Presented data demonstrate that TON2/FASS, which encodes the B′′ regulatory subunit of the protein phosphatase PP2A, regulates microtubule nucleation in interphase Arabidopsis cells. The key finding is that TON2 function regulates the balance of branched versus parallel microtubule-associated microtubule nucleation, thus revealing regulation of nucleation geometry.
Abstract
Organization of microtubules into ordered arrays involves spatial and temporal regulation of microtubule nucleation. Here, we show that acentrosomal microtubule nucleation in plant cells involves a previously unknown regulatory step that determines the geometry of microtubule nucleation. Dynamic imaging of interphase cortical microtubules revealed that the ratio of branching to in-bundle microtubule nucleation on cortical microtubules is regulated by the Arabidopsis thaliana B′′ subunit of protein phosphatase 2A, which is encoded by the TONNEAU2/FASS (TON2) gene. The probability of nucleation from γ-tubulin complexes localized at the cell cortex was not affected by a loss of TON2 function, suggesting a specific role of TON2 in regulating the nucleation geometry. Both loss of TON2 function and ectopic targeting of TON2 to the plasma membrane resulted in defects in cell shape, suggesting the importance of TON2-mediated regulation of the microtubule cytoskeleton in cell morphogenesis. Loss of TON2 function also resulted in an inability for cortical arrays to reorient in response to light stimulus, suggesting an essential role for TON2 and microtubule branching nucleation in reorganization of microtubule arrays. Our data establish TON2 as a regulator of interphase microtubule nucleation and provide experimental evidence for a novel regulatory step in the process of microtubule-dependent nucleation.
INTRODUCTION
Microtubule function in cell division, trafficking, and cell morphogenesis depends on the formation of specialized arrays. Organization of microtubules into arrays is regulated by the activity of distinct intracellular structures known as microtubule organizing centers (MTOCs; Pickett-Heaps, 1969), where new microtubules are assembled.
Well-studied MTOCs in animals and yeast are centralized structures, such as centrosomes and spindle pole bodies, which are responsible for organization of classic astral arrays in interphase and mitotic spindles during cell division. These bodies function in part by recruiting ring-shaped microtubule nucleation complexes, which contain γ-tubulin and GCP (for γ-tubulin complex protein)/Grip (for γ-tubulin ring protein) subunits. Assembled in the cytoplasm, γ-tubulin ring complexes (γ-TURCs) get recruited to centrosomes through interaction with anchoring proteins localized in the pericentriolar matrix (Takahashi et al., 2002; Zimmerman et al., 2004; Delgehyr et al., 2005).
Recruitment of γ-tubulin complexes to sites of microtubule nucleation is an important regulatory step in the formation of microtubule arrays. Studies on spindle formation in animal cells have shown that targeting of γ-TURCs to the centrosome and spindle microtubules is regulated during the cell cycle. The cascade of phosphorylation events triggered by cyclin-dependent kinase 1 (CDK1) and of polo-like kinase 1 (PLK1) has been demonstrated to promote binding of the γ-TURC targeting factor GCP-WD to spindle microtubules, facilitating the acentrosomal nucleation important during spindle formation (Lüders et al., 2006; Zhang et al., 2009; Johmura et al., 2011). In addition to CDK1 and PLK1, several other kinases and phosphatases were found to be involved in the regulation of microtubule nucleation at centrosomes (Fry et al., 1998; Horn et al., 2007; Kim et al., 2007; Sardon et al., 2008), highlighting the important role of protein phosphorylation in microtubule organization.
Despite the absence of the centrosome in plant cells, their microtubules are organized into ordered arrays. Rather than astral arrays, interphase plant cells feature a variety of array architectures with microtubules lying parallel to the plasma membrane at the cell cortex. The organization of these arrays is often associated with a growth pattern and shape of plant cells. For example, in jigsaw puzzle–like leaf pavement cells, the reiterating pattern of cell indentations and cell outgrowths is correlated with regions of high microtubule density spaced between regions with low microtubule density (Fu et al., 2005). Mitotic cells feature transformation of the interphase array to another striking cortical array, the preprophase band (PPB), which forms a hoop at the plane of future cytokinesis at the G2/M transition of the cell cycle (Gunning, 1982; Mineyuki et al., 1988; Granger and Cyr, 2000). As mitosis progresses, the PPB is rearranged into the mitotic spindle, which in turn is transformed at late telophase into the phragmoplast array, consisting of parallel microtubules oriented orthogonally to the cell division plane.
Recent studies have shed new light onto the nature of MTOCs in these acentrosomal arrays of higher plant cells. As in animals and yeast, microtubule nucleation is dependent on γ-TURC complexes. Biochemical isolations have shown that core proteins are assembled into a complex with stoichiometry similar to the γ-TURCs from yeast and animals (Nakamura et al., 2010). γ-TURC components γ-tubulin, GCP2, GCP4, and GCP-WD have been shown to play an essential role in organization of cortical microtubules, the spindle, and phragmoplast arrays in Arabidopsis thaliana (Binarová et al., 2006; Pastuglia et al., 2006; Nakamura and Hashimoto, 2009; Zeng et al., 2009; Kong et al., 2010).
Although the mechanisms targeting the γ-tubulin complexes to nucleation sites in plant cells are not known, imaging of the interphase cortical arrays gave valuable insights into the process generating acentrosomal microtubule arrays. It has been shown that the great majority of interphase cortical microtubules are nucleated from the walls of existing microtubules, with most new microtubules forming a branch at approximately a 40° angle (Murata et al., 2005; Chan et al., 2009; Nakamura et al., 2010). At a lower frequency, microtubules nucleated in parallel to the mother polymer and formed a bundle (Chan et al., 2009; Nakamura et al., 2010). More rarely, microtubules are also observed to nucleate from sites at the cell cortex not associated with existing microtubules (Shaw et al., 2003; Chan et al., 2009; Nakamura et al., 2010). Mechanisms involved in regulation of different nucleation types and their relative frequencies are not known.
Here, we show that microtubule branching nucleation is specifically promoted by the B′′ regulatory subunit of the Arabidopsis protein phosphatase 2A (PP2A), encoded by TONNEAU2/FASS (TON2). We demonstrate the important role of TON2 in cell morphogenesis and reorganization of cortical arrays in response to light. Our results uncover a regulatory step in the process of microtubule nucleation and suggest that the TON2-dependent PP2A signaling pathway plays an important role in the organization of interphase microtubule arrays in part through regulating nucleation geometry.
