Abstract
Chloroquine (CQ) is a 4-aminoquinoline drug used for the treatment of diverse diseases. It inhibits lysosomal acidification and therefore prevents autophagy by blocking autophagosome fusion and degradation. In cancer treatment, CQ is often used in combination with chemotherapeutic drugs and radiation because it has been shown to enhance the efficacy of tumor cell killing. Since CQ and its derivatives are the only inhibitors of autophagy that are available for use in the clinic, multiple ongoing clinical trials are currently using CQ or hydroxychloroquine (HCQ) for this purpose, either alone, or in combination with other anticancer drugs. Here we show that in the mouse breast cancer cell lines, 67NR and 4T1, autophagy is induced by the DNA damaging agent cisplatin or by drugs that selectively target autophagy regulation, the PtdIns3K inhibitor LY294002, and the mTOR inhibitor rapamycin. In combination with these drugs, CQ sensitized to these treatments, though this effect was more evident with LY294002 and rapamycin treatment. Surprisingly, however, in these experiments CQ sensitization occurred independent of autophagy inhibition, since sensitization was not mimicked by Atg12, Beclin 1 knockdown or bafilomycin treatment, and occurred even in the absence of Atg12. We therefore propose that although CQ might be helpful in combination with cancer therapeutic drugs, its sensitizing effects can occur independently of autophagy inhibition. Consequently, this possibility should be considered in the ongoing clinical trials where CQ or HCQ are used in the treatment of cancer, and caution is warranted when CQ treatment is used in cytotoxic assays in autophagy research.
Keywords: chloroquine, cisplatin, PtdIns3K, LY294002, mTOR, rapamycin, autophagy, breast cancer
Introduction
Chloroquine (CQ) has been used for the treatment of diverse diseases. When unprotonated, it can diffuse across cell membranes, get protonated and accumulate in acidic organelles such as the lysosomes.1 This lysosomotropic property results in the inhibition of lysosomal enzymes and makes it useful in the treatment of the malaria parasite.1 CQ has also been used for the treatment of rheumatoid arthritis, lupus erythematosus and amoebic hepatitis because of its anti-inflammatory and immunosuppressive effects, which have been attributed to lysosomal stabilization, suppression of antigen presentation, inhibition of prostaglandin and cytokine synthesis and the modulation of toll-like receptors.2 It has also been proposed to work as an antiviral agent by interfering with protein glycosylation necessary for viral function.3 More recently, the ability of CQ to block autophagy by inhibiting lysosomal proteases and autophagosome-lysosomal fusion events has generated further interest in this drug in other settings, including cancer treatment.1,4–6 Since autophagy is thought to act as a cell-survival pathway in cancer, CQ has been used in combination with diverse chemotherapeutic drugs and radiation and has been shown to enhance tumor cell killing. For instance, it has been suggested that CQ preferentially induces cell death of breast cancer cell lines compared with immortalized cells7 and to increase survival of mice implanted with the 4T1 breast cancer cell line when used as a single agent.8 CQ is perhaps the most widely used drug to inhibit autophagy in vitro and its effects on cell death when used together with other chemotherapeutic agents have been attributed to its inhibition of autophagy.9,10 Moreover, CQ and its derivatives are currently the only inhibitors of autophagy available for clinical treatment of patients. This widely accepted mechanism of autophagy inhibition together with its relatively low toxicity1 and well understood pharmacological properties has led to more than twenty clinical trials listed on the ClinicalTrials.gov website11 using CQ or hydroxychloroquine (HCQ) to test if inhibition of autophagy in a clinical setting can increase the effectiveness of cancer therapies.6
The phosphoinositide 3-kinases (PtdIns3Ks) have a key role in the regulation of cellular processes, including survival, proliferation and differentiation. PtdIns3Ks transduce signals from growth factors and cytokines by activating AKT, and subsequently activating the mTOR kinase, which has a crucial role in the regulation of cell growth and proliferation.12,13 Increased signaling through the PtdIns3K/mTOR pathway induces cellular transformation, as well as tumor progression and metastasis. Mutations in this pathway occur in up to one quarter of breast cancers and might cause resistance to upstream targeted inhibition like anti HER-2 agents and hormonal agents.14 Moreover, PtdIns3K, AKT and mTOR inhibitors are currently being used in clinical trials for treatment of a variety of cancers.12
In addition to its role in cell growth, the PtdIns3K/mTOR pathway is one of the main regulators of macroautophagy.15 Macroautophagy, referred to hereafter as autophagy, is a catabolic mechanism of intracellular turnover in which cells degrade their own cytoplasmic components to provide nutrients in response to stresses like starvation, growth factor deprivation or energetic requirements.4 Autophagy is regulated by the Atg (autophagy-related) proteins, which control a series of steps that culminate in the formation of the autophagosome, a double-membrane organelle that transports cytoplasmic material and damaged organelles to the lysosomes for their degradation. During nutrient availability, mTOR represses the activity of the Atg1/ULK1 complex and inhibits autophagy. Conversely, when amino acids are scarce or there is decreased signaling through growth factor receptors, mTOR is not activated and autophagy is induced.
Although autophagy has become a well-accepted survival pathway, it has also been suggested that when cellular damage is extensive, autophagy could be cytotoxic.16 Both mechanisms could be important during cancer treatment since many chemotherapeutic drugs as well as radiation therapy induce autophagy in a variety of cancer cell lines.16,17 One such widely used drug, cisplatin, causes tumor cell death and cell cycle arrest due to the formation of platinum-DNA adducts.18 Cisplatin and other platinum-based drugs have long been used for breast cancer treatment as single agents or in combination with other therapies,19 and recent evidence suggests that autophagy could be involved in resistance to cisplatin treatment in ovarian,20,21 skin22 and esophageal23 cancer cell lines.
We tested the idea that autophagy inhibition could sensitize to therapy using different anticancer agents. We found that cisplatin as well as the PtdIns3K pathway inhibitor LY294002 and mTOR inhibitor rapamycin all induced autophagy in two mouse breast cancer cell lines. CQ decreased the viability of cells treated with chemotherapy, and this effect was more pronounced when used together with LY294002 or rapamycin treatment. However, this effect could not be mimicked with bafilomycin A (Baf A), which is another lysosomotropic agent that inhibits autophagy or with Atg12 or Beclin 1 knockdown, which both inhibit early steps of the autophagy process. Moreover, CQ could still sensitize to treatment even when autophagy was blocked upstream of autophagosome formation. We therefore conclude that CQ-mediated chemosensitization to therapy is an autophagy-independent event in these cells and that, when treating humans with CQ along with anti-cancer agents, one should consider the possibility that CQ may be acting through mechanisms other than by inhibition of autophagy. Our study also suggests that autophagy researchers should be careful when interpreting experiments in which CQ treatment produces, or sensitizes to, a cytotoxic effect, since the effect may be mediated by mechanisms other than its inhibition of autophagy.
