Abstract
Oenocytes are a specialized cell type required for lipid processing, pheromone secretion, and developmental signaling. Their development has been well characterized in Drosophila melanogaster, but it remains unknown whether the developmental program is conserved in other insect species. In this study, we compare and contrast the specification and development of larval oenocytes between Drosophila and the red flour beetle, Tribolium castaneum. First, we identify several useful reagents to label larval oenocytes, including both a Tribolium GFP enhancer trap line and a simple flurophore-conjugated streptavidin staining method that recognizes oenocytes across insect species. Second, we use these tools to describe oenocyte development in Tribolium embryos, and our findings provide evidence for conserved roles of MAP-kinase signaling as well as the Spalt, Engrailed, HNF4, and Ventral veins lacking factors in producing abdominal-specific oenocyte cells. However, Tribolium embryos produce four-times as many oenocytes per abdominal segment as Drosophila, and unlike in Drosophila, these cells rapidly down-regulate the expression of the Spalt transcription factor. Thus, these results provide new insight into the molecular pathways regulating oenocyte specification across insect species.
Introduction
Oenocytes are a specialized cell type responsible for a host of important metabolic and behavioral roles in insects (Chapman, 1998; Snodgrass, 1993). Oenocyte function is perhaps best understood in Drosophila melanogaster, which has distinct populations of larval and adult oenocytes (Gould et al., 2001). Larval oenocytes initially form in the embryo and after maturation express a large number of different enzymes required for lipid metabolism (Gutierrez et al., 2007). In fact, the selective ablation of larval oenocytes results in the inability of larva to mobilize and process lipids from the fat body, ultimately causing severe growth defects and lethality prior to pupation (Gutierrez et al., 2007). The adult oenocytes develop independently from imaginal tissues during pupation, and recent papers have shown that they modulate courtship and aggression behavior in adult flies through the production of cuticular hydrocarbons that serve as pheromones (Billeter et al., 2009; Fernandez et al., 2010). Hence, larval oenocytes have been likened to vertebrate hepatocytes due to their critical role in lipid homeostasis, whereas adult oenocytes participate in animal communication through pheromone production. In this paper, we focus on comparing and contrasting the development of larval oenocytes between two different insect species.
Larval oenocytes in Drosophila melanogaster are an abdominal-specific cell type that form in clusters of three to nine cells within each hemisegment of the first seven abdomen segments (Gould et al., 2001). Elegant studies have shown that genetic interactions between the Abdominal-A (Abd-A) Hox factor, the Spalt transcription factors (Spalt-major (Salm) and Spalt-related (Salr)), and the epidermal growth factor receptor (EGFR) signaling pathway are required for oenocyte specification (Brodu et al., 2002, 2004; Elstob et al., 2001; Rusten et al., 2001). In this model, Abd-A induces the secretion of the Spitz EGF ligand from a specific abdominal sensory organ precursor cell that lies within the Spalt-expressing dorsal ectoderm. The neighboring cells that receive the EGF signal further up-regulate Spalt expression, delaminate into the embryo, and commit to a larval oenocyte cell fate. Since EGF behaves as a short-range morphogen in this context, only the three to four cells contacting the sensory precursor are specified at a time (Brodu et al., 2004; Shilo, 2005). However, once the first cells delaminate into the embryo, three to four new cells come into contact with the sensory precursor, receive the EGF signal, and repeat another cycle of oenocyte specification. As the sensory precursor secretes EGF for a limited period of time during embryogenesis, typically only two rounds of oenocyte recruitment occur to yield an average of six oenocytes per cluster (Brodu et al., 2004; Witt et al., 2010). After specification, the oenocytes mature and subsequently activate the expression of the Hepatocyte nuclear factor-4 (HNF4) transcription factor as well as numerous lipid-modifying enzymes required for lipid processing during larval growth (Gutzwiller et al., 2010; Palanker et al., 2009).
The experiments focused on Drosophila larval oenocyte production have provided a host of genetic and molecular oenocyte-specific markers. However, these reagents have so far been restricted to the study of D. melanogaster. In contrast, oenocytes in other species have only been described in larval/nymph stages using classical histological and anatomical approaches where they are identified by their large size, polytenic nuclei, and staining properties (Jackson and Locke, 1989; Snodgrass, 1993). Even without the benefit of molecular markers, these studies established a degree of diversity in the arrangement and number of oenocytes in different insect clades. While most species tend to group their oenocytes in small clusters of 6–10, Eastham described a parasitic wasp with >20 oenocytes/cluster (Eastham, 1929; Gould et al., 2001). Moreover, the distribution of oenocytes can vary greatly. Most clades have metameric clusters just under the integument of abdominal segments. However, some also disperse their oenocytes amongst the tissue of the fat body, while others such as Coleoptera (beetles) organize them in a continuous streak running the length of the abdomen (Chapman, 1998; Wheeler, 1892). There are even descriptions of grasshoppers and ants with oenocytes spreading to thoracic segments late in their respective larval/nymph stages (Wheeler, 1892; Zara and Caetano, 2004). Thus, the variation in final location and number of larval oenocytes between insect species provides a potentially useful model for performing comparative evolutionary-developmental biology studies.
Given the diversity of insects and the essential role of larval oenocytes in metabolic functions, we wanted to study oenocyte development in distantly related insect species. For comparison studies with Drosophila melanogaster, we chose the red flour beetle, Tribolium castaneum. T castaneum is an emerging model organism that has a sequenced genome, and unlike Drosophila, Tribolium is a short-germ band insect that adds segments sequentially during embryonic development (Richards et al., 2008; Roth and Hartenstein, 2008). Here, we identify several molecular markers that recognize larval oenocytes in Tribolium, including an oenoctye-specific enhancer trap line near the ventral-veins lacking (vvl) homologue, which is also expressed in Drosophila oenocytes (Inbal et al., 2003). In addition, we determined that fluorophore-conjugated streptavidin functions as a novel biomarker capable of detecting larval oenocytes within several insect species. Lastly, we use these biomarkers to identify both similarities and differences between the embryonic development of larval oenocytes within Tribolium compared to those in Drosophila. Our findings reveal that, similar to Drosophila, Tribolium larval oenocytes are abdominal-specific and are initially recruited from a Spalt-positive ectoderm that subsequently up-regulates the expression of the HNF4 factor. Unlike Drosophila, however, Tribolium oenocytes rapidly down-regulate Spalt expression, are recruited in much larger numbers (more than 20 oenocytes per hemisegment), and form a continuous streak of oenocytes between abdominal segments. The implications of the insect oenocyte staining methods and cross-species results are discussed.
