | 0.0% (0/147) | 81.9% (167/204) |
The results from bisulfite sequencing using the two primer sets (Figure 1) that span CpG 13–25 were used for the analysis. PosCtrl: Positive Control. NegCtrl=1738bp synthetic DNA negative control. Ss=subjects.
3. Results
Subjects
The total of sixty-nine MS subjects (twenty males and forty-nine females) and nineteen normal subjects (ten males and nine females) were included in the study (Tables 1 and 2). The average age of normal subjects was 33.4 and the average age of MS subjects studied was 42.2. The MS subjects included seven primary progressive type (PP), fifty relapsing/remitting type (RR) and twelve secondary progressive type (SP) MS.
Significant Difference in SHP-1 Promoter Methylation Between MS and Normal Subjects
Previous studies have indicated that abnormally high CpG DNA methylation of the SHP-1 gene promoter 2 (Figure 1) was associated with a reduction of SHP-1 expression in B and T cell lymphoma (Zhang et al., 2005, Koyama et al., 2003). Therefore, CpG methylation in this region in fifty-eight MS subjects (1,428 total clones) and nineteen normal subjects (568 total clones) was determined and a frequency distribution analysis at the subject level was performed. The analysis revealed that 39.7% of MS subjects possessed at least one clone with >30% methylation of the promoter compared with 10.5% of control subjects (Table 4A). An odds ratio (OR) and Fisher’s exact test was used to compare the relative numbers of subjects within the >30% methylation category. Overall, MS subjects were estimated to be nearly 5 times more likely to meet this criterion compared with control subjects (OR = 4.6, 95th confidence interval (CI), 1.1 – 19.2, p < 0.05). The results of the Fisher’s exact test confirmed a highly significant increase in the relative frequency of >30% promoter 2 methylation in the MS compared to normal control subjects (p = 0.015) (Table 4A). The differences between MS and control subjects were even more significant at the 40%, 50% and 60% methylation level, where the odds of an MS subject meeting these criteria were more than 20 times that of control subjects (Table 4A). Specifically, 38% of MS subjects (and 0% of normal subjects) had clones with 40% methylation of promoter 2, and 36% of MS patients contained clones with either 50% or 60% methylation of promoter 2 compared to 0% of normal subjects. In agreement with the odds ratio analysis, these differences were highly significant by Fisher’s exact test (40% cutoff p = 0.00055, 50% or 60% cutoff p = 0.00084) (Table 4A). Notably, 77% of the clones from the positive 293t cell control DNA sample and 0% of the negative control DNA sample had clones meeting the 30% methylation criteria. Thus, MS subjects possessed slightly more than one half the frequency of promoter 2 methylated clones seen in the hypermethylated 293t cells (Table 4B).
Significant methylation within the functional core of SHP-1 promoter 2
A previous analysis that related the extent of DNA methylation of individual promoter 2 clones from transformed lymphoblastoid cells to silencing of SHP-1 expression demonstrated that, though 30% methylation of CpG13–25 of SHP-1 promoter 2 was sufficient to suppress SHP-1 expression by 30% of normal, 60% methylation of this region was sufficient to substantially repress transcription of the SHP-1 gene to a third of normal levels (Nakase et al., 2009). To determine whether DNA methylation observed in clones of the MS subjects reached this functional level, percent DNA methylation within the CpG13–25 region of SHP-1 promoter 2 within the different groups was analyzed. This group analysis consisted of 870 clones from twenty-nine MS subjects, 518 clones from fourteen normal subjects, 204 clones of methylation positive control (293t cell DNA) and 147 clones of the synthetic DNA negative control. We observed rare clones (<1%) from the normal subject group having greater than 50% methylation (5 clones out of 518 clones examined) (Table 5A). In contrast, approximately 5% of clones from the MS subject groups had greater than 50%, or even 60% methylation, in contrast to the <1% from the normal subject group. The odds ratio analysis produced an OR for 50% cutoff of 5.3 (with 95th CI, 2.2–12.8, p <0.05), and an OR for 60% methylation cutoff level of 5.0 (with 95th CI, 2.1 – 12.3, p <0.05). Thus, MS subjects are 5 times more likely to have clones with at least 60% methylation at CpG13–25 compared with normal control subjects. The result of the odds ratio analysis was confirmed by Fisher’s exact test (p<0.00001 for 50% cutoff and p <0.00002 for 60% cutoff) (Table 5).
