Abstract
Background & Aims
The Helicobacter pylori toxin vacuolating cytotoxin (VacA) promotes gastric colonization and its presence (VacA+) is associated with more-severe disease. The exact mechanisms by which VacA contributes to infection are unclear. We previously found that limited exposure to VacA induces autophagy of gastric cells, which eliminates the toxin; we investigated whether autophagy serves as a defense mechanism against H pylori infection.
Methods
We investigated the effect of VacA on autophagy in human gastric epithelial cells (AGS) and primary gastric cells from mice. Expression of p62, a marker of autophagy, was also assessed in gastric tissues from patients infected with toxigenic (VacA+) or nontoxigenic strains. We analyzed the effect of VacA on autophagy in peripheral blood monocytes obtained from subjects with different genotypes of ATG16L1, which regulates autophagy. We performed genotyping for ATG16L1 in two cohorts of infected and uninfected subjects.
Results
Prolonged exposure of AGS and mouse gastric cells to VacA disrupted induction of autophagy in response to the toxin, because the cells lacked cathepsin-D in autophagosomes. Loss of autophagy resulted in the accumulation of p62 and reactive oxygen species. Gastric biopsies samples from patients infected with VacA+, but not nontoxigenic strains of H pylori, had increased levels of p62. Peripheral blood monocytes isolated from individuals with polymorphisms in ATG16L1 that increase susceptibility to Crohn's disease had reduced induction of autophagy in response to VacA+ compared to cells from individuals that did not have these polymorphisms. The presence of the ATG16L1 Crohn’s disease risk variant increased susceptibility to H pylori infection in 2 separate cohorts.
Conclusions
Autophagy protects against infection with H pylori; the toxin VacA disrupts autophagy to promote infection, which could contribute to inflammation and eventual carcinogenesis.
Keywords: stomach cancer, genetic, bacteria toxin, tumor
Introduction
Helicobacter pylori has been classified as a Class I carcinogen by the World Health Organization [1]. Chronic H. pylori infection is a risk for the onset of serious gastric disease, including peptic ulcer disease, mucosa lymphoid tissue (MALT) lymphoma and gastric cancer, the second leading cause of cancer deaths worldwide [1]. The mechanisms responsible for initial susceptibility to infection and subsequent chronic infection involve a complex interplay between host and bacterial factors. One specific virulence factor important for colonization and disease outcome is the vacuolating cytotoxin (VacA) [2]. The exact mechanisms by which VacA contributes to colonization and disease outcome remain unclear.
Autophagy is an evolutionarily conserved process that results in the sequestration of cytosolic components within double membrane compartments called autophagosomes. These compartments fuse with lysosomes to become autophagolysosomes, which degrade vesicle contents through the action of lysosomal hydrolases. Although initially recognized to occur in response to cellular stresses such as nutrient starvation, current evidence indicates that autophagy plays a critical role in modulating host immunity and inflammatory responses. Importantly autophagy is thought to serve as an innate defense mechanism against infection. Autophagy is stimulated in response to the invasion of intracellular pathogens and the presence of bacterial toxins [3]. In addition, studies in Caenorohabditis elegans, Drosophila and mice suggest that autophagy is a critical pathway for controlling infection. For example, inactivation of an autophagy dependent gene in C. elegans increases intracellular replication of Salmonella typhimurium [4]. In mice, disruption of a bacterial-selective autophagy pathway promotes intracellular replication of Shigella [5]. However, a number of pathogens evade or utilize the autophagy pathway for enhanced survival and persistence in host cells [6,7].
While an increasing number of pathogens have been demonstrated to subvert autophagic pathways to promote intracellular survival, much less is known about the potential consequences of pathogen-mediated disruption of autophagic pathways or, alternatively, the influence of autophagic defects within host cells for controlling infection in humans and influencing disease pathology. Recent studies from our lab revealed that within gastric epithelial cells in vitro, autophagic signaling is induced in response to infection with H. pylori in a VacA-dependent manner [8]. Furthermore, autophagy eliminates VacA. Here, we specifically evaluated the biologic significance of the autophagy pathway during H. pylori infection in vitro and in vivo in human subjects, and assessed the influence of both pathogen and host genetic factors.
Methods and Materials
Cells, bacteria and antibodies
Culture conditions for human gastric epithelial cells (AGS) and murine primary gastric cells were as described previously [9,10]. Growing conditions for wild-type H. pylori strain 60190 (ATCC 49503; cagA+ cagE+ VacA+) and its isogenic vacA mutant strain (provided by Dr. R. Peek) were as described previously [11,12]. Rabbit polyclonal p62 antibodies were from Santacruz Biotechnology, CA. Rabbit anti-H. pylori antibodies were from DAKO (Denmark). Mouse anti-human Lamp1 antibodies were purchased from (Developmental Studies Hybridoma Bank, Iowa City, IA). Cathepsin D antibodies were from Upstate (Lake Placid, NY). Rabbit anti-VacA antibodies were a kind gift from Dr. S. R. Blanke. All other reagents were obtained from Sigma-Aldrich (St. Louis, MO).
