Abstract
Autophagy regulates cell survival and cell death upon various cellular stresses, yet the molecular signaling events involved are not well defined. Here, we established the function of a proteolytic Cyclin E fragment (p18-CycE) in DNA damage-induced autophagy, apoptosis, and senescence. p18-CycE was identified in hematopoietic cells undergoing DNA damage-induced apoptosis. In epithelial cells exposed to DNA damage, chronic but not transient expression of p18-CycE leads to higher turnover of LC3 I/II and increased emergence of autophagosomes and autolysosomes. Levels of p18-CycE, which was generated by proteolytic cleavage of endogenous Cyclin E, were greatly increased by chloroquine and correlated with LC 3II conversion. Preventing p18-CycE genesis blocked conversion of LC3 I to LC3 II. Upon DNA damage, cytoplasmic ataxia-telangiectasia-mutated (ATM) was phosphorylated in p18-CycE-expressing cells resulting in sustained activation of the adenosine-mono-phosphate-dependent kinase (AMPK). These lead to sustained activation of mammalian autophagy-initiating kinase ULK1, which was abrogated upon inhibiting ATM and AMPK phosphorylation. Moreover, p18-CycE was degraded via autophagy followed by induction of senescence. Both autophagy and senescence were prevented by inhibiting autophagy, which leads to increased apoptosis in p18-CycE-expressing cells by stabilizing p18-CycE expression. Senescence was further associated with cytoplasmic co-localization and degradation of p18-CycE and Ku70. In brief, chronic p18-CycE expression-induced autophagy leads to clearance of p18-CycE following DNA damage and induction of senescence. Autophagy inhibition stabilized the cytoplasmic p18-CycE-Ku70 complex leading to apoptosis. Thus, our findings define how chronic apoptotic stress and DNA damage initiate autophagy and regulate cell survival through senescence and/or apoptosis.
Keywords: autophagy, DNA damage, senescence, p18-Cyclin E, AMPK, ULK1
Introduction
The roles of cell proliferation and apoptosis signaling pathways in the demise of tumor cells have been studied, yet remain poorly understood. Targeting these pathways therapeutically has met with limited success. Ionizing radiation (IR), a prototypical DNA-damaging agent is known to induce cell death; however, often tumors become resistant to treatment.1 Apoptosis is the most extensively studied cell death pathway; however, autophagy and necrosis have been recently explored as alternative mechanisms.2,3 Autophagy is a lysosomal-dependent self-digestion process of cytoplasmic content, such as damaged organelles or proteins, in response to nutrient starvation or other forms of cellular stress.2 Autophagy can play a dual role in mediating either cell survival or death in response to various stress stimuli.4,5
Autophagy signaling is regulated by autophagy-related genes (Atgs), which are evolutionarily conserved.2,6 The most widely studied autophagy protein is the microtubule associated protein 1 light chain 3 β (LC3b; referred as LC3) also known as ATG8. Upon induction of autophagy pro-LC3 gets cleaved at the C-terminus to expose Gly 120 (LC3 I) by a Cysteine protease ATG4b and is further lipidated to its phosphatidylethanolamine (PE)-conjugated form (LC3 II), which can then be localized to autophagosomal membranes.7–9 Other ATGs involved in LC3 modifications are ATG3 and ATG7 that act as E1 and E2 ubiquitin ligases for LC3b.8 ULK1, the mammalian homolog of yeast ATG1, complexes with the ATG13/FIP200 kinase to mediate mammalian target of rapamycin (mTOR) inhibition and autophagy initiation.10,11 The ubiquitin-binding receptor, p62 participates in autophagic protein degradation through interaction with ubiquitinated protein aggregates that it targets to autophagosomes.12 Autophagy can enhance cellular survival during nutrient starvation and other forms of cellular stress, such as DNA damage.13
Autophagy induced by nutrient starvation has been most extensively studied, however recently other stresses, such as DNA damage, ROS, hypoxia, and the unfolded protein response have been also implicated.13 Irradiation has been used in the therapy of many tumor types to take advantage of DNA damage-induced apoptosis in cancer cells. Yet, as it is often not effective, many chemotherapeutics have been utilized to radiosensitize tumors, for example by targeting DNA repair.14 Recently, many studies have established that irradiation induces autophagy and this in turn reduces the cell death effect of radiation through activation of cell survival pathways. Thus, inhibiting autophagy is now considered as a means to overcome the resistance induced by irradiation and to sensitize many tumor types, such as those of breast and glioma.13,15,16
p18-CycE was first identified in hematopoietic cells whereupon irradiation Cyclin E gets proteolytically cleaved at the c-terminus in a caspase-dependent manner.17 Once p18-CycE is generated, it interacts with Ku70 and dissociates the Bax-Ku70 complex, and thus provides the first step of Bax activation leading to apoptosis.18 Here, we have studied the role of chronic expression of an apoptosis inducer, p18-CycE in cell survival through regulation of DNA damage-induced autophagy and senescence. We showed that p18-CycE stable expression results in increased autophagy rather than apoptosis, and inhibiting autophagy sensitize cells to DNA damage-induced cell death. Endogenously generated p18-CycE, by proteolytic cleavage of Cyclin E, also correlates with increased LC3 II levels, and preventing its genesis blocks conversion of LC3 I to LC3 II. p18-CyE is regulated by autophagy-mediated degradation upon DNA damage, along with cytoplasmic Ku70 leading to induction of senescence and thus reduced long-term cell survival.
Results
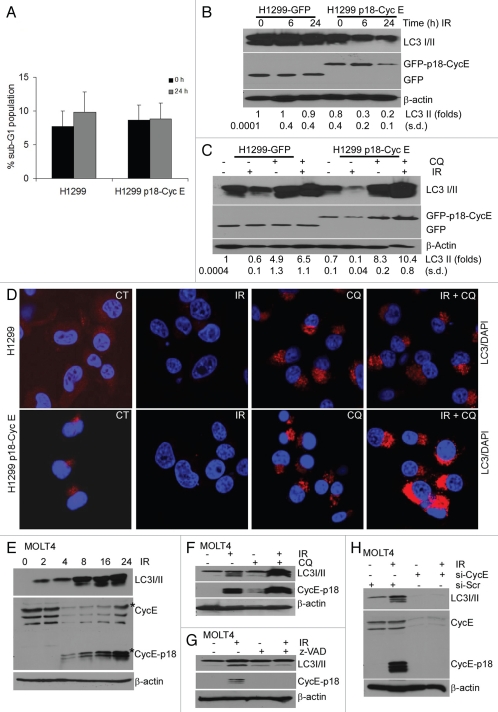
DNA damage-induced autophagy is enhanced in p18-CycE-expressing cells.
Since p18-CycE is not produced in epithelial cells, we generated NCI-H1299 cells stably expressing p18-CycE to study its function. There was no effect of p18-CycE on DNA damage-induced apoptosis in cells stably expressing p18-CycE (Fig. 1A).15 As autophagy has an important role in mediating cell survival upon apoptotic stress, we examined the possible role of p18-CycE in autophagy. Levels of LC3 I (delipidated form) and II (lipidated form) were reduced following irradiation. This change was much more extensive in p18-CycE-expressing cells (> 4-fold, as compared with no change w/o p18-CycE), suggesting either decreased LC3 expression or increased LC3 degradation (Fig. 1B). Quantitative real-time PCR indicated no significant changes in LC3 mRNA following irradiation, suggesting that LC3 transcription was not affected and that the reduced LC3 is likely to be due to its degradation (Fig. S1A). To determine the effect on steady-state levels of LC3, as opposed to accumulated LC3 due to a higher induction of autophagosome formation, experiments were performed in the presence of chloroquine, which inhibits autophagy-mediated LC3 degradation via inhibiting acidification of autophagolysosomes. Chloroquine addition prior to irradiation led to LC3 II accumulation, while another autophagosomeassociated ATG5-12 protein complex, known to be dissociated from the outer membrane after autophagosome formation19 remained unaffected (Fig. S1B). There was no significant difference in cell death following irradiation in the absence or presence of chloroquine, indicating that the doses used were not cytotoxic (Fig. S1C and S1D). Instead, it resulted in increased accumulation of LC3 II (up to 14-fold in p18-CycE-expressing as compared with only 6.5-fold in control cells), suggesting an increased induction of autophagy in p18-CycE-expressing cells (Fig. 1C and D).