Materials and Methods
Insect stocks
Beetles were reared in plastic bottles at 30°C in a Digitherm 47L incubator (Tritech Research, DT2-MP-38) humidified via a small tray of water. The KT817 (vvl-GFP) beetles were acquired from Sue Brown at the Kansas State University Tribolium stock center (http://www.geku-base.uni-goettingen.de/) (Trauner et al., 2009). The insertion site of the piggybac-XP3-GFP element was determined using inverse PCR (iPCR) on circularized genomic fragments resulting from Hha1 digestions. The PCR product was sequenced using primers PRF1 5’-ACCGATAAAAACACATGCGTCAAT-3’ and PRR1 5’-ATGCATTTGCCTTTCGCCTTAT-3’. The integration site and orientation were confirmed with PCR genotyping using locus specific primers (5’-AAGTCAGCCACCAATCATCG-3', 5’-GCTCAATTCGGGTGCTTGTT-3') paired with piggybac-specific primers (PRF1 and PLR1 5’-ACAGCGACGGATTCGCGCTAT-3') respectively. All Drosophila stocks were raised at 25°C and the Rho654-lacZ (Li-Kroeger et al., 2008) and svp-lacZ (Elstob et al., 2001) transgenic lines were previously described.
Antibody production
A Tribolium castaneum HNF4 bacterial expression vector was generated using PCR to amplify and clone the HNF4 cDNA encoding amino acids 162-494 in-frame with an N-terminal 6X-His tag (pET14b, Novagen). The expression plasmid was transformed into BL21-CodonPlus (DE3)-RP bacteria (Stratagene) and protein expression induced using 0.25 mM IPTG for 2 hours at 37°C. Bacteria w as lysed in 8M urea lysis buffer and purified using Ni-affinity chromatography as previously described (Gutzwiller et al., 2010). The purified protein was used to generate HNF4 antibodies in a rat using standard protocols (Cocalico Biologicals)
Embryo fixation and antibody staining
Tribolium embryos were separated from triple-sifted medium via sifting with a #50 sieve. Harvested embryos were dechorionated in 50% bleach solution for 8 min, rinsed in H2O, and fixed with shaking in 8% formaldehyde/heptane for 20 min. Embryos were extracted in methanol and triturated through a 20g needle with naked embryos removed after every ~10 passes. Drosophila embryos were collected with standard methods. Drosophila larvae and squash bug eggs were prepared similarly as Tribolium embryos but were instead treated with 100% bleach and fixed with shaking for 30 min.
For antibody stainings, samples were incubated overnight in a primary antibody cocktail in PBX (PBS plus 0.2%Triton-X) at 4°C, was hed 3 times 20 min in PBX, incubated for 2 hr at room temperature with the appropriate fluorophore conjugated secondary antibodies, washed 4 times 20 min in PBX, and mounted for microscopic analysis. Images were captured on either a Zeiss fluorescent microscope equipped with an apotome filter, or on a Leica confocal microscope. The following primary antibodies were used: Cut (mouse, 1:20, DSHB, #2B10), phospho-Histone 3 (mouse, 1:1000, Millipore, #05-806), Dm HNF4 (rat, 1:2000, (Gutzwiller et al., 2010)), Tc HNF4 (rat, 1:2000), Dm Spalt (rabbit, 1:1500, (Xie et al., 2007)), phospho-ERK (di-phosphorylated MAPK; Mouse, 1:50, Sigma, M-8159) Engrailed (mouse, 1:20, DSHB, #4D9), Beta-galactosidase (Chicken, 1:1000, Abcam) Dm Abd-A (guinea pig, 1:500, (Li-Kroeger et al., 2008)) and UbdA (Mouse, 1:5, DSHB FP6.87). For streptavidin detection of oenocytes, samples were incubated overnight with Cy5-conjugated streptavidin (1:1000), and washed 4 times 20 min with at least one additional wash overnight in PBX. Oenocyte counts were performed on 10 age-matched Drosophila embryos using Dm anti-HNF4 and 9 age-matched Tribolium embryos using the combination of vvl-GFP and Tc anti-HNF4. All results were analyzed by ANOVA using Excel.
Results
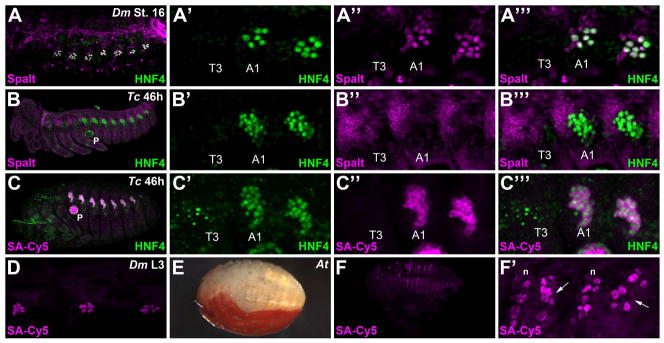
To conduct a comparative study of larval oenocyte formation between insect species, we first evaluated the ability of two antibodies that label Drosophila oenocytes to cross-react with conserved proteins in Tribolium embryos. In Drosophila embryos, the Spalt-major (Salm) and Hepatocyte nuclear factor 4 (HNF4) proteins are co-expressed in abdominal oenocytes in stage 16 Drosophila embryos (Fig 1A). Since Tribolium contains a single homologue of the HNF4 and Spalt genes and the Drosophila Salm and HNF4 antigens used to generate each antibody included highly conserved protein domains (not shown), we tested these reagents in Tribolium embryos (Gutzwiller et al., 2010; Xie et al., 2007). As shown in Figure 1B, we found that the antibody raised against the Drosophila Spalt protein, but not HNF4 (not shown), reveals a distinct expression pattern in Tribolium embryos. Consistent with the pattern seen in early Drosophila embryos, the Spalt antibody labels a broad swath of the dorsal ectoderm that includes a region from which oenocytes arise in Tribolium embryos (Fig 1, and see Figure 5 to compare early D. melanogaster embryo with early T. castaneum embryo) (Li-Kroeger et al., 2008; Rusten et al., 2001). In addition, we found that this Spalt antibody shows an expression pattern identical to the published Spalt in situ pattern in the Tribolium hindwing (not shown and (Tomoyasu et al., 2005)). However, unlike in Drosophila, no distinct Spalt-positive abdominal cell clusters were observed in mature embryos, suggesting this factor may not be maintained in Tribolium larval oenocytes.
Figure 1. Molecular markers of larval oenocytes within insect species.
A-A’’’) A lateral view of a stage 16 Drosophila melanogaster (Dm) embryo immunostained for Spalt (magenta) and HNF4 (green) reveals clusters of abdominal oenocytes. Close-up view of the third thoracic segment (T3) and first two abdominal segments (A1, A2 not labeled) highlights the abdominal specific formation of this cell type (A’-A’’’). Note, the additional nearby Spalt staining reveals cells of a sensory organ that forms part of the peripheral nervous system.
B-B’’’) Lateral view of a 46h Tribolium castaneum (Tc) embryo immunolabeled for HNF4 (green) and Spalt (magenta) reveals these two factors do not co-localize in oenocytes of the beetle embryo.
C-C’’’) Lateral view of a 46h Tribolium castaneum (Tc) embryo immunolabeled for HNF4 (green) and fluorophore-conjugated streptavidin (SA-Cy5) reveals clusters of oenocytes in the abdominal but not thoracic segments. Note, additional strong SA-Cy5 staining is observed in the pleuropod (P) in the first abdominal segment.
D) Close-up lateral view of three abdominal segments from a third instar Drosophila melanogaster larva stained with fluorophore-conjugated streptavidin (SA-Cy5) reveals clusters of approximately six oenocytes in each segment.
