Abstract
When Saccharomyces cerevisiae is starved of glucose, the gluconeogenic enzymes fructose-1,6-bisphosphatase (FBPase), malate dehydrogenase (MDH2), isocitrate lyase (Icl1) and phosphoenolpyruvate carboxykinase (Pck1) are induced. However, when glucose is added to prolonged starved cells, these enzymes are degraded in the vacuole via the vacuole import and degradation (Vid) pathway. Recent evidence suggests that the Vid pathway merges with the endocytic pathway at actin patches where endocytic vesicles are formed. The convergence of the Vid pathway with the endocytic pathway allows cells to remove intracellular and extracellular proteins simultaneously. However, the genes that regulate this step of the convergence have not been identified previously. Here we show that VID30 plays a critical role for the association of Vid vesicles and actin patches. Vid30 is constitutively expressed and interacts with Vid vesicle proteins Vid24 and Sec28 but not with the cargo protein FBPase. In the absence of SEC28 or VID24, Vid30 association with actin patches was prolonged. In cells lacking the VID30 gene, FBPase and Vid24 were not localized to actin patches, suggesting that Vid30 has a role in the association of Vid vesicles and actin patches. Vid30 contains a LisH and a CTLH domain, both of which are required for FBPase degradation. When these domains were deleted, FBPase trafficking to the vacuole was impaired. We suggest that Vid30 also has a role in the Vid pathway at a later step in a process that is mediated by the LisH and CTLH domains.
Keywords: vacuole import and degradation; fructose-1,6-bisphosphatase; malate dehydrogenase; isocitrate lyase; phosphoenolpyruvate carboxykinase; Vid vesicles; Vid pathway; autophagy
Introduction
Autophagy is a process by which a cell degrades its own components via the lysosome/vacuole and is important for many biological processes including normal development, life-span extension and cell survival during stress.1–14 Furthermore, disrupted autophagy has been associated with diseases such as cancer, neurodegeneration and aging.1,3,4,15,16 Multiple autophagy pathways including macroautophagy, microautophagy and chaperone-mediated autophagy have been characterized.1–14 The macroautophagy pathway is conserved from yeast to human. It is induced in Saccharomyces cerevisiae that have been starved of nitrogen.12,17–20 The starvation-induced macroautophagy pathway degrades proteins nonselectively and recycles amino acids during periods of starvation. This pathway requires ATG genes that are also involved in the Cvt pathway that targets aminopeptidase I from the cytoplasm to the vacuole and the pexophagy pathway that degrades peroxisomes.12,17–20 The nonselective macroautophagy pathway is inhibited by Tor1 and is induced by rapamycin even in the absence of nitrogen starvation. In animal models of aggregate prone diseases, rapamycin clears large aggregates and improves the performance of the affected animals.4,21–25 Hence, activation of the nonselective macroautophagy pathway may be beneficial for treating patients with aggregate-prone diseases.
This study focuses on a specialized autophagy pathway in Saccharomyces cerevisiae. When yeast cells are starved of glucose, gluconeogenic enzymes such as fructose-1,6-bisphosphatase (FBPase), malate dehydrogenase (MDH2), isocitrate lyase (Icl1) and phosphoenolpyruvate carboxykinase (Pck1) are induced.26–29 However, when glucose is added to glucose-starved cells, these enzymes are inactivated.28–35 This has been termed “catabolite inactivation” and has also been described for the high affinity glucose transporter,29 the galactose transporter36,37 and the maltose transporter.38,39 The inactivation and degradation of FBPase has been studied extensively. This protein is degraded in either the proteasome36,40–44 or in the vacuole.31,32,45–47 Interestingly, the site of FBPase degradation appears to depend on the duration of starvation.45 For example, when glucose is added to cells that have been starved for one d, FBPase is degraded in the proteasome. By contrast, when glucose is added to cells that are starved for three d, FBPase is degraded in the vacuole.45
The vacuole-dependent degradation pathway utilizes small vesicles known as Vid vesicles to sequester FBPase.48 This sequestration requires the heat shock protein Ssa2, cyclophilin A, and Vid22.49–51 The UBC1 gene is required for the formation of Vid vesicles.46 Vid24 is a peripheral protein on Vid vesicles52 and has been used to study the trafficking of Vid vesicles in response to glucose. Recent evidence suggests that the Vid pathway converges with the endocytic pathway.53 Using the Δvph1 mutant that blocks endocytosis at a late stage, FBPase is found in the lumen of endocytic compartments and colocalizes with the FM 4–64 dye.53 This dye is internalized from the plasma membrane and traffics to the endosomes and later to the vacuole.54 COPI coatomer proteins are present on Vid vesicles and they recruit Vid24 to Vid vesicles.53 Moreover, the coatomer subunit Sec28 also traffics to the FM-containing endosomes following the addition of glucose to glucose-starved wild-type cells.53
Recently, we reported that the Vid pathway utilizes the early steps of endocytosis to deliver cargo proteins to the vacuole.55 In yeast, actin polymerization is required for early steps of the endocytic process.56–59 Both Vid24 and Sec28 associate with actin patches during glucose starvation and following glucose re-feeding for up to 30 min.55 However, less colocalization is seen after the addition of glucose for 60 min.55 In addition, the cargo protein FBPase also associates with actin patches following glucose re-feeding and dissociates at the 60 min time point.55 The association of Vid vesicles and actin patches is critical for the integration of the Vid pathway with the endocytic pathway. It allows cells to remove intracellular and extracellular proteins simultaneously. However, the genes that regulate this step of convergence have not yet been identified.
In our current report, we characterized the role of the VID30 gene in the vacuole-dependent degradation of FBPase. We initially identified VID30 using a transposon library screen for genes that function in this degradation pathway. Interestingly, this gene also has a role in the proteasome pathway in which it forms a complex with Vid24 and functions as an E3 ligase in the ubiquitination of FBPase.60 VID30 is also required for the rapamycin-induced degradation of the high affinity hexose transporter Hxt761 and for the transcriptional regulation of multiple genes involved in nitrogen metabolism.62 However, the role of VID30 in the vacuole-dependent FBPase degradation pathway has not been examined. This may be due in part to the fact that the Vid pathway has not yet been fully characterized. Vid30 contains a LisH domain (lissencephaly type 1-like homology domain) and a CTLH domain (C-terminal to the LisH domain). The LIS1 gene is mutated in Miller-Dieker lissencephaly, a disorder that causes severe retardation and early death.63–66 The CTLH domain may be involved in microtubule function.67,68 However, the functions of these domains in the Vid pathway have not been elucidated. In our present study, we showed that Vid30 is distributed in multiple locations including Vid vesicles, actin patches and FM-containing compartments. Furthermore, this protein interacts with Vid vesicle proteins Vid24 and Sec28. In yeast cells lacking VID30, FBPase and Vid24 failed to associate with actin patches. This suggests an important role for VID30 in the association of Vid vesicles with actin patches. Vid30 lacking either the LisH or CTLH domains inhibited FBPase degradation and FBPase trafficking. Hence, we suggest that Vid30 also has a role in the trafficking of FBPase at a later step in a process mediated by the LisH and CTLH domains.
Results
VID30 is required for the degradation of FBPase in the vacuole.
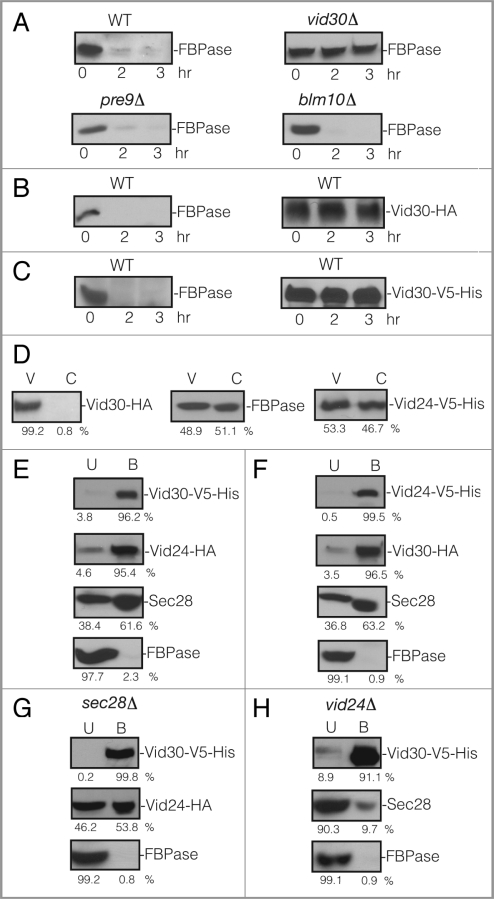
VID30 was previously identified in our laboratory using a transposon library screen for mutants that failed to degrade FBPase following glucose re-feeding. Interestingly, this gene is also involved in the proteasome-dependent degradation of FBPase.60 We have shown that FBPase is degraded in the vacuole when glucose is added to three d starved wild-type cells.45 To confirm the role of VID30 in the vacuole-dependent pathway, wild-type and vid30Δ cells were grown in low glucose medium for three d and then re-fed with fresh glucose. The wild-type cells degraded FBPase in response to glucose (Fig. 1A). However, FBPase degradation was impaired when glucose was added to vid30Δ cells that were starved of glucose for three d (Fig. 1A). This suggests that the VID30 gene is required for FBPase degradation in the vacuole. We also examined FBPase degradation in prolonged starved cells lacking the PRE9 gene and the BLM10 gene (Fig. 1A). PRE9 encodes that α 3 subunit of the 20S proteasome.69,70 BLM10 encodes a proteasome activator protein that binds to the core proteasome particle to activate proteolysis.71 FBPase was degraded normally in cells lacking either PRE9 or BLM10, suggesting that these genes are not involved in the degradation of FBPase in the vacuole in prolonged starved cells. In our current study, we focused on the function of Vid30 in the vacuole-dependent degradation of FBPase. Therefore, all subsequent experiments were performed using cells that were grown in low glucose medium for three d and then switched to medium containing high glucose for the indicated periods.
