1. Polymer-based gene delivery offers interesting and important scientific problems
In about 70% of over 1,400 gene therapy clinical trials that have been conducted to date worldwide, genetically-modified viruses have been the carrier of choice for delivery of therapeutic genetic material [1]. While the viruses promise both high efficiency of transfer and great protection of the therapeutic genes [2], this approach also carries a risk of causing adverse (inflammatory or immune) reactions [3, 4] or even cancer [5]. Non-viral systems, such as cationic lipids and synthetic polymers (in particular, polycations), have attracted the interest of a large number of researchers as safer alternatives [6]. In particular, polycations have become popular components of non-viral gene carriers because of the relative ease with which their chemical and physical properties can be engineered for specific applications. However, the polycation-based approach has been limited in its clinical application in large part due to the poor biological activities of synthetic polymers on both cellular and systemic levels. A major issue is the difficulty associated with target-cell-specific delivery of genetic materials in vivo [6, 7]. However, even the basic problem of achieving a sufficient efficiency in the transportation of therapeutic genes across various intracellular barriers also remains one of the leading challenges in the development of superior polycation-based gene delivery systems. In this regard, even the most effective polycation gene carrier (e.g., linear polyethylenimine or PEI for short) remains 105 times less efficient [8] than its viral counterpart [9]. Since the first demonstration of polycation-medicated gene transfection in 1987 [10], many polycation materials (both new and off-the-shelf) have been explored for gene delivery applications with the most intensively studied example being the PEI polycation (reviewed in Refs. [11–16]). An obvious reason for the great attention devoted to PEI is that this polycation affords the highest levels of in vitro gene transfection. It is believed that the high gene transfection efficiency observed with PEI is attributable to its unique ability to simultaneously overcome several key barriers to intracellular trafficking of the DNA particles (e.g., escape from endosomes [17, 18], protection of DNA from degradation by endonulceases [19], nuclear entry [17, 19, 20], DNA release and transcription [20]). Currently, however, the exact mechanisms of how PEI orchestrates the sequence of the intracellular processes required for effective expression of the transgene in the host cell, and the particular chemical/molecular attributes of PEI responsible for each event, remain largely unexplained, making it difficult to further improve the performances of the PEI-based carriers in other aspects of the delivery process. One recent example to improve the PEI-based delivery system is the incorporation of intracellularly degradable disulfide bonds in the backbone structure of the PEI molecule [21–24] to reduce the inherent cellular (and systemic) toxicity of the polycation [25–27]. While this modification improves the viability of the transfected cells, thereby enabling the use of the PEI chemistry at high molecular weight without causing cell death [22–24], this improvement accompanies an unwanted decrease in the overall gene transfection efficiency when the performances are compared at an identical PEI molecular weight [24]. Improved understanding of the polycation chemistry vs. performance mechanism relationships will provide useful insights to guide further (chemical and/or physical) modifications of this already useful polycation toward creating multipotent gene carriers that can accommodate all of the sophisticated functional requirements at various stages of the delivery process. In this article, we intend to identify and discuss several key areas which require further improvements in our molecular understanding of the cellular transport processes of polymer/DNA complexes (“polyplexes”).
2. The endocytosis-mediated delivery paradigm is in need of a breakthrough
Polyplexes can effectively be internalized by cells via the membrane invagination mechanism called “endocytosis” [28]. In particular, when the sizes of the polyplex particles are less than 200 nm [29, 30] and/or the cell uptake is mediated by certain cell surface receptors such as transferrin or low-density lipoprotein receptors [31–34], the internalization of the polyplexes is believed to occur by the so-called clathrin-mediated endocytosis (CME) mechanism [29, 30], which has been thought to be kinetically the most effective [35, 36] (and obviously the most commonly cited [28]) endocytic uptake pathway for various polyplexes. A caveat of utilizing this CME mechanism for non-viral gene delivery is that because of the eventual merger of the endosome compartments into lysosomes [37], the endocytosed cargo material (i.e., the therapeutic gene) is typically destined for acidic and/or enzymatic degradation at the final stage of the process (in the late endosome or lysosome) [38, 39]. Thus, the CME pathway is applicable to polyplex-based gene delivery applications, only when this pathway is used with a polymer material that is capable of inducing the release of the polyplexes from endosomes into the cell’s cytoplasm at a relatively early stage in the CME pathway. A recent study by Gabrielson and Pack [40] shows that for polymeric gene delivery, a different pathway called the caveolae-mediated endocytosis affords a more efficient means of gene transfection than the CME pathway, because the vesicles that result from the caveolar endocytosis do not develop into lysosomes. The authors also discovered that contrary to what has been reported for the caveolae-based internalization of viral fusogenic proteins [41], the caveolar endocytosis of PEI polyplexes involves a significant degree of acidification of the endosomal compartments. In light of these recent data, an optimized gene carrier should be one that (a) induces the caveolae-dependent internalization of the carrier, and (b) at the same time causes disruption of endosomal membranes selectively under acidic conditions. Focusing on the latter part of the requirement, many polymers have been identified or developed over the past two decades that possess the needed low-pH-activated endosomolytic property and are not detrimental to cell membranes under normal conditions (reviewed in Ref. [42]). Examples of such materials span both polycation and polyanion categories. For instance, such polycations as PEI [18], polyamidoamine (PAm) dendrimers [18] and imidazole-containing polymers [43] have been shown to be effective in endosomal escape of the associated polyplexes. This endosome escape property is related to the polycation’s proton buffering capability (“titratability”) [18, 44–48] such that the polycation molecules become more protonated, as the pH of the endosome decreases during the endocytosis pathway. It is believed that this process causes endosome rupture and release of contents to the cytosol because of (i) the osmotic overload produced by the increased concentrations of the H+ and Cl− ions within the endosome [49, 50], and/or (ii) the increased adsorption of the highly charged polycations to the inner surface of the endosome [51, 52]. The former hypothesis, termed “proton sponge effect”, has been a favorite explanation for the endosomal lysis ability of the polycations, although the validity of the osmotic lysis conjecture has not been rigorously tested (as will be discussed later in this article). Polyanions with tailored hydrophobicity and pH-dependent charge densities have also been developed and demonstrated to be a useful component for producing the desired effect of endosome disruption via low-pH-induced insolubilization of the polyanion molecules and subsequent fusion with cell membranes [53].