Results
67NR and 4T1 mouse breast cancer cells have functional autophagy that can be blocked by CQ treatment.
LC3, the mammalian homolog of Atg8, is cleaved and conjugated to phosphatidylethanolamine during autophagy. This modified form, termed LC3II, is involved in the elongation of the autophagosome. However, LC3II is also degraded inside the lysosomes or de-conjugated by Atg4.4,24 Therefore, in order to measure autophagic flux, lysosomal inhibitors (CQ, Baf A, pepstatin A/E64D) are used in order to inhibit LC3II degradation, since in the absence of these inhibitors, an autophagy-inducing treatment can result in a modest increase or even a decrease in the amount of LC3II.25 Another approach has been the use of a tandem-tagged LC3 fluorescent protein containing GFP and RFP or cherry. Because GFP signal is more sensitive to the acidic conditions in the lysosomal lumen,4,26 a solely red fluorescence signal indicates autophagolysosomes, while colocalizing red and green signals indicate autophagosomes that have not yet fused with the lysosomes.
We used two breast cancer cell lines with different metastatic abilities and different levels of basal autophagy. Although both cell lines were isolated from the same tumor, the 67NR cell line is highly tumorigenic but will rarely metastasize while the 4T1 cell line is highly metastatic.27 Autophagy is believed to be important for the normal recycling of cytoplasmic contents, and therefore a certain amount of autophagy occurs even in unstimulated cells (basal autophagy). Since LC3 can be conjugated and degraded during this process, LC3II accumulation and re-localization induced by lysosomal inhibition in complete medium can be used as a measure of basal autophagic flux.4,28,29 Thus, we evaluated basal autophagy levels in both cell lines by measuring LC3II accumulation after CQ or Baf A treatment for different time points, as well as the accumulation of p62, a protein degraded by autophagy (Fig. S1A and S1B). 4T1 cells showed accumulation of LC3II and p62 at earlier time points than 67NR cells after both CQ and Baf A treatment, indicating that the cell lines have different levels of basal autophagy and that these concentrations of CQ and Baf A are able to block basal autophagy in both cell lines.
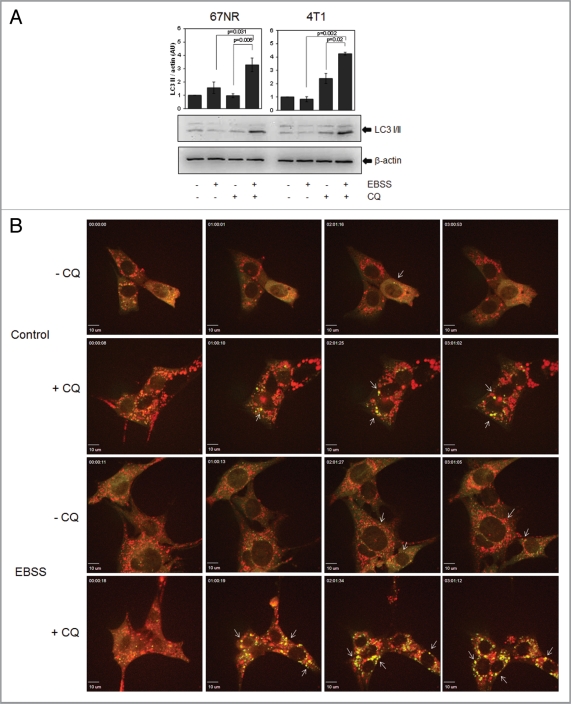
We then measured autophagic flux in 67NR and 4T1 cells upon starvation by EBSS (Earle's Balanced Salt Solution) treatment (Fig. 1A and B, Fig. S1D and E). CQ blocked LC3II degradation after starvation in EBSS medium and both cell lines underwent autophagic flux to similar extents as HeLa cells, a well-studied model of starvation-induced autophagy (Fig. 1A and Fig. S1C).5,25 Autophagic flux was also evident in 67NR GFP-cherry-LC3 expressing cells (Fig. 1B, Fig. S1D and S1E). In a time-lapse analysis, few yellow dots were observed under control conditions and green fluorescence showed mostly a uniform, cytoplasmic distribution. Red dots were primarily observed in these cells, indicative of autophagolysosomes under conditions of basal autophagy. CQ treatment induced colocalization of green and red dots. And although red vesicles were still observed, probably due to autophagolysosomes present before the addition of CQ, many vesicles were yellow (green and red co-localization). Upon starvation, yellow dots could be observed due to autophagosome formation and red vesicles accumulated beginning at early time points (1 h), indicating autophagosome formation and fusion with lysosomes where GFP fluorescence is quenched, i.e., autophagic flux. When CQ was added to starvation media, vesicles were largely yellow, indicating that EBSS-induced autophagic flux is effectively blocked by CQ treatment. Thus, 67NR and 4T1 cells undergo autophagy upon starvation and CQ decreases LC3II lysosomal degradation. Quantitation of GFP-cherry positive cells also indicated that autophagic flux was induced by EBSS treatment (Fig. S1D and S1E).
Figure 1.
67NR and 4T1 mouse breast cancer cells induce autophagy in response to starvation. 67NR and 4T1 cells were treated with EBSS ± CQ for 2 h to measure autophagy induction with an LC3 western blot (A). 67NR cells were also evaluated for starvation-induced autophagy with fluorescence microscopy using a GFP-cherry-LC3 construct. In (B), 67NR cells were starved for 3 h in EBSS ± 30 µM CQ and observed in a time-lapse confocal microscope. Pictures show frames of each movie at the indicated times (h). Arrows show yellow dots. Graphs in (A) show density analysis of the mean ± standard error of three independent experiments.