RESULTS
TON2 Function Is Required for Cell Morphogenesis
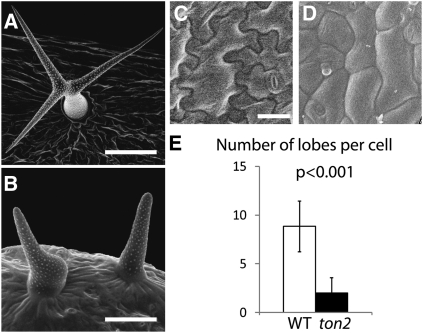
Previous research has established the requirement of the B′′ PP2A subunit TON2 for the formation of the PPB microtubule array, a cytoskeletal structure that plays an essential role in selecting the plane of cell division (Traas et al., 1995; Camilleri at al., 2002), and indicated that ton2 mutants display cell morphogenesis defects (Torres-Ruiz and Jürgens, 1994; Traas et al., 1995). ton2 trichomes fail to develop branches (Torres-Ruiz and Jürgens, 1994) and are often bloated (Figures 1A and 1B), and ton2 pavement cells have a less pronounced intercalating appearance (Torres-Ruiz and Jürgens, 1994) and form fewer lobes (Figures 1C to 1E). These observations indicated that TON2 may regulate the microtubule cytoskeleton both in dividing and in interphase cells, and we set out to investigate the role of TON2 in the interphase cortical microtubule cytoskeleton.
Figure 1.
Cell Morphogenesis Defects in ton2 Epidermal Cells.
(A) to (D) Leaf trichomes ([A] and [B]) and pavement cells ([C] and [D]) in the wild type ([A] and [C]) and in the ton2 mutant ([B] and [D]) imaged with a scanning electron microscope. Bars = 100 μm in (A), 50 μm in (B), and 25 μm in (C) and (D).
(E) Reduced number of lobes in ton2 pavement cells. Average number of lobes was 8.9 ± 2.6 in wild-type (WT) (n = 95) and 2.0 ± 1.5 in ton2 (n = 120) leaf pavement cells. P value calculated by the Student’s t test indicates a significant difference. Error bars show sd.
TON2 Regulates the Geometry of Microtubule Nucleation
As a first step to determine what properties of interphase microtubules may be regulated by TON2, we compared the cortical microtubule array organization in interphase cells of ton2-15 mutant and wild-type plants, both expressing a yellow fluorescent protein (YFP):TUA5 microtubule marker. Cortical arrays of the ton2 hypocotyl cells displayed a 27% reduction in density in the abaxial cell cortex as measured by the area occupied by microtubule label above a threshold level in confocal optical sections (Figures 2A and 2B; P < 0.001, single-tailed Student’s t test, n = 10 cells each genotype; see Methods for image processing details). This reduction in microtubule density was not restricted to hypocotyl cells; pavement cells in 7-d-old cotyledons displayed a 24% reduction in measured density (Figures 2A and 2B; P < 0.01 single-tailed Student’s t test, n = 10 cells each genotype). The reduction in cortical microtubule density suggested that TON2 function may be required for either microtubule stability at the cell cortex or for the formation of new cortical microtubules.
Figure 2.
Comparison of Microtubule Array Density in ton2 and Wild-Type Cells.
(A) Confocal reconstructions of upper hypocotyl cells of dark-grown seedlings expressing YFP:TUA5. Leaf epidermal pavement cells are shown below. Bar = 10 μm. wt, wild type.
(B) The percentage of the image area occupied by signal above a threshold value was measured as a rough estimate of microtubule array density (see Methods). The difference between paired means by cell type is significant at P < 0.01 (two-tailed Student’s t test, n = 10 cell for the wild type and ton2). Bars indicate se.
[See online article for color version of this figure.]
To investigate the mechanism of TON2 regulation, we analyzed microtubule dynamics and microtubule nucleation in the ton2-15 mutant background. We did not find large differences in the rates of polymerization, depolymerization, microtubule dynamicity, and microtubule rescue frequency in the epidermal cells of the ton2 mutant and the wild type (see Supplemental Table 1 online). However, the microtubule catastrophe rate was 2.4 times higher in the ton2 mutant, suggesting that the microtubule plus ends are less stable in the ton2 mutant.
To explore the role of TON2 in microtubule nucleation, we created ton2 heterozygous lines expressing GCP2:green fluorescent protein (GFP), a functional marker for γ-TURCs (Nakamura et al., 2010), together with mCherry:TUB5 to label microtubules (Figures 3A and 3B). Using these transgenic lines, we analyzed microtubule nucleation in a segregating population of seedlings expressing the wild-type or ton2 phenotype.
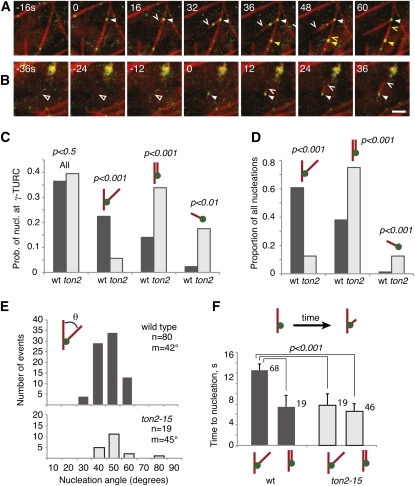
Figure 3.
TON2 Is Required to Regulate the Geometry of Microtubule-Associated Microtubule Nucleation.
Confocal time series at the cortex of ton2 pavement cells expressing 35S:mCherry:TUB5 (red channel) and GCP2:GFP (green channel) constructs.
(A) Recruitment of γ-tubulin complexes (arrowheads) to a cortical microtubule that results in a branching nucleation event (white arrowhead and carat) and a parallel nucleation event (yellow arrowhead and carat). GCP2:GFP also appears on the branching daughter microtubule but without evidence for nucleation from this location.
(B) Recruitment of a labeled nucleation complex (arrowhead) to a cortical site free of detectable microtubules (open arrowhead), resulting in nucleation of a new microtubule (carat). The green label at top right is autofluorescent background. Bars = 2 μm for (A) and (B).
(C) Probability of recruited nucleation (nucl.) complexes giving rise to nucleation events. Observation of stable localization of GCP2:GFP over at least three consecutive acquisition frames (~8 s) was defined as a cortical recruitment event. A total of 245 stabilized GCP2:GFP-labeled nucleation complexes were analyzed for wild-type (wt) cells and 163 for ton2 cells.
(D) Proportion of all observed nucleation events that were branched, parallel, or de novo. The proportion of branching nucleation dropped approximately fourfold in ton2 cells (n = 79 nucleation events in the wild type and 58 events in ton2; 10 cells from 10 plants for each).
(E) Distribution of branching nucleation angles in the wild type and ton2. The distributions are not significantly different from each other by either a Wilcoxon rank sum test or a two-tailed Student’s t test.
(F) Mean time from recruitment of GCP2:GFP-labeled γ-TURC complexes at cortical microtubules to the first detected nucleation as visualized by mCherry-TUA5. Measurements were acquired from 10 ton2-15 mutant cells and 11 wild-type cells. The numbers of measured events are indicated next to the bars.
In wild-type pavement cells, nucleation complexes were recruited to the cortex at a frequency of 57.7 ± 26.1 events per 100 μm2 per hour (n = 245 events in 11 cells), while in ton2 cells, the recruitment frequency was measured at 35.8 ± 23.9 events per 100 μm2 per hour (n = 163 events in 11 cells). Although the estimated recruitment rate in the mutant was nominally lower, the cell-to-cell variance was high, so the observed difference between the wild type and mutant is not significantly different in the observed samples. A similar proportion of recruitment events was observed to colocalize with microtubules in both wild-type and ton2 cells, at 87 and 76%, respectively.