Detection of reactive oxygen species and cell death using FACS
For the detection of reactive oxygen species, a redox-sensitive dye 5-(and-6)-chloromethyl-20,70-dichlorodihydrofluorescein diacetate, acetyl ester(CM-H2DCFDA) was used. Fluorescence was measured using a FACSCalibur flow cytometer.
Cell death analysis was performed using 3µM propidium iodide (Invitrogen) in PBS buffer and assessed by fluorescent-activated cell sorting FACS (FACSCalibur, BD Biosciences). The percentage of stained cells was determined and compared with appropriate negative controls.
Isolation of Peripheral blood monocytes (PBMC)
Venous blood was drawn from healthy genotyped volunteers in sterile EDTA coated tubes (BD Vacutainer,) diluted 1:1 with pyrogen-free saline and layered over Ficoll-Paque (GE Healthcare Sciences, Canada). Approval for the study was provided by the local ethics board (Approval No. MSH REB#02-0234-E).
Cells were spun at 400g for 30min to obtain the mononuclear cell fractionation via density centrifugation, washed and suspended in culture medium. Cells were counted and plated in 6-well plates where they were treated with various stimuli: muramyl dipeptide (MDP, 10µg/ml, InvivoGen, San Diego), and concentrated culture supernatants of H. pylori strain 60190 (200µl/ml) for 24 hours with bafilomycin (50µg/ml, Sigma) and rapamycin (100ng/ml, Enzo Life Sciences International, USA) used for the last 4 hours of incubation. Collected cells were then lysed and used for immunoblots.
Densitometric analysis was performed using FlourChem FCII software. All blots from each independent experiment were used. Densities of the LC3 II and actin bands were measured for each treatment and expressed as a ratio of LC3II/actin, normalized to each subjects control.
Statistical analysis
Analysis of variance (ANOVA), unpaired T test and other statistical analysis of the results were performed utilizing GraphPad Prism 4 for Macintosh V 4.0b.
Other assays
Immunofluorescence, immunohistochemistry, immunoblotting, bacterial survival, genotyping and other assays were performed as previously described and are outlined in detail in the supplementary materials section.
Results
Prolonged exposure to VacA toxin causes accumulation of defective autophagosomes
Previous studies have shown that limited exposure to H. pylori VacA for 6h triggers autophagy in AGS cells, resulting in degradation of the toxin in autophagosomes suggesting that autophagy serves as a protective host response during infection [8]. To further assess the biologic significance of the pathway during infection we exposed gastric cells to VacA toxin for prolonged periods to mimic the setting of chronic infection and utilized the LC3-eGFP-mRFP tandem construct to follow the maturation of autophagosomes [13]. After formation, autophagosomes fuse with lysosomes and acquire cathepsins and acid phosphatases to become mature autolysosomes [6,14]. Modified LC3 remains associated with autophagosomes until hydrolysis at the autolysosomal stage. During autophagosome maturation the mRFP-LC3 signal persists longer than the GFP-LC3 signal since mRFP is resistant to autolysosomal proteases and has a lower pKa [13]. As we had previously shown that VacA was necessary and sufficient to induce autophagy and that autophagy did not occur during infection with isogenic vacA− mutants we focused our experiments on VacA. When AGS cells transfected with the tandem construct were treated with culture supernatants from VacA+ H. pylori (CCMS), both the GFP-LC3 and mRFP-LC3 signals persisted (Figure 1B,D) in a comparable manner to cells incubated with vinblastine (Figure 1C,D), which prevents autophagosome-lysosome fusion. These findings were recapitulated in VacA+ CCMS treated primary murine gastric epithelial cells indicating the effect was not related to cancerous transformation (Supplementary figure 1).
Figure 1. Effects of prolonged VacA treatment on autophagy.
The confocal micrographs in panels A–C show accumulation of LC3-eGFP-mRFP in AGS cells incubated in nutrient starvation conditions (A), treated with wild type CCMS (B) or vinblastine (C) at different time points (2h, 6h and 24h). Panel D shows the comparison of the percent co-localization of the GFP and mRFP signals under the different conditions at 2h and 6h. * p< 0.001, as analyzed by Student’s T-test. Experiments were performed twice, each time in duplicate and at least 10 cells were used to calculate the Pearson’s co-localization coefficient using the Volocity acquisition software.
Panel E: The graph shows the quantitation of the hydrolysis of long half-life proteins in control, CCMS treated and rapamycin treated cells. AGS cells were grown for 72h in the presence of [3H]leucine and exposed to CCMS or rapamycin for 8h. [3H]leucine release due to protein hydrolysis was calculated as a percentage of the total radioactivity present in the medium normalized by control cell data. Panel F: Quantitation of cell viability using FACS analysis of propidium iodide staining of AGS cells incubated with CCMS (black bars) or standard culture media (white bars) for 12h prior to treatment with the indicated concentrations of rapamycin for an additional 12h. Graphs depict results (expressed as the mean± SEM) of 3 independent experiments performed in triplicate. * represents p<0.05
Next we measured the hydrolysis of long half-life proteins, whose turnover depends on autophagy, in VacA+ CCMS treated cells [15,16]. Figure 1E shows that the degradation of long half-life proteins in VacA+ CCMS treated cells was lower than in cells undergoing autophagy triggered by rapamycin treatment. Taken together, these findings suggest that the autophagosomes formed by VacA are defective and have reduced catalytic activity.