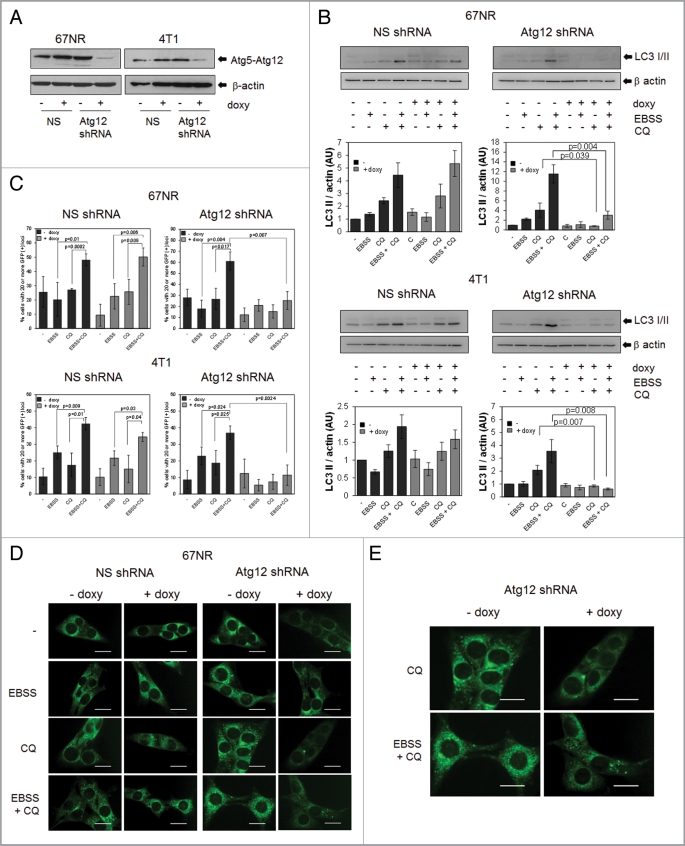
In order to genetically modulate autophagy, we established a doxycycline-inducible system to induce the expression of an Atg12 shRNA. Atg12 expression was substantially decreased after doxycycline treatment in both cell lines (Fig. 2A). Functional inhibition of autophagy by these knockdowns was demonstrated by a doxycycline-dependent reduction of starvation-induced LC3II formation and autophagic flux (Fig. 2B) as well as GFP-LC3 relocalization after starvation (Fig. 2C–E and Fig. S2) in both cell lines.
Figure 2.
Establishment of an inducible system to manipulate autophagy. 67NR and 4T1 cells were transduced with a lentivirus containing either an inducible Atg12 shRNA or a nonsilencing (NS) shRNA (see Materials and Methods). Atg12 was decreased after 72 h of doxycycline (doxy) treatment (A) as well as starvation-induced changes in LC3II (B) and re-distribution of LC3 (C–E). Pictures in (E) show a magnification of CQ and EBSS+CQ pictures in (D). Since doxycycline-treated cells showed less total green fluorescence than non-doxycycline treated cells, pictures in (E) for doxycycline-treated cells (+ doxy) were brightness-enhanced by 20% for a better comparison. Graphs in (B) show density analysis of three independent western blots; graphs in (C) show quantification of green dots from pictures shown in Figure 2D and E and Figure S2. All graphs show mean ± standard error of three independent experiments. Scale bars in (D and E) represent 20 µm.
Chemotherapeutic drugs induce autophagy in the 67NR and 4T1 cell lines but CQ sensitizes mainly to LY294002 or rapamycin treatment.
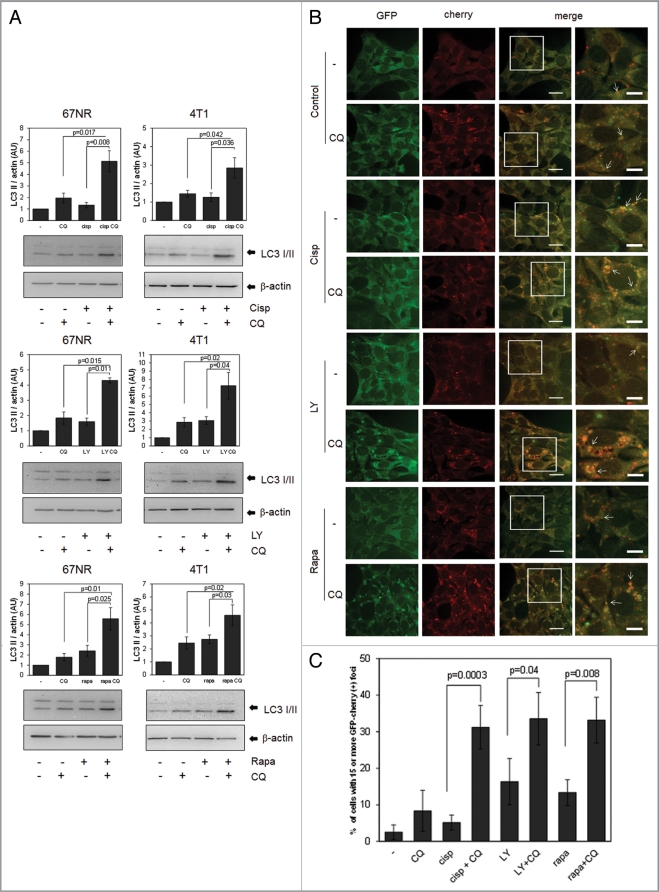
Many chemotherapeutic drugs are reported to induce autophagy in a variety of cell lines. In this work, we used cisplatin, a DNA damaging agent, and two drugs that target the PtdIns3K pathway, rapamycin, an mTOR inhibitor, and LY294002, a PtdIns3K inhibitor. PtdIns3K inhibitors such as LY294002 or wortmannin have been shown to either block autophagy4 or stimulate autophagy,30,31 depending on cell type or on the experimental context, since they inhibit both the class III PtdIns3K (Vps34), an important part of the autophagic nucleation process, and also the classical class I PtdIns3K, which negatively regulates autophagy by the downstream activation of mTOR by AKT.4 When both cell lines were treated with these drugs, we found an induction of autophagic flux by LY294002, as well as cisplatin and rapamycin, which was blocked by CQ as demonstrated using LC3 western blots (Fig. 3A) or fluorescence microscopy in 67NR GFP-cherry-LC3 expressing cells (Fig. 3B and C).
Figure 3.
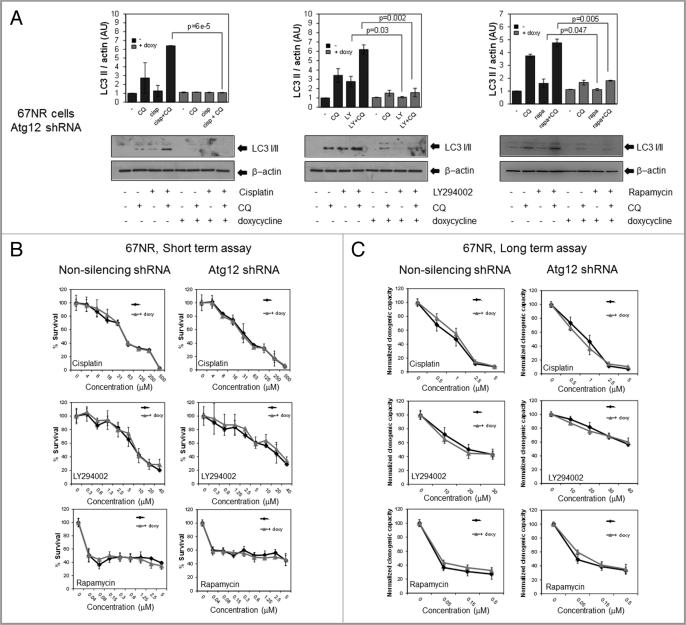
Chemotherapeutic drugs induce autophagy in the 67NR and 4T1 cell lines. Cells were treated with 1 mM cisplatin, (Cisp) for 6 h or with 30 µM LY294002 (LY) or 0.2 µM rapamycin (Rapa) for 8 h ± CQ and proteins were collected for WB (A). GFP-cherry-LC3 expressing 67NR cells grown in coverslips were treated with the same drug concentrations for 8 h, fixed and observed in a confocal microscope. The bar in merged images represents 20 µM and the one in the inset represents 10 µM. Quantification of yellow dots is shown in (C). Graph shows mean ± standard error of three independent experiments.