The pH-sensitive endosomolytic polymer-based approaches require cellular internalization of the polyplex particles by an endocytosis pathway which involves endosomal acidification. In this regard, the caveolar pathway [40] and other mechanisms such as the so-called macropinocytosis [18, 54] and micropinocytosis [54] pathways would all be desirable routes of polyplex internalization. On the other hand, the CME pathway is less desirable, because it involves lysosomal degradation of the DNA cargo [40]. Interestingly, it has been known that lipid-based gene carriers (“lipoplexes”) are normally taken up by cells via CME [55]. Only within past few years have studies questioned whether or not this is also the case for small-size polyplex particles. As recently reviewed by Midoux et al. [56], the results of these studies (fortunately) indicate that unlike the cases involving lipoplexes, polyplex particles can be internalized by different endocytic mechanisms, and the internalization mechanism can vary substantially depending on a number of factors, including polymer chemistry, cell type, cell polarization state, and cell division cycle. These findings appear to provide at least a partial explanation as to why some polycations with significant proton buffering capacities are not effective in gene transfection under certain conditions. That is, they are likely internalized predominantly by the CME pathway. The well-known PEI polyplexes appear to rely significantly on other internalization mechanisms such as the caveolae-dependent pathway. As illustrated in Figure 1, transfection of a gene delivered by a PEI vector is almost completely suppressed in HeLa cells when the caveolae pathway is blocked, whereas blocking the CME pathway reduces the transfection efficiency to only about 50%; also see the results of more extensive experiments by Gabrielson and Pack [40]. Therefore, it is not unreasonable to generalize that if one could force the polyplex uptake process to occur such that the interference of the CME pathway can be maximally suppressed, one would be able to achieve more improved efficiencies in the endosome escape processes regardless of the polymer type.
Figure 1.
(A) Effects of blocking specific endocytosis pathways on the gene transfection efficiency of PEI polyplexes in HeLa cells. The polyplexes were prepared using 11 kDa linear PEI and luciferase-encoding pDNA (pGL2) at an N:P ratio of 7.5 and a DNA concentration of 10 µg/ml. The cells were incubated either with chlorpromazine or with fillipin III (for an hour prior to the transfection with the PEI polyplexes and for another three hours after the transfection) in order to block, respectively, the clathrin-mediated endocytosis pathway or the caveolae pathway. The gene transfection efficiencies were quantified in terms of the luciferase expression levels of the pGL2 genes in the transfected cells (in relative light units) as measured by the standard luminescence assay; the luciferase expression levels were normalized by the total protein contents of the respectively treated cells. (B) The same data are shown in normalized forms relative to the gene transfection efficiency of the polyplexes obtained under no chlorpromazine or fillipin III conditions.
An important manifestation of the variability of the polyplex internalization mechanisms will be that a polymer formulation, optimized toward maximum endosomal lysis under a specific set of in vitro conditions, would not necessarily work equally well for cells under in vivo conditions, thus significantly hindering clinical applications. Therefore, the endocytosis-based gene delivery paradigm is in need of a breakthrough. Perhaps, the key to this will be the development of new polymer technologies that will allow precise and universal control of the polyplex endocytosis pathway. One possible approach will be to functionalize polyplex particles with receptor-specific ligand moieties (for instance, folic acid [57]) which, through binding to specific receptors, induce the caveolar endocytosis (or alternatively the macro/micropinocytosis) of the polyplex particles selectively over other endocytosis pathways, in particular the CME pathway. In this context, there are many unanswered fundamental questions that need to be addressed before this approach can proceed. One such question is whether a single ligand (e.g., folic acid) can activate the caveolar endocytic machinery invariably regardless of polyplex chemistry/size, cell type, cell polarization state, and cell division cycle. Another difficulty lies in the fact that the functionalization of polyplexes with caveolar-endocytosis-triggering ligands will likely increase unwanted interactions with non-targeted cells, which will provide additional difficulty to the already challenging task of systemically delivering polyplexes to specific tissues and cells [7].