E) Bright-field image showing a lateral view of a mature Hemipteran Anasa tristis embryo.
F-F’) Lateral view of a mature Hemipteran Anasa tristis embryo labeled with fluorophore-conjugated streptavidin (SA-Cy5) reveals clusters of oenocytes in the abdominal segments. A subset of neurons (n) are also labeled via streptavidin (F’)
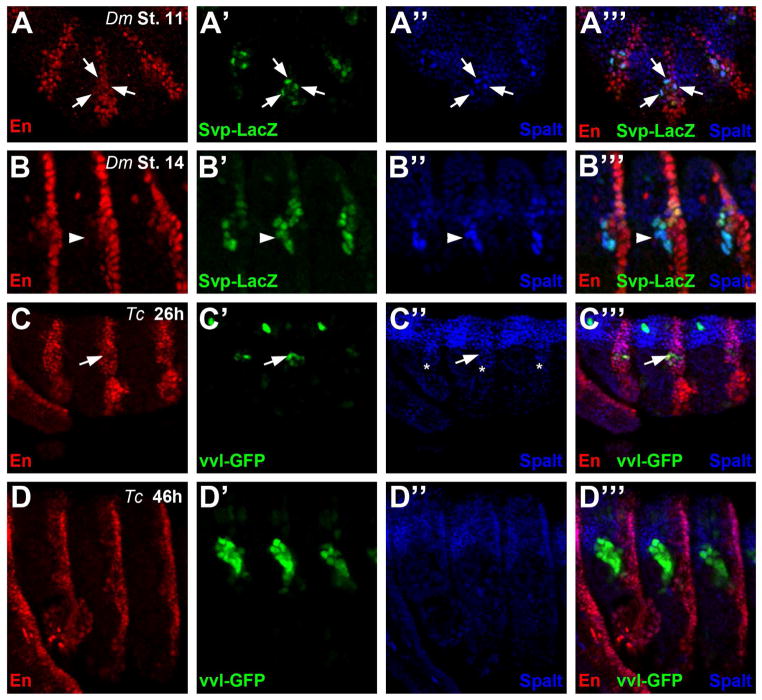
Figure 5. Oenocytes are derived from the engrailed stripe in both Drosophila and Tribolium.
A) Lateral view of three abdominal segments from a stage 11 svp-lacZ Drosophila embryo immunostained for Engrailed (En, red), β-galactosidase (β-gal, green), and Spalt (blue). svp-lacZ (β-gal) labels newly specified Drosophila oenocytes (arrows) that lie within the posterior compartment marked by En.
B) Lateral view of three abdominal segments from a stage 14 svp-lacZ Drosophila embryo immunostained for Engrailed (En, red), β-galactosidase (green), and Spalt (blue). At this stage, Drosophila oenocytes are still positive for svp-lacZ (β-gal) and Spalt, but they have downregulated En and lie anterior to the posterior compartment (arrowhead).
C) Lateral view of three abdominal segments from a 26h Tribolium embryo immunostained for En (red), vvl-GFP (green), and Spalt (blue). Note the early vvl-GFP positive cells (arrow) lie within the Spalt domain (C’’, asterisk) and overlaps with En expression in abdominal segments.
D) Lateral view of three abdominal segments from a 46h Tribolium embryo immunostained for En (red), vvl-GFP (green), and Spalt (blue). Note the GFP-positive oenocyte cells lie anterior to the En-positive posterior compartment cells and do not express Spalt.
Since the Drosophila HNF4 antibody failed to recognize a specific pattern in Tribolium embryos, we next generated antisera against the Tribolium HNF4 protein. As shown in Fig 1B, we detected an HNF4 expression pattern that labels a promising set of cell clusters that are restricted to abdominal segments in 46h embryos. Using optical sectioning, these cells are distinguishable from the underlying HNF4+ mesoderm by both the intensity of HNF4 staining and their lateral position. Consistent with these HNF4-positive cells being larval oenocytes, they also co-express a lipid-modifying enzyme (Alas, Suppl Fig 1) that is expressed in Drosophila oenocytes as well as several other oenocyte markers described below. Moreover, co-staining with the Spalt antibody confirmed that in contrast to the strong up-regulation seen in mature D. melanogaster oenocytes, oenocytes within older T. castaneum embryos lack Spalt expression (Fig 1B’’’).
A new biomarker for larval oenocytes
In the process of confirming that the HNF4-expressing abdominal-specific cells in Tribolium embryos are larval oenocytes, we serendipitously identified a new biomarker for oenocytes. While testing a host of antibodies for cross-reactivity in Tribolium (See Supplemental Table 1 for complete list of antibodies), we attempted to amplify the signal using the biotin-streptavidin labeling system. We instead found that whenever biotin-streptavidin was utilized, a consistent robust signal specifically labels the cytoplasm of the same abdominal cell clusters that express HNF4 as well as the pleuropod appendage of abdominal segment 1 (Fig 1C). The subsequent stepwise removal of both primary and secondary antibodies revealed that adding the fluorophore-conjugated streptavidin molecule alone results in this abdominal-specific pattern. The strong labeling of larval oenocytes by streptavidin is likely due to this cell type containing high levels of biotin, which serves as an essential co-enzyme for fatty acid metabolism (Fletcher and Myant, 1960). Thus, fluorescent conjugated streptavidin is a useful stain to recognize larval oenocytes in Tribolium embryos.
We next determined the general applicability of streptavidin as an oenocyte biomarker by testing the stain on two other insects. While streptavidin did not recognize larval oenocytes during D melanogaster embryogenesis, this stain was very effective at labeling oenocytes in third instar larva (Fig.1D). Moreover, the ability of streptavidin to recognize this cell type has the added advantage of not requiring larval dissections (see methods). To further test the applicability of the streptavidin stain to insects, we collected embryos of a Hemipteran species of the Anasa genus (often called squash bugs) from pumpkin squash in Southwestern Ohio. The egg-shell was manually dissected from each late-stage embryo (Fig. 1E), and the application of fluorescent-conjugated streptavidin revealed strong staining of abdominal regions consistent with oenocyte clusters (arrows, Fig 1F’). However, it should be pointed out that streptavidin staining in Anasa as well as in Drosophila and Tribolium also labeled a subset of other cells including neurons. Hence, additional markers should be used when possible to define streptavidin labeled cells as oenocytes. Nevertheless, these data suggest that streptavidin is likely to be a broadly applicable marker of larval oenocytes across many insect species.