Once a labeled nucleation complex was recruited, the probability of observing nucleation from that complex was nearly identical in wild-type and mutant cells, at 32 and 34%, respectively (total nucleation probability labeled as “All” in Figure 3C). Thus, while this analysis could not exclude that recruitment and nucleation frequency per unit area may be lower in the ton2 mutant, our measurements indicate that ton2-15 did not significantly affect the likelihood of nucleation once a γ-TURC complex was recruited to a stable position at the cell cortex.
By contrast, an examination of nucleation mode revealed a striking difference between wild-type and ton2 mutant cells. Plant cortical microtubules show three different modes of nucleation, with respect to existing cortical microtubules: microtubule-dependent branching nucleation, microtubule-dependent parallel nucleation, and de novo nucleation, where new microtubules form directly at the cell cortex or plasma membrane (see Supplemental Movies 1 to 3 and Supplemental Movie Legends 1 online; Shaw et al., 2003; Murata et al., 2005; Chan et al., 2009; Nakamura et al., 2010). In agreement with previous observations in hypocotyl cells, we found that branching nucleation was the predominant mode in wild-type pavement cells (48 events in 11 cells compared with 30 events of parallel nucleation, a ratio of 1.6). However, in ton2-15 cells, the ratio of branching to parallel nucleation was dramatically different, with only approximately one in eight microtubule-associated nucleation events being branched (Figure 3D, a ratio of 0.16, seven branching and 42 parallel events in 11 cells). If we estimate the probability of a stabilized γ-tubulin complex producing a branching nucleation, the likelihood was observed to drop fourfold compared with wild-type cells, from 22.4 to 5.6% (Figure 3C), a highly significant difference (P < 0.0001, Fisher’s exact test). The likelihood of parallel nucleation rose to make up for most of this drop, from 14.0% in the wild type to 33.9% in ton2. We also found a modest but significant increase in de novo nucleation events in ton2 mutant cells (Figures 3C and 3D). Taken together, these data suggest that TON2 acts as a regulator of microtubule nucleation geometry, increasing the frequency of branching nucleation and decreasing the frequency of parallel nucleation.
A mutation in GCP2, spr3, had been shown to affect the range of branched nucleation angles (Nakamura and Hashimoto, 2009). To ask if the branching angle was also affected in ton2-15, we measured this parameter in 10 mutant and 11 wild-type cells (Figure 3E). The low rate of branching nucleation in mutant cells limited measurement to 19 events, but these had a similar distribution and mean branching angle as measured in the larger number of events in wild-type cells, 44° and 42°, respectively (P > 0.5 by Student’s t test). Thus, the TON2 function appears to affect a switch between two modes of microtubule-associated microtubule nucleation, branched versus parallel, rather than the branching angle per se.
The analysis of nucleation likelihood following γ-TURC recruitment to cortical microtubules (Figure 3C) indicated that the nucleation activity of γ-TURCs was not diminished by ton2-15. To explore the activity state of the nucleation complex further, we asked if there might be a longer delay between the time from complex recruitment to the time when nucleation could be first observed (a measurement limited by the time required for enough labeled dimer to accumulate for confident detection). We found that the mean time to nucleation was not longer in ton2-15; rather, it was actually significantly shorter than in the wild type (see Supplemental Figure 1 online). Analysis by nucleation mode revealed that the difference between the wild type and ton2-15 is attributed only to branching nucleation events (Figure 3F). In the wild type, branching nucleations show a delay from recruitment to nucleation about twice as long on average as for parallel nucleations. The delay times for both nucleation modes in ton2-15 cells were similar as the parallel delay times in the wild type. The number of de novo events was relatively low, yielding large confidence intervals for statistical comparison, but the mean values of delay times in both wild-type and mutant cells were similar to the delay times for branching and parallel nucleation in ton2-15 and for parallel nucleation in the wild type (see Supplemental Figure 2 online). The reason for the longer delays for branched nucleation specifically when TON2 function is at wild-type levels is not known, but these results certainly indicate that loss of TON2 function does not result in longer delay times.
Since the ratio of parallel to branched nucleation was affected in ton2-15, we asked if cortical microtubules may be on balance more bundled in the mutant compared with the wild type. By quantifying the signal intensities of single microtubules and optical bundles (microtubule arrangements that appear as single structures by optical resolution), we did not observe evidence for an increase in bundling state in the ton2-15 mutant in the cortical arrays of growing hypocotyl cells. In fact, higher intensity structures were somewhat more common in the arrays of wild-type cells (see Supplemental Figure 3 online). Upon reflection, this may not be surprising in arrays with strong parallel order, such as those in growing hypocotyl cells, as branched microtubules can also be efficiently incorporated into bundles by interaction with neighboring microtubules driven by polymer treadmilling (Deinum et al., 2011).
Microtubule Reorganization in Response to Light Requires TON2
PPB formation involves reorganization of the interphase cortical microtubules into an equatorial array of parallel microtubules. As the PPB does not form in the ton2 mutant, we tested here if the TON2 function is important for reorganization of the cortical microtubule array in response to an external cue.
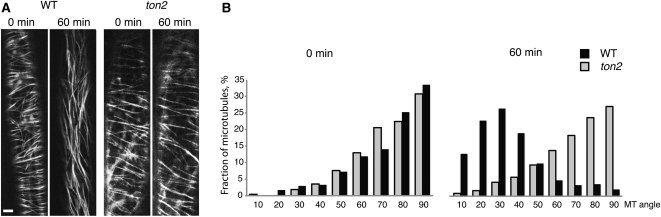
Previous studies showed that cortical microtubule arrays reorganize from transverse to longitudinal orientation after exposure of the dark-grown seedlings to blue light (Ueda and Matsuyama, 2000; Paredez et al., 2006; Sampathkumar et al., 2011). We assayed microtubule orientation in dark-grown wild-type and ton2 hypocotyl cells following light exposure and found that ton2 mutants were unable to reorganize their cortical microtubule arrays in response to light stimulation (Figure 4). Prior to blue light stimulation, the hypocotyl cells of dark-grown seedlings showed an average microtubule orientation of 69° both for the wild type (10 cells and 722 microtubules and bundles) and for the ton2 mutant (10 cells and 624 microtubules and bundles). After the 60-min light exposure, microtubules showed strong reorientation toward a more longitudinal orientation in wild-type cells (average angle of 30°) but remained largely unchanged in the mutant (average angle of 65°). Taken together, our data demonstrate an important role of TON2 in light-inducible microtubule reorganization and suggest that reordering of cortical arrays may depend on the balance between parallel and branching microtubule-bound nucleations regulated by TON2.
Figure 4.
Reorganization of Cortical Microtubule Arrays in Response to Light.
(A) Confocal reconstructions of YFP:TUA5-labeled microtubules acquired before and after exposure to light for 60 min. Bar = 5 μm.