Figure 1.
DNA damage-induced autophagy is enhanced in p18-CycE-expressing cells. (A) Parental and p18-CycE expressing NCI-H1299 cells were harvested, fixed, and stained with propidium iodide at 24 h following treatment with ionizing radiation (IR). Cell death is shown as percentage of cells with sub-G1 DNA content. (B and C) Cells were lysed at the indicated time following irradiation and/or chloroquine (100 µM) treatment and immunoblotted for LC3 I/II, GFP, GFP-p18-CycE and β-actin as a loading control. Fold change in LC3 II is indicated (D) Visualization of endogenous LC3 I/II at 24 h following IR in the absence or presence of chloroquine. Nuclei were stained with 4;6-diamidino-2-phenylindole (DAPI). (E) Western blot analysis for endogenous LC3 I/II and p18-CycE in MOLT4 cells at the indicated time following IR. (F and G) Western blot analysis for endogenous LC 3I/II and p18-CycE in MOLT4 cells at 16 h following IR in the absence or presence of chloroquine or z-VAD-fmk (10 µM). (H) Western blot analysis for endogenous LC3 I/II and p18-CycE in MOLT4 cells irradiated for 16 h after 24 h post-transfection with scrambled siRNA (si-Scr) or si-RNA against Cyclin E. β-actin served as loading control.
To rule out the possibility that the effect of p18-CycE expression on autophagy was because of unrelated events due to its ectopic expression, we examined the role of endogenous p18-CycE on autophagy signaling. Hematopoietic cell lines, such as MOLT4 and IM-9 were shown to generate p18-CycE following IR.17 p18-CycE was generated in MOLT4, IM-9 and Reh cells following IR in a time-dependent manner (Fig. 1E and Fig. S1E and S1F). LC3 II expression was correlated with p18-CycE generation in these cell lines (Fig. 1E and Fig. S1E and S1F). Chloroquine addition prior to IR resulted in accumulation of both LC3 II and p18-CycE in MOLT4 cells, again suggesting that p18-CycE expression correlates with autophagy induction (Fig. 1F). It has been shown that p18-CycE generation can be inhibited by z-VAD treatment or by depleting Cyclin E itself.17 z-VAD-fmk treatment inhibited p18-CycE as well as LC3 II formation in MOLT4 cells following IR (Fig. 1G). Using siRNA against Cyclin E also lead to diminished Cyclin E expression as well as p18-CycE generation following IR, compared with scrambled siRNA. LC3 II was also reduced upon depleting CyclinE and p18-CycE (Fig. 1H). These results suggest that p18-CycE expression is associated with autophagy and inhibiting p18-CycE generation also inhibit autophagy.
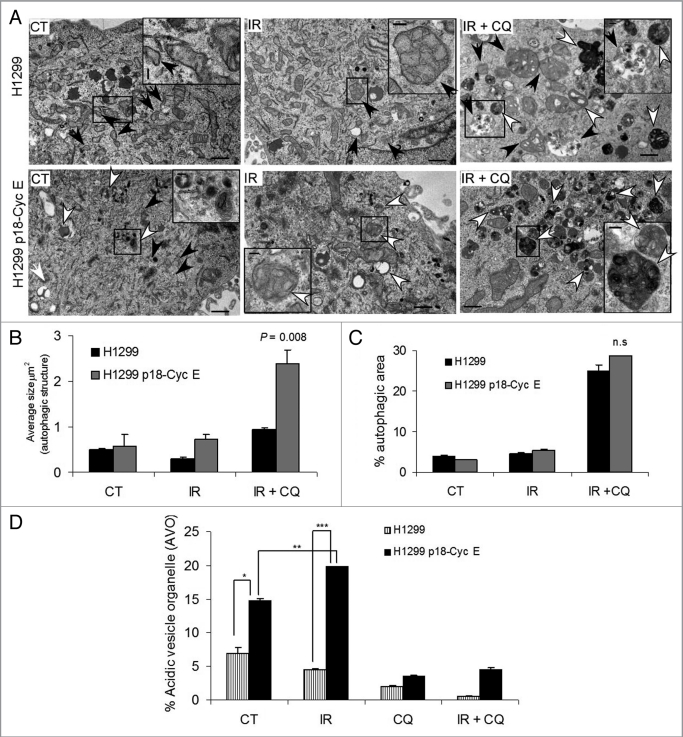
Transmission electron microscopy revealed a greater accumulation of autophagic structures in p18-CycE expressing cells (Fig. 2A). A more detailed quantitation showed an increased size of autophagic, double-membrane vesicles (autophagosomes) but not the total autophagic area in p18-CycE-expressing cells (Fig. 2B and C). Development of acidic vesicle organelles (AVOs), which is characteristic of autophagy, was determined by staining live cells with the lysosomo-tropic agent, acridine orange. When protonated, it accumulates in acidic compartments and fluoresce bright red. Cells constitutively-expressing p18-CycE showed increased number of AVOs, which were increased following irradiation in p18-CycE-expressing but not in parental cells. Upon addition of chloroquine, formation of AVOs was decreased in both parental and p18-CycE expressing cells (Fig. 2D). Altogether, these results indicate that DNA damage induces more autophagy in p18-CycE expressing cells.
Figure 2.
Autophagic area and flux is enhanced in p18-CycE-expressing cells following DNA-damage. (A) Transmission electron microscopic analysis of autophagic structures at 24 h post irradiation in the absence or presence of chloroquine. Early and late autophagic structures are indicated by black and white arrows, respectively, with black box regions enlarged in the inset. The scale bar in the original image represents 1 µm and the scale bar in the inset represents 200 nm. (B,C) Quantitative analysis for the average size of autophagic structures (*p = 0.008) and fraction of autophagic area normalized to cytoplasmic area (n.s). Data represent means ± s.e.m., with more than three different fields analyzed per sample. (D) Detection of acidic vesicular organelles (AVOs) by measuring red and green fluorescence of acridine orange-stained cells using FACS analysis in parental and p18-CycE expressing cells at 24 h following IR and/or chloroquine treatment (*p = 0.007, **p = 0.002, ***p < 0.001). Data represent means ± s.e.m., obtained from three independent triplicates.
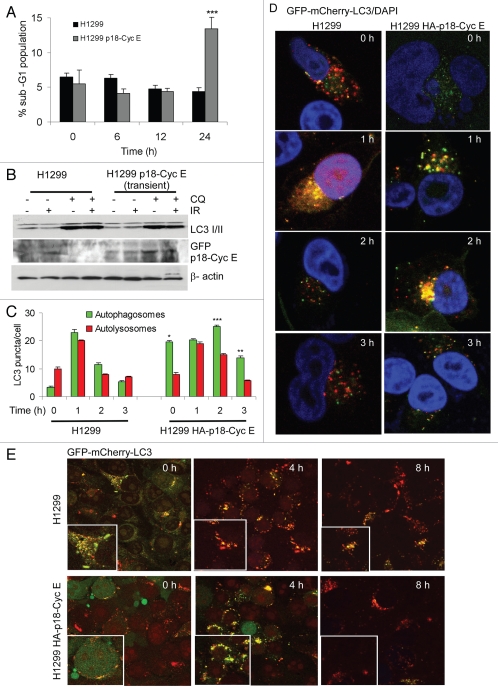
Stable, but not transient p18-CycE expression leads to increased autophagy.