An enhancer trap line for tracing oenocyte development in Tribolium
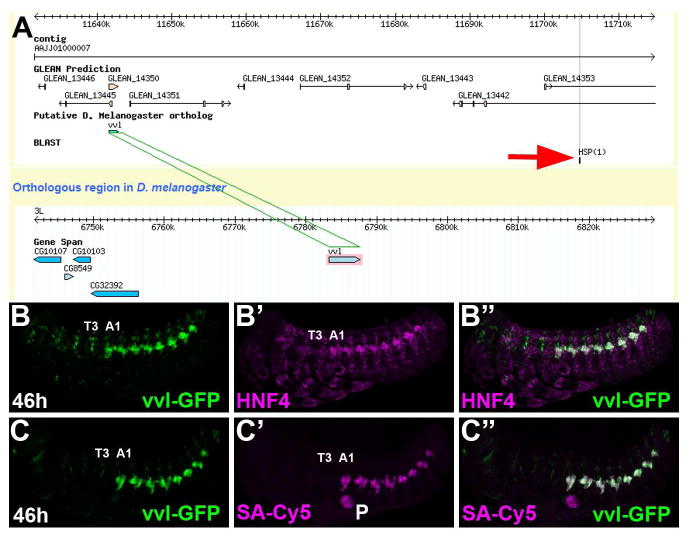
A powerful tool for labeling specific cell types in model organisms is the use of enhancer trap lines. The recent work of several research groups has made many such GFP enhancer trap lines in Tribolium castaneum available through the GEKU database (http://www.geku-base.uni-goettingen.de/) (Trauner et al., 2009). Scanning the available enhancer trap lines revealed several with interesting abdominal-specific expression patterns. One of these lines, KT817, was chosen for analysis and was found to express GFP in the same pattern as cells labeled by HNF4 (Fig. 2B) and streptavidin (Fig. 2C). (Note the additional weak GFP staining in each segment is due to non-insertion site-specific activity of the GFP construct). Using a iPCR protocol (see methods), we determined the piggybac-GFP integrated ~60kb away from the Tribolium homologue of the ventral veins lacking (vvl) gene, which is known to be expressed in the early larval oenocytes in D melanogaster (Inbal et al., 2003). As such, we believe the oenocyte-specific expression pattern is most likely due to an enhancer of Tribolium vvl, despite the presence of several other predicted gene models at the locus. This conclusion is made more the plausible by the relatively great distance between genes in the Tribolium genome as well as previous findings describing enhancers more than 100kb from their target genes (Levine, 2010). Hence, the KT817 enhancer trap is hereafter referred to as vvl-GFP.
Figure 2. Identification of the vvl-GFP oenocyte-specific enhancer trap.
A) Screen capture of a GBrowse view showing the integration site (Red Arrow) of the piggybac-GFP vector ~60kb downstream of the Tribolium ventral veinless (vvl) homologue.
B-C) Lateral view of a 46h vvl-GFP Tribolium embryo immunolabeled for GFP (green) and either HNF4 (B, magenta) or co-stained with streptavidin conjugated to Cy5 (SA-Cy5, magenta) reveals abdomen-specific GFP-positive cell clusters that co-label with HNF4 and SA-Cy5. The T3 and A1 segments are labeled.
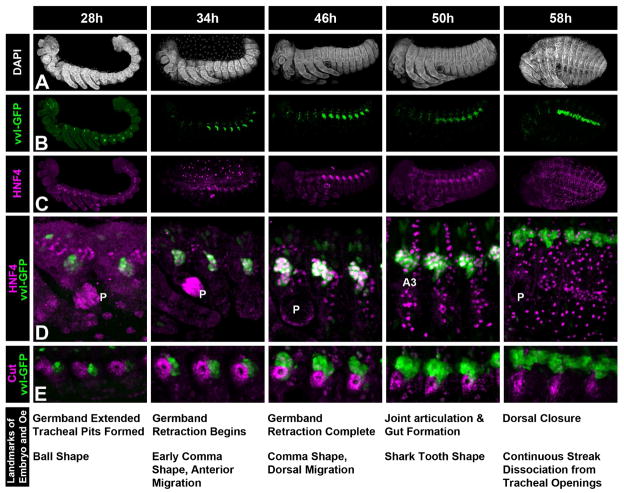
We next used the vvl-GFP and the HNF4 antibody as markers to study the development of Tribolium larval oenocytes during embryogenesis (Fig 3 and Suppl Fig 2). An antibody against the Cut transcription factor was also used to serve as a marker of the nearby tracheal pits (Fig 3E). For this analysis, we performed timed-embryo collections and defined several developmental time-points and landmarks that correlate with morphological transitions for oenocytes. First, the initial traces of vvl-GFP expression can be seen shortly after germ band extension is complete (26h), and this staining quickly strengthens to reveal a cluster of roughly 15 cells in each abdominal segment (Fig 3B at 28h). At this time, the GFP-positive cells lack HNF4 expression and the GFP signal has a slight but consistently stronger intensity in the anterior (A1-A3) versus posterior (A4-A8) abdominal segments (Fig 3B at 28h). This finding is congruent with Tribolium being a short germ band insect that adds new (younger) posterior abdominal segments during development (Schroder et al., 2008). By 34h, the A1 through A8 abdominal segments each have robust vvl-GFP expressing cells that co-express HNF4 (Fig 3B-D), and these cells form a crescent-shaped cluster of cells around the Cut-positive tracheal pit. At the completion of germ band retraction (46h), the number of GFP- and HNF4-positive oenocytes increases to greater than 20 cells per cluster in most abdominal segments, and these cells are present in a single cluster dorsal to the tracheal pit in each segment. By 50h, the joints in the legs articulate sharp boundaries, and the elongation of the gut tube can be seen in embryos tilted toward a dorsal view (not shown). At this stage, oenocyte clusters begin to spread both to the posterior and interiorly away from the integument resulting in the formation of a characteristic shark tooth shape. In addition, many non-oenocyte cells begin to express high levels of HNF4 but these cells are not recognized by streptavidin nor do they express vvl-GFP (Fig 3D at 50h and data not shown). By 58h, the gut tube forms a complete loop, and dorsal closure is complete. At this stage, the GFP-positive cells still stain strongly with streptavidin but HNF4 expression has greatly diminished. Moreover, the oenocytes form a continuous streak extending across segmental boundaries and have migrated dorsally, a morphology that matches the classical description of larval coleopteran oenocytes (Snodgrass, 1993; Wheeler, 1892).
Figure 3. Developmental time-course of oenocyte morphogenesis in Tribolium.
Embryos were stage matched and stained for general morphology (DAPI in A), oenocyte markers (vvl-GFP in B and D, HNF in C and D), and a Cut antibody that labels the developing tracheal pit (E). Oenocytes marked by vvl-GFP first appear as faint clusters in the posterior of the abdominal segments at 28h of development. GFP expression becomes more robust by 34h, the cells begin to express HNF4, and the oenocyte cluster stretches to the anterior and dorsal in each abdominal segment. At 46h, most oenocytes are positioned in a cluster dorsal to the tracheal pit with a small number of cells remaining just to the posterior resulting in the formation of a distinctive comma shape. By 50h, the GFP label reveals the oenocytes form a ‘shark tooth’ shaped cluster of cells in each abdominal segment and while the oenocytes maintain HNF4 expression, many additional cells also begin to express high levels of this protein. After dorsal closure (58h), the oenocytes form a continuous streak of cells and HNF4 expression has begun to fade. Moreover, the oenocytes have migrated interiorly and dorsally relative to the external openings of the tracheal pits.