VacA protects cells from rapamycin-mediated cell death
Autophagy is considered cytoprotective but in certain settings excessive autophagy can cause cell death [17,18]. For instance, overexpression of Atg1 in Drosophila melanogaster salivary glands triggers an autophagic phenotype followed by cell loss which is prevented by inhibiting autophagy [19]. Similarly prolonged treatment with rapamycin, which triggers autophagy, induces death of mammalian cells [20]. Therefore, we next assessed the effect of VacA mediated changes in autophagy on rapamycin-induced cell death. Similar to ATG12 siRNA treated cells, pre-incubation of cells with VacA + CCMS attenuated rapamycin-triggered cell death (Figure 1F, Supplementary figure 2). These findings indicate that VacA effects on autophagy can promote increased cell viability in response to rapamycin comparable to siRNA-mediated disruption of autophagy (Supplementary figure 2).
VacA induced autophagosomes lack Cathepsin-D
We next determined if VacA+CCMS-triggered autophagosomes failed to mature due to lack of fusion with lysosomes. VacA+CCMS-induced autophagosomes acquired the late endosomal lysosomal marker Lamp1 (Figure 2) and co localized with lysosensor probes (Supplementary figure 3) indicating that autophagosome-lysosome fusion and acidification were not affected.
Figure 2. VacA reduces Cathepsin D sorting to autophagosomes.
The micrographs in panels A–C show 3D projections of de-convolved confocal z-stack slices obtained each 0.25 µm from (A) control GFP-LC3 (green) expressing AGS cells, or GFP-LC3 (green) expressing AGS cells treated with (B) rapamycin, (C) starvation, (D) vinblastine or (E) with VacA +CCMS for12h. After treatment cells were immunolabeled for cathepsin D (blue) and Lamp-1 (Red). The insets in panels A–C show details of the framed area in the main panels. The top insets show the merge of GFP-LC3 and cathepsin D. The middle insets show the merge of GFP-LC3 and Lamp-1. The bottom insets show the merge of GFP-LC3, cathepsin D and Lamp-1. The scale bars indicate 10µm. Micrographs are representative of 100 cells observed in three independent experiments.
Previous reports indicate that VacA alters the degradative properties of the endocytic pathway by subverting the sorting and activation of cathepsin enzymes [11,21]. We had shown that in VacA positive but not isogenic VacA mutant infected cells, lysosomes lack cathepsin D, a key hydrolase, which in turn affects their degradative properties [11,21]. Therefore, we determined if cathepsin D was present in autophagosomes in VacA+CCMS treated cells. As shown in Figure 2 A–E and Supplementary figure 4, VacA+CCMS treated cells displayed a reduction in cathepsin D over time in comparison with control cells. Furthermore, in comparison with rapamycin, starvation, and vinblastine treated cells, negligible amounts of cathepsin D were found in Lamp1-labeled late endosomes in CCMS-treated cells. These findings indicate that VacA -induced autophagosomes fuse with lysosomes and acquire an acidic pH but lack cathepsin D and thus the catalytic activity required to complete the autophagic degradative process [22].
VacA induces formation of p62 aggregates in vitro and in vivo
Dysfunctional autophagy leads to an accumulation of p62, which has been implicated in promoting tumorigenesis [23,24]. Therefore, we determined the levels of p62 in AGS cells treated with VacA+CCMS. AGS cells treated with VacA+CCMS showed accumulation of p62, which increased over time and co-localized with GFP-LC3 (Figure 3A–E). The accumulation of p62 in VacA+CCMS-treated cells was similar to the p62 aggregates in cells where autophagy was suppressed using ATG12 siRNA (data not shown).
Figure 3. VacA induces p62 aggregation both in vitro and in vivo.
The accumulation of p62 in AGS cells (Panel A–C) and in gastric biopsies from patients infected with VacA (+) H. pylori or VacA(–) H. pylori is shown in these representative images (Panel D–E). Panel A shows control AGS cells transfected with GFP-LC3 and treated with CCMS either from wild type H. pylori (CCMS +) or VacA mutant H. pylori (CCMS−). Cells were fixed and immunostained for p62 (Red). Insets show the colocalization of GFP-LC3 and p62. Scale bars indicate 10µm.