Relationships between DNA methylation and subject parameters
Finally, we analyzed a potential correlation between frequency of highly methylated SHP-1 promoter clones in MS subjects with different MS disease type, years with MS, and expanded disability status scale (EDSS) score (Table 6). For each subject, we calculated % clone frequency (the number of clones that had >30% methylation / the total number of clones studied in a particular subject). From regression analysis of scatter plots (not shown), no association between the frequency of highly methylated clones in particular MS subjects with any subject parameter including EDSS (R2= 0.039), subject age (R2= 0.01), or years with MS (R2= 0.029) could be detected. Finally, there was no significant difference in the average frequency of clones with high promoter methylation (>30% methylation) between MS subjects with either active (n=6) or stable (n=36) relapsing-remitting MS analyzed by Student’s t-test (3.8% vs. 4.9%; p=0.79). Further, there was no significant difference in the proportion of subjects with either active or stable relapsing-remitting MS that displayed clones >30% versus <30% methylation (Active 1/5 vs. Stable 16/20; Fisher’s exact test p=0.37).
Table 6.
% DNA methylation from bisulfite sequencing and MS subject parameters.
| Specimen No. |
% freq of clones with >30% met |
Gender | Patient's age |
Yrs with MS |
Disease course |
Current EDSS |
|---|---|---|---|---|---|---|
| 65 | 33% | F | 27 | 0 | RR | 0 |
| 42 | 5% | F | 51 | 8 | RR | 0.5 |
| 47 | 0% | F | 45 | 7 | RR | 3 |
| 36 | 0% | F | 48 | 7 | RR | 2 |
| 29 | 0% | M | 39 | 8 | RR | 1.5 |
| 26 | 19% | F | 57 | 8 | RR | 3.5 |
| 46 | 0% | F | 31 | 3 | RR | 0.5 |
| 48 | 7% | F | 38 | 3 | RR | 0 |
| 21 | 0% | F | 50 | 4 | RR | 3.5 |
| 41 | 23% | F | 36 | 10 | RR | 1 |
| 63 | 0% | F | 30 | 1 | RR | 0 |
| 27 | 0% | F | 33 | 2 | RR | 0 |
| 59 | 5% | F | 27 | 1 | RR | 0 |
| 44 | 7% | F | 34 | 7 | RR | 0 |
| 23 | 0% | M | 37 | 3 | RR | 2.5 |
| 35 | 0% | F | 34 | 8 | SP | 4.5 |
| 30 | 10% | F | 36 | 8 | RR | 0 |
| 61 | 0% | F | 40 | 6 | RR | 0 |
| 43 | 0% | M | 32 | 1 | RR | 0 |
| 60 | 8% | M | 31 | 4 | RR | 1 |
| 69 | 0% | F | 71 | 8 | SP | 5 |
| 33 | 0% | F | 53 | 8 | RR | 3 |
| 28 | 8% | F | 35 | 2 | RR | 0 |
| 39 | 0% | F | 48 | 9 | RR | 1.5 |
| 31 | 0% | F | 41 | 2 | RR | 0 |
| 22 | 5% | M | 35 | 3 | RR | 0.5 |
| 62 | 19% | F | 52 | 1 | RR | 0 |
| 38 | 4% | M | 45 | 13 | SP | 7 |
| 45 | 0% | M | 48 | 15 | PP | 7 |
| 64 | 5% | F | 57 | 4 | PP | 6 |
| 7 | 0% | M | 27 | 7 | RR | 2 |
| 1 | 0% | F | 57 | 20 | SP | 7.5 |
| 6 | 0% | M | 33 | 4 | RR | 1 |
| 17 | 0% | F | 49 | 14 | SP | 5 |
| 25 | 0% | F | 33 | 6 | RR | 1.5 |
| 15 | 0% | F | 47 | 3 | RR | 2 |
| 34 | 0% | F | 54 | 19 | SP | 6.5 |
| 20 | 5% | M | 60 | 27 | PP | 6.5 |
| 5 | 0% | F | 35 | 3 | RR | 0.5 |
| 32 | 5% | F | 37 | 9 | RR | 5.5 |
| 14 | 0% | M | 48 | 8 | PP | 6 |
| 2 | 0% | F | 47 | 12 | RR | 1.5 |
| 13 | 0% | F | 27 | 0 | RR | 0.5 |
| 3 | 15% | M | 58 | 6 | RR | 4 |
| 12 | 2% | M | 42 | 10 | PP | 6.5 |
| 8 | 0% | F | 52 | 2 | RR | 0 |
| 11 | 2% | F | 54 | 8 | PP | 7 |
| 37 | 9% | F | 57 | 12 | SP | 4 |
| 19 | 0% | F | 30 | 11 | RR | 1 |
| 10 | 0% | F | 59 | 26 | SP | 4 |
| 9 | 0% | F | 57 | 5 | SP | 1 |
| 16 | 0% | M | 69 | 17 | SP | 4.5 |
| 18 | 2% | F | 39 | 5 | RR | 1 |
| 67 | 26% | F | 31 | 3 | RR | 0 |
| 4 | 0% | F | 30 | 5 | RR | 3 |
| 40 | 0% | M | 38 | 4 | RR | 0.5 |
| 66 | 4% | F | 35 | 6 | RR | 1 |
| 24 | 0% | M | 37 | 6 | RR | 1 |