(B) Distribution histogram of observed microtubule angles in relation to the longitudinal cell axis. Microtubules angles were grouped into bins of 10°. Microtubules perpendicular to the cell longitudinal axis were assigned the 90° angle. Measurements were taken from the same cells before and after light treatment. Note a change of microtubule orientation from predominantly transverse to longitudinal in the wild type but not in the ton2 mutant.
Plasma Membrane Targeting of TON2 Changes Cell Shape and Microtubule Organization
The observed microtubule defects in ton2 loss-of-function mutants indicate that TON2 is necessary for the organization of cortical microtubules. Here, we asked if TON2 functions as a regulator and is sufficient to induce changes in microtubule organization.
GFP:TON2 expressed from its own promoter was predominantly found in the cytoplasm, except at the late G2 phase, when we detected it at the cortical region corresponding to the PPB (see Supplemental Figure 4 online). To manipulate TON2 localization and distribution in the cell, we ectopically targeted TON2 to the plasma membrane by creating a fusion construct between the tdTomato fluorescent protein (Shaner et al., 2004), TON2, and the N-terminal part of the Ca2+-dependent protein kinase CPK34 containing the N-terminal myristoylation/palmitoylation signal (Myers et al., 2009). The resulting N-CPK34:tdTomato:TON2 (hereafter PM-TON2) construct was placed under control of the TON2 promoter and introduced into wild-type Arabidopsis plants.
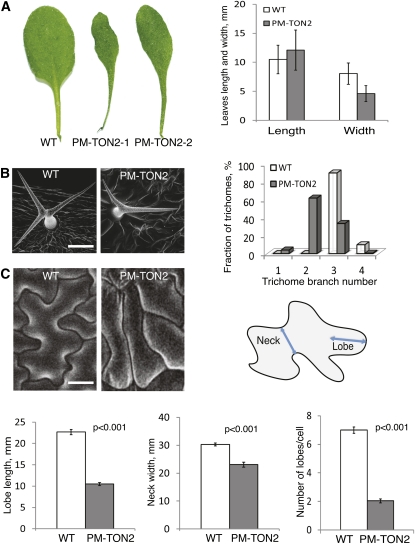
Transgenic PM-TON2 plants showed distinct signal at the plasma membrane (see Supplemental Figure 5 online) and displayed changes in plant morphology, with the prominent narrow leaves phenotype (Figure 5A). Morphometric analysis indicated that the narrow leaf phenotype was due to the reduction in the leaf width with no significant effect on the leaf length (Figure 5A). We did not find similar phenotypical changes in plants overexpressing TON2 under the control of the 35S promoter, indicating that the effect of the PM-TON2 construct is caused by the specific enrichment of TON2 at the plasma membrane.
Figure 5.
Ectopic Targeting of TON2 to the Plasma Membrane Causes Organ and Tissue Morphogenesis Defects.
(A) Leaf shape of the wild type (WT) and two different PM-TON2 plants. Average leaf width was reduced twofold in the PM-TON2 plants (P < 0.001) with no significant change in the leaf length (P = 0.4). The third and fourth mature rosette leaves were measured from 10 wild-type plants and nine PM-TON2 plants in the T1 generation. Error bars show sd.
(B) Scanning electron microscopy images of trichomes on wild-type and PM-TON2 leaves. Trichome branching data were collected from 10 wild-type plants and nine PM-TON2 plants in T1 generation (n = 676 for the wild type; n = 381 for PM-TON2).
(C) Pavement cells expressing PM-TON2 displayed less curved cell outlines with reduced number and length of lobes and necks. Data are presented as means ± se. P values in the diagrams show that differences in lobe length (n = 211 for the wild type; n = 123 for PM-TON2), neck width (n = 210 for the wild type; n = 123 for PM-TON2), and number of lobes (n = 80 for the wild type; n = 100 for PM-TON2) were significant.
Bars = 100 μm in (B) and 30 μm in (C).
[See online article for color version of this figure.]
On the cellular level, mistargeting of TON2 had no effect on the division planes and PPB formation in mitotic cells (see Supplemental Figure 6 online). Also, light-induced microtubule reorganization was not affected by PM-TON2 expression (see Supplemental Figure 7 online). However, interphase cells displayed changes in cell morphogenesis characteristic of microtubule organization defects. Wild-type trichomes of the Arabidopsis Columbia ecotype are stellar in appearance due to formation of three to four branches. Ninety percent of wild-type trichomes formed three branches, and 10% had four. TON2 targeting to the plasma membrane suppressed trichome branching. In the PM-TON2 plants, 4% of the trichomes did not branch, 62% branched one time, forming two branches, 33% had three branches, and 1% had four branches (Figure 5B).
Wild-type leaf pavement cells have a distinct puzzle-shaped morphology with regions of local cell outgrowth (lobes) between cell indentations (necks; Figure 5C). PM-TON2 pavement cells had lost the intercalating appearance of reiterating outgrowth and indentations (Figure 5C). The length of lobes and the width of necks were reduced in the PM-TON2 plants and cells were more elongated with straight outlines.
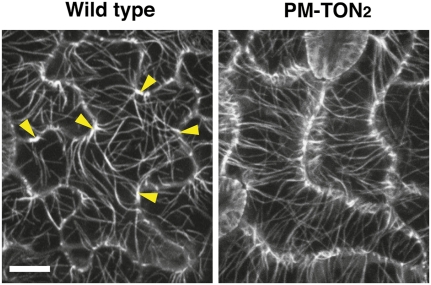
Reduced trichome branching and pavement cell morphogenesis defects have often been associated with microtubule cytoskeleton defects (Burk et al., 2001; Kirik et al., 2002a, 2002b; Fu et al., 2005, 2009). To test if the PM-TON2 construct caused changes in microtubule cytoskeleton organization, we introduced the YFP:TUA5 microtubule marker into PM-TON2 plants. In wild-type pavement cells, microtubules are concentrated at neck regions and are sparse in the lobes (Fu et al., 2005, 2009; Figure 6). By contrast, we found that in the PM-TON2 pavement cells, microtubule distribution along the cell periphery was uniform (Figure 6). In addition, microtubules were aligned in transverse orientation to the cell longitudinal axis. These observations indicate that altering the localization of TON2 by targeting it to the plasma membrane is sufficient to alter cytoskeletal organization and cell shape, suggesting that TON2 functions as a regulator of the cortical microtubule cytoskeleton.
Figure 6.
Plasma Membrane–Targeted TON2 Disrupts Microtubule Organization in Pavement Cells.
Confocal reconstructions of PM-TON2 and wild-type pavement cells expressing YFP:TUA5. Arrowheads point to cell indentations with dense microtubules in a wild-type pavement cell. Note that pavement cells of the PM-TON2 plants have a more uniform distribution of microtubules along the lateral cell outlines. Bar = 10 μm.
[See online article for color version of this figure.]