Panel B shows the accumulation of p62 in CCMS treated cells by Western blot with p62 antibody. Cells were left untreated or treated with CCMS from wild type H. pylori or VacA mutant H. pylori for 24 h. Lysates were then collected and Western blot was performed. β-actin was used as loading control. The blots shown are a representative of three independent experiments. Panel C shows the quantitation of p62 by densitometry for three different western blots. Cells treated with CCMS from wild type H. pylori showed significant accumulation of p62 compared to control cells. Statistical analysis was performed using a one way ANOVA on the Graph Pad Prism program. (P value=0.0133).
Panel D shows immunohistochemistry with p62 antibody on gastric biopsies from patients infected with cagA (+) vacA s1m1 and cagA(−) vacA s2m2 strains of H. pylori and quantitation in corresponding graph. Arrows indicate increased p62 staining in the gastric epithelial cells. Statistical analysis was performed using a Student’s T-test. * indicates P value = 0.0453
To extend our in vitro findings to the bacterial niche in humans, we next looked at p62 expression in human gastric biopsies. All H. pylori strains carry the VacA gene and can be divided into toxigenic, expressing a functional VacA toxin, or nontoxigenic based on their VacA genotype (s1m1 vs s2m2). Gastric tissue sections from patients infected with toxigenic s1m1 VacA-producing strains showed a significantly higher accumulation of p62 in the foveolar cells of the gastric epithelium when compared to tissue sections obtained from patients infected with a nonfunctional VacA genotype (s2m2) strain (Figure 3F–G). These in vivo observations were concordant with our in vitro findings. Furthermore, the expression level of p62 was independent of the degree of gastric inflammation indicating that p62 accumulation was likely due to a bacterial factor (data not shown)
In addition to increasing p62 aggregation, disrupted autophagy increases levels of reactive oxygen species (ROS) in tumor cells[23]. Therefore we next assessed the effect of VacA treatment on ROS expression in gastric cells by flow cytometry. As seen in Fig 4 (A–B), treatment with purified active VacA toxin increased the levels of ROS comparable to cells treated with hydrogen peroxide. In contrast levels of ROS in inactive VacA toxin-treated cells were comparable to untreated control cells. The VacA-mediated increased ROS was ameliorated by pre treatment with the anti-oxidant N- acetyl cysteine (Fig 4B). These findings indicate that VacA treatment increases ROS expression, which could be mediated by VacA’s effect on autophagy.
Figure 4. VacA increases production of reactive oxygen species.
AGS cells were incubated with activated VacA pure toxin or inactive toxin for 4h. Where indicated, the anti-oxidant NAC (5mM) was utilized alone, or with pure toxin. Cells incubated with H2O2 (200µM, 30min) served as a positive control. Panel A shows ROS production measured by using a redox-sensitive dye (CM-H2DCFDA) on live cells, followed by flow cytometry analysis. Overlaid curves indicate fluorescence curve for condition stated (green) versus control curve (red) with corresponding numbers representing the percentage of ROS + cells assessed within the gated region. In Panel B, the graph shows quantification of ROS experiments. Fold increase is expressed as ratio of the %ROS+ cells in each condition/control. Columns, means; bars, SE; *, P < 0.05, **, P< 0.01, using one-way ANOVA (n=3). The overall P value = 0.001.
Autophagy restricts the growth of intracellular VacA+ H. pylori
Autophagy can inhibit the growth of intracellular bacteria and our results suggest that prolonged exposure to VacA disarms autophagosomes thereby inhibiting their anti-bacterial function. To conclusively determine the effect of autophagy on intracellular survival of H. pylori, we employed wildtype and autophagy deficient Atg5−/− MEFs. Similar to mutant Shigella whose growth is restricted by autophagy[25], survival of VacA+ H. pylori was increased in autophagy disrupted Atg5−/− MEFs in comparison with wildtype MEFs (Figure 5). These findings indicate that autophagy restricts the growth of VacA+ H. pylori.
Figure 5. Disrupting autophagy during infection with VacA+ H. pylori enhances intracellular survival.
Panel A, B: The graphs show the quantitation of the intracellular survival of Shigella ΔiscB or VacA+ H. pylori in wildtype or Atg5−/− MEFs. Cells were infected with Shigella ΔiscB or VacA+ H. pylori under gentamicin assay conditions (see Materials and Methods). At the indicated invasion times, intracellular bacteria were retrieved from cells utilizing 1% saponin in PBS buffer and plated on Brucella agar (H. pylori) or LB agar (Shigella) for enumeration of colony formation units (CFU). Data is expressed as percent of bacteria quantitated at specificied time point compared to initial CFU of invasive bacteria detected at 6hrs for VacA+ H. pylori and 2hrs for Shigella ΔiscB. *P<0.001 Panel C shows wildtype and Atg5−/− MEF’s transfected with LC3-GFP and infected with wild type VacA+ H. pylori. Cells were fixed and observed under a spinning disk confocal microscope. Scale bars = 10µm.