% of colonies that had greater than 30% methylation from bisulfite sequencing analysis of CpG2–23 for each MS patient was tested for correlation with various MS subject parameters.
4. Discussion
DNA methylation is an epigenetic alteration that results in both acquired and heritable gene silencing and is critical for developmental progression, cell fate determination, and various diseases including hematopoietic cancers (Rush and Plass, 2002, Kanai, 2010, Issa, 2004, Egger et al., 2004, Tost, 2010, Vaissiere et al., 2008) and autoimmunity including MS (Burrell et al., 2011, Liggett et al., 2010, Meda et al., 2011, Brooks et al., 2010, Pedre et al., 2011, Casaccia-Bonnefil et al., 2008, Baranzini et al., 2010). Studies indicate that the extent of promoter methylation that inhibits gene expression differs depending on type of gene, ranging from 9% for RB1 gene (Ohtani-Fujita et al., 1997) to 84% for MLH1 gene in microsatellite-unstable colorectal tumor (Herman et al., 1998). In hematopoietic cancers, CpG methylation at the SHP-1 core promoter of approximately 60% substantially reduced SHP-1 gene expression to a third of normal levels (Oka et al., 2002, Koyama et al., 2003, Nakase et al., 2009, Zhang et al., 2005). The present studies have revealed a similar level of methylation of the SHP-1 promoter in a fraction of hematopoietic cells from MS subjects which may have functional relevance to disease.
Although SHP-1 is believed to be an important tumor suppressor in hematopoietic cells and transcriptional silencing promotes lymphoblastoid cell proliferation, the main function of SHP-1 determined from animal studies with null or hypomorphic mutations in the SHP-1 gene is to attenuate leukocyte activation and immune-mediated inflammation (Deng et al., 2002, Massa et al., 2002, Pao et al., 2007, Croker et al., 2008). As such, it was of particular interest that we previously described that PBMC of MS subjects had reduced levels of SHP-1 mRNA (specifically of promoter 2 transcripts) and protein relative to those of normal subjects (Christophi et al., 2009b, Christophi et al., 2008c). Consequently, it was of particular interest to determine whether DNA methylation of transcriptionally relevant CpG sites within the SHP-1 promoter 2 (Nakase et al., 2009) may be more common in MS compared to normal subjects. Statistical analysis of methylation with the CpG2–23 region performed at the subject level indicated that MS patients are nearly 5 times more likely to possess 30% or greater methylation within the SHP-1 promoter 2 compared to normal subjects. At more stringent levels, the group differences were even more robust with a highly-significant 20-fold elevation in the likelihood of having 40, 50, or 60% methylation in the SHP-1 promoter in MS subjects versus controls. Importantly, we also observed a highly-significant increase in percent DNA methylation in MS patients compared to normal subjects within the critical CpG13–25 region of the SHP-1 promoter 2. Of particular note, 60% methylation in this restricted region of the proximal promoter is sufficient to repress transcription from promoter 2 to a third of normal levels (Nakase et al., 2009). Assuming that individual clones of methylated genomic DNA are derived from a fraction of individual leukocytes in buffy coat samples from MS subjects, we propose that MS subjects are more likely to contain leukocytes in which SHP-1 is functionally repressed. Further examination of this question will require comparative analysis of percent methylation of promoter 2 in both silenced leukocytes and leukocytes with normal levels of SHP-1 expression within individual subjects.