Remarkably, transient but not stable expression of p18-CycE increased apoptosis, suggesting that autophagy might have evolved as an adaptive mechanism for cell survival (Figs. 1A and 3A). In cells transiently transfected with HA-p18CycE, the p18-CycE expression was slightly increased upon chloroquine treatment, when used either alone or in combination with irradiation. There was no difference, however, in LC3 I/II and p62 accumulation (Fig. 3B). These observations indicate that transient expression of p18-CycE is not sufficient to enhance autophagic signaling, with the autophagic response being rather a consequence of the cellular adaptation to chronic apoptosis stress in response to constitutive p18-CycE expression.
Figure 3.
Stable, but not transient p18-CycE expression leads to increased autophagy. (A) Cell death is shown as percentage of cells with sub-G1 DNA content in cells transiently transfected with p18-CycE following IR (*p < 0.001 compared for p18-CycE-expressing cells vs. parental cells at 24 h) Student's t-test. (B) Cells were lysed at 24 h post IR in the absence or presence of chloroquine and immunoblotted for LC3 I/II, GFP-p18-CycE, with β-actin as loading control. (C) Quantitation of autophagosomal (yellow) and autolysosomal (red) LC3 puncta following irradiation. **p = 0.017, *p = 0.0006, ***p = 0.03 Student's t-test. For all panels, values are mean ± s.e.m. with > 30 cells from different fields analyzed per sample. (D) Autophagic flux is shown by representative confocal microscopic images for cells stably-expressing GFP-mCherry-LC3 in cells without or those expressing HA-p18-CycE. (E) Representative confocal images are shown for cells stably expressing GFP-mCherry-LC3 without or with transient expression of HA-p18-CycE at the indicated times following IR.
Next, the autophagic flux was examined to distinguish between regulation of synthesis vs. degradation of LC3 I/II. GFP-mCherry-LC3 enabled simultaneous quantification of autophagosome induction and autolysosome maturation, since the GFP signal is sensitive to the acidic and proteolytic conditions of the lysosome, while mCherry is stable.20 The analysis revealed that the number of autophagosomes was only transiently increased after 1 h in parental cells, while p18-CycE-expressing cells had an increased number of autophagosomes, which was maintained at least until 3 h following irradiation, indicating pronounced induction of autophagy (Fig. 3C and D). Similarly the number of autolysosomes was increased at least until 2 h in p18-CycE-expressing cells compared with transient increase at 1 h in parental cells following irradiation suggesting that p18-Cyc E-expression has an effect on the rate of autophagy induction as well as autophagic flux (Fig. 3C). This finding also indicates that the total number of LC3 II puncta (autophagosomes + autolysosomes) were increased in p18-CycE-expressing cells compared with the parental cells. Both cell lines showed comparable GFP-mCherry-LC3 expression (Fig. S2A). Chloroquine addition prior to irradiation resulted in increased accumulation of autophagosomes in p18-CycE-expressing cells (Fig. S2B). Transient p18-CycE expression did not affect the LC3 flux following DNA damage (Fig. 3E). LC3 lipidation and delipidation are known to be dependent on ATG3 and ATG4b, respectively.19 ATG3 levels were high in p18-CycE-expressing cells, while ATG4b levels were lower, with no additional changes following irradiation, suggesting that p18-CycE expression favors steady-state LC3 lipidation. ATG4a expression was, however not affected (Fig. S2C).
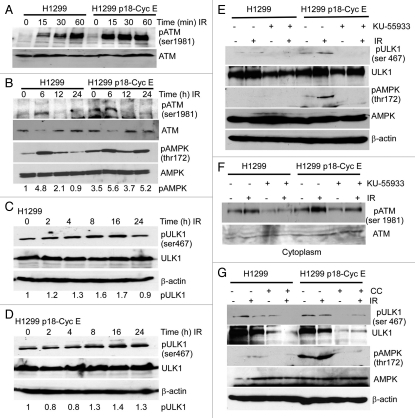
DNA damage induces sustained activation of AMPK and ULK1 in p18-CycE-expressing cells.
It is unclear how nuclear DNA damage initiates cytoplasmic signaling to regulate autophagy. Irradiation activates ataxia-telangiectasia mutated (ATM)-mediated signaling to induce a DNA damage response in the nucleus; however, ATM can also translocate to the cytoplasm.21–23 ATM has also been shown to activate AMP-activated kinase (AMPK), which triggers autophagy by inhibiting mTOR.24 There was a robust phosphorylation of cytoplasmic ATM on Ser1981 as early as 15 min, that lasted up to 6 h following irradiation in p18-CycE-expressing cells, while enhanced ATM activation in the nucleus was observed in parental but not p18-CycE-expressing cells (Fig. 4A> and B, and Fig. S3). Cytoplasmic AMPK phosphorylation on Thr172 was sustained (up to ∼5-fold) for at least 24 h in p18-CycE-expressing cells, however was only transiently detected at 6 h following irradiation in parental cells (Fig. 4B). mTOR is known to inhibit ULK1 activity through phosphorylation of its kinase domain, while AMPK activates ULK1 by phosphorylating its serine-proline rich region required to bind LC3 and activate autophagy.25–27 In p18-CycE-expressing cells ULK1 phosphorylation on Ser467 was sustained for at least 24 h following irradiation, while in parental cells ULK1 activation was increased only up to 16 h (Fig. 4C and D). Importantly, ULK1, AMPK, and ATM phosphorylation were all prevented by the specific ATM inhibitor, KU-55933 (Fig. 4E and F). Moreover, ULK1 Ser467 phosphorylation following irradiation was dependent on AMPK, since it was inhibited by Compound C, an AMPK inhibitor (Fig. 4G). Overall, these findings suggest that a DNA damage-induced phosphorylation cascade in the cytoplasm involved sequential ATM, AMPK, and ULK1 activation leading to autophagy initiation and which was sustained in the presence of p18-CycE.
Figure 4.
DNA damage induces sustained activation of AMPK and ULK1 in p18-CycE-expressing cells. (A and B) Cytoplasmic fractions of cells, at the indicated time following IR, were immunoblotted for ATM-ser1981, ATM, pAMPK-thr172, AMPK. β-actin was used as a loading control. (C and D) Total protein lysates of cells at the indicated time following IR were immunoblotted for pULK1-ser467, ULK1, and β-actin. (E) Total protein lysates of cells at 24 h following IR with or without 1 h pretreatment with the ATM inhibitor KU-55933 (10 µM) were immunoblotted for pULK1-ser467, ULK1, pAMPK-thr172, AMPK and β-actin. (F) Cytoplasmic fractions of cells at 6 h following IR with or without 1 h pretreatment with the ATM inhibitor KU-55933 (10 µM) were immunoblotted for ATM-ser1981 and ATM. (G) Total protein lysates of cells at 24 h following IR with or without 1 h pretreatment with the AMPK inhibitor Compound C (CC; 25 µM) were immunoblotted for pULK1-ser467, ULK1, pAMPK-thr172, AMPK and β-actin.
Autophagy inhibition increases apoptosis in p18-CycE-expressing cells.