3. An important gap exists in our understanding of the “proton sponge effect”
For the low-pH-specific endosome lysis activity of certain polycations, the most frequently employed explanation has been the proton sponge hypothesis. At the outset, it should be noted that despite popularity of this model, the theory has never been proven. There are two fundamental issues surrounding this concept. The first is that the exact chemical/molecular factors which impart proton absorbing (“sponging”) qualities to certain polycations are presently undetermined. In fact, the molecular origin of the proton sponge effect has been somewhat controversial and has been variably attributed to the retarded ionization of the tertiary [58, 59] or secondary [60, 61] amine group relative to the primary amine. Although this line of logic explains the “titratability” of branched and linear PEI and PAm dendrimers and the absence of such an ability in polylysine, this hypothesis contradicts the relative tendencies towards protonation among the different amine types. For instance, the pKa values of tri-, di- and mono-ethyl amines are approximately 10.8, 11.1 and 10.8, respectively. Instead, a more accurate picture is that the monomer pKa plays only part of the role in determination of the proton buffering capacity of a polycation, and it is the connectivity of the amine groups in a polycation chain that causes the retardation of the protonation of the amine groups relative to the same compounds in their monomeric state. This hypothesis is supported by the data shown in Figure 2 in which we compare the protonation behavior of two sets of polycation monomer and polymer combinations; i.e., poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA) vs. its monomer, and PEI vs. its copolymer derivative in which ethylenimine (EI) monomers are separated by spacer groups along the backbone. In both cases, at any value of the total added proton concentration (i.e., [H+]) the pH of the polymer solution is lower than the corresponding monomer solution, indicating that the polymer segments are always less protonated than the monomers under identical pH conditions. Additionally, our data demonstrate that as the amine spacing is increased by incorporation of hydrophilic spacer groups (i.e., in the polyethylenimine-co-poly(2-ethyl-2-oxazoline) (PEI-co-PEOz) case), the polycation protonation behavior tends asymptotically towards its monomeric behavior, further supporting the importance of the connectivity and small spacing between the amine groups. A self-consistent field theoretical study indicates that the electrostatic repulsion between adjacent charged groups is, at least partially, responsible for the suppression of the ionization of polycations [62]. Another factor that may retard the complete protonation of a polycation is the (quantum mechanical) electron delocalization effect. In this instance, the effect might be of importance in the PEI case in which the neighboring amine groups are only 3 atoms apart from one another, while the same effect is expected to be negligible for the PDMAEMA case where the adjacent tertiary amines are separated by a 12-atom distance. Overall, the results presented in Figure 2 suggest that within the intracellularly relevant pH range (5.0–7.4), PEI has a higher capacity in absorbing H+ ions than any other polymer tested (i.e., PDMAEMA and PEI-co-PEOz). With PDMAEMA or PEI-co-PEOz, it would take an increase in polymer material to achieve the same proton sponge effect as with PEI. Also of note, the proton buffering capacity of PDMAEMA decreases significantly when pH < 6.0, suggesting that in the PDMAEMA/DNA polyplex case, upon endocytosis the polyplexes have only a relatively small window of time to escape endosomes (i.e., during the early stages of the endosomal maturation pathway). The differences and trends observed in the proton buffering characteristics of these polycations are expected to be reflected in the differences in their relative abilities to enhance the endosomal escape of the corresponding polyplexes. The results of this example clearly illustrate that the interrelationships between the molecular characteristics of the polycation and the polycation’s proton buffering capability are far more complicated than one can simplistically extrapolate from the type of the polycation’s amine groups. At this time, we simply do not have sufficient data regarding what molecular factors determine the proton titratability of polycations to guide us, on a rational basis, towards new (or modified) polycation materials possessing better intracellular trafficking efficiencies.
Figure 2.
(A) Potentiometric titration curves of poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA127; here the subscript number denotes the degree of polymerization) and its monomer (DMAEMA), demonstrating the suppression of the protonation of the tertiary amine groups in the polymer chain relative to the monomeric amine groups. The tertiary amine group concentrations were the same for both the DMAEMA and PDMAEMA experiments: 7.5 mM. The solutions were initially prepared in deionized (DI) water. (B) Potentiometric titration curves of polyethylenimine-co-poly(2-ethyl-2-oxazoline) random copolymers (PEIx-co-PEOzy where x + y = 266) with varying monomer ratios (i.e., x:y = 9:1, 1:2, and 1:6), demonstrating the retarded protonation of the secondary amine groups in the copolymers with high EI monomer contents; this retardation effect is due to the connectivity and tight spacing between the amine groups, and becomes mitigated when hydrophilic spacers are incorporated along the chain (e.g., when x:y ≤ 1/2). The amine group concentrations were the same for all the samples: 10 mM.
The second issue regarding the validity of the proton sponge hypothesis is that it needs to be rigorously tested whether the osmotic stress produced by the proton sponge effect can, by itself, induce lysis of the endosomal membrane, or whether the endosome lysis process requires other mechanisms to be operative at the same time (e.g., hydrophobic and/or electrostatically-driven adsorption of polycation molecules to endosome membranes at low pH). To elucidate this issue, it is useful to calculate the osmotic pressure that is expected to be produced inside a polyplex-containing endosome vesicle when the pH of the endosome is shifted from 7.4 to 5.0. Given the size of a typical vesicle produced by CME (100 – 150 nm) [34, 54], it is reasonable to assume that each clathrin-coated vesicle will contain one polyplex particle on the order of 200 nm in diameter (see the next section). On the basis of estimates of the number of DNA molecules per polyplex particle in the literature (i.e., 1 to 7) [63–65], we will assume that each polyplex contains five plasmid DNA molecules (having a length of 5,000 base pairs). These dimensions give a value for the ethylenimine (EI) group concentration within the endosome of [N]o = 70 mM, where the N:P ratio (defined as the ratio of the number of amine (N) groups on PEI to the number of phosphate (P) groups on DNA) is equal to 7. As can be extracted from the data shown in Figure 2, a pH change from 7.4 to 5.0 in a PEI solution containing 10 mM EI monomers (i.e., [N]o = 10 mM) requires that the added proton concentration has to be increased by an amount of Δ[H+]o = 4.8 mM. Assuming that under the environment of the endosome compartment, the proton buffering behavior of PEI will be similar to that of the controlled experiment, and after correction for the small difference in [N]o between the two situations, we estimate that the same pH change in a polyplex-containing endosome will involve an influx of H+ (and Cl−) ions of an amount of Δ[H+]o = Δ[Cl−]o = 33 mM (= (4.8 mM/10 mM)×70 mM). Therefore, the osmotic pressure (due to the surplus amount of Cl− ions inside the endosome relative to the cytosol) is estimated to be π (≈ Δ[Cl−]o·RT where R is the gas constant and T is the temperature) ≈ 8.3 × 104 Pa. On the basis of the Young-Laplace relation (π = 2γ/r where γ and r denote the membrane tension and the vesicle radius, respectively) [66], and assuming the high-tension limit for the elastic response of the lipid membrane (i.e., γ = Ka·α where Ka is the area expansion modulus and α is the areal strain which is equal to [(r/ro)2-1] for a spherical vesicle with an initial radius ro at π = 0) [67], we estimate, by using a typical value of 180 mN/m for Ka [68], that the given amount of osmotic pressure will expand the membrane area only by 2.3% (i.e., α = 0.023). Interestingly, lipid vesicles can withstand area expansion up to 2 to 5% strain (i.e., αc ≈ 0.02 – 0.05) above which the membrane begins to lose its integrity [69]. Further, it should be noted that the above estimates of the osmotic pressure inside the endosomal vesicle (π) and the resultant degree of vesicle deformation (α) are their maximum likelihood values, because in real situations, the proton-absorbing capacity of polycations will be significantly reduced due to the presence of other electrolytes in the physiological medium and also due to the complexation of the polycations with DNA. Therefore, it is not unreasonable to argue that even under the influence of the polycation’s proton buffering reactions, the osmotic pressure built up during the acidification of the endosome is theoretically insufficient to cause endosome disruption, though it might be a significant contributory factor to the eventual disruption of the bilayer membrane. An important question that arises is then what are the other effects of polycation molecules that contribute to endosomal lysis? This is another key question that needs to be answered to better determine the direction of future developments of new polycation gene carriers.