Crohn’s disease autophagy risk allele reduces autophagy in response to VacA toxin
Our previous studies indicated that while autophagy targets VacA to eliminate the toxin and restrict the growth of the bacteria, during prolonged exposure to VacA the toxin can counteract this effect by disrupting autophagy. Therefore, we next asked whether alterations in the host genetic autophagy machinery could modulate infection in the human setting. We took advantage of the recent genome wide association studies that identified a SNP in the autophagy gene, ATG16L1 (ATG16L1T300A-rs2241880), as a causal risk variant for Crohn’s disease [26,27]. Although the functional relevance of this variant remains unclear, it is speculated that the ATG16L1 CD risk allele may result in an unstable ATG16L1 protein and lead to impaired cytokine responses and anti-microbial autophagy, though without affecting basal autophagy function [28–30].
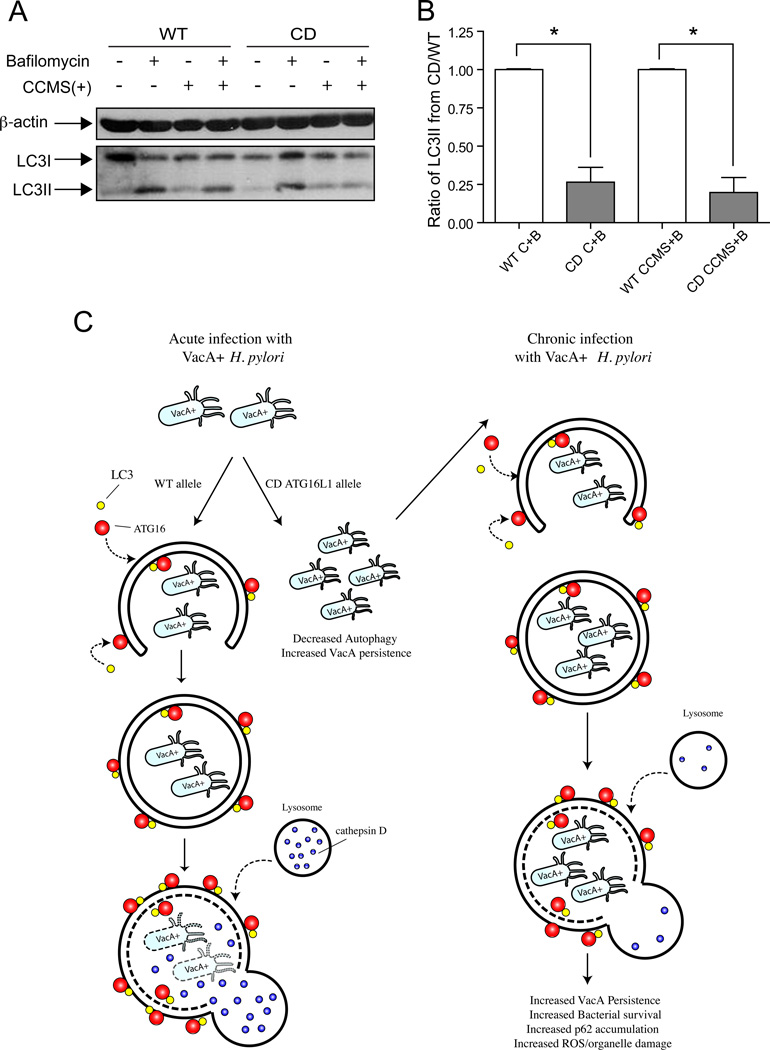
We first assessed if ATG16L1 was required for VacA-mediated autophagy. In ATG16L1 siRNA-treated cells a lack of LC3 II conversion in response to CCMS was detected indicating requirement for ATG16L1 (Supplementary figure 5). To determine if the ATG16L1 genotype influenced VacA mediated autophagy, we reconstituted the wildtype or CD allele in ATG16L1 knockdown cells. However, we could only rescue the autophagy phenotype by overexpressing ATG16L1 (Supplementary figure 5). Therefore, we assessed the induction of autophagy in response to VacA or CCMS in peripheral blood monocytes (PBMCs). As previous studies indicate the effects of the toxin may differ in immune cells compared to epithelial cells we first characterized the autophagic responses to VacA in PBMCs. The response to VacA in PBMCs recapitulated the findings observed in gastric epithelial cells as demonstrated by increased conversion of LC3II and p62 expression (Supplementary Figure 6). We then utilized PBMCs obtained from healthy volunteers who had been genotyped for ATG16L1. As we had shown previously that Nod2fs and ATG16L1 interact in the autophagic pathway we also genotyped these individuals for NOD2fs. None of the subjects harbored the NOD2fs mutation. Induction of autophagy and autophagic flux was determined by assessing conversion of LC3 I to LC3 II in control and CCMS-treated cells treated with bafilomycin. The LC3II/LC3I ratios of PBMC’s harboring the risk 300A allele were expressed as ratios of LC3II/LC3I in PBMC’s harboring the wild type protective 300T allele to indicate the difference between the two genotypes. In PBMCs with the protective 300T allele, VacA+ CCMS increased LC3 II conversion (Figure 6A,B) comparable to the Nod2 ligand MDP (data not shown), which is known to induce autophagy [31]. In PBMCs from individuals harboring the 300A “risk” allele, the response to VacA+CCMS was reduced when compared to the 300T allele-harboring PBMCs (Figure 6 A, B).