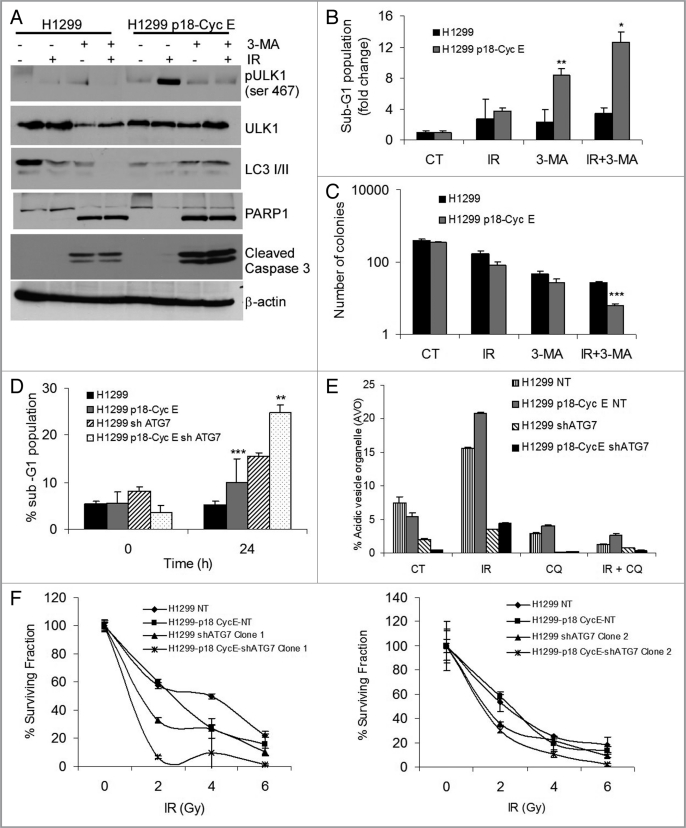
Autophagy inhibitors have been considered for overcoming radiation resistance in several tumor types.28 3-methyladenine (3-MA), a known pharmacological inhibitor of autophagy initiation, when used in combination with irradiation leads to reduced ULK1 phosphorylation on Ser467, reduced accumulation of LC3 II, and increased cleavage of caspase-3 and PARP1 (Fig. 5A). 3-MA treatment prior to irradiation increased the proportion of cells with sub-G1 DNA content in p18-CycE-expressing cells (Fig. 5B). In a long-term clonogenic survival assay, 3-MA treatment prior to irradiation reduced clonogenic survival (Fig. 5C). Next, pharmacological inhibition was complemented with a genetic approach using a lentiviral hairpin shRNA against ATG7. Cell lines were generated that stably expressed shATG7 using two different constructs targeting two different regions of ATG7 mRNA. Following shATG7 expression, the levels of ATG7 were reduced more than 2-fold and did not increase following irradiation (Fig. S4A). There was increased PARP1 cleavage and increased cell death in shATG7-expressing cells that was more extensive when p18-CycE was co-expressed (up to 8-fold as opposed to only 2-fold in parental cells), suggesting that autophagy inhibition sensitizes cells to irradiation, with p18-CycE expression enhancing the effect (Fig. S4A and S4B, and Fig. 5D). Endogenous LC3 II levels were decreased while those of LC3 I were increased in ATG7-knockdown cells, both constitutively as well as following irradiation, indicating that autophagy was inhibited in cells stably-expressing shATG7 (Fig. S4C and S4D). Moreover, the number of AVOs was reduced upon ATG7 knockdown in both parental and p18-CycE expressing cells (Fig. 5E and Fig. S4E). Long-term clonogenic survival was reduced in shATG7-expressing cells (Clone 1 and Clone 2) following IR in p18-CycE-expressing compared with the parental cells expressing nontarget shRNA (NT) control (Fig. 5F, left and right panels). These findings suggest that in spite of increased apoptosis upon autophagy inhibition, the long-term cell survival following DNA damage could also be affected by other factors that impact on cellular growth, as addressed below.
Figure 5.
Autophagy inhibition increases apoptosis in p18-CycE-expressing cells. (A) Cells harvested and lysed at 24 h post IR in the absence or presence of 3-MA (10 mM) were immunoblotted for pULK1-ser467, ULK1, LC3 I/II, PARP1, cleaved caspase-3, and β-actin. (B) Cell death at 24 h following IR in the absence or presence of 3-MA (10 mM) treatment is shown as percentage of cells with sub-G1 DNA content. **p = 0.02, *p = 0.03 Student's t-test. (C) Clonogenic assay following IR (5 Gy) in the absence or presence of 3-MA. *p < 0.001 Student's t-test. (D) Cell death is shown as percentage of cells with sub-G1 DNA content that stably express p18-CycE in cells without or with shATG7 expression (Clone 1) at 24 h following IR. *p < 0.001, **p = 0.001 Student's t-test. (E) Detection of acidic vesicular organelles (AVOs) by measuring red and green fluorescence of acridine orange-stained cells using FACS analysis in parental and p18-CycE expressing cells with or without shATG7 expression following IR and/or chloroquine (100 µM) treatment for 24 h. Data represent means ± s.e.m., obtained from three independent triplicates. (F) Clonogenic assay for cells stably-expressing p18-CycE in the absence or presence of shATG7 (Clone 1 in left panel, Clone 2 in right panel) following IR. p < 0.05 Student's t-test. For all panels, values are mean ± s.e.m. of three independent experiments performed in triplicates.
Expression of p18-CycE is regulated by autophagy.
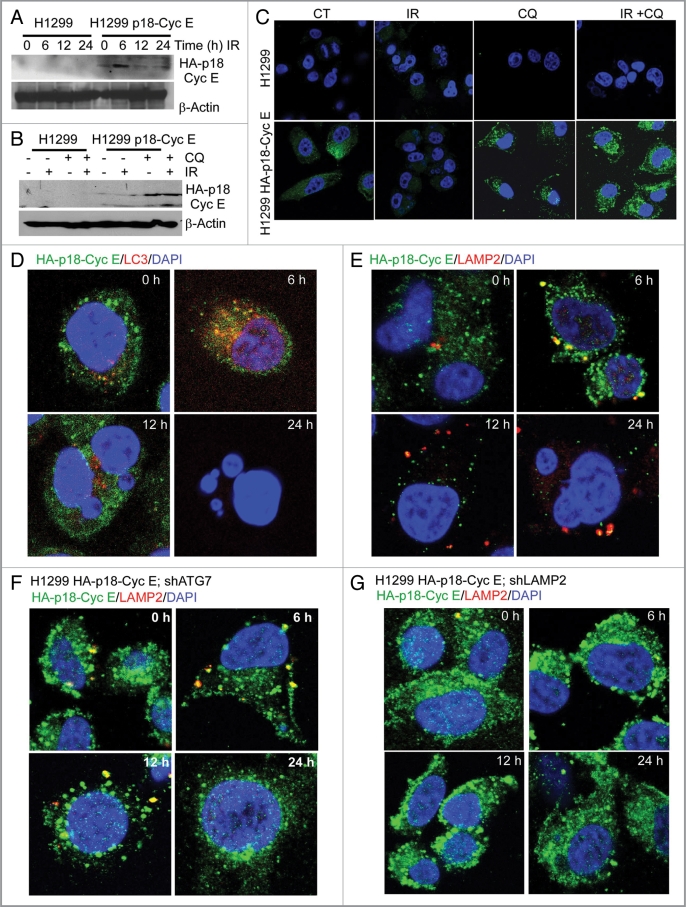
Since the long-term effect of irradiation was a decrease in clonogenic survival, it raised a question about the level of p18-CycE following irradiation. p18-CycE expression increased at 6 h, then decreased at 12–24 h following irradiation (Fig. 6A). Chloroquine treatment prior to irradiation stabilized p18-CycE levels, suggesting its autophagic degradation (Fig. 6B and D). Co-immunostaining for HA-p18-CycE with the autophagosomal marker LC3 and the lysosome-associated membrane protein 2 (LAMP2) revealed the presence of p18-CycE in autophagosomes and lysosomes, respectively at the earlier time-points, but not at 24 h (Fig. 6D and E). Interestingly, p18-CycE did not colocalize with p62, suggesting its p62-independent degradation (Fig. S5A). Moreover, p18-CycE expression was stabilized in shATG7-and shLAMP2-expressing cells, compared with its ∼3-fold degradation in p18-CycE-expressing cells with functional ATG7 and LAMP2 expression, indicating that inhibition of autophagy induction or degradation, respectively, stabilized p18-CycE following irradiation (Fig. 6F and G, and Fig. S5B and S5C). LAMP2 expression was efficiently reduced by shLAMP2 and was not changed upon irradiation (Fig. S5D). Thus, p18-CycE levels were decreased following irradiation by autophagy-mediated degradation. This finding further clarifies why there was no effect of irradiation on apoptosis in cells that stably express p18-CycE.