4. What is the ideal timing of DNA unloading, before or after nuclear entry?
After the endosomal escape, a desirable polyplex transport scenario is for the escaped polyplex particles to traffic towards and enter the nucleus of the cell to (at least partially) unload the DNA for transcription [70]. There is evidence that migration of polyplexes (or DNA) to the nucleus periphery through the cytoplasm is an active (not diffusive) transport process mediated by the microtubule network [71]. This process is typically not a rate-limiting step in intracellular trafficking of polyplexes (or DNA) [20]. However, the entry of polyplexes (or DNA) into the nucleus typically imposes a huge barrier to transgene expression [51, 72, 73] because of the small functional size of the nuclear pore complex in the nucleus (≈ 10 nm [74], or about 30 nm even under the inclusion of a nuclear localization signal/sequence [75]) of a non-dividing (“postmitotic”) cell. While experiments suggest that the nuclear entry of polyplexes (or naked DNA) is easier during the cell division (“mitosis”) period when the nuclear envelope becomes disintegrated [76], it is also well accepted that nuclear entry does not necessarily require a cell division event [72]. For naked DNA, the nuclear entry process during the postmitotic period involves specific “nuclear targeting” sequences (NTS) in the DNA which can activate the importin (or other nuclear import) machinery through mediating the formation of appropriate DNA/protein complexes [77, 78]. In contrast, for DNA molecules in the form of polyplexes, the sequence specificity of the nuclear entry has not been reported. Experiments based on microinjection of PEI/DNA polyplexes into the cytoplasm suggest that the complexation of DNA with certain polycations (such as PEI) significantly lowers the barrier for nuclear entry of the DNA, presumably due to the reduced size of the DNA upon complexation with polycations [20]. Recently, however, new evidence has emerged that DNA normally (at least partially) dissociates from the PEI/DNA complex upon escape from the endosome [79], which contradicts the above view regarding the role of PEI in enhancing gene transcription. There also has been a report that DNA complexes with an intracellularly degradable version of PEI (PEI with disulfide bonds in the backbone) that exhibits significant gene expression [24], although in this case cleavage of the disulfide bonds is expected to result in decondensation of DNA in the cytoplasm. Reduction of disulfide bonds may occur as early as during the endocytic stages of intracellular trafficking [80]. Currently, it remains a puzzling question how the observed “decompaction” of PEI/DNA polyplexes in the cytoplasm (which will likely cause an increase in the size of the polyplex particles) can contribute to the lowering of the barrier for nuclear entry of the polyplexes. This will remain an important question to be addressed in the future.
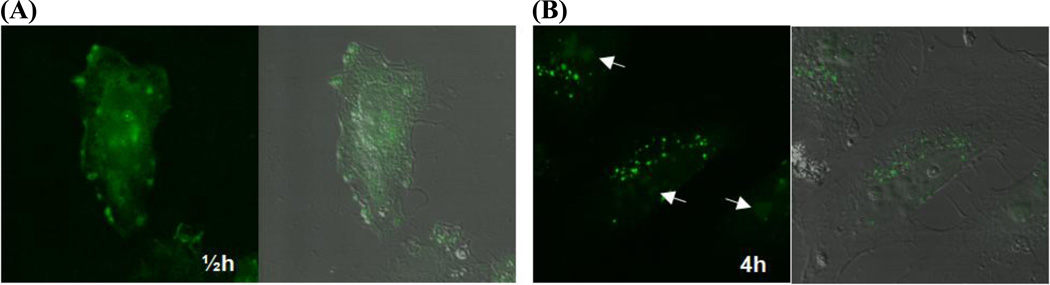
The compactness of the polyplex structure is expected to have competing effects on the performance of the polyplex. More compact structures afford better protection of DNA against nucleases and more efficient transportation of DNA through the cytoplasm (and controversially into the nucleus as well). On the other hand, stronger polycation-DNA binding that causes denser polyplex particles is detrimental to the timely release of DNA for transcription [81–83]. Therefore, an ideal gene carrier would be one which binds strongly to DNA during the earliest stages of intracellular transport but dissociates from DNA only upon arrival in the nucleus (i.e., right before transcription of the gene). Unfortunately, such a material has not been developed. Currently, PEI is considered to be one of the most effective cellular gene delivery polycations studied to date. We speculate that the key to this performance is the optimal binding affinity that PEI has for DNA. The puzzling aspect of the intracellular behavior of PEI polyplexes is demonstrated in the example presented in Figure 3, in which we present snapshots of the cytoplasmic and nuclear transport processes of the polyplexes prepared with 11 kDa PEI. At the N:P ratio used in this experiment (i.e., N:P = 7), the mean hydrodynamic diameter of the PEI-based polyplex particles is 229 nm. The confocal microscopy data revealed that the DNA molecules delivered by PEI to the nucleus (see Figure 3-B) do not exist in the form of PEI/DNA complexes (as indicated by the diffuse fluorescence in contrast to the punctuate appearance of the polyplexes in the endosome-confined state). This result further suggests that DNA dissociates from the PEI carrier prior to nuclear entry and enters the nucleus without relying on a sequence-specific nuclear import mechanism [77, 78], since the DNA used in the above experiment does not contain any of the known DNA nuclear localization sequences. Therefore, this observation poses an important question: What is the exact role that PEI plays in promoting the nuclear import of DNA? These and other results support the existence of an optimal binding level that gives the best compromise between nuclease protection vs. nuclear import/polyplex disintegration for gene transcription. Systematic and controlled studies of these issues will be required to establish precise polycation design requirements for vastly improved nuclear-targeted delivery of therapeutic genes.