Figure 6. Autophagy response is diminished in PBMC’s from individuals harboring the Crohn’s disease variant (T300A) of ATG16L1.
Panel A shows a representative western blot image using PBMC lysates. Lysates were run on an SDS-PAGE gel and Western blotting performed using anti-LC3 antibody. The blot shows the formation of LC3 II in PBMC lysates from individuals with the WT and CD allele in response to VacA+ CCMS. β-actin was used as loading control in both experiments. In Panel B, graphs depict densitometric analysis of LC3 II:LC3 I/actin ratio from PBMCs obtained from volunteers with either the WT or CD variant of ATG16L1. Cells were either untreated (C) or treated with CCMS for 24 hrs and with bafilomycin (B) added during the last 4 hrs of treatment. The ratios are normalized to the control or CCMS plus bafilomycin treated wild type control, respectively. Statistical analysis was performed using a one way ANOVA with a post test to compare each column to its control, * in Panel B indicates P value<0.0001.
Panel C shows the model depicting the effect of acute and chronic infection with H. pylori on the autophagy pathway in hosts with either wild type ATG16L1 or the CD risk allele.
Variants in the host autophagy gene, ATG16L1, increase susceptibility to infection
These in vitro findings suggested that induction of autophagy in response to VacA is inefficient in the presence of the CD ATG16L1 risk allele. Since our studies indicate that autophagy restricts the intracellular survival of VacA+ H. pylori we next determined if the presence of this ATG16L1 variant had any impact on susceptibility to infection in humans. We initially studied Scottish first degree relatives of patients with gastric cancer, to minimize potential confounders such as variants in dietary and environmental exposures, housing standards etc. since potential exposure to infection would be the same [32]. All subjects were genotyped for ATG16L1 and NOD2 and underwent upper endoscopy and biopsies, including culture for H. pylori, to diagnose and characterize the extent of H. pylori infection and related gastritis phenotypes. Genotyped population controls were derived locally from umbilical cord blood sampling. Odds ratios (OR) for H. pylori infection were then calculated. There was a significantly increased OR for H. pylori infection among subjects who were homozygous for the 300A allele (GG genotype) compared with those homozygous for the ‘protective’ 300T allele (AA genotype) (Table 1) but no correlation with the NOD2 genotype (Data not shown) Since induction of autophagy was entirely dependent on the presence of toxigenic VacA, we then assessed the correlation between VacA genotype of the infecting strain and ATG16L1 genotype [33]. Among patients harboring the toxigenic s1m1 strains, 38% had the GG genotype. In contrast, of those harboring the less toxigenic s1m2 strain, only 17% had the GG genotype. Taken together, these findings indicate that there was a positive correlation between the ATG16L1 genotype and susceptibility to infection with the toxigenic strain of H. pylori.
Table 1.
Association of the rs2241880 polymorphism with risk of H pylori infection in the German and Scottish studies. The Scottish study data were generated by comparing the two H pylori-infected subgroups (n=107) vs. the uninfected group (n=55); 4 samples (1 from infected and 3 from uninfected group) failed. The German study data were generated by comparing H pylori-infected subjects (n=273) vs. uninfected subjects (n=210); 3 samples failed (two from uninfected and 1 from infected group). Odds ratios, confidence intervals and p-values are given for per-genotype, dominant and recessive models.
| Number of Cases/Controls Odds Ratioa (95% Confidence Interval); p-value |
|||||
|---|---|---|---|---|---|
| AA | AG | GG | Recessive | Dominant | |
| German Study | 57/57 1.0 (ref) |
132/107 1.2 (0.8–1.9); 0.51 |
84/46 1.8 (1.1–3.1); 0.03 |
1.7 (1.0–2.8); 0.03 | 1.4 (0.9–2.1); 0.13 |
| Scottish Study | 26/18 1.0 (ref) |
39/24 1.2 (0.6–2.5); 0.67 |
42/13 2.3 (1.0–4.9); 0.05 |
2.3 (1.0–4.3); 0.05 | 1.5 (0.8–3.0); 0.18 |
For the Scottish study, odds ratios were adjusted for age, sex and within-family sampling. For the German study, odds ratios were adjusted for age and sex. The alleles at the rs2241880 polymorphic locus were in Hardy-Weinberg equilibrium, with non-significant χ2 values, in all control populations.
We replicated our study in a cohort of German subjects who similarly underwent genotyping and upper endoscopy and biopsies, including culture for H. pylori. Once again, subjects homozygous for the risk 300A allele had a significantly higher OR of H. pylori infection in comparison to those harboring the 300A allele (Table 1). These results confirm that the 300A allele in ATG16L1 confers a modest but significant host genetic risk of H. pylori infection. This is the first study to show that variants in an autophagy gene, ATG16L1, increase susceptibility to H. pylori acquisition and chronic infection in Caucasian populations.