Microtubule Cytoskeleton Genes MOR1 and TON1A Genetically Interact with TON2
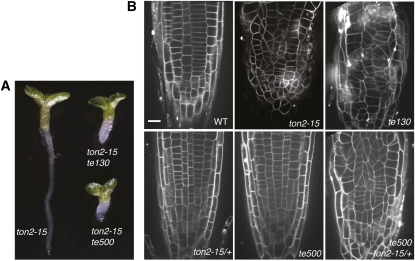
To identify other genes that work together with TON2 in microtubule cytoskeleton organization, we performed a genetic enhancer-suppressor screen of the ton2-15 mutant allele. ton2-15 is a weak ethyl methanesulfonate (EMS)-induced allele resulted from a nonconserved amino acid substitution in the C-terminal end of the TON2. Compared with stronger ton2 alleles, ton2-15 seedlings displayed milder growth phenotypes with less organ swelling (see Supplemental Figure 8 online). We mutagenized seeds of ton2-15 heterozygote plants with EMS and screened 2000 M2 seedlings progeny for phenotypes resembling strong ton2 mutants. Several mutants that enhanced the irregular cell division phenotype of ton2-15 were isolated. Two of the ton2-15 enhancers, te130 and te500 (Figure 7), showed segregation consistent with a single genetic locus and were selected for cloning by chromosome walking.
Figure 7.
te130 and te500 Mutations Enhance the ton2-15 Phenotype.
(A) Comparison of 7-d-old seedlings of the ton2-15 mutant with the te130 ton2-15 and te500 ton2-15 double mutants.
(B) Confocal sections of propidium iodide-stained root tips of the wild type (WT), ton2-15, te130, ton2-15 heterozygote (ton2-15/+), te500, and ton2-15 heterozygote homozygous for the te500 mutation (te500 ton2-13/+). Bar = 10 μm.
[See online article for color version of this figure.]
Double te130 ton2-15 mutants showed a pronounced cell division phenotype resembling strong ton2 alleles (Figure 7). Separated from the ton2-15 background, te130 mutants showed phenotypic traits similar to weak ton2 mutants, including smaller plant size, reduced trichome branching, and misaligned cell division planes in roots (Figure 7). Segregation analysis showed that te130 behaved as a single recessive mutation. Mapping of the recombination breakpoints placed the TE130 gene in an interval on chromosome 2, containing MOR1. MOR1 encodes a homolog of the MAP215 microtubule-associated protein, shown to regulate microtubule cytoskeleton organization and cell division in Arabidopsis (Whittington et al., 2001). Genetic crosses between te130 and mor1 showed no complementation among 57 F1 progeny seedlings, indicating that te130 is a novel mor1 allele. The additive phenotype of the te130 ton2-15 double mutant (Figure 7) combined with the similarity of single te130 and ton2 mutant phenotypes suggest that MOR1 and TON2 may regulate different microtubule properties, both of which are important for microtubule organization.
The te500 mutant enhanced the ton2-15 phenotype but did not show any phenotypic changes in the absence of the ton2-15 mutation. Intriguingly, ton2 heterozygous plants were haploinsufficient in the te500 mutant background and te500 ton2/+ plants showed misaligned division planes in roots (Figure 7). ton2 haploinsufficiency in the te500 mutant background indicates that the TON2-dependent pathway is hypersensitive to the activity of TE500. Conversely, since there are no detectable phenotypic changes in the te500 single mutant, the TE500-dependent pathway is also hypersensitive to TON2 levels.
We mapped TE500 to a 1-centimorgan (cM) interval on chromosome 3. This interval contains a tandem duplication of homologous genes TON1A and TON1B, which were shown previously to be required for PPB formation (Azimzadeh et al., 2008). A mutation disrupting both the TON1A and TON1B genes resulted in a severe cell division phenotype similar to ton2 (Azimzadeh et al., 2008). We sequenced the TON1A and TON1B coding regions in the te500 mutant background and found a premature stop codon in TON1A, terminating TON1A after 127 residues, thus suggesting that te500 is an allele of ton1a. Taken together, the reciprocal sensitivity between TON2 and TE500/TON1 provides genetic evidence suggesting that the two genes act in the same pathway regulating cortical microtubule arrays during cell division and morphogenesis.
DISCUSSION
Earlier studies suggested that the B′′ subunit of the PP2A phosphatase TON2 provides an essential function for the formation of the PPB microtubule array in cells entering mitosis and for array organization in the mitotic cells of roots. The specific microtubule properties regulated by TON2, however, were not known. Using live-cell imaging of interphase cortical arrays, we uncovered a role of TON2 in regulating microtubule nucleation and the dynamic organization of cortical arrays.
TON2 Is a Regulator of Microtubule Nucleation
Observations of microtubule nucleation in plant cell cortical arrays revealed that, initially, cytosolic γ-TURCs are recruited to existing cortical microtubules and to the cell cortex/plasma membrane, after which they initiate nucleation (Murata et al., 2005; Nakamura et al., 2010). Nucleation of new microtubules is geometrically constrained, with most initiating in a branching fashion in a narrow range of angles around 40° to the parent, forming branches, and the remainder growing parallel to the parent microtubule, thus forming a bundle (Figure 3; Murata et al., 2005; Chan et al., 2009; Nakamura et al., 2010). We found that the branching angle was unaffected in the ton2-15 mutant, but the frequency of microtubule branching nucleation from recruited nucleation complexes was reduced four times, whereas the frequency of the parallel nucleation increased 2.4-fold. The total frequency of nucleation from recruited nucleation complexes was unaffected, demonstrating that nucleation competence was not impaired by ton2-15. These results indicate that the balance between these two distinct nucleation geometries is specifically regulated by the TON2-dependent pathway, thus revealing a previously unknown regulatory event in microtubule nucleation.
What is the mechanism of the TON2-dependent regulation of microtubule nucleation geometry? In one scenario, the three-dimensional structure of the microtubule-bound γ-TURC may be important for fixing the orientation of the complex. Recent data provide indications that the conformation of γ-TURC subunits may be important in determining the branching nucleation angle. A missense mutation in GCP2 and downregulation of GCP4 have been shown to change the angle of microtubule branching in Arabidopsis (Nakamura and Hashimoto, 2009; Kong et al., 2010). Of note, human γ-TURC was shown to interact with subunits of the chaperonin containing T-complex protein, suggesting that conformation of the nucleation complex may be actively regulated (Teixidó-Travesa et al., 2010). TON2, as a putative B′′ subunit of the PP2A, may modulate the conformation of the γ-TURC through regulating the phosphorylation status of one or more structural subunits. Alternatively, TON2 may regulate interactions between the γ-TURC and yet to be determined microtubule-associated docking factors required for branching nucleation.
Interestingly, in addition to the change in branching nucleation mode, ton2 mutants showed a significant increase in the frequency of nucleation at cortical sites with no preexisting microtubules. The observed increase in de novo nucleation rate in the ton2 mutant may be an indirect effect caused by a higher concentration of free tubulin dimers in the ton2 cells due to a reduction in the total amount of polymerized tubulin. It is also possible that a TON2-dependent regulatory pathway may directly suppress de novo microtubule nucleation.