Figure 3.
Confocal microscopy and confocal-DIC overlay images (the left and right sides of each panel, respectively) of HeLa cells transfected for 2 h with PEI239-co-PEOz27 polyplexes containing YOYO-1-labeled (one per 300 bp) β-galactosidase plasmid (N:P = 7). Subsequently, the cells were incubated in polyplex-free medium, and the imaging measurements were performed at (A) ½ h and (B) 4 h after the transfection. The white arrows in (B) indicate the locations of the nuclei (particularly, the nucleoli). At ½ h the cell surfaces were densely covered with adsorbed polyplexes which gave diffuse fluorescence throughout the cell surfaces, and at 4 h large populations of the polyplexes were observed to be inside endosome/lysosome compartments (bright punctuate spots in (B)). A notable observation is that at 4 h, fluorescence is detected inside the nuclei of the cells transfected with the PEI polyplexes.
5. Concluding remarks
There is critical information missing in our understanding of the intracellular trafficking and transfection mechanisms of polymer-based gene carriers. The important scientific issues that need to be addressed include: (i) development of a method of controlling the cellular internalization mechanism of polyplex particles, irrespective of cell type, cell polarization state and cell division cycle; (ii) identifying the exact chemical and molecular factors responsible for the proton buffering behavior observed with certain polycations (such as polyethylenimine (PEI), currently one of the few most effective and versatile of all known synthetic gene carriers); (iii) defining the basic premise of the “proton sponge hypothesis” versus other possible effects of polycations contributing to the rupture of the endosome at low pH conditions; and (iv) understanding the exact mechanisms by which, for instance, PEI so effectively enhances the nuclear localization/release and transcription of the delivered DNA. A precise molecular-level understanding of the polyplex chemistry vs. performance relationships will provide a fundamental basis for developing new materials and strategies for vastly improved efficiencies of non-viral gene delivery systems. These approaches will help vitalize the gene therapy field towards realizing the full potential of the technology in both conventional and emerging areas of applications such as stem cell reprogramming [84–86].
Acknowledgment
The authors are grateful for financial support from the National Science Foundation (CBET-0828574). The authors would also like to thank Dr. Mini Thomas (Department of Chemistry, Purdue University), and Dana J. Gary and Hoyoung Lee (School of Chemical Engineering, Purdue University), for critical reading of this manuscript.
References
- 1.Gene Therapy Clinical Trials Worldwide (Provided by the Journal of Gene Medicine) http://www.wiley.co.uk/genetherapy/clinical/. [Google Scholar]
- 2.Mulligan RC. The Basic Science of Gene-Therapy. Science. 1993;260(5110):926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 3.Check E. Gene therapy: A tragic setback. Nature. 2002;420(6912):116–118. doi: 10.1038/420116a. [DOI] [PubMed] [Google Scholar]
- 4.Raper SE, Chirmule N, Lee FS, Wivel NA, Bagg A, Gao GP, Wilson JM, Batshaw ML. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Molecular Genetics and Metabolism. 2003;80(1–2):148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCcormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, Sorensen R, Forster A, Fraser P, Cohen JI, de Saint Basile G, Alexander I, Wintergerst U, Frebourg T, Aurias A, Stoppa-Lyonnet D, Romana S, Radford-Weiss I, Gross F, Valensi F, Delabesse E, Macintyre E, Sigaux F, Soulier J, Leiva LE, Wissler M, Prinz C, Rabbitts TH, Le Deist F, Fischer A, Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 6.Li SD, Huang L. Non-viral is superior to viral gene delivery. J. Control. Release. 2007;123(3):181–183. doi: 10.1016/j.jconrel.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Bae YH. Drug targeting and tumor heterogeneity. J. Control. Release. 2009;133(1):2–3. doi: 10.1016/j.jconrel.2008.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lechardeur D, Verkman AS, Lukacs GL. Intracellular routing of plasmid DNA during non-viral gene transfer. Advanced Drug Delivery Reviews. 2005;57(5):755–767. doi: 10.1016/j.addr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Seisenberger G, Ried MU, Endress T, Buning H, Hallek M, Brauchle C. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science. 2001;294(5548):1929–1932. doi: 10.1126/science.1064103. [DOI] [PubMed] [Google Scholar]
- 10.Wu GY, Wu CH. Receptor-Mediated Invitro Gene Transformation by a Soluble DNA Carrier System. Journal of Biological Chemistry. 1987;262(10):4429–4432. [PubMed] [Google Scholar]
- 11.Furgeson DY, Kim SW. Polymeric Drug Delivery I: Particulate Drug Carriers. 2006;Vol. 923:182–197. [Google Scholar]
- 12.Godbey WT, Barry MA, Saggau P, Wu KK, Mikos AG. Poly(ethylenimine)-mediated transfection: A new paradigm for gene delivery. Journal of Biomedical Materials Research. 2000;51(3):321–328. doi: 10.1002/1097-4636(20000905)51:3<321::aid-jbm5>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 13.Godbey WT, Mikos AG. Recent progress in gene delivery using non-viral transfer complexes. J. Control. Release. 2001;72(1–3):115–125. doi: 10.1016/s0168-3659(01)00267-x. [DOI] [PubMed] [Google Scholar]