Figure 6.
Expression of p18-CycE is regulated by autophagy. (A) Total protein lysates were immunoblotted for HA-p18-CycE and β-actin at the indicated time following IR. (B) Cells were lysed at the indicated time following IR and/or chloroquine (100 µM) treatment and immunoblotted for HA-p18-CycE using anti-HA antibody and β-actin as loading control. (C) Confocal co-immunostaining for HA-p18-CycE in parental and p18-CycE-expressing cells at 24 h following IR with or without chloroquine. (D and E) Confocal co-immunostaining for HA-p18-CycE/LC3I/II and HA-p18-CycE/LAMP2 in cells stably-expressing p18-CycE at the indicated time following IR. (F and G) Confocal co-immunostaining for HA-p18-CycE/LAMP2 in p18-CycE-expressing cells with ATG7 or LAMP2 knockdown at the indicated time following IR. Nuclei were stained with DAPI.
DNA damage induces autophagy-dependent senescence in p18-CycE-expressing cells.
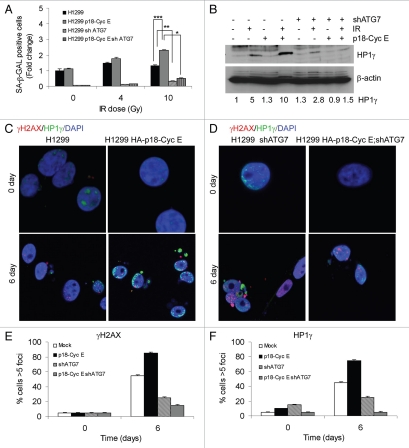
Induction of autophagy in p18-CycE-expressing cells reduced both apoptosis and long-term clonogenic survival. As these findings suggested that other factors may be involved, we examined senescence, a potent tumor suppressor mechanism, that has recently been associated with autophagy.29 Examining SA-β-galactosidase-positive cells showed that the number of senescent cells was increased more than 2-fold in p18-CycE-expressing cells (Fig. 7A). Representative images for SA-β-galactosidase staining are shown (Fig. S6A and S6B). Importantly, in cells with stable shATG7 expression, senescence was significantly inhibited, indicating its dependence on autophagy (Fig. 7A). Expression of another senescence marker, HP1γ was induced up to 10-fold in p18-CycE-expressing cells compared with 5-fold in parental cells. There was no increase in HP1γ in p18-CycE-expressing cells while a slight increase (∼2-fold) noticed in parental cells with ATG7 knockdown (Fig. 7B). Senescence-associated heterochromatin foci (SAHF), which were marked with HP1γ, were increased in number and size in p18-CycE-expressing cells following irradiation, but reduced instead in shATG7-expressing cells (Fig. 7C and D). These HP1γ-associated SAHF were distinct from the residual DNA damage foci marked by γH2AX. Quantitation of both γH2AX and HP1γ foci revealed increased number in p18-CycE-expressing cells, which were decreased to a larger extent upon ATG7 knockdown, compared with parental cells (Fig. 7E and F). Overall, these observations suggest that DNA damage-induced autophagy mediates senescence, which is enhanced in p18-CycE-expressing cells, resulting in decreased long-term clonogenic survival, whereas apoptosis being responsible for reduced cell survival in autophagy-compromised cells.
Figure 7.
DNA damage induces autophagy-dependent senescence in p18-CycE-expressing cells. (A) SA-β-Gal-positive, p18-CycE in the absence or presence of shATG7-expressing cells at 6 d post IR. ***p = 0.003, **p = 0.005, *p < 0.001 Student's t-test. Data represent means ± s.e.m. of three independent experiments done in triplicates. (B) Expression of HP1γ and β-actin in parental and p18-CycE expressing cells in the absence or presence of shATG7 at 6 d post IR. Fold change in HP1γ is indicated. (C and D) Visualization of endogenous HP1γ and γH2AX-associated foci by confocal immunostaining in parental and p18-CycE-expressing cells in the absence or presence of shATG7 expression following IR. (E and F) Percentage of cells having at least 5 foci per cell for γH2AX and HP1γ in parental and p18-CycE-expressing cells in the absence or presence of shATG7 expression following IR.
Coordinate cytoplasmic Ku70 and p18-CycE degradation leads to senescence.
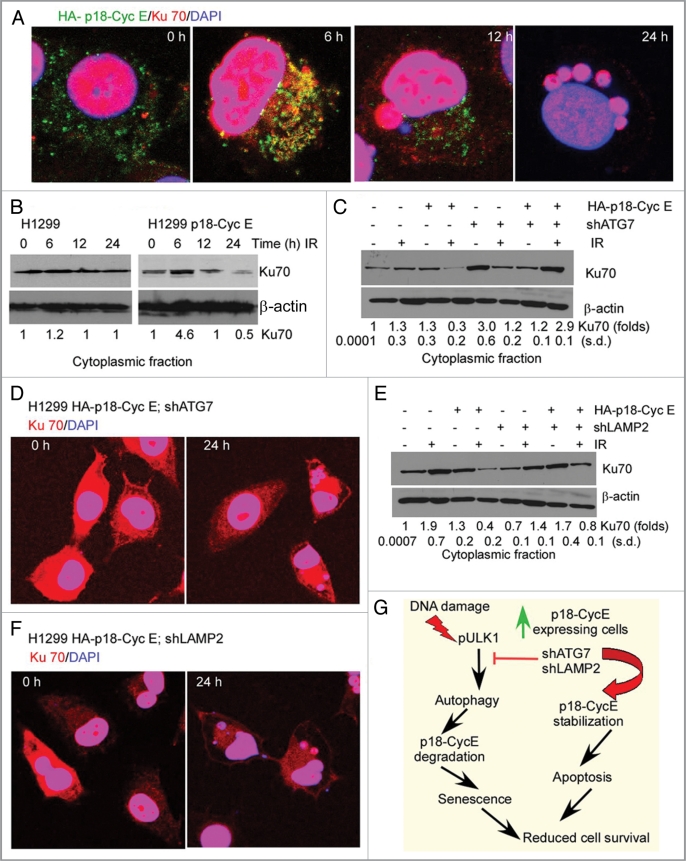
Since p18-CycE expression was inversely correlated with induction of senescence, we next examined whether there was a direct link between autophagy-mediated p18-CycE degradation and senescence. Our previous findings have established that p18-CycE interacts with cytosolic Ku70, causing the release of Bax and its activation leading to apoptosis.30 Similar to Ku70, the p18-CycE-Ku70 protein complex degradation was dependent on hdm2-dependent ubiquitination in cells expressing functional p53.30,31 Recently, depletion of cytosolic Ku70 has been correlated with development of cellular senescence.32,33 Since p18-CycE degradation leads to senescence, we next examined whether Ku70 was involved in this process. Co-immunostaining for p18-CycE and Ku70 revealed their colocalization at 6 h and degradation of only cytoplasmic Ku70 at 12 and 24 h (up to 2-fold) following irradiation in p18-CycE-expressing cells (Fig. 8A and B, and Fig. S7A). Degradation of cytoplasmic Ku70 was prevented in both ATG7-and LAMP2-depleted p18-CycE-expressing cells (Fig. 8C–F), while nuclear Ku70 levels remained unchanged (Fig. S7B and S7C). These observations indicate that following irradiation, p18-CycE and Ku70 co-localize in the cytoplasm where they are then degraded in a coordinated manner. Moreover, cytoplasmic Ku70 degradation is facilitated in the presence of p18-CycE and requires autophagy signaling, which in turn leads to induction of senescence and reduced cell proliferation.