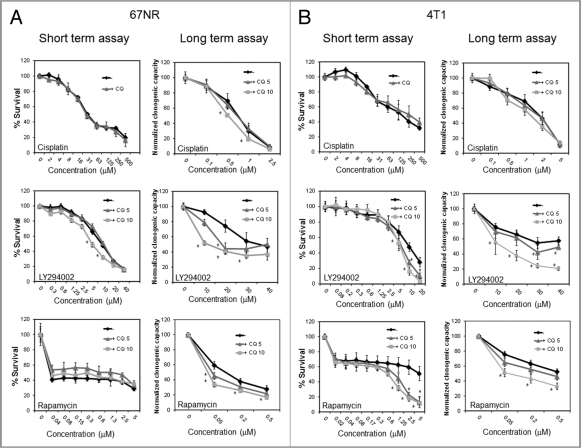
In order to test if autophagy induced by the drugs had an effect on cell survival, we performed short- (MTS) and long-term (clonogenic) survival assays in cells treated with the different drugs together with CQ in order to block autophagy in the 67NR (Fig. 4A) and 4T1 cell lines (Fig. 4B). CQ decreased viability of the cisplatin-treated 67NR cells in long-term but not in short-term assays as determined by the cisplatin dose-response curve and did not significantly affect viability in cisplatin-treated 4T1 cells in either short- or long-term assays. However, when used together with LY294002, CQ decreased viability in shortand long-term assays in both cell lines, this effect being more striking in long-term clonogenic assays. Similarly, CQ sensitized to rapamycin treatment in long-term clonogenic assays in the 67NR cell line and in both short- and long-term assays in 4T1 cells. Thus, CQ blocks autophagy in response to all three treatments, but chemosensitizes preferentially in response to PtdIns3K or mTOR inhibition, i.e., the pathway that directly regulates autophagy.
Figure 4.
CQ sensitizes to LY294002 and rapamycin treatment but has a minimal effect on sensitization to cisplatin treatment. 67NR (A) and 4T1 (B) cells were treated with cisplatin, LY294002 or rapamycin at the indicated doses ± CQ. For short-term (MTS) assays, cells were treated for 24 h with cisplatin or for 48 h with LY294002 or rapamycin (67NR cells) or for 72 h with rapamycin (4T1 cells) with or without 5 mM (CQ 5) or 10 mM CQ (CQ 10). When not indicated, CQ concentration was 10 µM. For long-term (clonogenic) assays, cells were treated for 24 h ± CQ, the treatment was washed and cells were allowed to recover for 5 d. Graphs are normalized to 100% per treatment and show mean ± standard error of three independent experiments done in triplicate. * = statistical difference, p = 0.05.
CQ sensitization to LY294002 or rapamycin treatment is independent of autophagy.
To test if the observed effects were due to autophagy inhibition, we used our inducible system to decrease Atg12 protein expression in 67NR cells. Atg12 or non-silencing shRNA cells without doxycycline showed a similar induction of autophagy after treatment with the drugs as that seen in the parental cell line (Fig. 5A and Fig. S3A). When doxycycline was administered, cisplatin-, LY294002- and rapamycin-induced autophagy was almost completely blocked when measured by changes in LC3II (Fig. 5A). Surprisingly, however, no significant differences in the dose-response curves to the drugs were observed when autophagy was blocked (Fig. 5B and C) by the addition of doxycycline in short- (Fig. 5B) or long-term clonogenic assays (Fig. 5C). Treatment of 4T1 cells containing the inducible Atg12 shRNA gave comparable results, as no differences in viability were observed after drug treatment in short-term MTS or long-term clonogenic experiments (Fig. S3B and S3C) in the presence or absence of doxycycline to induce knockdown of Atg12. Similar results were also observed in the 4T1 cell line using Becn1 siRNAs to decrease the expression of another Atg protein that is required for induction of autophagy (Fig. S4A and S4B).
Figure 5.
Autophagy inhibition by Atg12 knockdown has no effect on cell survival in 67NR cells treated with cisplatin, LY294002 or rapamycin. 67NR cells expressing an inducible Atg12 shRNA or a nonsilencing shRNA were treated with doxycycline and then with 1 µM cisplatin, 30 µM LY294002 or 0.2 µM rapamycin (A) or at the indicated concentrations (B and C). In (A), protein was collected for western blot after an 8 h treatment ± CQ. In (B), viability was measured with MTS reagent (short-term assay) after 24 h (cisplatin) or 48 h (LY294002 or rapamycin). In (C), cells grown in the presence of doxycycline were treated for 24 h, the treatment was washed, replaced with fresh medium ± doxycycline (doxy) and cells were allowed to recover for 4 d (long-term, clonogenic assays). Graphs are normalized to 100% per treatment and show mean ± standard error from three independent experiments performed in triplicate.
These results suggest that reduction in autophagy does not affect cisplatin, LY294002 or rapamycin toxicity. Since this is the case, it is unlikely that CQ mediated sensitization to these drugs is due to its ability to block the completion of the autophagic process since sensitization cannot be mimicked by Atg12 or Beclin 1 knockdown. However, since knockdown of these proteins prevents autophagy at an earlier time point in the process, the possibility remains that CQ blockage of autophagy at a later point could lead to sensitization.