- 14.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J. Control. Release. 1999;60(2–3):149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 15.Neu M, Fischer D, Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. J. Gene. Med. 2005;7(8):992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- 16.Thomas M, Klibanov AM. Non-viral gene therapy: polycation-mediated DNA delivery. Applied Microbiology and Biotechnology. 2003;62(1):27–34. doi: 10.1007/s00253-003-1321-8. [DOI] [PubMed] [Google Scholar]
- 17.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in-Vivo - Polyethylenimine. Proc. Natl. Acad. Sci. U. S. A. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonawane ND, Szoka FC, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. Journal of Biological Chemistry. 2003;278(45):44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 19.Oh YK, Suh D, Kim JM, Choi HG, Shin K, Ko JJ. Polyethylenimine-mediated cellular uptake, nucleus trafficking and expression of cytokine plasmid DNA. Gene Therapy. 2002;9(23):1627–1632. doi: 10.1038/sj.gt.3301735. [DOI] [PubMed] [Google Scholar]
- 20.Pollard H, Remy JS, Loussouarn G, Demolombe S, Behr JP, Escande D. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. Journal of Biological Chemistry. 1998;273(13):7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 21.Choi S, Lee KD. Enhanced gene delivery using disulfide-crosslinked low molecular weight polyethylenimine with listeriolysin o-polyethylenimine disulfide conjugate. J. Control. Release. 2008;131(1):70–76. doi: 10.1016/j.jconrel.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forrest ML, Koerber JT, Pack DW. A degradable polyethylenimine derivative with low toxicity for highly efficient gene delivery. Bioconjugate Chem. 2003;14(5):934–940. doi: 10.1021/bc034014g. [DOI] [PubMed] [Google Scholar]
- 23.Kloeckner J, Wagner E, Ogris M. Degradable gene carriers based on oligomerized polyamines. European Journal of Pharmaceutical Sciences. 2006;29(5):414–425. doi: 10.1016/j.ejps.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y, Mo H, Koo H, Park JY, Cho MY, Jin GW, Park JS. Visualization of the degradation of a disulfide polymer, linear poly(ethylenimine sulfide), for gene delivery. Bioconjugate Chem. 2007;18(1):13–18. doi: 10.1021/bc060113t. [DOI] [PubMed] [Google Scholar]
- 25.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J. Gene. Med. 2004;6(10):1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 26.Chollet P, Favrot MC, Hurbin A, Coll JL. Side-effects of a systemic injection of linear polyethylenimine-DNA complexes. J. Gene. Med. 2002;4(1):84–91. doi: 10.1002/jgm.237. [DOI] [PubMed] [Google Scholar]
- 27.Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Advanced Drug Delivery Reviews. 2006;58(14):1523–1531. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacological Reviews. 2006;58(1):32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 29.Grosse S, Aron Y, Thevenot G, Francois D, Monsigny M, Fajac I. Potocytosis and cellular exit of complexes as cellular pathways for gene delivery by polycations. J. Gene. Med. 2005;7(10):1275–1286. doi: 10.1002/jgm.772. [DOI] [PubMed] [Google Scholar]
- 30.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin-and caveolae-mediated endocytosis. Biochem. J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dautryvarsat A, Ciechanover A, Lodish HF. Ph and the Recycling of Transferrin During Receptor-Mediated Endocytosis. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1983;80(8):2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: Formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- 33.Schmid SL. Clathrin-coated vesicle formation and protein sorting: An integrated process. Annu. Rev. Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- 34.Takei K, Haucke V. Clathrin-mediated endocytosis: membrane factors pull the trigger. Trends Cell Biol. 2001;11(9):385–391. doi: 10.1016/s0962-8924(01)02082-7. [DOI] [PubMed] [Google Scholar]
- 35.Goncalves C, Mennesson E, Fuchs R, Gorvel JP, Midoux P, Pichon C. Macropinocytosis of polyplexes and recycling of plasmid from clathrin-dependent pathway impair the transfection efficiency into human hepatocarcinoma cells. Mol. Ther. 2004;9:S317–S317. doi: 10.1016/j.ymthe.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 36.Zhou XH, Huang L. DNA Transfection Mediated by Cationic Liposomes Containing Lipopolylysine - Characterization and Mechanism of Action. Biochimica Et Biophysica Acta-Biomembranes. 1994;1189(2):195–203. doi: 10.1016/0005-2736(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 37.Sonawane ND, Verkman AS. Determinants of [Cl−] in recycling and late endosomes and Golgi complex measured using fluorescent ligands. J. Cell Biol. 2003;160(7):1129–1138. doi: 10.1083/jcb.200211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein JL, Brown MS, Anderson RGW, Russell DW, Schneider WJ. Receptor-Mediated Endocytosis - Concepts Emerging from the Ldl Receptor System. Annual Review of Cell Biology. 1985;1:1–39. doi: 10.1146/annurev.cb.01.110185.000245. [DOI] [PubMed] [Google Scholar]