Figure 1.
VID30 is required for FBPase degradation in 3 d glucose starved cells. (A) Wild-type, vid30Δ, pre9D and blm10Δ mutant cells were starved for three d and transferred to medium containing fresh glucose for 0, 2 and 3 h. FBPase degradation was then determined. (B) Vid30 was tagged with HA and expressed in wild-type cells. Levels of FBPase and Vid30-HA in response to glucose re-feeding were determined. (C) Vid30 was tagged with V5-His and expressed in wild-type cells. FBPase degradation and Vid30-V5-His levels were examined. (D) Vid30-HA cells were glucose starved and shifted to glucose for 20 min. Levels of Vid30-HA, FBPase and Vid24-V5-His in the Vid vesicle enriched (V) and the cytosolic (C) fractions were examined. (E) Wild-type cells co-expressing Vid30-V5-His and Vid24-HA were glucose starved and shifted to glucose for 20 min. Vid30-V5-His was pulled down and the distribution of Vid30-V5-His, Vid24-HA, Sec28 and FBPase in the unbound (U) vs. bound (B) fractions was then determined. (F) Wild-type cells that co-expressed Vid24-V5-His and Vid30-HA were glucose starved and re-fed with glucose for 20 min and Vid24-V5-His was pulled down from total lysates. The distribution of Vid24-V5-His, Vid30-HA, Sec28 and FBPase in the unbound vs. bound fractions was examined. (G) Vid30-V5-His was co-expressed with Vid24-HA in the sec28D mutant that was starved for three d and then transferred to medium containing glucose for 20 min. Vid30-V5-His was pulled down and the presence of Vid24-HA and FBPase in the unbound vs. bound fractions was determined. (H) Vid30-V5-His was expressed in the vid24Δ mutant that was glucose starved and shifted to glucose for 20 min. Cells were lysed and Vid30-V5-His was pulled down. The presence of Vid30-V5-His, Sec28 and FBPase in the unbound and bound fractions was then determined. Relative ratios of protein levels were quantitated using ImageJ software.
Vid30 is constitutively expressed.
It has been previously reported that the expression of VID30 mRNA increases in response to low ammonia.62 We thus examined the expression of Vid30 under our experimental conditions. Vid30 was tagged with HA at the C-terminus and integrated into its chromosomal locus in wild-type cells. This allowed for the expression of Vid30-HA under the control of its own promoter. FBPase was degraded in Vid30-HA tagged cells (Fig. 1B, left panel), suggesting that HA tagging does not interfere with Vid30 function. Vid30-HA was present during glucose starvation and its levels did not increase significantly following glucose re-feeding for 3 h (Fig. 1B, right panel). In some experiments, Vid30 was tagged with V5-His and FBPase degradation was examined. FBPase was also degraded in cells containing this tag (Fig. 1C, left panel), suggesting that Vid30-V5-His is functional. Furthermore, levels of Vid30-V5-His did not change significantly whether cells were starved or replenished with glucose (Fig. 1C, right panel).
We examined whether Vid30 was localized to Vid vesicles. Wild-type cells expressing Vid30-HA were starved and re-fed with glucose for 20 min. The cells were lysed and total extracts were subjected to differential centrifugation to separate cytosol from the Vid vesicle enriched fraction. Most of the Vid30-HA was detected in the Vid vesicle enriched fraction (Fig. 1D). FBPase and Vid24-V5-His were distributed in both the Vid vesicle and the cytosolic fractions (Fig. 1D).
Vid30 interacts with Vid24 and Sec28.
In the proteasomal pathway, Vid30 forms a complex with Vid24 and FBPase.60 However, it has not been examined whether Vid30 forms a complex with Vid24 and FBPase during the vacuole-dependent degradation process. To investigate this, wild-type cells co-expressing Vid30-V5-His and Vid24-HA were starved for three d and transferred to high glucose containing medium for 20 min. Vid30-V5-His was pulled down and samples were separated into unbound and bound fractions. Proteins were then immunoblotted with antibodies directed against V5 and HA to detect Vid30 and Vid24, respectively (Fig. 1E). As expected, the majority of Vid30 was pulled down in the bound fraction. Most of the Vid24 was also in the bound fraction, suggesting that Vid30 and Vid24 interact. Consistent with our previous report that Sec28 and Vid24 interact,60 Sec28 was also present in the bound fraction. As coatomer proteins are involved in other protein trafficking pathways, it is not surprising that a fraction of Sec28 was also present in the unbound fraction. However, in contrast to a previous report on the proteasomal pathway,60 the majority of FBPase was detected in the unbound fraction in our experiment.
Similar findings were obtained using wild-type cells that co-expressed Vid24-V5-His and Vid30-HA. In these experiments, Vid24-V5-His was pulled down and the bound and unbound fractions were immunoblotted to detect Vid24-V5-His and Vid30-HA (Fig. 1F). The majority of Vid24 and Vid30 were in the bound fractions. Sec28 was also in the bound fraction while most of FBPase was in the unbound fraction. These results suggest that Vid30 interacts with Vid24 and Sec28 during the vacuole-dependent degradation pathway. However, FBPase was not identified in the bound fraction, suggesting that FBPase does not form a complex with Vid30 and Vid24.
Given that Vid30, Vid24 and Sec28 interact, we next determined whether the interaction of Vid30 with Vid24 was affected in cells lacking SEC28 (Fig. 1G) and whether the interaction of Vid30 with Sec28 was affected in cells lacking the VID24 gene (Fig. 1H). To study the role of SEC28 in the Vid30 and Vid24 interaction, Vid30-V5-His and Vid24-HA were co-expressed in the sec28Δ mutant that was glucose starved and then re-fed with glucose for 20 min. Vid30-V5-His was pulled down and the presence of Vid24 in the unbound and bound fractions was then determined (Fig. 1G). In this strain, the majority of Vid30-V5-His was pulled down in the bound fraction. However, the level of Vid24 in the bound fraction was reduced in this strain when compared with that seen in wild-type cells (see Fig. 1E). Most of the FBPase remained in the unbound fraction. Thus, the absence of Sec28 appears to impair the interaction of Vid30 with Vid24.
The interaction of Vid30 with Sec28 was also examined in cells lacking the VID24 gene (Fig. 1H). Vid30-V5-His was expressed in the vid24Δ mutant that was glucose starved and then transferred to medium containing fresh glucose for 20 min. Vid30 was pulled down and the presence of Sec28 in the bound and unbound fractions was then determined (Fig. 1H). The majority of Vid30-V5-His was pulled down in the bound fraction. However, the level of Sec28 in the bound fraction was reduced. In this strain, FBPase also remained in the unbound fraction. Thus, the interaction of Vid30 with Sec28 was affected in the absence of Vid24.
Vid30 associates with actin patches.
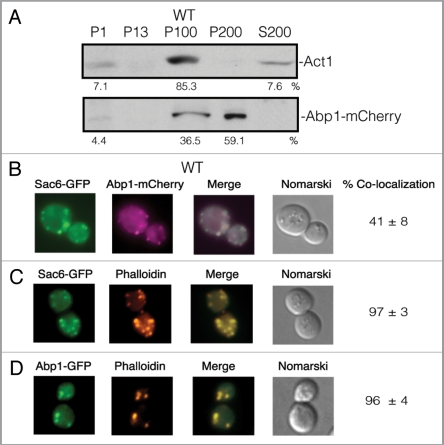
Both Vid24 and Sec28 localize to actin patches during glucose starvation and also at 30 min following glucose re-feeding.55 Since Vid30 interacts with both Vid24 and Sec28, it may also localize to actin patches. Abp1 has been used to mark actin patches in cells that are grown in rich medium. Because we used prolonged starvation conditions, we first determined whether or not Abp1 colocalizes with actin (Act1) under our growth conditions. We produced an Abp1-mCherry construct and transformed it into wild-type cells. Cells were glucose starved for three d and lysates were fractionated by differential centrifugation. The presence of Abp1-mCherry and Act1 in the P1, P13, P100, P200 and S200 fractions was then examined (Fig. 2A). Act1 was detected in multiple locations in the P1, P100 and S200 fractions and the majority of Act1 was distributed in the P100 fractions. In a similar manner, Abp1-mCherry was also distributed in multiple locations in the P1, P100 and P200 fractions. However, more than 50% of the Abp1-mCherry was in the P200 fraction that contained minimal amounts of actin. Based on these fractionation results, it appears that less than 50% of Abp1-mCherry colocalized with actin.
Figure 2.
Abp1 and actin (Act1) are distributed in multiple locations. (A) Wild-type cells expressing Abp1-mCherry were glucose starved for 3 d. Cells were fractionated by differential centrifugation. The distribution of Act1 and Abp1-mCherry in the P1, P13, P100, P200 and S200 fractions was determined. (B) Abp1-mCherry was transformed into wild-type cells that expressed Sac6-GFP. The distribution of Abp1-mCherry and Sac6-GFP was determined. (C) Wild-type cells expressing Sac6-GFP was processed and actin patches were visualized with phalloidin conjugated with rhodamine. (D) Wild-type cells expressing Abp1-GFP was processed and actin patches were visualized with phalloidin conjugated with rhodamine.