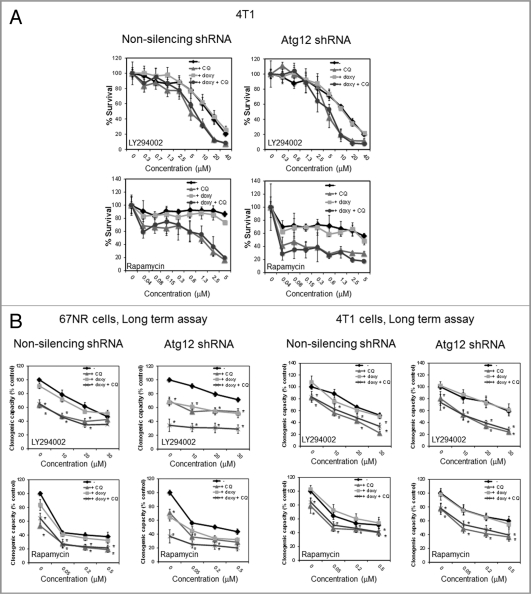
If CQ sensitization to these three drugs is not due to its effect on autophagy, one would expect that CQ should be able to chemosensitize even if autophagy is blocked earlier in the process. To test whether this is the case, we blocked autophagy using the inducible Atg12 shRNA and then treated with CQ and the anticancer drugs. In support of our hypothesis, we found that CQ sensitized 4T1 cells to LY294002 or rapamycin treatment even in the absence of Atg12 in short- (Fig. 6A) or long-term assays (Fig. 6B). 67NR cells expressing the inducible Atg12 shRNA in the presence of doxycycline grew slower than the non-doxycycline-treated cells and this effect was mimicked by CQ treatment in long-term assays, suggesting that autophagy is required for efficient growth of these cells. Interestingly, this was not the case in 4T1 cells, which had similar cell numbers in the presence or absence of doxycycline in long-term assays (Fig. 6B, untreated controls). We are currently investigating the autophagic-dependent growth of 67NR cells. However, despite this autophagy requirement for normal growth in the 67NR cells, both cell lines were sensitive to CQ in the absence of Atg12 after treatment with LY294002 or rapamycin. Similar results were found with cisplatin (Fig. S5A) or with Becn1 knockdown in the 4T1 cells (Fig. S4B).
Figure 6.
CQ induced sensitization to LY294002 and rapamycin treatment is not mimicked by Atg12 knockdown and CQ sensitizes even in the absence of Atg12. In (A), 4T1 cells were treated with LY294002 for 48 h or with rapamycin for 72 h ± doxycycline (doxy) or CQ and viability was evaluated in a short-term MTS assay. In (B), cells were grown in the absence or presence of doxycycline for 72 h, treated for 24 h ± doxycycline or CQ, the treatment was washed and replaced with fresh medium ± doxycycline and the cells were allowed to recover for 4 d for long-term (clonogenic) assays. Graphs in (A) show mean ± standard deviation of one representative experiment from three independent experiments performed in triplicate. Graphs in (B) show mean ± standard error of three independent experiments performed in triplicate. In (B), all the treatments were normalized to untreated controls.
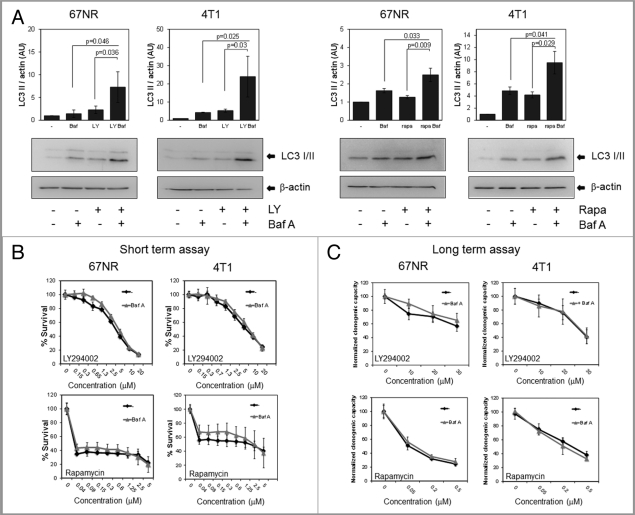
As further evidence that CQ was inducing chemosensitization independent of autophagy, we treated cells with bafilomycin A (Baf A), a lysosomotropic compound that blocks the autophagic process at a similar point as CQ. While Baf A could block the degradation of LC3II in 67NR and 4T1 cells after LY294002 or rapamycin treatment similarly to the effect of CQ (Fig. 7A), it had no effect on the dose-response curves when used together with LY294002 or rapamycin in short- (Fig. 7B) or long-term assays (Fig. 7C). Thus, CQ chemosensitizes but Baf A does not under these conditions despite the fact that both CQ and Baf A are equally effective at blocking autophagy and LC3-II degradation.
Figure 7.
Autophagy inhibition with bafilomycin A1 (Baf A) does not have the same effect as CQ. 67NR and 4T1 cells were treated with 20 µM LY294002 (4 h) or 0.2 µM rapamycin (6 h) ± 1 nM Baf A for 4 h and protein was collected for western blot (A). Cells were treated with LY294002 (48 h) or rapamycin (48 h for 67NR and 72 h for 4T1) ± 1 nM Baf A. Viability was measured with MTS reagent for short-term assays (B) and with colony formation assays for long-term assays after a 24 h treatment with the drugs (C). Graphs show mean ± standard deviation of one representative experiment from three independent experiments performed in triplicate.
Finally, since autophagy seems to have no role in the induction or protection from cisplatin, LY294002 or rapamycin-induced cell death, we tested the role of apoptosis in these treatments (Fig. S5B). When treating 4T1 cells with cisplatin together with the caspase inhibitor zVAD, we found a slight decrease in cell death with cisplatin treatment and no changes in cell viability were observed when treating cells with LY294002 or rapamycin in the presence of zVAD. Moreover, zVAD did not increase survival when used together with the drugs and CQ, suggesting that the CQ-induced decrease in cell viability involves a caspase-independent mechanism.
Discussion
The role of autophagy during cancer treatment is controversial. Although it is widely accepted that many anticancer agents induce autophagy in cancer cells, it is not yet clear if this autophagy represents a survival mechanism activated in response to stress induced by the treatment, if it is a cell death pathway activated when apoptosis is disabled, if it has no effect on tumor cell viability at all, or if all three effects arise in different contexts. In breast cancer, tamoxifen,32,33 trastuzumab,34 camptothecin,35 2-deoxyglucose,36 radiation37 and rapamycin38 have all been shown to induce autophagy in cancer cell lines. Studies suggest that autophagy is involved in the resistance to tamoxifen,33 trastuzumab,34 camptothecin35 and a BH3 mimetic,39 suggesting a protective role of autophagy during treatment with these drugs. On the other hand, induction of autophagy by rapamycin treatment has been suggested to decrease survival of irradiated cells38 and silencing of Bcl-2 has been reported to induce autophagic cell death that can sensitize cells to doxorubicin treatment,40 suggesting that autophagy plays a role in the promotion of breast cancer cell death with these treatments.