Figure 8.
Coordinate cytoplasmic Ku70 and p18-CycE degradation leads to senescence. (A) Confocal co-immunostaining for HA-p18-CycE and Ku70 at 6–24 h following IR in HA-p18-CycE-expressing cells. (B) Cytoplasmic fractions of parental and p18-CycE expressing cells following IR at the indicated time were immunoblotted for Ku70 and β-actin. (C) Cytoplasmic fractions of p18-CycE-expressing cells with or without shATG7 expression at the indicated time following IR were immunoblotted for Ku70 and β-actin. (D) Confocal co-immunostaining for Ku70 in HA-p18-CycE-expressing cells in the absence or presence of shATG7 at 24 h following IR. Nuclei were stained with DAPI. (E) Cytoplasmic fractions of p18-CycE-expressing cells with or without shLAMP2 expression at the indicated time following IR were immunoblotted for Ku70 and β-actin. (F) Confocal co-immunostaining for Ku70 in HA-p18-CycE-expressing cells in the absence or presence of shLAMP2 at 24 h following IR. Nuclei were stained with DAPI. (G) Model for cell survival regulation by p18-CycE via autophagy, apoptosis, and senescence.
Discussion
Here we established that DNA damage-induced autophagy is enhanced by stable p18-CycE expression in contrast to apoptosis, which is induced by transient expression of p18-CycE. DNA damage lead to increased LC3 II levels in cells with stable, but not transient p18-CycE expression. This observation was confirmed by the autophagic structures detected by transmission electron microscopy. Moreover, autophagic flux, determined with the GFP-mCherry-LC3 fusion protein, was higher in cells with stable but not transient expression of p18-CycE. These findings clearly indicate that autophagy emerged as an adaptive mechanism mediating cell survival upon chronic apoptotic stress, in this case provided by stable expression of p18-CycE. Autophagic signaling has been reported to be initiated by activation of AMPK and ULK1 kinase complexes, however the link between DNA damage and autophagy is still not clear.10,11 Here we showed that DNA damage caused by ionizing radiation led to phosphorylation of ATM at Ser1981, primarily in the nucleus in parental cells. Interestingly, in p18-CycE-expressing ATM phosphorylation was predominant in the cytoplasm and not the nucleus. Increased cytoplasmic ATM activation directly impacted on increased AMPK and ULK1 activation, thus providing evidence that, indeed, nuclear DNA damage signaling can be transduced to the cytoplasm and can activate autophagy to mediate cell survival. Interestingly, ATM and AMPK inhibitor as well as 3-MA inhibited ULK1 phosphorylation, but at the same time also led to reduced ULK1 expression indicating that ULK1 Ser467 could also regulate the total protein of ULK1 which needs to be explored.
Since autophagy can mediate cell survival, inhibiting autophagy can sensitize cells to DNA damage-induced cell death. Inhibition of autophagy by pharmacological inhibitors or by genetically reducing ATG7 and LAMP2 expression using shRNA-mediated gene silencing lead to increased apoptosis and reduced clonogenic survival in p18-expressing cells. Moreover, autophagy inhibition stabilized p18-CycE levels, indicating that its expression can be regulated via autophagic degradation. p18-CycE co-localization with LC3 and LAMP2 was also observed following DNA damage directly implicating its involvement in autophagy.
Since inhibition of autophagy sensitized cells to DNA damage-induced cell death, increased autophagy resulted in cell survival but with a limited effect on long-term clonogenic survival, indicating that there are other factors involved in regulation of overall cell survival. Recently, senescence has been associated with autophagy and autophagy-mediated tumor suppression by inhibiting cellular proliferation.29,34 DNA damage-induced senescence was also increased in p18-CycE expressing cells and was inhibited upon autophagy inhibition, suggesting that autophagy can lead to induction of senescence and thus can have cell survival as well as growth inhibitory function. Moreover, p18-CycE colocalizes with Ku70 in the cytoplasm upon DNA damage followed by its degradation, which were prevented upon autophagy inhibition. These findings provide more insight into the mechanism of senescence induction since recent reports indicate that reduced cytosolic levels of Ku70 are correlated with the senescence phenotype.32,33 Therefore, acute expression of p18CycE induced apoptosis while its chronic expression facilitated senescence via autophagy; in both cases p18-CycE reduced cancer cell survival following DNA damage (Fig. 8E).
Recently, autophagy signaling has gained a broad interest based on its emerging critical role in mediating cell survival and cell death outcomes in response to antitumor therapies in various human cancer model systems. Cancer cells were recently shown to acquire oncogene-induced autophagy as an adaptation for cell survival under various stress conditions, such as hypoxia, nutrient and growth factor deprivation, and therefore inhibiting autophagy can have growth-inhibitory as well as apoptosis-inducing effects.34,35 It is thus important to define the role of autophagy in mediating cell survival in tumors and how modulating proteins regulated by autophagy, such as p18-CycE, can induce apoptosis and/or senescence to reduce tumor growth. We hereby defined how chronic apoptotic stress can modulate DNA damage-induced autophagy and other growth inhibitory processes, such as senescence that could help in developing better strategies for inducing cell death in cancer cells. Our findings may be relevant to cellular adaptation to other apoptosis cues encountered during tumor development or therapeutic response that constitute a new area of investigation for future studies.
Materials and Methods
Reagents and plasmids.
HA-p18-CycE and GFP P18-Cyclin E were generated as described previously.30 The GFP-mCherry-LC3 and pCL10 plasmids were a kind gift from Dr. Jayant Debnath (University of California San Francisco). The lentiviral packaging plasmids pVSVG and dr 8.7 were from Invitrogen. pLKO.1-puro control vector, shATG7 (SHCLNGNM_006395), and shLAMP2 (SHCLNG-NM_002294) Mission shRNAs were from Sigma Aldrich. Following reagents were purchased: chloroquine, 3-methyladenine (3-MA), AMPK inhibitor Compound-C (Sigma Aldrich, C6628, M9281, P5499); ATM inhibitor KU-55933 (Selleck Chemicals LLC, S1092); z-VAD-fmk (EMD Biosciences); Cyclin E and scrambled siRNA (Dharmacon).
Cell culture, lentiviral transduction for p18-CycE, transfections, and treatments.