As an additional test, we transformed Abp1-mCherry into wildtype cells that expressed Sac6-GFP. Both proteins are localized to actin-containing structures as reported in the Saccharomyces Genome Database. In wild-type cells that co-expressed Sac6-GFP and Abp1-mCherry, less than 50% of Abp1-mCherry colocalized with Sac6-GFP (Fig. 2B). We next utilized a different protocol to examine the colocalization of GFP and actin-related structures.55 The cells that expressed GFP fusion proteins were fixed and permeabilized, and actin was stained with rhodamine-conjugated phalloidin. Using this method, GFP-tagged proteins and actin can be visualized simultaneously. However, the overall morphology of the cell was somewhat changed using this protocol. For example, when the vacuole and endosomes were pre-labeled with FM and then treated with this protocol, these organelles were no longer observed following the fixation and permeabilization. This may account for the diminished GFP signals as GFP fusion proteins move to these compartments. Nevertheless, more than 90% of Sac6-GFP showed colocalization with actin that was stained with phalloidin (Fig. 2C). Furthermore, more than 90% of Abp1-GFP also showed colocalization with actin stained with phalloidin (Fig. 2D). One explanation for these results is that the phalloidin protocol preserves the actin-containing structures at the internalization sites, whereas other actin-related structures were not preserved. Since our study focused on the localization of Vid vesicles at the internalization sites and since the phalloidin protocol gave consistent and reliable results under our experimental conditions, we used the phalloidin protocol to examine colocalization of various GFP fusion proteins with actin in this study. We have shown that Vid24, Sec28 and cargo proteins all associate with actin patches by the 30 min time point and less association is seen by the 60 min time point following glucose re-feeding.55 Therefore, all subsequent actin studies were performed in cells that were shifted to glucose for up to 60 min.
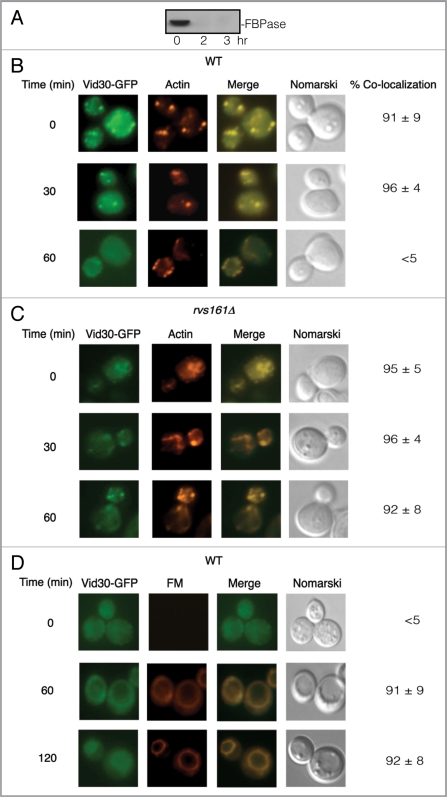
To examine whether Vid30 was distributed to actin patches, Vid30 was fused with GFP and integrated into the VID30 chromosomal locus in wild-type cells. FBPase was degraded in the wild-type cells expressing Vid30-GFP following glucose refeeding (Fig. 3A), indicating that GFP tagging does not interfere with FBPase degradation. A portion of Vid30-GFP colocalized with actin patches during glucose starvation and after the addition of glucose for up to 30 min (Fig. 3B). However, less colocalization was seen following the addition of glucose for 60 min, suggesting that Vid30 and actin patches dissociate by this time point.
Figure 3.
Vid30-GFP is associated with actin patches and FM-containing compartments. (A) Wild-type cells expressing Vid30-GFP were glucose starved and shifted to glucose for 0, 2 and 3 h. FBPase degradation was then examined. (B) The distribution of Vid30-GFP and actin patches was determined. (C) rvs161Δ cells expressing Vid30-GFP were re-fed with glucose and the distribution of Vid30-GFP and actin patches was examined. (D) Wild-type cells expressing Vid30-GFP were transferred to glucose-containing medium in the presence of FM for the indicated periods. The distribution of Vid30-GFP and FM was visualized using fluorescence microscopy.
Vid30 association with actin patches is prolonged in the Δrvs161 mutant.
In yeast, Rvs161 interacts with Rvs167 and both are required for the scission of endocytic vesicles from the plasma membrane.56–59 In order to determine whether RVS161 functions in Vid30 dissociation from actin patches, we expressed Vid30-GFP in rvs161Δ cells. In this strain, the majority of Vid30 was associated with actin patches during glucose starvation and this persisted following the addition of glucose for up to the 60 min time point (Fig. 3C). Thus, the association of Vid30-GFP and actin patches appears to be prolonged in the Δrvs161 mutant.
Vid30 traffics to FM-containing compartments in response to glucose.
We next examined if Vid30 traffics to the FM-containing endosomes and to the vacuole. Vid30-GFP was expressed in wild-type cells that were grown in low-glucose medium and were re-fed with glucose in the presence of FM 4–64 dye. In wild-type cells, the majority of FM reaches the vacuole by the 120 min time point.53 Therefore, all FM studies were conducted in cells that were shifted to glucose for up to 120 min. During glucose starvation, a portion of Vid30 was observed in punctate structures (Fig. 3D). By 120 min, a fraction of Vid30 was observed on the vacuole membrane. Hence, a percentage of Vid30 appears to traffic to the FM-containing compartments in response to glucose re-addition.
Vid30 association with actin patches was prolonged in cells lacking SEC28 or VID24.
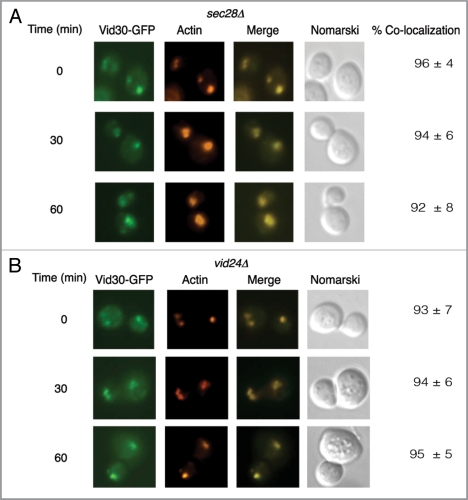
Our interaction data suggest that Vid30 interacts with Vid24 and Sec28. Therefore, we next determined whether SEC28 or VID24 have some roles in the association of Vid30 with actin patches. For these studies, we expressed Vid30-GFP in cells lacking either SEC28 or VID24 and examined the distribution of Vid30. In cells lacking SEC28, a significant amount of Vid30-GFP was expressed and colocalized with actin patches during glucose starvation (Fig. 4A). High levels of Vid30-GFP remained associated with actin patches at the 30 min and the 60 min time points (Fig. 4A). Therefore, the absence of SEC28 appears to prolong the association of Vid30 with actin patches.
Figure 4.
Vid30 association with actin patches is prolonged in the sec28Δ mutant and the vid24Δ mutant. (A) Vid30-GFP was expressed in the sec28Δ mutant that was glucose starved and then transferred to medium containing fresh glucose for the indicated time points. The distribution of Vid30-GFP with actin patches was determined. (B) Vid30-GFP was expressed in the vid24Δ mutant that was transferred from low to high glucose medium for the indicated time points. The distribution of Vid30-GFP with actin patches was examined.
We investigated whether the VID24 gene also has a role in Vid30 association with actin patches (Fig. 4B). Vid30-GFP was expressed in the vid24Δ strain and the association with actin patches was then determined (Fig. 4B). In this strain, a percentage of Vid30 was present as punctate structures and showed colocalization with actin patches during glucose starvation. Vid30 was associated with actin patches following the addition of glucose for 60 min. Thus, the association of Vid30 with actin patches appears to be prolonged in the absence of the VID24 gene.
FBPase and Vid24 are present in the Vid vesicle-enriched fraction in cells lacking the VID30 gene.
As shown above, Vid30 is distributed in multiple compartments including Vid vesicles, actin patches and FM-positive compartments. Therefore, we sought to determine which step of the FBPase trafficking pathway requires the VID30 gene. If VID30 is needed for FBPase association with Vid vesicles, then the FBPase levels in the Vid vesicle enriched fraction should be reduced in the vid30Δ mutant. In contrast, if VID30 is needed after FBPase is associated with Vid vesicles, then FBPase should be detected in the Vid vesicle fraction of the vid30Δ mutant. To investigate this, wild-type and vid30Δ mutant cells were shifted to glucose for 20 min and homogenized. Total lysates were subjected to differential centrifugation. In wild-type cells, FBPase was present in both the Vid vesicle and the cytosolic fractions (Fig. 5A, left panel). In the vid30Δ mutant, FBPase was also detectable in the Vid vesicle and the cytosolic fractions (Fig. 5A, right panel).
Figure 5.
FBPase and Vid24 are present in the Vid vesicle enriched fraction in vid30Δ mutant cells. (A) Wild-type and the vid30Δ mutant cells were transferred to high glucose-containing medium for 20 min. The presence of FBPase in the Vid vesicle and cytosolic fractions was then determined. (B) Wild-type and vid30Δ cells expressing Vid24-HA were re-fed with glucose for 20 min. The distribution of Vid24 in the Vid vesicle and the cytosolic fractions was then examined. Protein levels were quantitated using ImageJ software.
The presence of FBPase in the Vid vesicle fraction suggests that these vesicles are formed in the vid30Δ mutant. Therefore, Vid24 that resides on Vid vesicles should also be detectable in the Vid vesicle fraction for this mutant. To examine this, wild-type and vid30Δ mutant cells expressing Vid24-HA were glucose starved for three d and replenished with fresh glucose for 20 min. Cells were homogenized and total lysates were subjected to differential centrifugation. In wild-type cells, Vid24 was in both the Vid vesicle enriched fraction and the cytosolic fraction (Fig. 5B, left panel). In the vid30Δ mutant, the Vid24 levels in the cytosolic fraction were minimal and most of the Vid24 was in the Vid vesicle fraction (Fig. 5B, right panel), suggesting that Vid vesicles are formed in the vid30Δ mutant.
FBPase fails to associate with actin patches in cells lacking the VID30 gene.