Because CQ and its analogs block autophagy, together with the fact that they have a relatively low toxicity to humans, CQ or HCQ are widely used to inhibit autophagy in both in vitro and in vivo studies, as well as in current clinical trials in cancer patients. As expected, we found that CQ effectively blocked autophagy at the lysosomal degradation step in two mouse breast cancer cell lines (Fig. 1). We also found that cisplatin, a DNA damaging agent, and two agents that target an autophagy regulatory pathway, LY294002 and rapamycin, which target PtdIns3K and mTOR kinases respectively, all induced autophagy similarly in both breast cancer cell lines (Fig. 3). However, when used in combination treatment with CQ, CQ sensitized to LY294002 and rapamycin treatment in both cell lines (Fig. 4), and this effect was more striking in long-term clonogenic assays than in short-term MTS assays, indicating that the treatments are not only altering the kinetics of cell death but actually decreasing the number of cells that survive the treatment and that are subsequently able to proliferate and form colonies after the treatment is removed.
It should be noted that although DNA damaging agents induce autophagy in different cancer cell lines,35,41,42 and that rapamycin is a widely used inducer of autophagy,4 PtdIns3K inhibitors like LY294002 or wortmannin have sometimes been used to block autophagy since they also inhibit the class III PtdIns3K (Vps34), an important part of the autophagic nucleation process.4 However in our experiments, the net effect of all three agents was similar: LY294002 as well as cisplatin and rapamycin all induced autophagic flux as measured by changes in LC3II and by a GFP-cherry-LC3 construct flux assay (Fig. 3) and autophagy was blocked by induction of the Atg12 shRNA (Fig. 5A). Previous studies have shown that PtdIns3K inhibitors like 3MA induce autophagy when used in full medium and block autophagy when used together with an autophagy-inducing stimulus like starvation.31 We hypothesize that something similar is happening with LY294002 in these cells, and that therefore LY294002 induces autophagy, as is seen in glioma cells.30 Since LY294002 and 3MA are not able to discriminate between PtdIns3K family isoforms,31,43 LY294002 could be inhibiting class I PtdIns3K in normal, full-medium conditions, where it would be more active than class III PtdIns3K (Vps34). On the other hand, it could preferentially inhibit class III PtdIns3K (Vps34) under conditions that promote its activation, i.e., starvation or autophagic-inducing stimuli. Moreover, both LY294002 and wortmannin have been shown to inhibit mTOR at concentrations similar to those needed for inhibition of mammalian PI3-kinases,44 suggesting another mechanism through which this drug could be promoting autophagy.
Because all the treatments we employed induced autophagy to similar levels, we hypothesized that the cells might need autophagy for survival during treatment with DNA damaging drugs and targeted therapies that directly regulate the core autophagy pathway. Previous reports have shown that the combination of PtdIns3K pathway inhibitors with CQ or other lysosomotropic agents results in enhanced cancer cell killing in breast,7 prostate9 and glioma9,10 cancer cell lines. In these studies, PtdIns3K pathway inhibition decreased tumor growth and cell proliferation in vitro but apoptosis was only induced after the addition of lysosomotropic agents. These effects were attributed to the inhibitory effect of CQ in autophagy induced by PtdIns3K pathway inhibition. Other recent studies have proposed a role of autophagy in cisplatin resistance in ovarian cancer cell lines,20 where it has been suggested that autophagy protects from cisplatin treatment through the removal of ubiquitinated proteins and reduction of ER stress.21
Similarly to these previous studies, we found that PtdIns3K pathway inhibition by LY294002 or rapamycin treatment induced autophagic flux and CQ sensitized to both treatments. We also found autophagy induction and CQ sensitization to cisplatin treatment in the 67NR cells. However, these sensitization effects were independent of autophagy for the following reasons. First, an inducible Atg12 knockdown system that effectively blocked starvation, cisplatin, LY294002 and rapamycin-induced autophagy (Figs. 2 and 5), or Beclin 1 knockdown (Fig. S4) did not cause any differences in survival in response to the drugs in either cell line in short- or long-term assays (Fig. 5B and C, Fig. S3B, S3C and Fig. S4B). Second, CQ treatment caused a similar increase in chemosensitivity whether or not Atg proteins were present (Fig. 6, Fig. S4C and Fig. S5A), indicating that CQ is capable of increasing sensitivity to the drugs independent of Atg12 or Beclin 1. These data also suggest that the observed toxicity is not due to increased numbers of “blocked” autophagosomes, which should not accumulate when autophagosome formation is inhibited. Finally, Baf A, a lysosomal ATPase inhibitor, which inhibits autophagy at the same stage that CQ does, decreased LY294002 and rapamycin induced LC3II degradation in both cell lines but did not sensitize to the drugs in short- or long-term assays (Fig. 7). These three separate findings all lead to the same conclusion—CQ sensitization to cisplatin and PtdIns3K pathway inhibitors occurs in an autophagy-independent manner. The mechanism by which CQ could be inducing cell death seems to be independent of apoptosis since it was not decreased by caspase inhibition (Fig. S5B) and could possibly involve other functions of CQ independent of autophagy that are less well characterized, such as its ability to intercalate into DNA45 or to activate ATM and p53.46
We believe this work denotes a potentially important finding since active clinical trials in breast cancer patients and people with other types of cancers are using CQ or HCQ together with other therapies in order to block autophagy induced by cancer treatment.11 In addition, CQ is often used in the field to substantiate that autophagy is involved in a process, cell death or otherwise. Although we do not exclude the possibility that autophagy might have protective effects in other types of cancers or in cells with a specific mechanism of transformation, we propose that autophagy induced by cancer treatment is not necessarily a general mechanism of tumor cell chemoresistance and, even if clinical trials using CQ combined with other agents show success, this might not be due to CQ effects on autophagy induced by chemotherapy. It will be necessary to better understand under which circumstances autophagy is or is not chemo-protective and to develop more selective autophagy inhibitors if we are to maximize the benefits of autophagy manipulation during cancer therapy.
Materials and Methods
Cell lines and cell culture.