Because we can detect on average a 5% frequency of highly methylated clones (>60% methylation) in the MS subject group (Table 5), we may hypothesize that SHP-1 is functionally reduced in approximately 5% of the leukocytes in the blood of those with MS. In assessing the functional significance of this methylation, whether these leukocytes constitute autoreactive cells with increased inflammatory potential would be particularly important (Deng et al., 2003, Stefanova et al., 2003). For instance, the frequency of myelin reactive T cells in MS to any particular antigenic epitope is generally less than 1 in 10,000 of total T cells in the blood (0. 01%) (Hellings et al., 2001, de Rosbo and Ben-Nun, 1998). As a consequence, functional repression of SHP-1 in 5% of the peripheral leukocytes may potentially accommodate increased inflammatory activity in multiple lineages of pathogenic T cells directed towards hundreds of different myelin antigenic epitopes that could potentially be sufficient to mediate inflammatory demyelinating disease in MS. In future studies, it will be important to determine whether SHP-1-deficient leukocytes significantly overlap with these pathogenic populations of lymphocytes in the MS population.
We observed that DNA methylation seen in MS subjects includes substantial methylation within the critical CpG13–25 region containing the binding sites for various transcription factors, such as Sp1, Oct-1, NF-κB, PU.1 and CREB-1 important for the expression of SHP-1 (Cheng et al., 2007, Nakase et al., 2009, Wlodarski et al., 2007). Because many of these CpG sites are either adjacent or overlapping these transcriptional elements, increased methylation blocks factor binding and interferes with transcriptional activation of the SHP-1 gene (Nakase et al., 2009). Consistent with this hypothesis, the CpG13–25 region is critical for HTLV-1 Tax-mediated suppression of SHP-1 expression which acted at a NF-κB site to recruit NF-κB, Tax, histone deacetylase (HDAC1), and DNA methyltransferase (DNMT1) to increase methylation of promoter 2 (Nakase et al., 2009, Cheng et al., 2007). These studies suggest similar pathways for acquired DNA methylation at the SHP-1 promoter 2 that may lead to gene repression during development of inflammatory diseases. We propose that such acquired SHP-1 deficiency in leukocytes may impart susceptibility to autoimmunity as recently described in the mouse (Croker et al., 2008) rather than being directly related to changes in disease activity as suggested in Table 6. The latter possibility is supported by a lack of significant difference in high promoter methylation between active and stable relapsing-remitting MS subjects suggesting that SHP-1 promoter methylation may be independent of active disease in this group. However, we hesitate to draw any firm conclusions from this exploratory analysis due to the small number of active relapsing-remitting MS subjects included in our study.