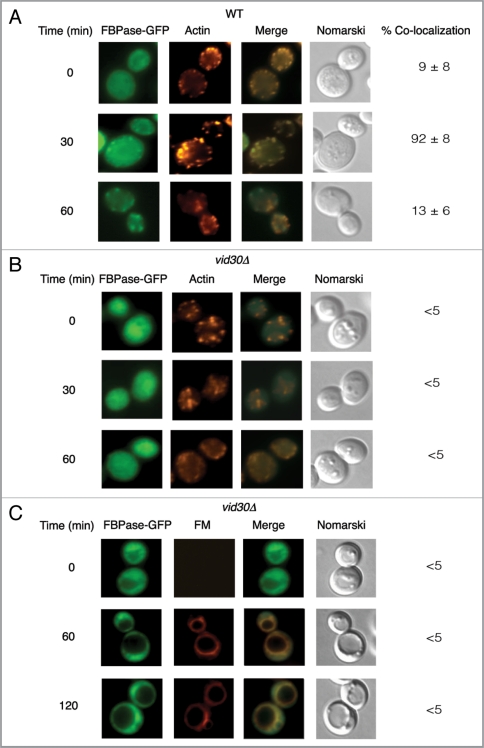
We next assessed whether VID30 plays a role in the association of FBPase with actin patches. FBPase-GFP was expressed in wild-type and the vid30Δ mutant cells that were glucose starved and transferred to medium containing fresh glucose. The distribution of FBPase and actin patches was then examined. In wild-type cells, FBPase associated with actin patches following the addition of glucose for 30 min. However, less colocalization was seen by the 60 min time point (Fig. 6A). In the vid30Δ strain, FBPase-GFP distribution was uneven but FBPase association with actin patches was not detected before or after the addition of glucose (Fig. 6B). This suggests that FBPase fails to associate with actin patches in cells lacking VID30.
Figure 6.
FBPase does not associate with actin patches in vid30Δ mutant cells. Wild-type (A) and vid30Δ mutant cells (B) expressing FBPase-GFP were glucose starved and re-fed with glucose for up to 60 min and examined by fluorescence microscopy for the distribution of FBPase-GFP and actin patches. (C) The vid30Δ mutant cells expressing FBPase-GFP were re-fed with glucose in the presence of FM for up to 120 min. The distribution of FBPase-GFP and FM was examined.
Since FBPase fails to associate with actin patches in the vid30Δ mutant, targeting of this protein to the FM-containing endosomes or to the vacuole may also be affected in the absence of the VID30 gene. To investigate this, the vid30Δ strain expressing FBPase-GFP was grown in low-glucose medium and transferred to high-glucose medium in the presence of the FM dye for various time points (Fig. 6C). In the vid30Δ mutant, FBPase-GFP did not show clear colocalization with FM, suggesting that FBPase is not targeted to these compartments.
Vid24 fails to associate with actin patches in the vid30Δ mutant.
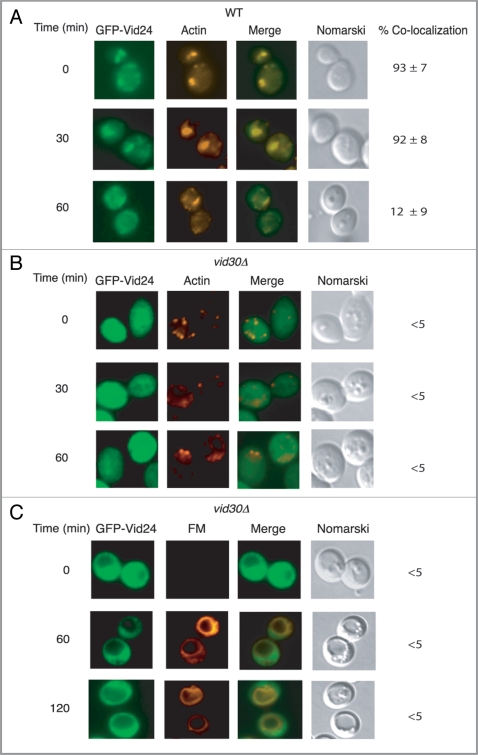
Because FBPase did not show colocalization with actin patches in cells lacking VID30, we next studied whether the association of Vid vesicle protein Vid24 was affected in the absence of this gene. GFP-Vid24 was expressed in wild-type and the vid30Δ mutant strain that were grown in low glucose medium for three d and then re-fed with high glucose for various periods. In wild-type cells, GFP-Vid24 was localized to actin patches during glucose starvation and following the addition of glucose up to 30 min. However, less colocalization of GFP-Vid24 with actin patches was seen at the 60 min time point (Fig. 7A). In cells lacking the VID30 gene, GFP-Vid24 did not colocalize with actin patches before or after the addition of glucose (Fig. 7B). This suggests that GFP-Vid24 does not associate with actin patches in the absence of the VID30 gene.
Figure 7.
Vid24 does not associate with actin patches in cells lacking VID30. Wild-type (A) and vid30Δ mutant cells (B) expressing GFP-Vid24 were starved and replenished with glucose for up to 60 min and examined by fluorescence microscopy for the localization of GFP-Vid24 and actin patches. (C) The vid30Δ cell was re-fed with glucose and FM for up to 120 min and examined for the distribution of GFP-Vid24 and FM.
Since Vid24 fails to associate with actin patches, this protein may not be targeted to FM-containing endosomes or to the vacuole. To investigate this, we expressed GFP-Vid24 in cells lacking VID30 and examined the localization of GFP-Vid24 in cells that were starved for three d and re-fed with glucose in the presence of the FM dye (Fig. 7C). GFP-Vid24 signal was not uniform in the vid30Δ mutant. However, there was no obvious colocalization of GFP-Vid24 with FM-containing structures in this strain (Fig. 7C). These results suggest that Vid24 fails to traffic to FM-containing compartments in the absence of the VID30 gene.
The LisH domain of Vid30 is required for the degradation of FBPase.
Vid30 contains a LisH domain and a CTLH domain (Fig. 8A). In order to study the functions of these domains, we attempted to produce random mutations by PCR based mutagenesis. Unfortunately, we did not obtain mutants that were defective in FBPase degradation. As an alternative, we produced Vid30 mutant proteins lacking either one of the two domains using site directed mutagenesis. Vid30 lacking either the LisH domain or the CTLH domain was expressed. Furthermore, these proteins were localized to Vid vesicle fraction. Moreover, they showed association with actin patches initially and less association in response to glucose like the full-length Vid30 (see Figs. 8–11). Therefore, these mutant proteins were used to study whether or not the absence of these domains affected the Vid pathway.
Figure 8.
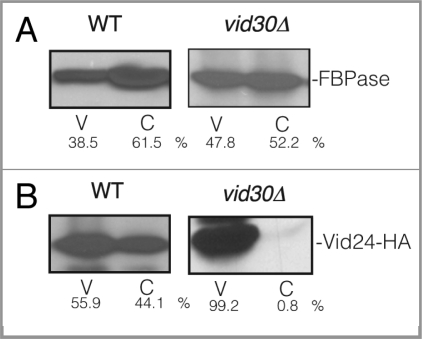
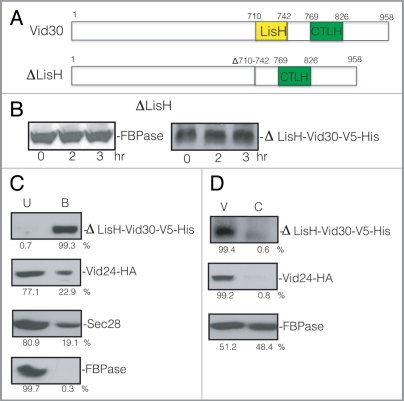
The LisH domain is required for FBPase degradation. (A) Schematic illustration of the position of the LisH and CTLH domains in Vid30 and the position of the LisH domain that was deleted in the ΔLisH mutant. (B) ΔLisH mutant cells were glucose starved and re-fed with glucose for up to 3 h. Levels of FBPase and ΔLisH-Vid30-V5-His were examined. (C) Cells co-expressing ΔLisH-Vid30-V5-His and Vid24-HA were starved and shifted to glucose for 20 min. ΔLisH-Vid30-V5-His was pulled down and the levels of Vid24-HA, Sec28 and FBPase in the bound vs. unbound fractions were then determined. (D) The ΔLisH mutant cells were shifted to glucose for 20 min. The distribution of ΔLisH-Vid30-V5-His, Vid24-HA and FBPase in the Vid vesicle and cytosolic fractions was examined. Relative ratios of proteins were quantitated using ImageJ software.
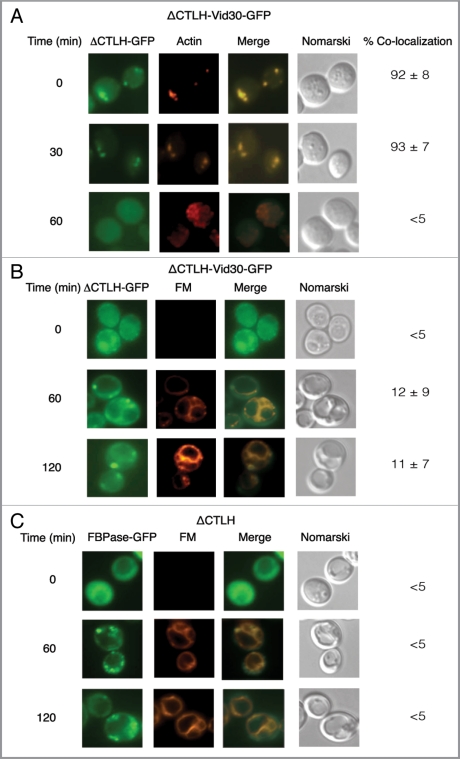
Figure 11.
ΔCTLH-Vid30-GFP and FBPase accumulate in punctate compartments following glucose re-feeding. (A) ΔCTLH-Vid30-GFP was expressed in yeast cells that were starved and re-fed with glucose for the indicated times. ΔCTLH-Vid30-GFP and actin patches were visualized by fluorescence microscopy. (B) The distribution of ΔCTLH-Vid30-GFP and FM was examined. (C) FBPase-GFP was expressed in ΔCTLH mutant cells. The localization of FBPase-GFP and FM was then determined.