MOLT4, IM-9 and Reh hematopoietic cells were maintained in RPMI medium while NCI-H1299 human lung carcinoma cells were maintained in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum (Atlanta Biologicals, S11050), L-glutamine (Invitrogen, 25030-081), and 100 unit/ml penicillin-streptomycin (Invitrogen, 15140-155) in a humidified incubator at 37°C, 5% CO2. NCI-H1299 cells stably-expressing HA-p18-CycE and only GFP vector control were generated by infecting cells with a lentivirus carrying HA-p18-CycE and enhanced-GFP separated by an IRES2 sequence under the control of an EF1α promoter as described previously.30 For all immunostaining experiments HA-p18-CycE expressing stable cells was generated by transfecting HA-p18-CycE without IRES2-GFP. Respective plasmids were transfected using the Fugene transfection reagent (Promega, E2311), following the manufacturer's protocol and infected or transfected cell population were selected with either Puromycin (0.5 µg/ml) or G418 (1 mM) as required for the specific constructs. For control cells indicated as parental cells, only vector-transfected or infected cells were used following antibiotic selection. Cells were irradiated with 10 Gy, unless otherwise mentioned, at 25°C, using a Mark I Irradiator (J.C. Shepherd & Associates) with a 137Cs source emitting at a fixed dose-rate of 2.0 Gy/min, as described previously.36
Lentiviral transduction of shRNA.
shRNA target sequences were: ATG7 3′UTR, 5′-CCGGGCCTGCTGAGGAGCTC TCCATCTCGAGATGGAGAGCTCCTCAGCAGGCTTTT-3′ (Clone1); ATG7 CDS, 5′-CCGGCCAGAGAGTTTACCTCTC ATTCTCGAGAATGAGAGGTAAACTCTCTGGTTTTT-3′ (Clone2) and LAMP2 CDS-5′-CCGGGCCATCAGAATTCCAT TGAATCTCGAGATTCAATGGAATTCTGATGGCTTTTT-3′ (Clone 1); LAMP2 CDS, 5′-CCGGGTACGCTATGAAACTACAAATCTCGAGATTTGTAGTTTCATAGCGTACTTTTT-3′ (Clone 2). 293T cells were transfected with these shRNAs together with the pVSVG and dr8.2 plasmids in a 3:1:1 ratio using the Fugene transfection reagent. Media containing viral particles was then collected after 24 h of transfection, passed through 0.8-micron filter, and added to NCI-H1299 parental and p18-CycE-expressing cells along with polybrene (10 µg/ml). After overnight incubation at 37°C, the media was replenished and cells were selected for puromycin resistance (0.5 µg/ml) for 6 d, after which knockdown was further validated.
Cell death analysis.
Cell viability was determined by staining with propidium iodide (PI; 1 µg/ml) and flow cytometric analysis on a FACScan with a 488nm Argon laser (BD Biosciences). Quantitation was performed by scoring cells for sub-G1 DNA content using the ModFit software.
Cell fractionation and immunoblotting.
Cells were lysed following the respective treatments in cell lysis buffer containing 50 mM Tris pH 7.4, 120 mM NaCl, 5 mM EDTA, 0.5% NP40, and 1 mM DTT supplemented with the phosphatase inhibitor cocktail I, II (Sigma, P5726) and complete protease inhibitor tablet (Roche Diagnostics, 11836170001). Cytoplasmic and nuclear fractions were obtained as described previously.37 PARP1 and β-tubulin served as nuclear and cytoplasmic markers, respectively. Western blots were quantified using the Image J program. Background signal was subtracted from the band intensities, which were then normalized to the β-actin controls. The differences in the band intensities were compared with the untreated control and were plotted as fold change.
The following primary antibodies were used: anti-LC3, anti-pAMPK (Thr172), anti-AMPK, anti-pULK1 (Ser467), anti-ULK1, anti-PARP1, anti-cleaved caspase-3, anti-γH2AX, anti-ATG7, anti-ATG3, and anti-pATM (Ser1981) (Cell Signaling Technologies, 2775, 2535, 2532, 4634, 4776, 9542, 9661, 2577, 2631, 3415, 4526); anti-p62, anti-LAMP2, anti-Ku70, anti-GFP, anti-cyclin E (Santa Cruz Biotechnologies, sc-48402, sc-5571, sc-17789, sc-9996, sc-198); anti-ATG4a and anti-ATG4b (Abcam, ab64738, ab64732); anti-HA and β-actin (Sigma, H9658, A5441); anti-ATM (Calbiochem, PC116); anti-HP1γ and secondary anti-mouse HRP (Millipore, 05-690, AP127P); secondary anti-rabbit HRP (Fisher Scientific, 31460).
Confocal immunostaining.
Cells were plated at 2 × 105) cells/cm2 on 22 × 22 mm coverslips in 35-mm culture dishes. Following the respective treatments, cells were fixed with 2.0% paraformaldehyde/PBS for 15 min at room temperature, washed 3X for 10 min each, permeabilized with 0.1% Triton X-100 in PBS for 5 min, and blocked in 10% fetal bovine serum in PBS for 1 h. The coverslips were then immunostained using the antibodies diluted in blocking buffer, followed by fluorescently conjugated secondary antibody. DAPI was added before penultimate washing to stain nuclei. They were then mounted in Vectashield (Vector Laboratories, H-1000). Images were collected using an HCX Plan Apo 63X/1.4N.A. oil immersion objective lens on a Leica TCS-SP2 confocal microscope (Leica Microsystems AG). Quantification was based on data observed from at least 50 cells.
Analysis of GFP-mCherry-LC3 puncta.
Cells with stable expression of GFP-mCherry-LC3 were grown overnight before irradiation and fixation with 4% paraformaldehyde at the indicated times, washed several times with PBS, mounted using Vectashield, and analyzed using an HCX Plan Apo 63x/1.4N.A. oil immersion objective lens on a Leica TCS-SP2 confocal microscope (Leica Microsystems AG). Quantification of LC3 puncta was performed using the Red and Green Puncta Colocalization Macro with Image J program, as described.20,38,39
Electron microscopic analyses.
Cells were immediately fixed in 2.5% glutaraldehyde/4% formaldehyde in 0.1M Cacodylate buffer, pH 7.3, for 24 h, followed by post fixation with 1% osmium tetroxide for 1 h. After en bloc staining and dehydration with ethanol, the samples were embedded with eponate 12 medium (Tell Pella, Inc.). Thin sections (85-nm) were cut with a diamond knife, double-stained with uranyl acetate and lead citrate, and analyzed using a PhilipsCM12 electron microscope (FEI Company) operated at 60 kv. Cells with more than 10 vacoules were scored as autophagy positive. The autophagic area was quantitated in at least 10 cells per sample and was normalized to the total cytoplasmic area.
Supravital cell staining with acridine orange for autophagy detection.
Acridine orange stains cytoplasm and nucleolus fluorescent bright green and dim red, respectively whereas the acidic compartments fluoresce bright red.40 The intensity of red is directly proportional to the degree of acidification of cellular compartments and thus the volume of the cellular acidic compartments can be quantified using acridine orange staining. After the indicated treatments, live cells were stained with Acridine orange (Sigma-Aldrich, 318337) according to published protocols.40 Briefly, the cells were stained at a final concentration of 1 µg/ml in Phenol red free medium for 20 min. Images were captured on a Leica DMIRB fluorescence microscope equipped with a Retiga SRV cooled CCD camera (QImaging) and running ImagePro Plus capture software (Media Cybernetics). The microscope was configured with a 490-nm band-pass blue excitation filters, a 500-nm dichroic mirror and a 515-nm long-pass barrier filter. For quantitation, green (510–530 nm) and red (650 nm) fluorescence emission from 1 × 104 cells illuminated with blue (488 nm) excitation light was measured with FACScan (BD Biosciences).