The 67NR cell line was a gift from Dr. H. Ford at UCD and 4T1 cells were obtained from ATCC. Both cell lines were maintained in DMEM (Cellgro, 10-013-CV) + 10% fetal bovine serum (FBS, Sigma, F6178) at 37C and 5% CO2. Atg12 mouse GIPZ shRNAmir (clone V2LMM_72549, RMM4431-99212717) was obtained from Open Biosystems. The shRNA was cloned into the pTRIPZ inducible vector (Open Biosystems, RHS4750) according to manufacturer's protocols. Lentiviruses containing the pTRIPZ vector were made in HEK293FT cells transfected with Trans-Lentiviral Packaging System (Open Biosystems, TLP4606), pTRIPZ plasmids and Arrest-In transfection reagent (Open Biosystems, ATR1740). Both cell lines were transduced with lentiviruses containing pTRIPZ Atg12 shRNA or nonsilencing shRNA (Open Biosystems, RHS4743) using polybrene (Sigma, H9268, 8 µg/ml). After puromycin (Sigma, P8833) selection (2 µg/ml), cells were maintained in medium containing 1 µg/ml puromycin and tetracycline free FBS (Hyclone, SH30070.03T). Clones were isolated and validated for Atg12 knockdown and LC3II decrease after EBSS (Hyclone, SH30029) treatment ± CQ. For shRNA induction, cells were treated with 1 µg/ml doxycycline (Clontech, 8634-1) for 72 h replacing doxycycline every 24 h.
Retroviruses expressing pBabe GFP-cherry-LC3 (construct was a gift from Dr. Debnath's lab at UCSF) were made using GP2-293 cells transfected with pBabe GFP-cherry-LC3 and pVSV-G plasmids (Clontech, Retro-X Universal Retroviral Expression System, 631530) using TransIT-LT1 (Mirus, MIR2300) transfection reagent. After puromycin selection, clones were isolated and individually screened for green and red fluorescence.
Unless otherwise specified, cells were treated with 10 µM CQ (Sigma, C6628) or 1 nM Baf A (Sigma, B1793). For western blot experiments longer than 4 h, CQ or Baf A were added for the last 4 h of treatment.
Viability assays.
For short-term MTS assays, cells were plated in 96 well plates and 24 h later, they were treated with cisplatin (Sigma, P4394), rapamycin (Sigma, R0395) or LY294002 (Calbiochem, 440202) ± CQ or Baf A and incubated for 24 h (cisplatin), 48 h (LY294002 and rapamycin, 67NR cells) or 72 h (rapamycin, 4T1 cells). Cells were treated with MTS reagent (Promega, G111A) according to manufacturer's instructions and read at 490 nm.
For long-term clonogenic assays, cells were plated in 12 well plates and 24 h later, they were pre-treated with CQ or Baf A for 1 h and then treated with the different drugs ± CQ or Baf A for 24 h. The treatment was washed and replaced with regular media. Cells were allowed to recover and grow for 4–5 d, fixed and stained with crystal violet (BD, 212525). Stain was solubilized with 30% acetic acid and absorbance was measured at 540 nm.
Protein isolation and western blots.
After treatment, cells were washed with PBS and lysed with RIPA buffer with protease inhibitors (Complete, Roche, 04693132001). Protein was quantitated using Bradford reagent (Bio Rad, 500-0006).15 µg (for LC3) or 20–30 µg (for Atg12 or Beclin 1) of protein were loaded in a SDS-PAGE and PVDF membranes (Millipore, IPVH07850) were probed with anti-LC3 (Novus-Biologicals, NB100-2220), Atg12 (Cell Signaling, D88H11), Beclin 1 (Cell Signaling, 3738) or actin antibodies (Sigma, A5441). Membranes were imaged in a Bio-Rad Universal Hood II system using Quantity-One 4.5.0. Software. Density analysis was performed using ImageJ 1.43m (NIH) and graphs show normalized arbitrary units (AU) from three independent experiments. Since LC3II signal was higher than LC3I, overexposed images of all the LC3 blots included in the paper are shown in Figure S6 for a better assessment of LC3I.
Fluorescence microscopy.
Cells were plated on coverslips and after treatment, fixed with 4% formaldehyde/ PBS, washed and mounted on slides with gelvatol mounting medium (16% Airvol 205 Air Products Ltd, 140 mM NaCl, 10 mM Na3PO4, 33% glycerol, pH = 7). Slides were observed in an Olympus spinning disk confocal microscope using a 60° magnification and Slidebook 5.0 software (Intelligent Imaging Innovations, Inc.). For time-lapse movies, 67NR GFP-cherry-LC3 expressing cells were plated in glass bottom dishes with compartments (Greiner Bio-one, 627870), treated and visualized during 3 h in a 31 Marianas Spinning Disk Zeiss Axio Observer Z1 microscope with Okolab cage incubator with temperature, CO2, air and humidity control. Quantification of yellow dots was done using particle analysis plug-in of Image J 1.43m (NIH). A cell was considered GFP-cherry-LC3 positive when it had more than 15 or 20 yellow dots/cell. Graphs show the quantification of three independent experiments and a total of 100 cells were counted per treatment. CQ concentrations were increased in time-lapse experiments (30 µM) to counter the strong effects of EBSS, which has been shown to significantly decrease lysosomal pH in contrast to other autophagy-inducing stimuli.47
GFP-LC3 redistribution evaluation.
4T1 and 67NR cells that had been previously transduced with the pTRIPZ lentiviral vector containing an Atg12 or a nonsilencing shRNA were transfected with Bgl-II linearized GFP-LC3 pcDNA3.1 hygro (+) (Invitrogen, V790-20) using TransIT-LT1 (Mirus) transfection reagent. After hygromycin selection (Cellgro, 30-240-CR, 500 µg/ml), cells were plated on coverslips, treated and fixed for imaging. Quantification of green dots was done using particle analysis plug-in of Image J 1.43m (NIH). A cell was considered GFP-LC3 positive when it had 20 or more green dots per cell. Graphs show the quantification of two independent experiments performed in duplicate and a total of 100 cells were counted per treatment.
siRNA experiments.
4T1 cells were transfected with Dharmacon BECN-1 ON-TARGETplus SMARTpool siRNAs (L-055895-00) or with ON-TARGETplus non-targeting siRNA #1 (D-001810-02-20) using Dharmafect-2 transfection reagent (Thermo Fisher Scientific, T-2002-03). Twenty-four hours after transfection, cells were trypsinized and plated for experiments. Protein knockdown was evaluated 72 h after transfection.
Statistical analysis.
Data was analyzed using one-way ANOVA followed by a Tukey test for survival experiments, or Student's t-test for mean comparison in density analysis or GFP-cherry-LC3/GFP-LC3 dot quantification, using OriginPro 8.0 Software, Origin Lab Corporation.
Acknowledgments
Supported by NIH grant CA150925. Imaging experiments were performed in the University of Colorado Anschutz Medical Campus Advanced Light Microscopy Core supported in part by NIH/NCRR Colorado CTSI Grant Number UL1 RR025780. Contents are the authors' sole responsibility and do not necessarily represent official NIH views.