MT Reorganization in Response to Light
Experimental and theoretical studies suggest that organization of plant cortical microtubules into ordered arrays depends on intrinsic microtubule behaviors, such as polymer treadmilling, encounters between microtubules with angle-dependent outcomes, microtubule-dependent microtubule nucleation, and alteration of essential properties, such as plus-end stability in specific cell domains (Shaw et al., 2003; Dixit and Cyr, 2004; Baulin et al., 2007; Kawamura and Wasteneys, 2008; Allard et al., 2010; Eren et al., 2010; Hawkins et al., 2010; Nakamura et al., 2010; Shi and Ma, 2010; Tindemans et al., 2010; Ambrose et al., 2011; Deinum et al., 2011). While clues to how arrays might attain parallel organization are arising from these studies, outstanding questions are how particular array orientations are obtained, how arrays are dynamically reorganized by developmental and environmental cues, and what microtubule properties and behaviors are important for array reorganization.
Blue light stimulates a dramatic reorientation of transverse cortical arrays in dark-grown hypocotyl cells to a longitudinal orientation (Ueda and Matsuyama, 2000; Paredez et al., 2006). During this transition, the old organization needs to be dismantled and the new organization built. It has been speculated that the angle of nucleation relative to existing microtubules could play a role in array reorganization, as parallel nucleation would tend to maintain the order of the existing array, while branching nucleation would produce new microtubules at a wider range of angles, supporting transitions in array organization (Ehrhardt, 2008). A recent theoretical study, where different modes of microtubule-dependent nucleation were simulated, has shown that the proportion of parallel microtubule nucleation can play a role in promoting ordering of microtubule arrays in simulations that also included a background of isotropic de novo nucleation (Deinum et al., 2011).
Strong reduction in the frequency of microtubule branching nucleation concomitant with the relative increase in parallel nucleations was a prominent microtubule cytoskeleton defect found in the ton2-15 mutant (Figure 3), providing a valuable opportunity to address the significance of nucleation modes for reorganization of microtubule arrays. Transverse microtubule arrays in ton2-15 mutants did not reorient in response to light exposure, indicating that TON2 function is required for efficient array reorientation and thus suggesting that the balance of nucleation modes may be important for dynamic reorganization. We suggest that TON2 functions in microtubule reorientation by promoting a higher ratio of branching to parallel nucleation, thus allowing a greater proportion of polymers to be generated at discordant angles to promote the transition of array order. It will be important to determine whether the TON2 activity in regulating the balance between different nucleation types is regulated by light signaling.
Plasma Membrane Targeting of TON2 Induces Changes in Cell Shape and Microtubule Cytoskeleton Organization
Expressed from the native TON2 promoter, GFP:TON2 was predominantly found in the cytoplasm, except at late G2 when we observed accumulation at the cortical region corresponding to the PPB. The specific enrichment of GFP:TON2 at the PPB site is consistent with the immunolocalization data for the TON2 homologs genes ATERNATIVE DISCORDIA1 and DISCORDIA1 from maize (Zea mays; Wright et al., 2009). Targeting of TON2 to the plasma membrane by the PM-TON2 construct disrupted this spatial and temporal regulation of TON2 localization and caused changes in microtubule cytoskeleton organization and cell shape. Microtubules in leaf pavement cells had strong transverse ordering and cells failed to develop their characteristic puzzle piece morphology, suggesting that TON2 functions as a regulator of microtubule cytoskeleton organization.
TON2 may regulate the phosphorylation status of cell cortex/plasma membrane proteins, important for microtubule cytoskeleton organization and cell morphogenesis. Of note, overexpression of the ROP GTPase pathway components, ROP6 (for Rho of plants 6) and RIC1 (for Rop-interacting CRIB-containing protein 1), caused changes in pavement cell morphology and microtubule organization that strongly resemble the PM-TON2 phenotype (Fu et al., 2009). Plasma membrane–localized ROP6 is suggested to activate the RIC1 effector by promoting its localization to cortical microtubules in specific domains, thus increasing microtubule density in pavement cell indentations. We can envision that TON2 and ROP6/RIC1 may function in the same pathway, and ectopic localization of TON2 at the plasma membrane may ectopically activate the ROP6/RIC1 pathway. This scenario would predict that the ROP GTPase pathway is also involved in the regulation of microtubule nucleation. Alternatively, TON2 and ROP6/RIC1 may work in separate pathways regulating different mechanisms of microtubule organization. It will be important to test in future studies the involvement of TON2 and ROP GTPase pathways in the hypothesized cortical microtubule organization mechanisms directed by regional regulation of microtubule behaviors, such as nucleation rate or stability (Dixit et al., 2006; Ehrhardt and Shaw, 2006; Ambrose et al., 2011).
Role of TON2 in Cortical Array Organization
In this study, we document (1) a significant defect in the ability of cortical microtubule arrays to dynamically reorient in response to light, (2) that ectopic targeting of TON2 can drive a dramatic reorganization of cortical arrays in pavement cells, and (3) that cortical arrays in both hypocotyl and pavement cells in the ton2-15 mutant are less dense than in the wild type. In the original description of FASS/TON2 (Traas et al., 1995), cortical arrays in root meristem cells were observed to be highly disordered, a phenotype that we did not observe in the hypocotyl or cotyledon. The lines we used to mark microtubules did not yield sufficient signal quality to clearly discern microtubule organization in the root meristem, so we could not verify this observation. In our analyses of microtubule nucleation and dynamics, we found that microtubules in ton2-15 had higher catastrophe rates and that the proportion of parallel to branching nucleation was dramatically altered. We hypothesize that the altered mode of nucleation geometry could be a principle cause of the reorientation defect, whereas increased catastrophe rates could play a significant role in the observed effects on array density. As cortical array ordering is hypothesized to be the result of integrating many cortical microtubule behaviors and interactions, the relative importance of these component activities for creating specific array organizations could vary from cell type to cell type. Thus, a loss of TON2 function could lead to disorganization in some arrays but not in others. It is also important to point out that the ton2-15 allele used in this study, although resulting in severe stunting of plant growth, sterility, and a complete lack of preprophase bands, is not null in activity. It is possible that a complete loss of function could result in more severe switches in nucleation mode or larger changes in other microtubule properties, such as nucleation complex recruitment rate or polymerization catastrophe rate.
Genetic Interaction between TON2 and TON1A
So far, four genes have been identified that are required for PPB formation in Arabidopsis: TON2, TON1A, TON1B, and MOR1 (Traas et al., 1995; Camilleri et al., 2002; Kawamura et al., 2006; Azimzadeh et al., 2008). Two of them, TON1A and MOR1, were identified here as genetic enhancers of the weak ton2-15 allele.
Haploinsufficiency of TON2 in the te500/ton1a mutant background, together with the similar mutant phenotypes of ton2 and ton1a ton1b double mutants, suggest that TON2 and TON1 function in the same process. TON1A and TON1B were reported to be phosphorylated (Benschop et al., 2007; Sugiyama et al., 2008), making them possible targets for TON2 mediated dephosphorylation.