When LisH was deleted, FBPase degradation was retarded (Fig. 8B, left panel). When ΔLisH-Vid30 was tagged with V5-His, the resulting protein was expressed and its level did not decrease following the addition of glucose (Fig. 8B, right panel), suggesting that this mutant protein is stable under our experimental conditions.
Since full-length Vid30 interacts with Vid24 and Sec28, we determined whether the LisH mutation affected Vid30 interaction with Vid24 or Sec28. Cells co-expressing ΔLisH-Vid30-V5-His and Vid24-HA were glucose starved and transferred to medium containing fresh glucose for 20 min. ΔLisH-Vid30-V5-His was then pulled down and the presence of Vid24-HA in the bound and unbound fractions was examined (Fig. 8C). The majority of the ΔLisH-Vid30-V5-His was pulled down in the bound fraction. However, the levels of Vid24-HA and Sec28 in the bound fraction were reduced when compared with that detected for full-length Vid30 (see Fig. 1E). Under these conditions, most of the FBPase remained in the unbound fraction. As such, the absence of the LisH domain reduces the Vid24 and Sec28 levels in the bound fraction.
We next assessed whether the ΔLisH mutant protein was present in the Vid vesicle enriched fraction. The ΔLisH mutant cells were glucose starved for three d and transferred to medium containing fresh glucose for 20 min. The distribution of the mutant protein in the cytosolic and Vid vesicle fractions was then examined (Fig. 8D). In the ΔLisH mutant cells, the majority of ΔLisH-Vid30-V5-His and Vid24 were detectable in the Vid vesicle enriched fraction. FBPase was found in both the Vid vesicle and the cytosolic fractions. Thus, levels of FBPase and Vid24 in the Vid vesicle fraction were not reduced in the ΔLisH mutant.
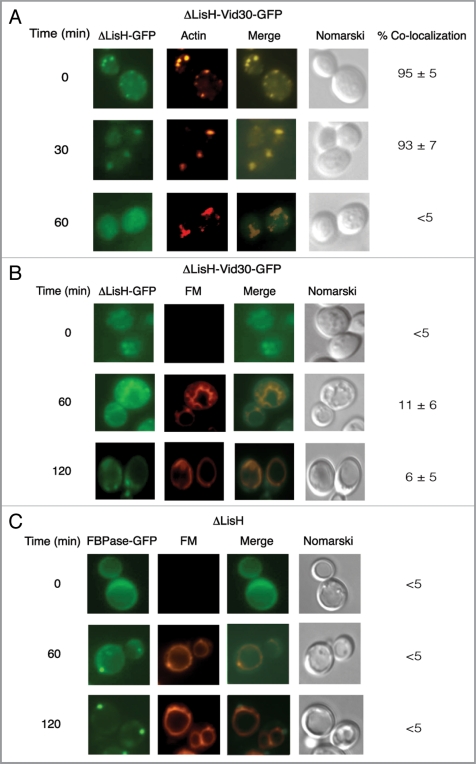
We investigated whether the ΔLisH mutant protein was targeted to actin patches using a yeast strain that expressed ΔLisH-Vid30-GFP. High levels of ΔLisH-Vid30-GFP colocalized with actin patches during glucose starvation. However, less colocalization was seen at 60 min (Fig. 9A). This suggests that ΔLisH-Vid30-GFP and actin patches associate and then dissociate with kinetics similar to that seen for the full-length Vid30 (see Fig. 3B).
Figure 9.
The ΔLisH mutant protein and FBPase accumulate in punctate structures in response to glucose. (A) ΔLisH-Vid30-GFP was expressed in yeast cells that were starved and re-fed with glucose for the indicated time points. The distribution of ΔLisH-Vid30-GFP and actin patches was determined by fluorescence microscopy. (B) The distribution of ΔLisH-Vid30-GFP and FM was examined. (C) FBPase-GFP was expressed in the ΔLisH mutant strain that was starved and re-fed with glucose. FBPase-GFP and FM was visualized by fluorescence microscopy.
We next determined whether the ΔLisH mutant protein was targeted to FM-containing compartments (Fig. 9B). The ΔLisH-Vid30-GFP strain was starved of glucose for three d and replenished with glucose in the presence of FM for the indicated time points (Fig. 9B). Following glucose addition for 120 min, the majority of ΔLisH-Vid30-GFP was present in punctate structures, while most of the FM was on the vacuole membrane.
We examined whether or not the trafficking of FBPase was affected when the LisH domain was deleted (Fig. 9C). In the ΔLisH cells, the trafficking of FBPase-GFP was affected when the LisH domain was deleted (Fig. 9C). At the 120 min time point, high levels of FBPase were observed in punctate structures. We have attempted to study the colocalization of FBPase with ΔLisH-Vid30 by producing FBPase that was fused with YFP, CFP, venus, cerulean and mCherry. Unfortunately, none of these fusion proteins were functional and they inhibited FBPase degradation to different degrees. We have also produced Vid30-mCherry, ΔLisH-Vid30-mCherry and DCTLH-Vid30-mCherry and transformed them into wild-type cells that expressed FBPase-GFP. Unfortunately, all the Vid30-mCherry constructs were mislocalized to the vacuole. Therefore, further colocalization experiments were not pursued in this study.
The CTLH domain of Vid30 is required for FBPase degradation.
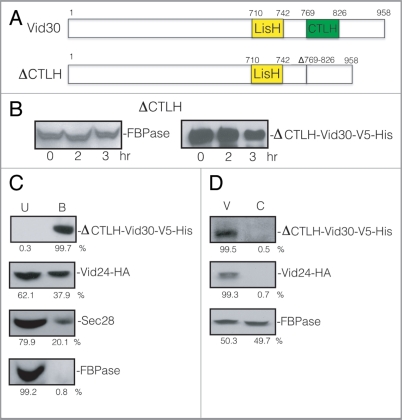
We next investigated whether the lack of the CTLH domain affects the FBPase degradation pathway (Fig. 10A). FBPase degradation was inhibited in the ΔCTLH strain (Fig. 10B, left panel), suggesting that the CTLH domain has a role in the FBPase degradation pathway. The ΔCTLH mutant protein was detectable up to 3 h after glucose addition (Fig. 10B, right panel).
Figure 10.
FBPase degradation is inhibited in the ΔCTLH mutant. (A) Schematic illustration of the position of the LisH and CTLH domains in Vid30 and the CTLH domain that was deleted to produce the CTLH domain mutant. (B) The CTLH domain of Vid30 was deleted and the mutant protein was tagged with V5-His. FBPase degradation was examined. The levels of ΔCTLH-Vid30-V5-His were determined. (C) Cells that co-expressed ΔCTLH-Vid30-V5-His and Vid24-HA were shifted to glucose for 20 min. Total lysates were prepared and ΔCTLH-Vid30-V5-His was pulled down. The presence of ΔCTLH-Vid30-V5-His, Vid24-HA, Sec28 and FBPase in the unbound and bound fractions was determined. (D) Cells that co-expressed ΔCTLH-Vid30-V5-His and Vid24-HA were glucose starved for three d and re-fed with glucose for 20 min. The distribution of ΔCTLH-Vid30-V5-His, Vid24-HA and FBPase in the Vid vesicle and cytosolic enriched fractions was examined and quantitated using ImageJ software.
We determined whether the ΔCTLH mutant affected the interaction of Vid30 with Vid24. Cells co-expressing ΔCTLH-Vid30-V5-His and Vid24-HA were glucose starved and re-fed with fresh glucose for 20 min. ΔCTLH-Vid30-V5-His was pulled down from total lysates and the presence of ΔCTLH-Vid30-V5-His and Vid24-HA in the bound and unbound fractions was determined (Fig. 10C). Most of the ΔCTLH-Vid30-V5-His was precipitated in the bound fraction but levels of Vid24-HA and Sec28 in the bound fraction were reduced in this mutant. The majority of FBPase was in the unbound fraction. Thus, the absence of the CTLH domain appears to reduce the levels of Vid24 and Sec28 in the bound fraction.
Next, we examined whether or not the ΔCTLH mutant protein was distributed to the Vid vesicle enriched fraction. Cells expressing ΔCTLH-Vid30-V5-His and Vid24-HA were glucose starved and re-fed with glucose for 20 min. Cell lysates were fractionated by differential centrifugation and the presence of ΔCTLH-Vid30-V5-His and Vid24 was determined (Fig. 10D). The majority of ΔCTLH-Vid30-V5-His was in the Vid vesicle fraction. Similarly, most of the Vid24 was in the Vid vesicle fraction in this strain. FBPase was distributed in both the Vid vesicle and the cytosolic fractions. These results suggest that FBPase and Vid24 are localized to the Vid vesicle fraction when the CTLH domain is deleted.
We investigated whether the ΔCTLH-Vid30 mutant protein associated with actin patches. Cells expressing the ΔCTLH-Vid30-GFP fusion protein were glucose starved and transferred to medium containing fresh glucose for the indicated time points (Fig. 11A). The mutant protein colocalized with actin patches during glucose starvation. However, less colocalization was seen at 60 min, suggesting that the CTLH mutant protein and actin associate and then dissociate in response to glucose re-feeding with kinetics similar to that shown for the full-length Vid30 (see Fig. 3B).
We determined whether or not the ΔCTLH-Vid30 protein traffics to FM-containing compartments. Cells expressing ΔCTLH-Vid30-GFP were glucose starved and shifted to glucose in the presence of FM for various periods (Fig. 11B). At the 120 min time point, a fraction of the ΔCTLH-Vid30-GFP protein was observed in punctate structures and did not show colocalization with FM-containing compartments.
Finally, we determined whether or not the trafficking of FBPase was affected when the CTLH domain was deleted. FBPase-GFP was expressed in a strain that contained the CTLH deletion, which was then starved of glucose and re-fed with glucose in the presence of FM for the indicated time points (Fig. 11C). By 120 min, a high percentage of FBPase-GFP was in punctate structures and did not show colocalization with FM-containing structures (Fig. 11C), suggesting that FBPase trafficking is impaired when the CTLH domain is deleted.