Comparison of oenocyte specification between D. melanogaster and T. castaneum
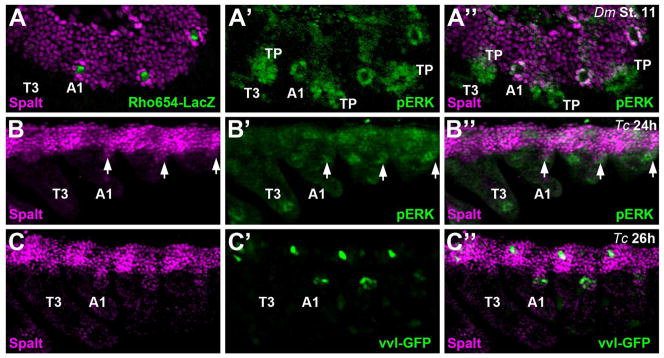
Drosophila oenocytes are initially specified in stage 11 embryos when dorsal ectoderm cells that express Spalt receive a transient EGF signal (Elstob et al., 2001; Rusten et al., 2001). The EGF sending cell, a specific sensory organ precursor (SOP), can be labeled using a transgenic reporter line taken from the rhomboid locus: Rho654-LacZ, and the EGF receiving cells can be visualized using a phospho-ERK antibody that recognizes the activated MAP-kinase signaling pathway (Fig 4A) (Gabay et al., 1997; Li-Kroeger et al., 2008; Witt et al., 2010). The EGF receiving cells subsequently up-regulate and maintain Spalt expression during oenocyte maturation. In contrast, we found that the mature Tribolium oenocytes (as marked by HNF4 and streptavidin) do not maintain Spalt expression (Fig 1B). However, it is possible that Tribolium oenocytes are initially recruited from Spalt-expressing dorsal ectoderm cells and subsequently down-regulate this factor during maturation. To test this idea, we carefully analyzed the early expression of Spalt, phospo-ERK, and vvl-GFP in beetle embryos. As shown in Fig 4B, a small ventral outcropping of Spalt-positive cells are weakly, but consistently, stained using the phospho-ERK antibody. While these embryos do not express the vvl-GFP transgene at this stage of development, the earliest vvl-GFP expression is detected within the Spalt-positive outcropping that is restricted to the abdominal segments (Fig 4C). However, unlike in Drosophila, Tribolium Spalt expression is rapidly lost in these cells with no protein detected in GFP-positive cells by 34h of development.
Figure 4. Spalt and pERK signaling are present in oenocyte precursors in both Drosophila and Tribolium.
A) Lateral view of a stage 11 Rho654-LacZ Drosophila embryo immunostained for Spalt (magenta in A and A’’), β-galactosidase (green in A), and phospho-ERK (pERK, green in A’ and A’’). Rho654-lacZ specifically marks the C1 sensory precursor cell that secretes the EGF ligand to activate the MAP-kinase pathway (p-ERK) in the neighboring oenocyte precursors that subsequently up-regulate Spalt. Note, pERK activity also marks developing tracheal pits (TP).
B) Lateral view of a 24h Tribolium embryo immunostained for Spalt (magenta in B and B’’), and phospho-ERK (pERK, green in B’ and B’’). Note the presence of a small outcropping of Spalt expression that overlaps with pERK activity in the posterior of each abdominal segment (arrow).
C) Lateral view of a 26h Tribolium embryo immunostained for Spalt (magenta in C and C’’), and vvl-GFP (green in C’ and C’’). Note the small outcropping of Spalt expression now overlaps with the early vvl-GFP expression in abdominal segments.
Another useful marker relevant to Drosophila oenocyte specification is the Engrailed (En) homeodomain protein that specifically labels posterior compartment cells within each segment (Kornberg, 1981). As shown in Fig 5A, oenocytes, which are co-labeled with the Seven-up-lacZ (Svp-lacZ) enhancer trap line and the Spalt antibody, arise in groups of three to four cells within, or in very close proximity to, the En-positive posterior compartment (Gebelein and Mann, 2007). Oenocyte recruitment in Tribolium proceeds along a similar trajectory. At 26h of development, the earliest recruited oenocytes, as marked by vvl-GFP and the Spalt out-cropping, are contained within the En-positive posterior compartment cells of abdominal segments (Fig 5C). As Tribolium oenocytes mature and begin to express HNF4 and co-label with streptavidin, however, they lose En expression, move to the anterior, and reside outside of the posterior compartment (Fig 5D). Drosophila oenocytes similarly down-regulate En-expression in older embryos and reside outside of the posterior compartment (Fig 5B). Altogether, these studies suggest Tribolium oenocytes similarly arise from posterior compartment cells that activate the MAP-kinase signaling pathway and express Spalt, but unlike in Drosophila, Spalt expression is rapidly lost in Tribolium oenocytes suggesting this factor is not required for their maturation.
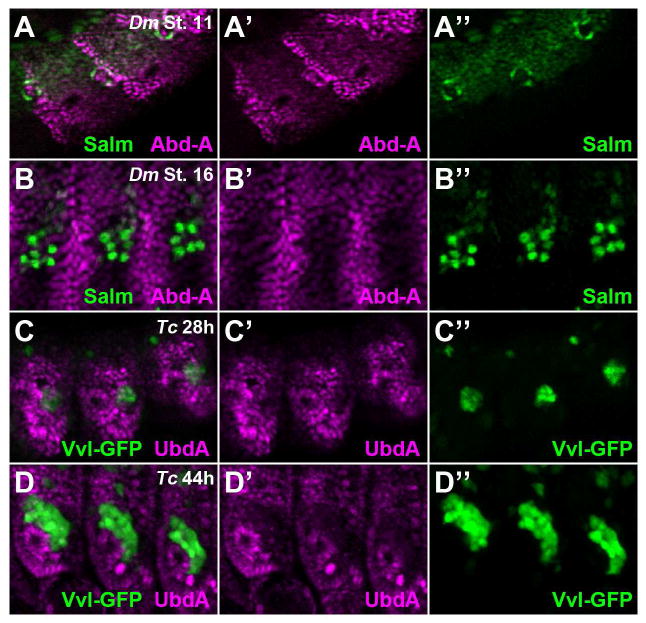
We next analyzed which abdominal segments of Drosophila and Tribolium embryos produce oenocytes. In D melanogaster embryos, oenocytes form in the first seven abdominal segments that express the Abd-A Hox factor (Brodu et al., 2002). Abd-A is most highly expressed in the En-positive cells of the posterior compartment that produce oenocytes in stage 11 Drosophila embryos (Fig 6A) (Gebelein and Mann, 2007). After their maturation, however, Drosophila oenocytes express relatively low levels of Abd-A compared to neighboring abdominal cells (Fig 6B). Previous expression analysis of Tribolium embryos revealed that strong abd-A expression is detected in the posterior compartments of the first eight abdominal segments (Shippy et al., 1998). In this study, we demonstrated that all eight of these Tribolium segments produce oenocytes and that these cells arise from the posterior compartment. Unfortunately, our specific Drosophila Abd-A antibodies failed to recognize Tribolium Abd-A. However, we were able to determine if the Tribolium oenocytes express either the Abd-A or Utx (the Ultrabithorax homologue in Tribolium) proteins using the mouse-UbdA antibody (mAb FP6.87) that cross-reacts with both in Tribolium embryos. As shown in Fig 6, we found that the UbdA antibody detects abdominal Hox factors during early oenocyte specification (Fig 6C) and that, similar to Drosophila oenocytes, the Tribolium oenocytes express relatively low levels of abdominal Hox factors upon maturation (Fig 6D).