Abbreviations
- CQ
chloroquine
- HCQ
hydroxychloroquine
- PtdIns3K
phosphatidyl inositol 3-kinase
- mTOR
mammalian target of rapamycin
- Atg
autophagy related
- EBSS
Earle's Balanced Salt Solution
- Baf A
bafilomycin A
- doxy
doxycyline
- zVAD
carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 2.Lesiak A, Narbutt J, Sysa-Jedrzejowska A, Lukamowicz J, McCauliffe DP, Wozniacka A. Effect of chloroquine phosphate treatment on serum MMP-9 and TIMP-1 levels in patients with systemic lupus erythematosus. Lupus. 2010;19:683–688. doi: 10.1177/0961203309356455. [DOI] [PubMed] [Google Scholar]
- 3.Savarino A, Di Trani L, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6:67–69. doi: 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, et al. Inhibition of macro-autophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, et al. Principles and Current Strategies for Targeting Autophagy for Cancer Treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu C, Solomon VR, Ulibarri G, Lee H. The efficacy and selectivity of tumor cell killing by Akt inhibitors are substantially increased by chloroquine. Bioorg Med Chem. 2008;16:7888–7893. doi: 10.1016/j.bmc.2008.07.076. [DOI] [PubMed] [Google Scholar]
- 8.Jiang PD, Zhao YL, Deng XQ, Mao YQ, Shi W, Tang QQ, et al. Antitumor and antimetastatic activities of chloroquine diphosphate in a murine model of breast cancer. Biomed Pharmacother. 2010;64:609–614. doi: 10.1016/j.biopha.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101–116. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan QW, Cheng C, Hackett C, Feldman M, Houseman BT, Nicolaides T, et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal. 2010;3:ra81. doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Institutes of Health (U.S.), National Library of Medicine (U.S.), United States, author. Food and Drug Administration. ClinicalTrials.gov.
- 12.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 14.Baselga J. Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer. Oncologist. 2011;16(Suppl 1):12–19. doi: 10.1634/theoncologist.2011-S1-12. [DOI] [PubMed] [Google Scholar]
- 15.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maycotte P, Thorburn A. Autophagy and cancer therapy. Cancer Biol Ther. 2011;11:127–137. doi: 10.4161/cbt.11.2.14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen N, Karantza V. Autophagy as a therapeutic target in cancer. Cancer Biol Ther. 2011;11:157–168. doi: 10.4161/cbt.11.2.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 19.Martín M. Platinum compounds in the treatment of advanced breast cancer. Clin Breast Cancer. 2001;2:190–208. doi: 10.3816/CBC.2001.n.022. discussion 9. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Cheng Y, Ren X, Zhang L, Yap KL, Wu H, et al. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene. 2011 doi: 10.1038/onc.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H, Su J, Xu Y, Kang J, Li H, Zhang L, et al. p62/SQSTM1 involved in cisplatin resistance in human ovarian cancer cells by clearing ubiquitinated proteins. Eur J Cancer. 2011;47:1585–1594. doi: 10.1016/j.ejca.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Claerhout S, Verschooten L, Van Kelst S, De Vos R, Proby C, Agostinis P, et al. Concomitant inhibition of AKT and autophagy is required for efficient cisplatin-induced apoptosis of metastatic skin carcinoma. Int J Cancer. 2010;127:2790–2803. doi: 10.1002/ijc.25300. [DOI] [PubMed] [Google Scholar]
- 23.O'Donovan TR, O'Sullivan GC, McKenna SL. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy. 2011;7:509–524. doi: 10.4161/auto.7.5.15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 25.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 27.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 28.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkhitko A, Myachina F, Morrison TA, Hindi KM, Auricchio N, Karbowniczek M, et al. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc Natl Acad Sci USA. 2011;108:12455–12460. doi: 10.1073/pnas.1104361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, et al. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 31.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M, et al. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–1607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- 33.Samaddar JS, Gaddy VT, Duplantier J, Thandavan SP, Shah M, Smith MJ, et al. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol Cancer Ther. 2008;7:2977–2987. doi: 10.1158/1535-7163.MCT-08-0447. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. PLoS ONE. 2009;4:e6251. doi: 10.1371/journal.pone.0006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 36.Xi H, Kurtoglu M, Liu H, Wangpaichitr M, You M, Liu X, et al. 2-Deoxy-D-glucose activates autophagy via endoplasmic reticulum stress rather than ATP depletion. Cancer Chemother Pharmacol. 2011;67:899–910. doi: 10.1007/s00280-010-1391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 38.Paglin S, Lee NY, Nakar C, Fitzgerald M, Plotkin J, Deuel B, et al. Rapamycin-sensitive pathway regulates mitochondrial membrane potential, autophagy, and survival in irradiated MCF-7 cells. Cancer Res. 2005;65:11061–11070. doi: 10.1158/0008-5472.CAN-05-1083. [DOI] [PubMed] [Google Scholar]
- 39.Gao P, Bauvy C, Souquere S, Tonelli G, Liu L, Zhu Y, et al. The Bcl-2 homology domain 3 mimetic gossypol induces both Beclin 1-dependent and Beclin 1-independent cytoprotective autophagy in cancer cells. J Biol Chem. 2010;285:25570–25581. doi: 10.1074/jbc.M110.118125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akar U, Chaves-Reyez A, Barria M, Tari A, Sanguino A, Kondo Y, et al. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy. 2008;4:669–679. doi: 10.4161/auto.6083. [DOI] [PubMed] [Google Scholar]
- 41.Katayama M, Kawaguchi T, Berger MS, Pieper RO. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14:548–558. doi: 10.1038/sj.cdd.4402030. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Hou N, Faried A, Tsutsumi S, Kuwano H. Inhibition of autophagy augments 5-fluorouracil chemotherapy in human colon cancer in vitro and in vivo model. Eur J Cancer. 2010;46:1900–1909. doi: 10.1016/j.ejca.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 43.Crabbe T, Welham MJ, Ward SG. The PtdIns3K inhibitor arsenal: choose your weapon! Trends Biochem Sci. 2007;32:450–456. doi: 10.1016/j.tibs.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Brunn GJ, Williams J, Sabers C, Wiederrecht G, Lawrence JC, Jr, Abraham RT. Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J. 1996;15:5256–5267. [PMC free article] [PubMed] [Google Scholar]
- 45.Wenzel NI, Chavain N, Wang Y, Friebolin W, Maes L, Pradines B, et al. Antimalarial versus cytotoxic properties of dual drugs derived from 4-aminoquinolines and Mannich bases: interaction with DNA. J Med Chem. 2010;53:3214–3226. doi: 10.1021/jm9018383. [DOI] [PubMed] [Google Scholar]
- 46.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 47.Ni HM, Bockus A, Wozniak AL, Jones K, Weinman S, Yin XM, et al. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011;7:188–204. doi: 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.