Quantitative real-time (RT)-PCR analysis.
PCR amplification reactions were comprised of 1 X iQ SYBR Green Supermix (Bio-Rad, 1708882) and 4 µM each of sense and antisense primer. RT-PCR primers for LC3 (QT01673007) were purchased from Qiagen. The GAPDH primers: forward 5′-TCACCATCTTCCAGGAG-3′ and reverse 5′-GCTTCACCACCTTCTTG-3′ were synthesized by Integrated DNA Technologies. Reactions were performed in a 96-well optical reaction plate with optical caps (Denville Scientific, C18096-10) in an iCycler iQ Real-Time PCR Detection System spectrofluorometric thermal cycler (Bio-Rad) with an initial 3 min incubation at 95°C followed by 40 cycles of amplification: 95°C for 15 sec and 60°C for 1 min.
Clonogenic cell survival assay.
Cells were trypsinized after 8 h following irradiation and drug treatment as indicated, counted and plated in 6-well plates in triplicate. Cells were then allowed to grow for 10 d, fixed and stained in methanol:acetic acid (75:25, v/v) containing 0.5% crystal violet (w/v) to visualize colonies of at least 50 cells.
SA-β-Galactosidase assay.
Senescent cells were detected using an SA-β-Galactosidase staining kit available from Cell Signaling Technology (9860). Cells were fixed following treatment in 20% formaldehyde, 2% glutaraldehyde solution in 1 × PBS and stained with staining solution containing X-gal supplemented with potassium ferrocyanide and potassium ferricyanide following the manufacturer's protocol. Images were obtained using bright phase microscopy at 20 X magnification and percentage of SA-β-Gal positive cells was plotted against total cell number.
Statistical analyses.
Cell populations with sub-G1 DNA content, SA-β-Gal positive staining, and mRNA levels were expressed as percentages or fold change. The size of autophagic structures was represented as relative area values calculated by selecting specific areas using the Canvas 11 software. The autophagic area was calculated as percentage fraction normalized to the total cytoplasmic area. In clonogenic assays the absolute number of colonies was plotted. Data were obtained from at least three independent experiments performed in triplicate with the error bars denoting standard error of the mean. p values were determined by Student's t-test (Microsoft Excel).
Acknowledgments
We thank Drs. J. Debnath (University of California San Francisco) for GFP-mCherry-LC3, Dr. B.P. Rubin for insightful suggestions, and M. Yin, J. DeVecchio, C. Shemo, J. Alharbi, P. Chatterjee, G, Choudhary, and A. Sharma (Cleveland Clinic) for technical assistance. This work was supported by National Cancer Institute (CA127264) to A.A., NIH (AG031903) to S.M, and US. Department of Defense (PC094405) Post-doctoral Training Award to K.S. All authors designed and analyzed experiments. K.S. and J.D.A. performed the experiments; K.S. and A.A. wrote the paper. A.A. supervised the project.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Chen Q, Gong B, Almasan A. Distinct stages of cytochrome c release from mitochondria: evidence for a feedback amplification loop linking caspase activation to mitochondrial dysfunction in genotoxic stress induced apoptosis. Cell Death Differ. 2000;7:227–233. doi: 10.1038/sj.cdd.4400629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 6.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 7.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanida I, Sou YS, Minematsu-Ikeguchi N, Ueno T, Kominami E. Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. FEBS J. 2006;273:2553–2562. doi: 10.1111/j.1742-4658.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- 9.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalmers AJ, Lakshman M, Chan N, Bristow RG. Poly (ADP-ribose) polymerase inhibition as a model for synthetic lethality in developing radiation oncology targets. Semin Radiat Oncol. 2010;20:274–281. doi: 10.1016/j.semradonc.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Gewirtz DA, Hilliker ML, Wilson EN. Promotion of autophagy as a mechanism for radiation sensitization of breast tumor cells. Radiother Oncol. 2009;92:323–328. doi: 10.1016/j.radonc.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/00085472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 17.Mazumder S, Gong B, Chen Q, Drazba JA, Buchsbaum JC, Almasan A. Proteolytic cleavage of cyclin E leads to inactivation of associated kinase activity and amplification of apoptosis in hematopoietic cells. Mol Cell Biol. 2002;22:2398–2409. doi: 10.1128/MCB.22.7.23982409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazumder S, Plesca D, Kinter M, Almasan A. Interaction of a cyclin E fragment with Ku70 regulates Bax-mediated apoptosis. Mol Cell Biol. 2007;27:3511–3520. doi: 10.1128/MCB.01448-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitinlike conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta A, Roy S, Lazar AJ, Wang WL, McAuliffe JC, Reynoso D, et al. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST) Proc Natl Acad Sci USA. 2010;107:14333–14338. doi: 10.1073/pnas.1000248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinz M, Stilmann M, Arslan SC, Khanna KK, Dittmar G, Scheidereit C. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. Mol Cell. 2010;40:63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 23.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM-and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci USA. 2010;107:4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PLoS ONE. 2010;5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, et al. Principles and Current Strategies for Targeting Autophagy for Cancer Treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plesca D, Mazumder S, Gama V, Matsuyama S, Almasan A. A C-terminal fragment of Cyclin E, generated by caspase-mediated cleavage, is degraded in the absence of a recognizable phosphodegron. J Biol Chem. 2008;283:30796–30803. doi: 10.1074/jbc.M804642200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gama V, Gomez JA, Mayo LD, Jackson MW, Danielpour D, Song K, et al. Hdm2 is a ubiquitin ligase of Ku70-Akt promotes cell survival by inhibiting Hdm2-dependent Ku70 destabilization. Cell Death Differ. 2009;16:758–769. doi: 10.1038/cdd.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seluanov A, Danek J, Hause N, Gorbunova V. Changes in the level and distribution of Ku proteins during cellular senescence. DNA Repair (Amst) 2007;6:1740–1748. doi: 10.1016/j.dnarep.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ju YJ, Lee KH, Park JE, Yi YS, Yun MY, Ham YH, et al. Decreased expression of DNA repair proteins Ku70 and Mre11 is associated with aging and may contribute to the cellular senescence. Exp Mol Med. 2006;38:686–693. doi: 10.1038/emm.2006.81. [DOI] [PubMed] [Google Scholar]
- 34.Di Micco R, Sulli G, Dobreva M, Liontos M, Botrugno OA, Gargiulo G, et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol. 2011;13:292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Q, Chai YC, Mazumder S, Jiang C, Macklis RM, Chisolm GM, et al. The late increase in intracellular free radical oxygen species during apoptosis is associated with cytochrome c release, caspase activation, and mitochondrial dysfunction. Cell Death Differ. 2003;10:323–334. doi: 10.1038/sj.cdd.4401148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh K, Sinha S, Malonia SK, Bist P, Tergaonkar V, Chattopadhyay S. Tumor suppressor SMAR1 represses IkappaBalpha expression and inhibits p65 transactivation through matrix attachment regions. J Biol Chem. 2009;284:1267–1278. doi: 10.1074/jbc.M801088200. [DOI] [PubMed] [Google Scholar]
- 38.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 39.Cherra SJ, 3rd, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, et al. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Traganos F, Darzynkiewicz Z. Lysosomal proton pump activity: supravital cell staining with acridine orange differentiates leukocyte subpopulations. Methods Cell Biol. 1994;41:185–194. doi: 10.1016/S0091-679X(08)61717-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.