- 39.Maxfield FR, McGraw TE. Endocytic recycling. Nature Reviews Molecular Cell Biology. 2004;5(2):121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 40.Gabrielson NP, Pack DW. Efficient Polyethylenimine-Mediated Gene Delivery Proceeds via a Caveolar Pathway in HeLa Cells. J. Control. Release. 2009;136(1):54–61. doi: 10.1016/j.jconrel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Ferrari A, Pellegrini V, Arcangeli C, Fittipaldi A, Giacca M, Beltram F. Caveolae-mediated internalization of extracellular HIV-1 tat fusion proteins visualized in real time. Mol. Ther. 2003;8(2):284–294. doi: 10.1016/s1525-0016(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 42.Cho YW, Kim JD, Park K. Pollycation gene delivery systems: escape from endosomes to cytosol. J. Pharm. Pharmacol. 2003;55(6):721–734. doi: 10.1211/002235703765951311. [DOI] [PubMed] [Google Scholar]
- 43.Pack DW, Putnam D, Langer R. Design of imidazole-containing endosomolytic biopolymers for gene delivery. Biotechnol. Bioeng. 2000;67(2):217–223. [PubMed] [Google Scholar]
- 44.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene. Med. 2005;7(5):657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 45.Behr JP. The proton sponge: A trick to enter cells the viruses did not exploit. Chimia. 1997;51(1〓2):34–36. [Google Scholar]
- 46.Kichler A, Leborgne C, Coeytaux E, Danos O. Polyethylenimine-mediated gene delivery: a mechanistic study. J. Gene. Med. 2001;3(2):135–144. doi: 10.1002/jgm.173. [DOI] [PubMed] [Google Scholar]
- 47.Merdan T, Kunath K, Fischer D, Kopecek J, Kissel T. Intracellular processing of poly(ethylene imine)/ribozyme complexes can be observed in living cells by using confocal laser scanning microscopy and inhibitor experiments. Pharmaceutical Research. 2002;19(2):140–146. doi: 10.1023/a:1014212630566. [DOI] [PubMed] [Google Scholar]
- 48.Thomas M, Klibanov AM. Enhancing polyethylenimine's delivery of plasmid DNA into mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2002;99(23):14640–14645. doi: 10.1073/pnas.192581499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Behr JP. Gene-Transfer with Synthetic Cationic Amphiphiles - Prospects for Gene-Therapy. Bioconjugate Chem. 1994;5(5):382–389. doi: 10.1021/bc00029a002. [DOI] [PubMed] [Google Scholar]
- 50.Haensler J, Szoka FC. Polyamidoamine Cascade Polymers Mediate Efficient Transfection of Cells in Culture. Bioconjugate Chem. 1993;4(5):372–379. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
- 51.Bieber T, Meissner W, Kostin S, Niemann A, Elsasser HP. Intracellular route and transcriptional competence of polyethylenimine-DNA complexes. J. Control. Release. 2002;82(2–3):441–454. doi: 10.1016/s0168-3659(02)00129-3. [DOI] [PubMed] [Google Scholar]
- 52.Pichon C, Roufai MB, Monsigny M, Midoux P. Histidylated oligolysines increase the transmembrane passage and the biological activity of antisense oligonucleotides. Nucleic Acids Res. 2000;28(2):504–512. doi: 10.1093/nar/28.2.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones RA, Cheung CY, Black FE, Zia JK, Stayton PS, Hoffman AS, Wilson MR. Poly(2-alkylacrylic acid) polymers deliver molecules to the cytosol by pH-sensitive disruption of endosomal vesicles. Biochem. J. 2003;372:65–75. doi: 10.1042/BJ20021945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bishop NE. An update on non-clathrin-coated endocytosis. Rev. Med. Virol. 1997;7(4):199–209. doi: 10.1002/(sici)1099-1654(199712)7:4<199::aid-rmv203>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 55.Wasungu L, Hoekstra D. Cationic lipids, lipoplexes and intracellular delivery of genes. J. Control. Release. 2006;116(2):255–264. doi: 10.1016/j.jconrel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 56.Midoux P, Breuzard G, Gomez JP, Pichon C. Polymer-Based Gene Delivery: A Current Review on the Uptake and Intracellular Trafficking of Polyplexes. Curr. Gene Ther. 2008;8(5):335–352. doi: 10.2174/156652308786071014. [DOI] [PubMed] [Google Scholar]
- 57.Pelkmans L, Helenius A. Endocytosis via caveolae. Traffic. 2002;3(5):311–320. doi: 10.1034/j.1600-0854.2002.30501.x. [DOI] [PubMed] [Google Scholar]
- 58.Lim YB, Kim SM, Lee Y, Lee WK, Yang TG, Lee MJ, Suh H, Park JS. Cationic hyperbranched poly(amino ester): A novel class of DNA condensing molecule with cationic surface, biodegradable three-dimensional structure, and tertiary amine groups in the interior. Journal of the American Chemical Society. 2001;123(10):2460–2461. doi: 10.1021/ja005715g. [DOI] [PubMed] [Google Scholar]
- 59.von Harpe A, Petersen H, Li YX, Kissel T. Characterization of commercially available and synthesized polyethylenimines for gene delivery. J. Control. Release. 2000;69(2):309–322. doi: 10.1016/s0168-3659(00)00317-5. [DOI] [PubMed] [Google Scholar]
- 60.Kasturi SP, Sachaphibulkij K, Roy K. Covalent conjugation of polyethyleneimine on biodegradable microparticles for delivery of plasmid DNA vaccines. Biomaterials. 2005;26(32):6375–6385. doi: 10.1016/j.biomaterials.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 61.Zhong ZY, Song Y, Engbersen JFJ, Lok MC, Hennink WE, Feijen J. A versatile family of degradable non-viral gene carriers based on hyperbranched poly(ester amine)s. J. Control. Release. 2005;109(1–3):317–329. doi: 10.1016/j.jconrel.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 62.Witte KN, Kim S, Won YY. Self-Consistent Field Theory Study of the Effect of Grafting Density on the Height of a Weak Polyelectrolyte Brush. Journal of Physical Chemistry B. 2009 doi: 10.1021/jp809814j. revision under review. [DOI] [PubMed] [Google Scholar]