Figure 6. Abdominal Hox expression in Drosophila and Tribolium oenocytes.
A) Lateral view of three abdominal segments from a stage 11 Drosophila embryo immunostained for Abd-A (magenta) and Spalt (green). High spalt levels labels the newly specified Drosophila oenocytes that co-express Abd-A.
B) Lateral view of three abdominal segments from a stage 16 Drosophila embryo immunostained for Abd-A (magenta) and Spalt (green). At this stage, Drosophila oenocytes are still positive for Spalt, but express less Abd-A.
C) Lateral view of three abdominal segments from a 28h Tribolium embryo immunostained for UbdA (magenta) and vvl-GFP (green). Note the early vvl-GFP positive cells express an abdominal Hox factor as detected by the UbdA antibody.
D) Lateral view of three abdominal segments from a 46h Tribolium embryo immunostained for UbdA (magenta) and vvl-GFP (green). Note the GFP-positive oenocyte cells express low levels of abdominal Hox factors.
Oenocyte enumeration in Tribolium
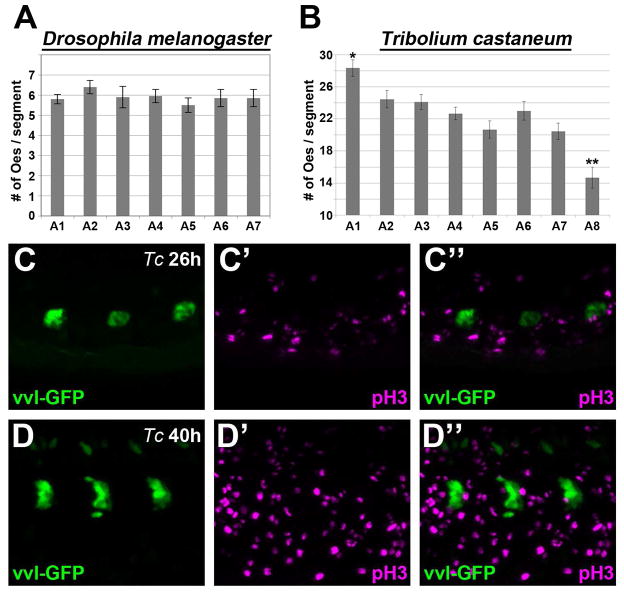
Previous publications have demonstrated that Drosophila melanogaster embryos have abdominal oenocyte clusters averaging approximately six cells in each hemi-segment (Brodu et al., 2002, 2004; Gould et al., 2001). Using HNF4 as a marker, we found similar numbers of oenocytes in stage 16 D melanogaster embryos (5.79 ± 0.92 per cluster), and we also determined that the number of oenocytes do not significantly differ between the abdominal segments along the anterior-posterior axis (Fig 7A). In contrast, the visualization of larval oenocytes in Tribolium reveals much larger cell clusters in abdominal segments. To assess the number of oenocytes that form per cluster in Tribolium, we selected to analyze embryos labeled for HNF4 and vvl-GFP at a time point (46h) in which strong nuclear HNF staining is easily detected (see Fig 4). Oenocyte numbers were tabulated for each abdominal segment of nine age-matched embryos, which revealed an overall average of 22.3 ± 0.6. However, the number of oenocytes that form between abdominal segments varied with position along the anterior-posterior axis at this developmental time-point. As shown in Fig 7B, the anterior-most A1 segment averaged 28.3 ± 1.0 oenocytes, whereas the posterior-most A8 segment averaged 14.7 ± 1.3 cells per cluster. Unfortunately, we were unable to ascertain oenocyte numbers in older embryos because oenocytes from distinct segments come into close contact with each other (see 58 to 65h time points in Fig 3 and Suppl Fig 2), and the nuclear HNF4 staining used to count these cells fades during development. However, this difference in oenocyte number along the anterior-posterior axis is likely a transient event as the size of vvl-GFP-positive clusters in older Tribolium embryos and early larvae are similar within all abdominal segments (see Suppl Fig 2). Thus, unlike in the long-germ band Drosophila embryo that simultaneously produces similar numbers of oenocytes within all abdominal segments, the short-germ band Tribolium embryo produces oenocytes in a gradient from high to low along the anterior-posterior axis of the abdomen.
Figure 7. Oenocyte enumeration in Tribolium.
A-B) Comparisons of oenocyte numbers per cluster in each abdominal segment of Drosophila (A) and Tribolium (B) embryos. Graph shows the mean and standard error for the number of oenocytes that form within each abdominal segment. Drosophila embryos average approximately 6 oenocytes per cluster with no statistical differences observed between the A1 through A7 abdominal segments. In contrast, Tribolium embryos average greater than 20 oenocytes per cluster and the A1 (*) and A8 (**) segments are statistically different from all other abdominal segments (p<0.05).
C-D) Lateral view of three abdominal segments from either a 26h (C) or 40h (D) Tribolium embryo immunostained for vvl-GFP (green) and phospho-histone3 (pH3, magenta) reveal that cells expressing GFP fail to colocalize with the pH3 G2-M marker.
The higher number of oenocytes produced in Tribolium could be due to either the recruitment of more oenocytes from a pool of competent cells or the subsequent expansion of oenocytes through cell division. To distinguish between these possibilities, we stained vvl-GFP embryos with the mitotic marker phospho-Histone3 (pH3) (Mahadevan et al., 1991). As shown in Figure 7, pH3 labeled numerous cells within the Tribolium embryo. However, we did not detect co-labeling of GFP-positive cells with pH3 at any stage of embryogenesis (Fig 7C-D). These findings are consistent with previous studies in Drosophila that showed oenocytes are post-mitotic following their initial specification (Gould et al., 2001). Thus, the expanded numbers of vvl-GFP-positive oenocytes in Tribolium is not through proliferation after specification and is likely due to increased cell recruitment of this cell type.
Discussion
Larval oenocytes constitute a cell type found throughout insects, and functional studies in Drosophila melanogaster revealed they are essential for proper lipid metabolism and larval growth (Chapman, 1998; Gutierrez et al., 2007). In this study, we compare and contrast the specification and development of larval oenocytes between D melanogaster and the red flour beetle, Tribolium castaneum. Our findings have two important implications: 1) We identified a general tool (fluorophore-conjugated streptavidin) that can be used to recognize oenocytes in other insect species as well as several useful markers for studying oenocytes in Tribolium. 2) We applied these tools to describe the specification of oenocytes in Tribolium embryos, and our findings show both similarities and differences in their development compared to Drosophila.