Even if SHP-1 is not a primary susceptibility gene for MS, SHP-1 could play a subsequent role in some aspects of MS disease because SHP-1 controls the expression of multiple proinflammatory pathways in both lymphocytes and myeloid cells (Kruger et al., 2000, Zhou et al., 2010, Pao et al., 2007, Christophi et al., 2009b, Sathish et al., 2007, Johnson et al., 1999). Epigenetic alteration is potentially a reversible process, and some epigenetic drugs are being tested in clinical trials for the treatment of hematopoietic malignancies (Egger et al., 2004). Studies demonstated that the activity of proinflammatory signaling molecules, such as NF-κB and STATs, which are negatively regulated by SHP-1, are increased in MS leukocytes and CNS lesions (Christophi et al., 2009b, Frisullo et al., 2006, Gobin et al., 2001). Even though such repression may be limited to a subset of MS patients, if MS patients were screened for epigenetic modification and specific ways to reverse epigenetic changes were discovered, it may be possible to alter the inflammatory disease process in some MS subjects.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baranzini SE, Mudge J, Van Velkinburgh JC, Khankhanian P, Khrebtukova I, Miller NA, Zhang L, Farmer AD, Bell CJ, Kim RW, May GD, Woodward JE, Caillier SJ, Mcelroy JP, Gomez R, Pando MJ, Clendenen LE, Ganusova EE, Schilkey FD, Ramaraj T, Khan OA, Huntley JJ, Luo S, Kwok PY, Wu TD, Schroth GP, Oksenberg JR, Hauser SL, Kingsmore SF. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464:1351–1356. doi: 10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks WH, Le Dantec C, Pers JO, Youinou P, Renaudineau Y. Epigenetics and autoimmunity. J Autoimmun. 2010;34:J207–J219. doi: 10.1016/j.jaut.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Burrell AM, Handel AE, Ramagopalan SV, Ebers GC, Morahan JM. Epigenetic mechanisms in multiple sclerosis and the major histocompatibility complex (MHC) Discov Med. 2011;11:187–196. [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Pandozy G, Mastronardi F. Evaluating epigenetic landmarks in the brain of multiple sclerosis patients: a contribution to the current debate on disease pathogenesis. Prog Neurobiol. 2008;86:368–378. doi: 10.1016/j.pneurobio.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kydd AR, Nakase K, Noonan KM, Murakami A, Tao H, Dwyer M, Xu C, Zhu Q, Marasco WA. Negative regulation of the SH2-homology-containing protein-tyrosine phosphatase-1 (SHP-1) P2 promoter by the HTLV-1 tax oncoprotein. Blood. 2007 doi: 10.1182/blood-2006-11-058388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophi GP, Gruber RC, Panos M, Christophi RL, Jubelt B, Massa PT. Interleukin-33 upregulation in peripheral leukocytes and CNS of multiple sclerosis patients. Clin Immunol. 2011 doi: 10.1016/j.clim.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophi GP, Hudson CA, Gruber R, Christophi CP, Massa PT. Promoter-specific induction of the phosphatase SHP-1 by viral infection and cytokines in CNS glia. J Neurochem. 2008a;105:2511–2523. doi: 10.1111/j.1471-4159.2008.05337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophi GP, Hudson CA, Gruber RC, Christophi CP, Mihai C, Mejico LJ, Jubelt B, Massa PT. SHP-1 deficiency and increased inflammatory gene expression in PBMCs of multiple sclerosis patients. Lab Invest. 2008b;88:243–255. doi: 10.1038/labinvest.3700720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophi GP, Hudson CA, Gruber RC, Christophi CP, Mihai C, Mejico LJ, Jubelt B, Massa PT. SHP-1 deficiency and increased inflammatory gene expression in PBMCs of multiple sclerosis patients. Lab Invest. 