- 63.Arscott PG, Li AZ, Bloomfield VA. Condensation of DNA by Trivalent Cations. 1 Effects of DNA Length and Topology on the Size and Shape of Condensed Particles. Biopolymers. 1990;30(5–6):619–630. doi: 10.1002/bip.360300514. [DOI] [PubMed] [Google Scholar]
- 64.Golan R, Pietrasanta LI, Hsieh W, Hansma HG. DNA toroids: Stages in condensation. Biochemistry. 1999;38(42):14069–14076. doi: 10.1021/bi990901o. [DOI] [PubMed] [Google Scholar]
- 65.Lai E, van Zanten JH. Monitoring DNA/poly-L-lysine polyplex formation with time-resolved multiangle laser light scattering. Biophys. J. 2001;80(2):864–873. doi: 10.1016/S0006-3495(01)76065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evans DF, Wennerstrom H. The Colloidal Domain: Where Physics, Chemistry, Biology, and Technology Meet. New York, NY: VCH; 1994. [Google Scholar]
- 67.Evans E, Rawicz W. Entropy-Driven Tension and Bending Elasticity in Condensed-Fluid Membranes. Phys. Rev. Lett. 1990;64(17):2094–2097. doi: 10.1103/PhysRevLett.64.2094. [DOI] [PubMed] [Google Scholar]
- 68.Discher BM, Won YY, Ege DS, Lee JCM, Bates FS, Discher DE, Hammer DA. Polymersomes: Tough vesicles made from diblock copolymers. Science. 1999;284(5417):1143–1146. doi: 10.1126/science.284.5417.1143. [DOI] [PubMed] [Google Scholar]
- 69.Needham D, Nunn RS. Elastic-Deformation and Failure of Lipid Bilayer-Membranes Containing Cholesterol. Biophys. J. 1990;58(4):997–1009. doi: 10.1016/S0006-3495(90)82444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gebhart CL, Kabanov AV. Perspectives on Polymeric Gene Delivery. Journal of Bioactive and Compatible Polymers. 2003;18:147–166. [Google Scholar]
- 71.Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc. Natl. Acad. Sci. U. S. A. 2003;100(7):3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dean DA, Strong DD, Zimmer WE. Nuclear entry of nonviral vectors. Gene Therapy. 2005;12(11):881–890. doi: 10.1038/sj.gt.3302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grosse S, Thevenot G, Aron Y, Duverger E, Abdelkarim M, Roche AC, Monsigny M, Fajac I. In vivo gene delivery in the mouse lung with lactosylated polyethylenimine, questioning the relevance of in vitro experiments. J. Control. Release. 2008;132(2):105–112. doi: 10.1016/j.jconrel.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 74.Melchior F, Gerace L. Mechanisms of Nuclear-Protein Import. Current Opinion in Cell Biology. 1995;7(3):310–318. doi: 10.1016/0955-0674(95)80084-0. [DOI] [PubMed] [Google Scholar]
- 75.Dworetzky SI, Lanford RE, Feldherr CM. The Effects of Variations in the Number and Sequence of Targeting Signals on Nuclear Uptake. J. Cell Biol. 1988;107(4):1279–1287. doi: 10.1083/jcb.107.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brunner S, Furtbauer E, Sauer T, Kursa M, Wagner E. Overcoming the nuclear barrier: Cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Mol. Ther. 2002;5(1):80–86. doi: 10.1006/mthe.2001.0509. [DOI] [PubMed] [Google Scholar]
- 77.Dean DA, Dean BS, Muller S, Smith LC. Sequence requirements for plasmid nuclear import. Experimental Cell Research. 1999;253(2):713–722. doi: 10.1006/excr.1999.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gasiorowski JZ, Dean DA. Mechanisms of nuclear transport and interventions. Advanced Drug Delivery Reviews. 2003;55(6):703–716. doi: 10.1016/s0169-409x(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 79.Itaka K, Harada A, Yamasaki Y, Nakamura K, Kawaguchi H, Kataoka K. In situ single cell observation by fluorescence resonance energy transfer reveals fast intra-cytoplasmic delivery and easy release of plasmid DNA complexed with linear polyethylenimine. J. Gene. Med. 2004;6(1):76–84. doi: 10.1002/jgm.470. [DOI] [PubMed] [Google Scholar]
- 80.Yang J, Chen H, Vlahov IR, Cheng JX, Low PS. Evaluation of disulfide reduction during receptor-mediated endocytosis by using FRET imaging. Proc. Natl. Acad. Sci. U. S. A. 2006;103(37):13872–13877. doi: 10.1073/pnas.0601455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol. Bioeng. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 82.Cohen RN, van der Aa M, Macaraeg N, Lee AP, Szoka FC. Quantification of plasmid DNA copies in the nucleus after lipoplex and polyplex transfection. J. Control. Release. 2009;135(2):166–174. doi: 10.1016/j.jconrel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Honore I, Grosse S, Frison N, Favatier F, Monsigny M, Fajac I. Transcription of plasmid DNA: Influence of plasmid DNA/polyethylenimine complex formation. J. Control. Release. 2005;107(3):537–546. doi: 10.1016/j.jconrel.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 84.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 85.Vogel G. Reprogramming Cells. Science. 2008;322(5909):1766–1767. doi: 10.1126/science.322.5909.1766. [DOI] [PubMed] [Google Scholar]
- 86.Yu JY, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin, Thomson JA., II Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]