Streptavidin is a useful marker of oenocyte fate in insects
Due to its relatively small size and high affinity for streptavidin, biotin has long been a favorite molecule for labeling and purifying biological proteins. However, biotin is also an essential coenzyme required for numerous cellular reactions including proper fatty acid synthesis and metabolism (Fletcher and Myant, 1960). Hence, it is not surprising that larval oenocytes, which express many lipid-modifying enzymes required for fatty acid metabolism, have relatively high concentrations of biotin that can be labeled using fluorophore-conjugated streptavidin. We showed the utility of streptavidin staining in species found within Diptera, Coleoptera, and Hemiptera, suggesting this oenocyte labeling method is likely to be broadly applicable to many insect species. However, two important caveats should be kept in mind when using streptavidin to label cells. First, the ability of streptavidin staining to strongly label larval oenocytes is stage dependent. For example, streptavidin staining recognized oenocytes in the Drosophila melanogaster third instar larva but did not label these cells in the embryo. In contrast, streptavidin strongly labeled Tribolium oenocytes shortly after their specification in the beetle embryo. This differential labeling of larval oenocytes by streptavidin suggests each organism may have distinct requirements for concentrating biotin for enzymatic reactions within oenocytes at different developmental time points. Second, since biotin plays an essential role in the citric acid cycle, streptavidin is likely to label most cells, and thus, is not an oenocyte-specific stain. However, cells such as oenocytes that require a high concentration of biotin relative to their neighbors will be easily detected by streptavidin. Additional cell types including a subset of neuronal cells are labeled by streptavidin in Drosophila, Tribolium and Anasa embryos and/or larvae (not shown) (Ziegler et al., 1995). Thus, other markers and/or morphological characteristics such as cell size and position within the organism may be required to definitively identify cells as larval oenocytes. Nevertheless, the ease of obtaining and using fluorophore-conjugated streptavidin to label larval oenocytes makes it a powerful tool for the study of this cell type.
The comparative development of larval oenocytes between Tribolium and Drosophila
Oenocyte development can be divided into two key steps; cell specification and cell maturation. In Drosophila, larval oenocyte specification requires the activation of EGF signaling in dorsal ectoderm cells that express the Spalt transcription factor. The EGF signal originates from a specific sensory organ precursor cell within the posterior compartment of abdominal segments. An average of six neighboring cells per abdominal hemi-segment receive sufficient EGF signaling to further up-regulate Spalt expression and commit to oenocyte cell fate (Brodu et al., 2002, 2004). At the time of cell specification, oenocytes also express several other transcription factors including the Ventral-veinless (Vvl) and Seven-up (Svp) proteins (Brodu et al., 2002; Inbal et al., 2003). After oenocyte specification, these cells are post-mitotic, remain clustered, and eventually activate high levels of the HNF4 transcription factor as well as numerous lipid-modifying enzymes through largely unknown mechanisms during maturation (Gutierrez et al., 2007; Palanker et al., 2009).
In this study, we generated and used several markers to study the development of larval oenocytes in Tribolium castaneum, and our findings reveal similarities and differences in oenocyte development between these two species. Like in Drosophila, Tribolium larval oenocytes are abdominal-specific (although oenocytes form in the first 8 abdominal segments in Tribolium versus the first 7 abdominal segments in Drosophila), and these cells are post-mitotic after cell specification. Moreover, the oenocytes arise from Spalt-expressing dorsal ectoderm cells within the posterior compartment, and these cells also label with the phospho-ERK antibody. However, unlike in Drosophila, Tribolium oenocytes do not up-regulate Spalt and its expression is not maintained in mature oenocytes. These findings suggest that Spalt may play an essential role in the specification process, but not for oenocyte maturation. Instead, maturing larval oenocytes in both Tribolium and Drosophila express the HNF4 transcription factor. In addition, our developmental time course analysis of vvl-GFP is consistent with Tribolium oenocytes engaging in an extensive pattern of migration that is not seen in Drosophila. After specification in the posterior compartment, Tribolium oenocytes become localized around the forming trachea that lies in a more anterior-dorsal region of each abdominal segment. The Tribolium clusters subsequently form a continuous streak of oenocytes between abdominal segments, consistent with the pattern described in classical studies (Snodgrass, 1993; Wheeler, 1892).
An additional difference between Drosophila and Tribolium is oenocyte enumeration. Both species form clusters of oenocytes, but the Tribolium clusters are roughly 4 times larger with 22 cells/cluster versus 6 in Drosophila clusters. This difference may be a result of a more extensive timeframe during which oenocytes are recruited in Tribolium. Like Drosophila, most Tribolium oenocytes are recruited in an initial pulse, which happens at 24-26h of beetle development. However, additional vvl-GFP-positive cells (Tribolium oenocytes) continue to arise from the Engrailed stripe in small numbers until 46h of development. The continued production in vvl-GFP-positive cells is not due to cell division, as we detected no co-labeling with the phopho-Histone-3 marker of cell proliferation. Hence, larval oenocytes are post-mitotic after specification in both Drosophila and Tribolium. In addition, we found that anterior abdominal segments produce more oenocytes per segment than posterior abdominal segments in Tribolium but not Drosophila. This finding suggests that short-germ band insects (Tribolium) produce fewer oenocytes within the posterior segments that are added later in development, whereas long-germ band insects (Drosophila) that form all abdominal segments simultaneously produce similar oenocyte numbers in each segment.
An unanswered question our findings raise is what is the source of the phospho-ERK signal in Tribolium and is it EGF signaling dependent? Unlike in Drosophila, there does not seem to be a central singular signaling cell, as broad swathes of phospho-ERK activity that cover more than 10 cells at a time are observed in Tribolium. Moreover, phospho-ERK activity may represent the activation of several major tyrosine kinase-dependent signaling pathways, and thus, phospho-ERK staining does not conclusively demonstrate EGF signaling is involved in oenocyte specification in Tribolium. In Drosophila, the EGF-secreting cells can be identified by analyzing the expression of the rhomboid serine-proteases that processes EGF (Elstob et al., 2001; Lage et al., 1997). We analyzed the embryonic expression of the rhomboid genes in Tribolium embryos and found that the only rhomboid orthologue with a discernible expression pattern is rhomboid-A (the root orthologue of rhomboids1-3 from D. melanogaster). However, while we detected rhomboid-A expression in the area of trachea formation (not shown), we did not detect expression relevant to the region of oenocyte recruitment. This finding could indicate that either rhomboid is not the source of phospho-ERK signaling to induce oenocytes or that the levels of rhomboid expressed in this region are below our detection threshold. Consistent with the later possibility, the expression of Drosophila rhomboid in the abdominal sensory cells that induce oenocytes is much lower than the expression detected within the Drosophila tracheal pits. Hence, future loss-of-function studies on the rhomboid genes and the EGF pathway are required to definitively ascertain their role in Tribolium oenocyte specification.
Supplementary Material
Acknowledgments
We thank Alex Gould, GEKU, the Bloomington Drosophila Stock Center, and the Developmental Studies Hybridoma Bank (Univ of Iowa) for reagents. We thank Tingjia Lao and Padmapriyadarshini Ravisankar for technical assistance. This work was supported by an NIH grant GM079428A to B.G and an NSF grant (IOS0950964) to Y. T.