2008c;88:243–255. doi: 10.1038/labinvest.3700720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophi GP, Hudson CA, Panos M, Gruber R, Massa PT. Modulation of macrophage infiltration and inflammatory activity by the phosphatase SHP-1 in virus-induced demyelinating disease. J Virol. 2009a;83:522–539. doi: 10.1128/JVI.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophi GP, Panos M, Hudson CA, Christophi RL, Gruber RC, Mersich AT, Blystone SD, Jubelt B, Massa PT. Macrophages of multiple sclerosis patients display deficient SHP-1 expression and enhanced inflammatory phenotype. Lab Invest. 2009b;89:742–759. doi: 10.1038/labinvest.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker BA, Lawson BR, Berger M, Eidenschenk C, Blasius AL, Moresco EM, Sovath S, Cengia L, Shultz LD, Theofilopoulos AN, Pettersson S, Beutler BA. Inflammation and autoimmunity caused by a SHP1 mutation depend on IL-1, MyD88, and a microbial trigger. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0806619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker BA, Lewis RS, Babon JJ, Mintern JD, Jenne DE, Metcalf D, Zhang JG, Cengia LH, O'Donnell JA, Roberts AW. Neutrophils require SHP1 to regulate IL-1beta production and prevent inflammatory skin disease. J Immunol. 2011;186:1131–1139. doi: 10.4049/jimmunol.1002702. [DOI] [PubMed] [Google Scholar]
- David M, Chen HE, Goelz S, Larner AC, Neel BG. Differential regulation of the alpha/beta interferon-stimulated Jak/Stat pathway by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol.Cell.Biol. 1995;15:7050–7058. doi: 10.1128/mcb.15.12.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosbo NK, Ben-Nun A. T-cell responses to myelin antigens in multiple sclerosis; relevance of the predominant autoimmune reactivity to myelin oligodendrocyte glycoprotein. J Autoimmun. 1998;11:287–299. doi: 10.1006/jaut.1998.0202. [DOI] [PubMed] [Google Scholar]
- Deng C, Minguela A, Hussain RZ, Lovett-Racke AE, Radu C, Ward ES, Racke MK. Expression of the tyrosine phosphatase SRC homology 2 domain-containing protein tyrosine phosphatase 1 determines T cell activation threshold and severity of experimental autoimmune encephalomyelitis. J Immunol. 2002;168:4511–4518. doi: 10.4049/jimmunol.168.9.4511. [DOI] [PubMed] [Google Scholar]
- Deng C, Wu B, Yang H, Hussain RZ, Lovett-Racke AE, Christadoss P, Racke MK. Decreased expression of Src homology 2 domain-containing protein tyrosine phosphatase 1 reduces T cell activation threshold but not the severity of experimental autoimmune myasthenia gravis. J Neuroimmunol. 2003;138:76–82. doi: 10.1016/s0165-5728(03)00119-x. [DOI] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Feng X, Petraglia A, Chen M, Byskosh P, Boos M, Reder A. Low expression of interferon-stimulated genes in active multiple sclerosis is linked to subnormal phosphorylation of STAT1. J Neuroimmunol. 2002;129:205. doi: 10.1016/s0165-5728(02)00182-0. [DOI] [PubMed] [Google Scholar]
- Frisullo G, Angelucci F, Caggiula M, Nociti V, Iorio R, Patanella AK, Sancricca C, Mirabella M, Tonali PA, Batocchi AP. pSTAT1, pSTAT3, and T-bet expression in peripheral blood mononuclear cells from relapsing-remitting multiple sclerosis patients correlates with disease activity. J Neurosci Res. 2006;84:1027–1036. doi: 10.1002/jnr.20995. [DOI] [PubMed] [Google Scholar]
- Gobin SJ, Montagne L, Van Zutphen M, Van Der Valk P, Van Den Elsen PJ, De Groot CJ. Upregulation of transcription factors controlling MHC expression in multiple sclerosis lesions. Glia. 2001;36:68–77. doi: 10.1002/glia.1096. [DOI] [PubMed] [Google Scholar]