Based on homology to human centrosomal proteins FOP and OFD1 and interaction with an Arabidopsis centrin homolog, CEN1, it was suggested that TON1A is involved in centrosome-related functions important for the formation of microtubule arrays at the plant cell cortex (Azimzadeh et al., 2008). Observed microtubule nucleation defects in the ton2 mutant, together with the similarity of TON1A to centrosomal proteins and the genetic interaction between ton1a and ton2 mutants, suggest that the TON2 and TON1 proteins are the founding components of a pathway that regulates microtubule nucleation geometry and, possibly, recruitment or number of nucleation complexes. Obvious future experiments are to determine if ton1 mutants have similar microtubule nucleation defects and to identify the upstream and downstream targets of both proteins.
Conclusions and Perspectives
Our data reveal the existence of a regulatory pathway that regulates the geometry of the microtubule nucleation. The key finding presented here is that TON2 plays a role in determining the geometry of the microtubule nucleation at the cortex but does not affect the probability of nucleation. The TON2-dependent signal transduction pathway offers an opportunity to investigate how different types of microtubule nucleation are regulated and how they contribute to formation of different microtubule arrays and their reorganization. Although plant cells do not have centrosomes, many homologs of the centrosomal genes important for microtubule nucleation can be found in plant genomes. Addressing functions of the plant genes homologous to animal centrosomal proteins may provide valuable insights into the evolutionarily conserved mechanisms important for the formation of noncentrosomal microtubule arrays.
METHODS
Plant Material, Growth Conditions, and Position-Based Cloning of ton2 Enhancer Mutants
All plants were grown at 22 to 25°C in light cabinets or in the greenhouse in a 16-h/8-h day/night light period. Columbia-0 was used as the wild type in this study. Agrobacterium tumefaciens strains GV3101 and pMK90RK were used to transform Arabidopsis thaliana plants.
The ton2-15 mutant was obtained from the ABRC (CS84613 EMS line), and a missense mutation in the last exon of TON2 was identified by sequencing. Two thousand seeds from ton2-15 heterozygous plants were mutagenized in 0.3% EMS solution. Staining with propidium iodide was used to identify mutants which enhanced the irregular cell division phenotype of the ton2-15. Positional mapping of the ton2 enhancer mutants te130 and te500 was done in the F2 generation from crosses with Landsberg erecta ecotype as described by Lukowitz et al. (2000). The position of TE130 was found to be in the interval between the markers M323 (67.97 cM) and VE01 (69.14 cM) on chromosome 2. TE500 was mapped between the AFC1 (73.97 cM) and BGL1 (75.23 cM) markers on chromosome 3.
Molecular Cloning
TON2pro:GFP:TON2, TON2pro:tdTOMATO:TON2, and TON2pro:PM-tdTOMATO:TON2 constructs were created by modifying the pMDC43 vectors (Curtis and Grossniklaus, 2003). Using PmeI and Acc65I sites, the 35S promoter of pMDC43 was exchanged with the 1991-bp fragment of the 5′ regulatory region before the TON2 start codon. The full-length TON2 coding region was PCR amplified from genomic DNA using FAS1-S1 (5′-ACAATGTATAGCGGATCTAGCGATG-3′) and TON-AS-noSTOP (5′-CTGAGACTCTTCCTCAGGTGGTTC-3′) and inserted into the modified pMDC43 vectors using Invitrogen Gateway cloning technology. The mCherry:TUB5 microtubule marker construct was created by introducing the Arabidopsis tubulin beta-5 coding sequence into the modified pEarleyGate104 vector, in which YFP was substituted for mCherry (Earley et al., 2006). For plasma membrane targeting of TON2, we fused TON2 with of the N-terminal fragment (amino acids 1 to 76) of CPK34, which contains a myristoylation/palmitoylation site (Myers et al., 2009). The corresponding DNA fragment was inserted into the Acc65I site of TON2pro:tdTOMATO:TON2, resulting in the TON2pro:PM-tdTOMATO:TON2 (PM-TON2) construct.
Microscopy and Image Analysis
For microscopy, seeds were germinated on 0.5× Murashige and Skoog agar at 22°C.
Imaging was performed on a spinning-disk confocal microscope setup using a Leica DM6000 inverted microscope, Leica ×100 oil objective, Yokogawa CSUX scanner, and Photometrics Evolve 512 camera. GFP/YFP and tdTomato were excited at 488 and 561 nm, respectively. Images were acquired using Slidebook 5 software (Intelligent Imaging Innovations). Image processing was performed with ImageJ (W. Rasband, National Institutes of Health, Bethesda, MD), Imaris (Bitplane Scientific Software), and Photoshop (Adobe Systems) software packages.
For identification and tracking of GCP2:GFP-labeled γ-TURCs, time series images of 200 to 300 s in length (4-s intervals) were registered using ImageJ plugins: StackReg45 and MultiStackReg (http://www.stanford.edu/∼bbusse/work/downloads.html). Images were background subtracted using the rolling ball function of ImageJ with a 15-pixel diameter. Subregions in each image series were selected for further analysis based on the quality of focus in both channels throughout the selected region (the cells have complex three-dimensional geometry and the images are optical sections). The cropped images were then loaded into Imaris, and the Spots function was employed to identify labeled puncta in the GCP2 channel, using a 5-pixel target diameter. The quality score was adjusted to detect the labeled complexes as verified by eye throughout a given time series. Identified spots were tracked using the autoregressive motion algorithm with a 2-pixel max distance, a 2-frame max gap size, and a requirement for at least a 3-frame track duration. All identified spots/tracks were then analyzed for microtubule colocalization and nucleation by examining the microtubule channel. Spots present in frame 1 and at the extreme margin of the analyzed region were discarded.
To systematically identify nucleation events, we first examined the GCP2 channel for evidence of nucleation complex recruitment to the cell cortex, an event that we defined as GCP2:GFP label being detected at the same location in the confocal section for at least three consecutive camera acquisitions (~8 s). We then examined each of these recruitment events in the microtubule channel for evidence of new microtubule nucleation.
Microtubule array density in wild-type and mutant cells was estimated as follows. Confocal optical sections were processed in ImageJ by background subtracting with a 15-pixel diameter rolling ball filter, passing through a Fourier band-pass filter with an upper limit of 4 pixels and a lower limit of 3 pixels, then blindly thresholding using the Otsu algorithm. The percentage of the analyzed area occupied by pixels above threshold was used as the estimator of array density. This value represents optical coverage in the image and does not take into account true microtubule dimensions nor does it attempt to account for the state of microtubule bundling (i.e., a bundle of two microtubules would yield about the same value as a single microtubule if the signal for a single polymer was well above background).
For the microtubule reorientation assay, 3-d-old dark-grown wild-type and ton2 seedlings were exposed to light using the 100-W incandescent light source of the microscope at full power, aligned for Kohler illumination. To ensure that undamaged hypocotyl cells were imaged, cells with transverse and dynamic microtubules were selected for the imaging.