Discussion
Current evidence indicates that autophagy can be induced in cells as an innate defense mechanism to eliminate bacteria or toxins during infection [6,7]. However, a number of pathogens are able to subvert this pathway as a strategy for their increased intracellular survival [6,34]. Here we show that although autophagy is initially induced in gastric epithelial cells (AGS) in response to VacA and serves a cytoprotective role, prolonged exposure to VacA causes disrupted autophagy, as determined by i) persistence of both the GFP and RFP signal when the tandem LC3-eGFP-mRFP construct was used, ii) reduced degradation of long-lived proteins, accumulation of p62 and elevated ROS levels iii) protection of cells from rapamycin-induced autophagic cell death and iv) lack of cathepsin D within autophagosomes in VacA treated cells. In addition, our in vitro findings were recapitulated in the natural setting of infection in human subjects as evidenced by higher levels of endogenous p62 in gastric tissues from patients infected with toxigenic H. pylori.
Our discovery that H. pylori infection is increased in hosts harboring the ATG16L1 300A polymorphism provides support for the importance of the pathway in mediating resistance to infection in humans. The 300A polymorphism is relatively common in Caucasian populations and the implications of research to date in Crohn’s disease would suggest that it facilitates chronic inflammation. That the resultant protein may result in reduced autophagic responses to VacA and increased susceptibility to infection with an enteric microbe in humans is intriguing. We propose a model whereby during initial exposure to infection where bacterial load and therefore levels of VacA may be low, host cell autophagy plays an important role in reducing the effects of the toxin, and clearance of bacteria (Figure 6C). In individuals with the ATG16L1 risk allele, a reduced host autophagic response could result in enhanced toxigenic effects, thereby increasing susceptibility to acquisition of infection. In contrast to the setting of initial exposure to infection, during chronic infection, persistent exposure of VacA could disrupt the autophagic pathway thereby resulting in a failure to clear bacteria and accumulation of genotoxic material in cells (p62, ROS). Since H. pylori possesses a variety of antioxidant strategies [35] accumulation of ROS would likely not impact bacterial survival. However increased ROS and genotoxic materials in epithelial cells may ultimately promote carcinogenesis (Figure 6C).
Autophagy plays dual roles, both as a tumor suppressor and tumor enhancer. The inability to eliminate protein aggregates and damaged organelles that produce genotoxic free radicals could increase the mutation rate within the genome, promoting cellular changes associated with oncogenesis. Tumor cells induce autophagy to handle metabolic stress and promote survival. However, defects in autophagy within tumor cells could also lead to cell death, inflammation and genetic instability. This in turn might create a microenvironment that predisposes to cancer [36,37]. Mice lacking Beclin 1 accumulate p62, ROS and DNA damage in the liver leading to tissue damage, inflammation and ultimately hepatocellular carcinoma [23,24,37]. Thus our finding that VacA-mediated disrupted autophagy increases known risk factors for tumorigenesis including ROS in vitro and p62 in vivo in human subjects suggests that alterations in this pathway may be highly relevant for disease outcome during H. pylori infection. Thus infection with H. pylori may serve as a unique model system to determine the role of autophagy disruption in microbial-mediated carcinogenesis.
To our knowledge this is the first study to provide evidence that the Crohn’s disease variant of ATG16L1 alters susceptibility to infection with an enteric microbe in human subjects at the population level. These findings support a role for altered autophagy in regulating the host response to enteric microbes in Crohn’s disease pathogenesis. Of interest recent studies indicate that infection with H. pylori may actually serve a protective role against inflammatory bowel disease [38,39]. It is interesting to speculate that due to increased susceptibility to infection, early exposure and acquisition of H. pylori in individuals with the ATG16L risk allele may decrease their risk for the subsequent development of inflammatory bowel disease. However, further studies are needed to confirm this contention.
In summary, our studies have uncovered previously unappreciated roles of autophagy in host susceptibility to H. pylori infection. We are the first group to identify an autophagy gene, ATG16L1, as a host candidate gene for susceptibility to H. pylori infection. Our novel in vitro and in vivo translational studies delineate mechanisms by which H. pylori usurps the autophagy pathway which in turn leads to accumulation of ROS and p62. Rather than eliminating the bacteria and the toxin, this sequence of events predisposes to chronic H. pylori infection, and ultimately may promote gastric cancer. Thus, the relationship between host autophagy genotype and VacA toxin-mediated induction of disarmed autophagosomes is an elegant example of the complex manner in which host factors and bacterial virulence factors modulate host cell responses and disease susceptibility.
Supplementary Material
Acknowledgments
Grant Support:
We would like to acknowledge the various funding agencies that supported this study. NLJ is supported by CIHR (No. 178886) and by the Crohn’s and Colitis Foundation of Canada. SH was funded by a Canadian Association of Gastroenterology/ CCFC Fellowship. MRT was funded by NSERC discovery grant. SRB was supported by a grant from the NIH (R01 AI045928). RMP was supported by NIH (DK 58587, CA77955 and CA116037). EK was funded by RUB/M 122/13. GH and EEO were supported by Cancer research UK (C8969/A6657).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare no conflict of interest.