Discussion
In our current study, we characterized the role of the VID30 gene in the Vid pathway. We have shown that Vid30 is expressed in yeast cells that are glucose starved for 3 d. The expression of this protein does not increase when the cells are transferred to medium containing high glucose. In addition, Vid30 interacts with Vid24 and Sec28. We found, however, that FBPase was not coprecipitated with either Vid30 or with Vid24. This suggests that FBPase does not associate with the Vid24/Vid30 complex. This finding is consistent with the idea that FBPase and Vid24 exist in topologically distinct environments. The majority of FBPase is sequestered inside Vid vesicles48 whereas Vid24 is localized on the Vid vesicle membrane as a peripheral protein.52 The interaction of Vid30 with Vid24 was reduced in the absence of Sec28. Likewise, the interaction of Vid30 with Sec28 was also affected in cells lacking Vid24. Furthermore, the association of Vid30 with actin patches was prolonged in cells lacking either the VID24 gene or the SEC28 gene, suggesting that these genes have some roles in the dissociation of Vid30 and actin patches.
Since Vid30 interacts with Vid24 and Sec28, it was not surprising to find that Vid30 showed similar distribution patterns to those described for Vid24 and Sec28 during the vacuole-dependent degradation process.53,55 For instance, Vid30 is localized to Vid vesicles, to actin patches and to the FM-containing compartments. Although Vid30 is distributed to multiple compartments, the complete absence of this protein does not affect the levels of Vid24 and FBPase in the Vid vesicle fraction. However, Vid24 and FBPase did not colocalize with actin patches in cells lacking Vid30. These results suggest that Vid30 plays an important role in the association of Vid vesicles and actin patches.
Vid30 contains a LisH domain and a CTLH domain, and both are required for FBPase degradation. In either the ΔLisH or the ΔCTLH mutant, FBPase degradation was inhibited. Levels of Vid24 and Sec28 in the bound fraction were also reduced in the absence of these domains. Vid30 lacking either the LisH or CTLH domain was present in Vid vesicles and actin patches. However, these mutant proteins accumulated in punctate structures. In a similar manner, when either the LisH or CTLH domain was deleted, FBPase-GFP was also localized to punctate structures. Therefore, we suggest that the LisH and CTLH domains have some role in the Vid pathway at a later step.
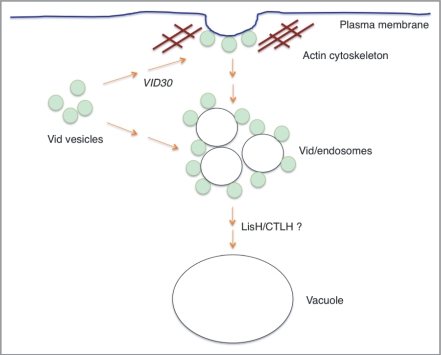
Based on our current data, we propose the following model for VID30 functions in the Vid pathway (Fig. 12). Vid30 is constitutively expressed and interacts with Vid24 and Sec28. Vid30, Vid24 and Sec28 are present on Vid vesicles and associate with actin patches. In the absence of Vid30, FBPase and Vid24 fail to associate with actin patches, suggesting that VID30 is needed for the association of Vid vesicle and actin patches. At present, it is not known how Vid vesicles and actin patches associate. Actin patches may mark the sites for Vid vesicles to cluster on the plasma membrane. Vid30 may be involved in the targeting of Vid vesicles to actin patches on the plasma membrane. Alternatively, Vid vesicles may cluster in the cytoplasm and actin associates with Vid vesicle clusters in the cytoplasm. Therefore, Vid30 may have a role in Vid vesicle clustering in the cytoplasm. Future experiments will be needed to sort out these possibilities. Following glucose addition, Vid/endosomes are released into the cytoplasm. The END3, SLA1 and RVS167 genes are involved in various steps of the actin assembly process and are required for the Vid pathway.55 These genes may be involved in the formation of Vid/endosomes or in the release of these organelles from the plasma membrane. When the LisH or CTLH domain is deleted, FBPase is localized in punctate structures. This distribution pattern is different from that seen in cells lacking the VID30 gene. Hence, we suggest that Vid30 also has a role in the Vid pathway at a later step that is mediated by the LisH and CTLH domains.
Figure 12.
A model for VID30 in the Vid pathway. Vid vesicles associate with actin patches on the plasma membrane. Vid30 is present in Vid vesicles, actin patches, FM-containing endosomes and the vacuole. In the absence of Vid30, Vid vesicles fail to associate with actin patches. This suggests that Vid30 has a role in the association of Vid vesicles and actin patches. The absence of LisH or CTLH domain results in the distribution of these mutant proteins in punctate structures. When either the LisH or the CTLH domain is deleted, FBPase is localized in punctate structures. Because the patterns of FBPase distribution in the LisH and CTLH mutants are distinct from those seen in the complete absence of the VID30 gene, we suggest that these domains have a role in the Vid pathway at a later step.
In conclusion, the convergence of the Vid pathway with the endocytic pathway enables cells to degrade both intracellular proteins and extracellular proteins. Therefore, the association of Vid vesicles and actin patches is critical for the Vid pathway to be integrated into the endocytic pathway. The VID30 gene is the first gene that is required for the association of Vid vesicles and actin patches.
Materials and Methods
Strains, media and antibodies.
The yeast strains used in the study are listed in Table 1 and the primers are listed in Table 2. Cells were grown in YPD (1% yeast extracts, 2% peptone and 2% glucose) medium for high glucose conditions or YPKG (1% yeast extracts, 2% peptone, 1% potassium acetate and 0.5% glucose) medium for low glucose conditions. Wild-type cells that expressed Sac6-GFP and Abp1-GFP were purchased from Invitrogen (95700). Antibodies raised against HA were obtained from Roche (11583816001) and antibodies against V5 were purchased from Invitrogen (R96025). Mouse monoclonal anti-DsRed was purchased from BD PharMingen (51-8115GR). FM 4–64 dye (T3166) and rhodamine-conjugated phalloidin (R415) were purchased from Invitrogen.
Table 1.
Strains used in this study
| Strain | Genotype |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| HLY635 | MATα ura3-52 LEU2 trp1Δ63 his3Δ200 GAL2 |
| vid30Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vid30::kanMX4 |
| pre9Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pre9::kanMX4 |
| blm10Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 blm10::kanMX4 |
| HLY1840 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 VID30-HA-HIS3 |
| HLY1841 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vid24::kanMX4 VID30-HA-HIS3 |
| HLY1987 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 VID30-V5-His-URA3 |
| HLY2254 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 VID30-V5-His-URA3 VID24-HA-HIS3 |
| HLY2207 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 VID24-V5-His-URA3 VID30-HA-HIS3 |
| HLY2851 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sec28::kanMX4 VID30-V5-His-URA3 VID24-HA-HIS3 |
| HLY2884 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ABP1-mCherry-LEU2 |
| HLY2890 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 SAC6-GFP-HIS3 ABP1-mCherry-LEU2 |
| HLY2588 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 VID30-GFP-HIS3 |
| HLY1418 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 FBP1-GFP-HIS3 |
| HLY2586 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 GFP-VID24-GFP-URA3 |
| HLY2594 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sec28::kanMX4 VID30-GFP-HIS3 |
| HLY2676 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vid24::kanMX4 VID30-GFP-HIS3 |
| HLY1844 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vid30::kanMX4 VID24-HA-HIS3 |
| HLY2164 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vid30::kanMX4 FBP1-GFP-HIS3 |
| HLY1655 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vid30::kanMX4 GFP-VID24-HIS3 |
| HLY2751 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ΔLisH-VID30-V5-His-URA3 VID24-HA-HIS3 |
| HLY2691 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ΔLisH-VID30-GFP-HIS3 |
| HLY2257 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 FBP1-GFP-HIS3 ΔLisH-VID30-V5-His-URA3 |
| HLY2700 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ΔCTLH-VID30-V5-His-URA3 VID24-HA-HIS3 |
| HLY2693 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ΔCTLH-VID30-GFP-HIS3 |
| HLY2258 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 FBP1-GFP-HIS3 ΔCTLH-VID30-V5-His-URA3 |
Table 2.
Primers used in this study
| Abp1-GFP | |
| P204F | GACCCACGTCTTGTTCAAAAGCCAACCGCTGCTGGTTCCAAGATTGATCCT |
| P202R | GACGTCTTTGTATAGTTCATCCATGCCATGTGTAATCCCAGCAGCTGTTAC |
| Vid30-HA and Vid30-GFP | |
| P79 F | AACTCTTCAGATCCAAGATATTACAAAGCTATTAACTTCGACGAAGATGTGTTGAATTTACGGATCCCCGGGTTAATTAA |
| P79 R | GGTGTCAAGTAAATTTCATTATATAAACGGTTTAAGACATATTAATATGCGATTTTTGGGGAATTCGAGCTCGTTTAAAC |
| Vid30-V5-His | |
| P171 | CCCGGGGACGTCGACGTATCCTACACCTCTACTTCGACCATCACCACAACC |
| P172 | GGATCCGTTAATTAATAAATTCAACACATCTTCGTCGAAGTTAATAGCTTT |
| Vid24-HA | |
| P116F | CATCTTTGAAAAATAAAGTCGAGTCCAGTGATTGTTCTTTTGAGTTTGCTCGGATCCCCGGGTTAATTAA |
| P116R | TAGACATAGACATGCTGTTATCATACCAAATAGAAAAGTGTACAGTCTTTGAATTCGAGCTCGTTTAAAC |
| FBPase-GFP | |
| P121F | ATTTGGTTGGGTTCTTCAGGTGAAATTGACAAATTTTTAGACCATATTGGCAAGTCACAGCGGATCCCCGGGTTAATTAA |
| P121R | CCATCCCATTCCATTCGCTACTTCCTTTCTCTTTTCCTAAGAATTTTCATTATTAGAAGGGAATTCGAGCTCGTTTAAAC |
| LisH deletion | |
| P188 | TGCCCATTGACATTAACTGAACCGTCATCCACGC |
| P189 | GCGTGGATGACGGTTCAGTTAATGTCAATGGGCA |
| CTLH deletion | |
| P186 | TCAAAATTAAATTTTCAATTTCGTCATCACGTTCTTCTTTCATGATCTGTCTTTC |
| P187 | GAAAGACAGATCATGAAAGAAGAACGTGATGACGAAATTGAAAATTTAATTTTGA |
Differential centrifugation, FBPase degradation and His Pull Down.