- Hellings N, Baree M, Verhoeven C, D'Hooghe MB, Medaer R, Bernard CC, Raus J, Stinissen P. T-cell reactivity to multiple myelin antigens in multiple sclerosis patients and healthy controls. J Neurosci Res. 2001;63:290–302. doi: 10.1002/1097-4547(20010201)63:3<290::AID-JNR1023>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BW, Herrick TM, Hildebrand JD, Zhang Y, Cooper JA. Dab1 tyrosine phosphorylation sites relay positional signals during mouse brain development. Curr Biol. 2000;10:877–885. doi: 10.1016/s0960-9822(00)00608-4. [DOI] [PubMed] [Google Scholar]
- Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- Jiao H, Berrada K, Yang W, Tabrizi M, Platanias LC, Yi T. Direct association with and dephosphorylation of Jak2 kinase by the SH2-domain-containing protein tyrosine phosphatase SHP-1. Mol Cell Biol. 1996;16:6985–6992. doi: 10.1128/mcb.16.12.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KG, Leroy FG, Borysiewicz LK, Matthews RJ. TCR signaling thresholds regulating T cell development and activation are dependent upon SHP-1. J Immunol. 1999;162:3802–3813. [PubMed] [Google Scholar]
- Kanai Y. Genome-wide DNA methylation profiles in precancerous conditions and cancers. Cancer Sci. 2010;101:36–45. doi: 10.1111/j.1349-7006.2009.01383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornek B, Lassmann H. Neuropathology of multiple sclerosis-new concepts. Brain Res Bull. 2003;61:321–326. doi: 10.1016/s0361-9230(03)00095-9. [DOI] [PubMed] [Google Scholar]
- Koyama M, Oka T, Ouchida M, Nakatani Y, Nishiuchi R, Yoshino T, Hayashi K, Akagi T, Seino Y. Activated proliferation of B-cell lymphomas/leukemias with the SHP1 gene silencing by aberrant CpG methylation. Lab Invest. 2003;83:1849–1858. doi: 10.1097/01.lab.0000106503.65258.2b. [DOI] [PubMed] [Google Scholar]
- Kruger J, Butler JR, Cherapanov V, Dong Q, Ginzberg H, Govindarajan A, Grinstein S, Siminovitch KA, Downey GP. Deficiency of Src homology 2-containing phosphatase 1 results in abnormalities in murine neutrophil function: studies in motheaten mice. J Immunol. 2000;165:5847–5859. doi: 10.4049/jimmunol.165.10.5847. [DOI] [PubMed] [Google Scholar]
- Liggett T, Melnikov A, Tilwalli S, Yi Q, Chen H, Replogle C, Feng X, Reder A, Stefoski D, Balabanov R, Levenson V. Methylation patterns of cell-free plasma DNA in relapsing-remitting multiple sclerosis. J Neurol Sci. 2010;290:16–21. doi: 10.1016/j.jns.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa PT, Ropka SL, Saha S, Fecenko KL, Beuler KL. Critical role for protein tyrosine phosphatase SHP-1 in controlling infection of central nervous system glia and demyelination by Theiler's murine encephalomyelitis virus. J Virol. 2002;76:8335–8346. doi: 10.1128/JVI.76.16.8335-8346.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, Mcfarland HF, Paty DW, Polman CH, Reingold SC, Sandberg-Wollheim M, Sibley W, Thompson A, Van Den Noort S, Weinshenker BY, Wolinsky JS. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50:121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- Meda F, Folci M, Baccarelli A, Selmi C. The epigenetics of autoimmunity. Cell Mol Immunol. 2011;8:226–236. doi: 10.1038/cmi.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase K, Cheng J, Zhu Q, Marasco WA. Mechanisms of SHP-1 P2 promoter regulation in hematopoietic cells and its silencing in HTLV-1-transformed T cells. J Leukoc Biol. 2009;85:165–174. doi: 10.1189/jlb.0608383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani-Fujita N, Dryja TP, Rapaport JM, Fujita T, Matsumura S, Ozasa K, Watanabe Y, Hayashi K, Maeda K, Kinoshita S, Matsumura T, Ohnishi Y, Hotta Y, Takahashi R, Kato MV, Ishizaki K, Sasaki MS, Horsthemke B, Minoda K, Sakai T. Hypermethylation in the retinoblastoma gene is associated with unilateral, sporadic retinoblastoma. Cancer Genet Cytogenet. 1997;98:43–49. doi: 10.1016/s0165-4608(96)00395-0. [DOI] [PubMed] [Google Scholar]