References
- Billeter JC, Atallah J, Krupp JJ, Millar JG, Levine JD. Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature. 2009;461:987–991. doi: 10.1038/nature08495. [DOI] [PubMed] [Google Scholar]
- Brodu V, Elstob PR, Gould AP. abdominal A specifies one cell type in Drosophila by regulating one principal target gene. Development. 2002;129:2957–2963. doi: 10.1242/dev.129.12.2957. [DOI] [PubMed] [Google Scholar]
- Brodu V, Elstob PR, Gould AP. EGF receptor signaling regulates pulses of cell delamination from the Drosophila ectoderm. Dev Cell. 2004;7:885–895. doi: 10.1016/j.devcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Chapman RF. The Insects: Structure and Function. 4. Cambridge University Press; 1998. [Google Scholar]
- Eastham LES. The Post-embryonic Development of Phaenoserphus viator Hal. (Proctotrypoidea), a Parasite of the Larva of Pterostichus niger (Carabidae), with Notes on the Anatomy of the Larva. Parasitology. 1929;21:1–21. [Google Scholar]
- Elstob PR, Brodu V, Gould AP. spalt-dependent switching between two cell fates that are induced by the Drosophila EGF receptor. Development. 2001;128:723–732. doi: 10.1242/dev.128.5.723. [DOI] [PubMed] [Google Scholar]
- Fernandez MP, Chan YB, Yew JY, Billeter JC, Dreisewerd K, Levine JD, Kravitz EA. Pheromonal and behavioral cues trigger male-to-female aggression in Drosophila. PLoS Biol. 2010;8:e1000541. doi: 10.1371/journal.pbio.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher K, Myant NB. Biotin in the synthesis of fatty acid and cholesterol by mammalian liver. Nature. 1960;188:585. doi: 10.1038/188585a0. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Gebelein B, Mann RS. Compartmental modulation of abdominal Hox expression by engrailed and sloppy-paired patterns the fly ectoderm. Developmental biology. 2007;308:593–605. doi: 10.1016/j.ydbio.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould AP, Elstob PR, Brodu V. Insect oenocytes: a model system for studying cell-fate specification by Hox genes. J Anat. 2001;199:25–33. doi: 10.1046/j.1469-7580.2001.19910025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- Gutzwiller LM, Witt LM, Gresser AL, Burns KA, Cook TA, Gebelein B. Proneural and abdominal Hox inputs synergize to promote sensory organ formation in the Drosophila abdomen. Dev Biol. 2010;348:231–243. doi: 10.1016/j.ydbio.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbal A, Levanon D, Salzberg A. Multiple roles for u-turn/ventral veinless in the development of Drosophila PNS. Development. 2003;130:2467–2478. doi: 10.1242/dev.00475. [DOI] [PubMed] [Google Scholar]
- Jackson A, Locke M. The formation of plasma membrane reticular systems in the oenocytes of an insect. Tissue Cell. 1989;21:463–473. doi: 10.1016/0040-8166(89)90059-1. [DOI] [PubMed] [Google Scholar]
- Kornberg T. Engrailed: a gene controlling compartment and segment formation in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:1095–1099. doi: 10.1073/pnas.78.2.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage P, Jan YN, Jarman AP. Requirement for EGF receptor signalling in neural recruitment during formation of Drosophila chordotonal sense organ clusters. Current biology : CB. 1997;7:166–175. doi: 10.1016/s0960-9822(97)70087-3. [DOI] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Current biology : CB. 2010;20:R754–763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Kroeger D, Witt LM, Grimes HL, Cook TA, Gebelein B. Hox and senseless antagonism functions as a molecular switch to regulate EGF secretion in the Drosophila PNS. Dev Cell. 2008;15:298–308. doi: 10.1016/j.devcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan LC, Willis AC, Barratt MJ. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell. 1991;65:775–783. doi: 10.1016/0092-8674(91)90385-c. [DOI] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 2009;9:228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Bucher G, Friedrich M, Grimmelikhuijzen CJ, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Roth S, Hartenstein V. Development of Tribolium castaneum. Dev Genes Evol. 2008;218:115–118. doi: 10.1007/s00427-008-0215-2. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Cantera R, Urban J, Technau G, Kafatos FC, Barrio R. Spalt modifies EGFR-mediated induction of chordotonal precursors in the embryonic PNS of Drosophila promoting the development of oenocytes. Development. 2001;128:711–722. doi: 10.1242/dev.128.5.711. [DOI] [PubMed] [Google Scholar]
- Schroder R, Beermann A, Wittkopp N, Lutz R. From development to biodiversity--Tribolium castaneum, an insect model organism for short germband development. Dev Genes Evol. 2008;218:119–126. doi: 10.1007/s00427-008-0214-3. [DOI] [PubMed] [Google Scholar]
- Shilo BZ. Regulating the dynamics of EGF receptor signaling in space and time. Development. 2005;132:4017–4027. doi: 10.1242/dev.02006. [DOI] [PubMed] [Google Scholar]
- Shippy TD, Brown SJ, Denell RE. Molecular characterization of the Tribolium abdominal-A ortholog and implications for the products of the Drosophila gene. Development genes and evolution. 1998;207:446–452. doi: 10.1007/s004270050135. [DOI] [PubMed] [Google Scholar]
- Snodgrass RE. Principles of insect morphology. Ithaca: Cornell University Press; 1993. [Google Scholar]
- Tomoyasu Y, Wheeler SR, Denell RE. Ultrabithorax is required for membranous wing identity in the beetle Tribolium castaneum. Nature. 2005;433:643–647. doi: 10.1038/nature03272. [DOI] [PubMed] [Google Scholar]
- Trauner J, Schinko J, Lorenzen MD, Shippy TD, Wimmer EA, Beeman RW, Klingler M, Bucher G, Brown SJ. Large-scale insertional mutagenesis of a coleopteran stored grain pest, the red flour beetle Tribolium castaneum, identifies embryonic lethal mutations and enhancer traps. BMC Biol. 2009;7:73. doi: 10.1186/1741-7007-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler WM. Concerning the blood tissue of insects. Psyche. 1892;6:216–258. [Google Scholar]
- Witt LM, Gutzwiller LM, Gresser AL, Li-Kroeger D, Cook TA, Gebelein B. Atonal, Senseless, and Abdominal-A regulate rhomboid enhancer activity in abdominal sensory organ precursors. Dev Biol. 2010;344:1060–1070. doi: 10.1016/j.ydbio.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B, Charlton-Perkins M, McDonald E, Gebelein B, Cook T. Senseless functions as a molecular switch for color photoreceptor differentiation in Drosophila. Development. 2007;134:4243–4253. doi: 10.1242/dev.012781. [DOI] [PubMed] [Google Scholar]
- Zara FJ, Caetano FH. Ultramorphology and histochemistry of fat body cells from last instar larval of the Pachycondyla (=Neoponera) villosa (Fabricius) (Formicidae: Ponerinae) Braz J Biol. 2004;64:725–735. doi: 10.1590/s1519-69842004000400022. [DOI] [PubMed] [Google Scholar]
- Ziegler R, Engler DL, Davis NT. Biotin-containing proteins of the insect nervous system, a potential source of interference with immunocytochemical localization procedures. Insect biochemistry and molecular biology. 1995;25:569–574. doi: 10.1016/0965-1748(94)00095-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.