FBPase degradation and differential centrifugation were performed as described previously.51,71 The preparation for Vid vesicle enriched fraction was performed as described.51 For differential centrifugation, wild-type cells expressing Abp1-mCherry were glucose starved for three d and harvested. Cell lysates were centrifuged sequentially at 1,000 × g, 13,000 × g, 100,000 × g and 200,000 × g. Pellet fractions from 1,000 × g (P1), 13,000 × g (P13), 100,000 × g (P100) and 200,000 × g (P200) and the cytosolic fraction (S200) were resolved by SDS-PAGE. The distribution of Abp1-mCherry and actin (Act1) was examined by western blotting. For His pull-down experiments, cells with various tags were glucose starved and shifted to glucose for 20 min. Cells were harvested and solubilized in buffer containing 2% Triton X-100. His pull-downs were performed according to the manufacturer's instructions (Qiagen, 30210). After pull-down, the samples were separated into unbound and bound fractions, which were then immuno-blotted with anti-FBPase antibodies and anti-Sec28 antibodies. Vid24-HA and Vid30-HA were detected with HA antibodies, while Vid24-V5-His and Vid30-V5-His were detected with anti-V5 antibodies.
Fluorescence microscopy.
For actin staining, yeast cells were grown under starvation conditions for three d in 2 ml of YPKG. At each time-point, samples of the cells (300 µl) were taken and fixed with 22 µl of formaldehyde for 5 min at room temperature. The cells were harvested by centrifugation at 1,500 g for 2.5 min. Following the removal of the supernatant, the cells were washed in 400 µl of PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). After further centrifugation at 1,500 g for 2.5 min, the supernatants were removed and −20°C acetone (800 ml) was added dropwise while vortexing the sample. The cells were then incubated for 5 min at −20°C. Cells were washed in 400 µl of PBS and re-suspended in 80 µl of PBS. The cells were finally stained with 1 µl of rhodamine-conjugated phalloidin at 0.2 U/µl in methanol and incubated for 30 min in the dark at room temperature. GFP and actin were visualized by fluorescence microscopy at 26°C with FLUAR 100X objective lens (1.30 NA) using FITC and rhodamine filters respectively. The cells were imaged using a Zeiss Axiovert S100 inverted microscope with an Axiocam MRm CCD camera and Axiovision v. 4.5 Software.
For FM staining studies, yeast cells were grown under starvation conditions for three d in 1 ml YPKG. At the 0 min time point, 100 µl was taken directly from the YPKG culture and 2.5 µl of 100 mM NaN3 was added. After centrifugation, cells were re-suspended in 1 ml YPD and 1 µl FM dye (16 mg/ml in DMSO) was added. At each designated time point thereafter, 100 µl aliquots of cells were taken from the YPD culture and 2.5 µl of 100 mM NaN3 was added. GFP and FM signals were visualized by fluorescence microscopy. The percent colocalization of GFP fusion proteins with either FM signal or actin was determined from cells from at least three images and was represented as mean and standard deviation.
Abp1-mCherry, LisH and CTLH domain mutation.
Abp1-GFP was PCR amplified with P202 and P204 from cells that expressed Abp1-GFP (Invitrogen) and cloned into TOPO plasmid (B557). The plasmid was digested with PshA1 and Pac1 and ligated with the fragment produced by Swa1 and Pac1 digestion from a plasmid harboring the mCherry and the LEU2 selection (B508). The resulting plasmid (B556) contained the fusion of Abp1 in frame with mCherry. The expression of Abp1-mCherry was confirmed by fluorescence microscopy. To produce the LisH and CTLH domain mutation, the VID30 gene was amplified by PCR reaction using P171 and P172 (Table 2) as the forward and reverse primers, respectively. The VID30 gene was cloned into a TOPO plasmid (Invitrogen) to produce B487. Vid30-GFP (B506) was generated by digesting the VID30-TOPO plasmid (B487) with PacI and ZraI, and this was ligated to a fragment that was produced by PacI and SwaI digestion of the FBPase-GFP-HIS3 plasmid (B430). Site directed mutagenesis was performed according to the manufacturer's suggestions (Strategene, 200518). The ΔCTLH mutation was generated by PCR using the P186 and P187 (Table 2) forward and reverse primers, respectively. The ΔLisH mutation was produced using the P188 and P189 (Table 2) primers. ΔLisH-Vid30-GFP (B523) was generated by site directed mutagenesis using a Vid30-GFP (B506) template and the P188 and P189 forward and reverse primers. ΔCTLH-Vid30-GFP (B525) was produced by PCR using the Vid30-GFP (B506) as the template and P186 and P187 as the forward and reverse primers, respectively. The resulting mutations were confirmed by DNA sequencing at the Core Facility of the Penn State University College of Medicine.
Acknowledgments
We thank Ryan Lucas for generating many of the reagents and plasmids used in this study. Primers were synthesized at the Core Facility of the Penn State University College of Medicine. This study was sponsored by an NIH grant R01 GM59480 and a Tobacco Settlement Fund to Hui-Ling Chiang.
Abbreviations
- FBPase
fructose-1,6-bisphosphatase
- MDH2
malate dehydrogenase
- Vid
vacuole import and degradation
- FM
N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide
References
- 1.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Platini F, Perez-Tomas R, Ambrosio S, Tessitore L. Understanding autophagy in cell death control. Curr Pharm Des. 2010;16:101–113. doi: 10.2174/138161210789941810. [DOI] [PubMed] [Google Scholar]
- 3.Cherra SJ, Dagda RK, Chu CT. Autophagy and Neurodegeneration: Survival at a cost? Neuropathol Appl Neurobiol. 2010;36:125–132. doi: 10.1111/j.1365-2990.2010.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Arencibia M, Hochfeld WE, Toh PP, Rubinsztein DC. Autophagy, a guardian against neurodegeneration. Semin Cell Dev Biol. 2010;21:691–698. doi: 10.1016/j.semcdb.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: Molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010;21:719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang RC, Levine B. Autophagy in cellular growth control. FEBS Lett. 2010;584:1417–1426. doi: 10.1016/j.febslet.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monastyrska I, Rieter E, Klionsky DJ, Reggiori F. Multiple roles of the cytoskeleton in autophagy. Biol Rev Camb Philos Soc. 2009;84:431–448. doi: 10.1111/j.1469-185X.2009.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 11.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 13.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang DS, Lee JH, Nixon RA. Monitoring autophagy in Alzheimer's disease and related neurodegenerative diseases. Methods Enzymol. 2009;453:111–144. doi: 10.1016/S0076-6879(08)04006-8. [DOI] [PubMed] [Google Scholar]
- 16.Yang F, Yang YP, Mao CJ, Cao BY, Cai ZL, Shi JJ, et al. Role of autophagy and proteasome degradation pathways in apoptosis of PC12 cells overexpressing human alpha-synuclein. Neurosci Lett. 2009;454:203–208. doi: 10.1016/j.neulet.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 18.Mijaljica D, Prescott M, Klionsky DJ, Devenish RJ. Autophagy and vacuole homeostasis: a case for self-degradation? Autophagy. 2007;3:417–421. doi: 10.4161/auto.4441. [DOI] [PubMed] [Google Scholar]
- 19.Matsushita M, Suzuki NN, Fujioka Y, Ohsumi Y, Inagaki F. Expression, purification and crystallization of the Atg5-Atg16 complex essential for autophagy. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:1021–1023. doi: 10.1107/S1744309106036232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsushita M, Suzuki NN, Obara K, Fujioka Y, Ohsumi Y, Inagaki F. Structure of Atg5.Atg16, a complex essential for autophagy. J Biol Chem. 2007;282:6763–6772. doi: 10.1074/jbc.M609876200. [DOI] [PubMed] [Google Scholar]
- 21.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 22.Renna M, Jimenez-Sanchez M, Sarkar S, Rubinsztein DC. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem. 2010;285:11061–11067. doi: 10.1074/jbc.R109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng S, Clabough EB, Sarkar S, Futter M, Rubinsztein DC, Zeitlin SO. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010;6:e1000838. doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menzies FM, Huebener J, Renna M, Bonin M, Riess O, Rubinsztein DC. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2010;133:93–104. doi: 10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubinsztein DC. Autophagy: where next? EMBO Rep. 2010;11:3. doi: 10.1038/embor.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minard KI, McAlister-Henn L. Glucose-induced degradation of the MDH2 isozyme of malate dehydrogenase in yeast. J Biol Chem. 1992;267:17458–17464. [PubMed] [Google Scholar]
- 27.Minard KI, McAlister-Henn L. Glucose-induced phosphorylation of the MDH2 isozyme of malate dehydrogenase in Saccharomyces cerevisiae. Arch Biochem Biophys. 1994;315:302–309. doi: 10.1006/abbi.1994.1504. [DOI] [PubMed] [Google Scholar]
- 28.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 29.Holzer H. Proteolytic catabolite inactivation in Saccharomyces cerevisiae. Revis Biol Celular. 1989;21:305–319. [PubMed] [Google Scholar]
- 30.Toyoda Y, Fujii H, Miwa I, Okuda J, Sy J. Anomeric specificity of glucose effect on cAMP, fructose 1,6-bisphosphatase, and trehalase in yeast. Biochem Biophys Res Commun. 1987;143:212–217. doi: 10.1016/0006-291X(87)90652-8. [DOI] [PubMed] [Google Scholar]