- Oka T, Ouchida M, Koyama M, Ogama Y, Takada S, Nakatani Y, Tanaka T, Yoshino T, Hayashi K, Ohara N, Kondo E, Takahashi K, Tsuchiyama J, Tanimoto M, Shimizu K, Akagi T. Gene silencing of the tyrosine phosphatase SHP1 gene by aberrant methylation in leukemias/lymphomas. Cancer Res. 2002;62:6390–6394. [PubMed] [Google Scholar]
- Oka T, Yoshino T, Hayashi K, Ohara N, Nakanishi T, Yamaai Y, Hiraki A, Sogawa CA, Kondo E, Teramoto N, Takahashi K, Tsuchiyama J, Akagi T. Reduction of hematopoietic cell-specific tyrosine phosphatase SHP-1 gene expression in natural killer cell lymphoma and various types of lymphomas/leukemias : combination analysis with cDNA expression array and tissue microarray. Am J Pathol. 2001;159:1495–1505. doi: 10.1016/S0002-9440(10)62535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao LI, Lam KP, Henderson JM, Kutok JL, Alimzhanov M, Nitschke L, Thomas ML, Neel BG, Rajewsky K. B cell-specific deletion of protein-tyrosine phosphatase Shp1 promotes B-1a cell development and causes systemic autoimmunity. Immunity. 2007;27:35–48. doi: 10.1016/j.immuni.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Pedre X, Mastronardi F, Bruck W, Lopez-Rodas G, Kuhlmann T, Casaccia P. Changed histone acetylation patterns in normal-appearing white matter and early multiple sclerosis lesions. J Neurosci. 2011;31:3435–3445. doi: 10.1523/JNEUROSCI.4507-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchusatsawat K, Wongpiyabovorn J, Shuangshoti S, Hirankarn N, Mutirangura A. SHP-1 promoter 2 methylation in normal epithelial tissues and demethylation in psoriasis. J Mol Med. 2006;84:175–182. doi: 10.1007/s00109-005-0020-6. [DOI] [PubMed] [Google Scholar]
- Rush LJ, Plass C. Alterations of DNA methylation in hematologic malignancies. Cancer Lett. 2002;185:1–12. doi: 10.1016/s0304-3835(02)00288-4. [DOI] [PubMed] [Google Scholar]
- Sathish JG, Dolton G, Leroy FG, Matthews RJ. Loss of Src homology region 2 domain-containing protein tyrosine phosphatase-1 increases CD8+ T cell-APC conjugate formation and is associated with enhanced in vivo CTL function. J Immunol. 2007;178:330–337. doi: 10.4049/jimmunol.178.1.330. [DOI] [PubMed] [Google Scholar]
- Stefanova II, Hemmer B, Vergelli M, Martin R, Biddison WE, Germain RN. TCR ligand discrimination is enforced by competing ERK positive and SHP-1 negative feedback pathways. Nat Immunol. 2003 doi: 10.1038/ni895. [DOI] [PubMed] [Google Scholar]
- Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol Biotechnol. 2010;44:71–81. doi: 10.1007/s12033-009-9216-2. [DOI] [PubMed] [Google Scholar]
- Tsui HW, Hasselblatt K, Martin A, Mok SC, Tsui FW. Molecular mechanisms underlying SHP-1 gene expression. Eur J Biochem. 2002;269:3057–3064. doi: 10.1046/j.1432-1033.2002.02986.x. [DOI] [PubMed] [Google Scholar]
- Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Wlodarski P, Zhang Q, Liu X, Kasprzycka M, Marzec M, Wasik MA. PU.1 activates transcription of SHP-1 gene in hematopoietic cells. J Biol Chem. 2007;282:6316–6323. doi: 10.1074/jbc.M607526200. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Somani AK, Siminovitch KA. Roles of the SHP-1 tyrosine phosphatase in the negative regulation of cell signalling. Sem.Immunol. 2000;12:361–378. doi: 10.1006/smim.2000.0223. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci U S A. 2005;102:6948–6953. doi: 10.1073/pnas.0501959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Collins CA, Wu P, Brown EJ. Protein tyrosine phosphatase SHP-1 positively regulates TLR-induced IL-12p40 production in macrophages through inhibition of phosphatidylinositol 3-kinase. J Leukoc Biol. 2010 doi: 10.1189/jlb.0409289. [DOI] [PubMed] [Google Scholar]