Author Contributions:
DR, MRT and MA performed experiments and data analysis. MR, MS, EGM, HM, JRG and GH performed experiments and sample preparation.
VG, SRB, TY, RMP, PC, MSS provided reagents. CS helped with data analysis. NLJ, DR, MT, SH planned the experiments. NLJ, DR, SH, MT, RMP, EEO, SG, DP wrote the manuscript. NLJ conceived of and supervised the study.
References
- 1.Polk DB, Peek RM., Jr Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzalez CA, Figueiredo C, Lic CB, et al. Helicobacter pylori cagA and vacA genotypes as predictors of progression of gastric preneoplastic lesions: a long-term follow-up in a high-risk area in Spain. Am J Gastroenterol. 106:867–874. doi: 10.1038/ajg.2011.1. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia K, Thomas C, Akbar M, et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogawa M, Yoshikawa Y, Kobayashi T, et al. A tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe. 9:376–389. doi: 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orvedahl A, Levine B. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ. 2008 doi: 10.1038/cdd.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terebiznik MR, Raju D, Vazquez CL, et al. Effect of Helicobacter pylori's vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy. 2009;5:370–379. doi: 10.4161/auto.5.3.7663. [DOI] [PubMed] [Google Scholar]
- 9.Amieva MR, Salama NR, Tompkins LS, Falkow S. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell Microbiol. 2002;4:677–690. doi: 10.1046/j.1462-5822.2002.00222.x. [DOI] [PubMed] [Google Scholar]
- 10.Franco AT, Israel DA, Washington MK, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci U S A. 2005;102:10646–10651. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terebiznik MR, Vazquez CL, Torbicki K, et al. Helicobacter pylori VacA toxin promotes bacterial intracellular survival in gastric epithelial cells. Infect Immun. 2006;74:6599–6614. doi: 10.1128/IAI.01085-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng PY, Jones NL. Helicobacter pylori strains expressing the vacuolating cytotoxin interrupt phagosome maturation in macrophages by recruiting and retaining TACO (coronin 1) protein. Cell Microbiol. 2003;5:25–40. doi: 10.1046/j.1462-5822.2003.00250.x. [DOI] [PubMed] [Google Scholar]
- 13.Kimura S, Noda T, Yoshimori T. Dissection of the Autophagosome Maturation Process by a Novel Reporter Protein, Tandem Fluorescent-Tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 14.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 16.Mortimore GE, Poso AR. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu Rev Nutr. 1987;7:539–564. doi: 10.1146/annurev.nu.07.070187.002543. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimori T. Autophagy: paying Charon's toll. Cell. 2007;128:833–836. doi: 10.1016/j.cell.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 18.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 19.Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwamaru A, Kondo Y, Iwado E, et al. Silencing mammalian target of rapamycin signaling by small interfering RNA enhances rapamycin-induced autophagy in malignant glioma cells. Oncogene. 2007;26:1840–1851. doi: 10.1038/sj.onc.1209992. [DOI] [PubMed] [Google Scholar]
- 21.Satin B, Norais N, Telford J, et al. Effect of helicobacter pylori vacuolating toxin on maturation and extracellular release of procathepsin D and on epidermal growth factor degradation. J Biol Chem. 1997;272:25022–25028. doi: 10.1074/jbc.272.40.25022. [DOI] [PubMed] [Google Scholar]
- 22.Zheng X, Chu F, Mirkin BL, et al. Role of the proteolytic hierarchy between cathepsin L, cathepsin D and caspase-3 in regulation of cellular susceptibility to apoptosis and autophagy. Biochim Biophys Acta. 2008;1783:2294–2300. doi: 10.1016/j.bbamcr.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 23.Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogawa M, Yoshimori T, Suzuki T, et al. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 26.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 29.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn's disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 12:99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 32.El-Omar EM, Carrington M, Chow WH, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 33.Argent RH, Thomas RJ, Aviles-Jimenez F, et al. Toxigenic Helicobacter pylori infection precedes gastric hypochlorhydria in cancer relatives, and H. pylori virulence evolves in these families. Clin Cancer Res. 2008;14:2227–2235. doi: 10.1158/1078-0432.CCR-07-2022. [DOI] [PubMed] [Google Scholar]
- 34.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006;61:847–860. doi: 10.1111/j.1365-2958.2006.05302.x. [DOI] [PubMed] [Google Scholar]
- 36.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol. 22:212–217. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luther J, Dave M, Higgins PD, Kao JY. Association between Helicobacter pylori infection and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Inflamm Bowel Dis. 16:1077–1084. doi: 10.1002/ibd.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luther J, Owyang SY, Takeuchi T, et al. Helicobacter pylori DNA decreases pro-inflammatory cytokine production by dendritic cells and attenuates dextran sodium sulphate-induced colitis. Gut. 60:1479–1486. doi: 10.1136/gut.2010.220087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.