- 31.Chiang HL, Schekman R. Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature. 1991;350:313–318. doi: 10.1038/350313a0. [DOI] [PubMed] [Google Scholar]
- 32.Chiang HL, Schekman R, Hamamoto S. Selective uptake of cytosolic, peroxisomal, and plasma membrane proteins into the yeast lysosome for degradation. J Biol Chem. 1996;271:9934–9941. doi: 10.1074/jbc.271.17.9934. [DOI] [PubMed] [Google Scholar]
- 33.Gancedo C. Inactivation of fructose-1,6-diphosphatase by glucose in yeast. J Bacteriol. 1971;107:401–405. doi: 10.1128/jb.107.2.401-405.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gamo FJ, Navas MA, Blazquez MA, Gancedo C, Gancedo JM. Catabolite inactivation of heterologous fructose-1,6-bisphosphatases and fructose-1,6-bisphosphatase-beta- galactosidase fusion proteins in Saccharomyces cerevisiae. Eur J Biochem. 1994;222:879–884. doi: 10.1111/j.1432-1033.1994.tb18935.x. [DOI] [PubMed] [Google Scholar]
- 36.Horak J, Regelmann J, Wolf DH. Two distinct proteolytic systems responsible for glucose-i nduced degradation of fructose-1,6-bisphosphatase and the Gal2p transporter in the yeast Saccharomyces cerevisiae share the same protein components of the glucose signaling pathway. J Biol Chem. 2002;277:8248–8254. doi: 10.1074/jbc.M107255200. [DOI] [PubMed] [Google Scholar]
- 37.Horak J, Wolf DH. The ubiquitin ligase SCF(Grr1) is required for Gal2p degradation in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2005;335:1185–1190. doi: 10.1016/j.bbrc.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Riballo E, Herweijer M, Wolf DH, Lagunas R. Catabolite inactivation of the yeast maltose transporter occurs in the vacuole after internalization by endocytosis. J Bacteriol. 1995;177:5622–5627. doi: 10.1128/jb.177.19.5622-5627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gadura N, Michels CA. Sequences in the N-terminal cytoplasmic domain of Saccharomyces cerevisiae maltose permease are required for vacuolar degradation but not glucose-induced internalization. Curr Genet. 2006;50:101–114. doi: 10.1007/s00294-006-0080-3. [DOI] [PubMed] [Google Scholar]
- 40.Regelmann J, Schule T, Josupeit FS, Horak J, Rose M, Entian KD, et al. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol Biol Cell. 2003;14:1652–1663. doi: 10.1091/mbc.E02-08-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schüle T, Rose M, Entian KD, Thumm M, Wolf DH. Ubc8p functions in catabolite degradation of fructose-1, 6-bisphosphatase in yeast. EMBO J. 2000;19:2161–2167. doi: 10.1093/emboj/19.10.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schork SM, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J Biol Chem. 1995;270:26446–26450. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- 43.Schork SM, Bee G, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase in yeast is mediated by the proteasome. FEBS Lett. 1994;349:270–274. doi: 10.1016/0014-5793(94)00668-7. [DOI] [PubMed] [Google Scholar]
- 44.Hämmerle M, Bauer J, Rose M, Szallies A, Thumm M, Dusterhus S, et al. Proteins of newly isolated mutants and the amino-terminal proline are essential for ubiquitin-proteasome- catalyzed catabolite degradation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. J Biol Chem. 1998;273:25000–25005. doi: 10.1074/jbc.273.39.25000. [DOI] [PubMed] [Google Scholar]
- 45.Hung GC, Brown CR, Wolfe AB, Liu J, Chiang HL. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J Biol Chem. 2004;279:49138–49150. doi: 10.1074/jbc.M404544200. [DOI] [PubMed] [Google Scholar]
- 46.Shieh HL, Chen Y, Brown CR, Chiang HL. Biochemical analysis of fructose-1,6-bisphosphatase import into vacuole import and degradation vesicles reveals a role for UBC1 in vesicle biogenesis. J Biol Chem. 2001;276:10398–10406. doi: 10.1074/jbc.M001767200. [DOI] [PubMed] [Google Scholar]
- 47.Shieh HL, Chiang HL. In vitro reconstitution of glucose-induced targeting of fructose-1, 6- bisphosphatase into the vacuole in semi-intact yeast cells. J Biol Chem. 1998;273:3381–3387. doi: 10.1074/jbc.273.6.3381. [DOI] [PubMed] [Google Scholar]
- 48.Huang PH, Chiang HL. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J Cell Biol. 1997;136:803–810. doi: 10.1083/jcb.136.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown CR, McCann JA, Hung GG, Elco CP, Chiang HL. Vid22p, a novel plasma membrane protein, is required for the fructose-1,6-bisphosphatase degradation pathway. J Cell Sci. 2002;115:655–666. doi: 10.1242/jcs.115.3.655. [DOI] [PubMed] [Google Scholar]
- 50.Brown CR, Cui DY, Hung GG, Chiang HL. Cyclophilin A mediates Vid22p function in the import of fructose-1,6-bisphosphatase into Vid vesicles. J Biol Chem. 2001;276:48017–48026. doi: 10.1074/jbc.M109222200. [DOI] [PubMed] [Google Scholar]
- 51.Brown CR, McCann JA, Chiang HL. The heat shock protein Ssa2p is required for import of fructose-1, 6-bisphosphatase into Vid vesicles. J Cell Biol. 2000;150:65–76. doi: 10.1083/jcb.150.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiang MC, Chiang HL. Vid24p, a novel protein localized to the fructose-1, 6-bisphosphatase-containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J Cell Biol. 1998;140:1347–1356. doi: 10.1083/jcb.140.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown CR, Wolfe AB, Cui D, Chiang HL. The vacuolar import and degradation pathway merges with the endocytic pathway to deliver fructose-1,6-bisphosphatase to the vacuole for degradation. J Biol Chem. 2008;283:26116–26127. doi: 10.1074/jbc.M709922200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown CR, Dunton D, Chiang HL. The vacuole import and degradation pathway utilizes early steps of endocytosis and actin polymerization to deliver cargo proteins to the vacuole for degradation. J Biol Chem. 2010;285:1516–1528. doi: 10.1074/jbc.M109.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galletta BJ, Cooper JA. Actin and endocytosis: mechanisms and phylogeny. Curr Opin Cell Biol. 2009;21:20–27. doi: 10.1016/j.ceb.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/S0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 58.Sekiya-Kawasaki M, Groen AC, Cope MJ, Kaksonen M, Watson HA, Zhang C, et al. Dynamic phosphoregulation of the cortical actin cytoskeleton and endocytic machinery revealed by real- time chemical genetic analysis. J Cell Biol. 2003;162:765–772. doi: 10.1083/jcb.200305077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 60.Santt O, Pfirrmann T, Braun B, Juretschke J, Kimmig P, Scheel H, et al. The yeast GID complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism. Mol Biol Cell. 2008;19:3323–3333. doi: 10.1091/mbc.E08-03-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snowdon C, Hlynialuk C, van der Merwe G. Components of the Vid30c are needed for the rapamycin-induced degradation of the high-affinity hexose transporter Hxt7p in Saccharomyces cerevisiae. FEMS Yeast Res. 2008;8:204–216. doi: 10.1111/j.1567-1364.2007.00327.x. [DOI] [PubMed] [Google Scholar]
- 62.van der Merwe GK, Cooper TG, van Vuuren HJ. Ammonia regulates VID30 expression and Vid30p function shifts nitrogen metabolism toward glutamate formation especially when Saccharomyces cerevisiae is grown in low concentrations of ammonia. J Biol Chem. 2001;276:28659–28666. doi: 10.1074/jbc.M102280200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mizuguchi M, Takashima S, Kakita A, Yamada M, Ikeda K. Lissencephaly gene product. Localization in the central nervous system and loss of immunoreactivity in Miller-Dieker syndrome. Am J Pathol. 1995;147:1142–1151. [PMC free article] [PubMed] [Google Scholar]
- 64.Chong SS, Pack SD, Roschke AV, Tanigami A, Carrozzo R, Smith AC, et al. A revision of the lissencephaly and Miller-Dieker syndrome critical regions in chromosome 17p13.3. Hum Mol Genet. 1997;6:147–155. doi: 10.1093/hmg/6.2.147. [DOI] [PubMed] [Google Scholar]
- 65.Lo Nigro C, Chong CS, Smith AC, Dobyns WB, Carrozzo R, Ledbetter DH. Point mutations and an intragenic deletion in LIS1, the lissencephaly causative gene in isolated lissencephaly sequence and Miller-Dieker syndrome. Hum Mol Genet. 1997;6:157–164. doi: 10.1093/hmg/6.2.157. [DOI] [PubMed] [Google Scholar]
- 66.Dobyns WB, Stratton RF, Greenberg F. Syndromes with lissencephaly. I: Miller-Dieker and Norman-Roberts syndromes and isolated lissencephaly. Am J Med Genet. 1984;18:509–526. doi: 10.1002/ajmg.1320180320. [DOI] [PubMed] [Google Scholar]
- 67.Kobayashi N, Yang J, Ueda A, Suzuki T, Tomaru K, Takeno M, et al. RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8-alpha and ARMC8beta are components of the CTLH complex. Gene. 2007;396:236–247. doi: 10.1016/j.gene.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 68.Tomaru K, Ueda A, Suzuki T, Kobayashi N, Yang J, Yamamoto M, et al. Armadillo Repeat Containing 8alpha Binds to HRS and Promotes HRS Interaction with Ubiquitinated Proteins. Open Biochem J. 2010;4:1–8. doi: 10.2174/1874091X01004010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kusmierczyk AR, Hochstrasser M. Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol Chem. 2008;389:1143–1151. doi: 10.1515/BC.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 71.Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Finley D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]