To ask if the degree of bundling might differ between wild-type and ton2-15 mutant cells, the signal intensities of microtubule structures were sampled as follows. Longitudinal transects were drawn on images of cortical arrays in hypocotyl cells (expressing 35S:mCherry:TUA5), and the point of intersection with microtubule structures was then measured for cross sectional brightness by drawing a second 5-pixel-wide line at a right angle to the microtubule axis at each point of intersection using the segmented line toll in ImageJ. Plot profile was then used to collect an intensity profile. The peaks corresponding to microtubules were identified by aid of a derivative filter on the profile values and cross-checked by visual inspection. The image was inspected for microtubules that were most likely singles based on inspection of polymer dynamics in an image time series bracketing the measured frame. After background subtraction, the values from the intensity profile corresponding to these reference peaks were averaged and used to normalize the values for all peaks. This method cannot distinguish between true microtubule bundles and polymers that may simply be more closely spaced than can be resolved optically.
Microtubule dynamics measurements were performed using ImageJ tools as described (DeBolt et al., 2007), and a nonparametric Mann-Whitney test was used to calculate P values. To measure frequencies of rescue and catastrophe events, we only measured those microtubules that did not touch other microtubules during their polymerization. This was done to eliminate the effect of microtubule density (which is different in the wild type and ton2) on the microtubule plus-end behavior.
For morphometric measurements of pavement cells, lobes were defined as cellular outgrowths if they were coupled with corresponding indentations in neighboring cells. Fully developed rosette leaves of the second pair were used to image pavement cells using a scanning electron microscope (FEI Quanta 450). Images were analyzed with ImageJ.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT1G20010 (TUB5), AT5G18580 (FASS/TON2), and AT5G19360 (CPK34).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Distribution of the Time Intervals between γ-TURC Recruitment to Cortical Microtubules and the Following Nucleation Event.
Supplemental Figure 2. Time from Nucleation Complex Recruitment to Detection of Nucleation, Including de Novo Nucleation Events.
Supplemental Figure 3. Distribution of Microtubule Structure Intensities.
Supplemental Figure 4. GFP:TON2 Localizes the PPB and in the cytoplasm.
Supplemental Figure 5. N-Terminal Myristoylation/Palmitoylation Signal of Calcium-Dependent Protein Kinase CPK34 Localizes TON2 Protein to the Plasma Membrane.
Supplemental Figure 6. Plasma Membrane–Targeted TON2 Does Not Interfere with Division Plane Selection and PPB Formation.
Supplemental Figure 7. Plasma Membrane–Targeted TON2 Does Not Interfere with Light-Inducible Reorganization of Cortical Microtubule Arrays.
Supplemental Figure 8. Seedling Phenotype of ton2 Mutants.
Supplemental Table 1. In Vivo Measurements of Microtubule Growth Phases and Transition Frequencies in Hypocotyl Cells of Light-Grown 7-d-Old Seedlings.
Supplemental Movie 1. Microtubule Branching Nucleation Visualized with Cherry:TUB5 (Red Channel) and GCP2:GFP (Green Channel) Fluorescent Markers.
Supplemental Movie 2. Microtubule Parallel Nucleation.
Supplemental Movie 3. Microtubule de Novo Nucleation.
Supplemental Movie Legends 1. Legends for Supplemental Movies 1, 2, and 3.
Acknowledgments
We thank Jeffrey Harper for sharing a plasmid containing the calcium-dependent protein kinase CPK34 cDNA and Masayoshi Nakamura and Takashi Hashimoto for GCP2:GFP seeds. We thank the ABRC for providing the ton2-15 mutant. Work with the scanning electron microscope was supported by National Science Foundation Instrument Grant DBI-0923448. The work was also supported by the Illinois State University (A.K. and V.K.) and by National Science Foundation Grant 0524355 (D.W.E.).
AUTHOR CONTRIBUTIONS
A.K., D.W.E., and V.K. designed the research, analyzed data, and wrote the article. A.K. and V.K. performed the research.
References
- Allard J.F., Wasteneys G.O., Cytrynbaum E.N. (2010). Mechanisms of self-organization of cortical microtubules in plants revealed by computational simulations. Mol. Biol. Cell 21: 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C., Allard J.F., Cytrynbaum E.N., Wasteneys G.O. (2011). A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat. Commun. 2: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J., Nacry P., Christodoulidou A., Drevensek S., Camilleri C., Amiour N., Parcy F., Pastuglia M., Bouchez D. (2008). Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 20: 2146–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulin V.A., Marques C.M., Thalmann F. (2007). Collision induced spatial organization of microtubules. Biophys. Chem. 128: 231–244 [DOI] [PubMed] [Google Scholar]
- Benschop J.J., Mohammed S., O’Flaherty M., Heck A.J., Slijper M., Menke F.L. (2007). Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Binarová P., Cenklová V., Procházková J., Doskocilová A., Volc J., Vrlík M., Bögre L. (2006). Gamma-tubulin is essential for acentrosomal microtubule nucleation and coordination of late mitotic events in Arabidopsis. Plant Cell 18: 1199–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk D.H., Liu B., Zhong R., Morrison W.H., Ye Z.H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827 [PMC free article] [PubMed] [Google Scholar]
- Camilleri C., Azimzadeh J., Pastuglia M., Bellini C., Grandjean O., Bouchez D. (2002). The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Sambade A., Calder G., Lloyd C. (2009). Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell 21: 2298–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S., Gutierrez R., Ehrhardt D.W., Melo C.V., Ross L., Cutler S.R., Somerville C., Bonetta D. (2007). Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc. Natl. Acad. Sci. USA 104: 5854–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinum E.E., Tindemans S.H., Mulder B.M. (2011). Taking directions: The role of microtubule-bound nucleation in the self-organization of the plant cortical array. Phys. Biol. 8: 056002. [DOI] [PubMed] [Google Scholar]
- Delgehyr N., Sillibourne J., Bornens M. (2005). Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118: 1565–1575 [DOI] [PubMed] [Google Scholar]
- Dixit R., Chang E., Cyr R. (2006). Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Mol. Biol. Cell 17: 1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., Cyr R. (2004). Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell 16: 3274–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W. (2008). Straighten up and fly right: Microtubule dynamics and organization of non-centrosomal arrays in higher plants. Curr. Opin. Cell Biol. 20: 107–116 [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W., Shaw S.L. (2006). Microtubule dynamics and organization in the plant cortical array. Annu. Rev. Plant Biol. 57: 859–875 [DOI] [PubMed] [Google Scholar]
- Eren E.C., Dixit R., Gautam N. (2010). A three-dimensional computer simulation model reveals the mechanisms for self-organization of plant cortical microtubules into oblique arrays. Mol. Biol. Cell 21: 2674–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Meraldi P., Nigg E.A. (1998). A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 17: 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Gu Y., Zheng Z., Wasteneys G., Yang Z. (2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Fu Y., Xu T., Zhu L., Wen M., Yang Z. (2009). A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr. Biol. 19: 1827–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger C.L., Cyr R.J. (2000). Microtubule reorganization in tobacco BY-2 cells stably expressing GFP-MBD. Planta 210: 502–509 [DOI] [PubMed] [Google Scholar]
- Gunning B.E.S. (1982). The cytokinetic apparatus: Its development and spatial regulation. In The Cytoskeleton in Plant Growth and Development, C.W. Lloyd, ed (New York: Academic Press), pp. 229–292
- Hawkins R.J., Tindemans S.H., Mulder B.M. (2010). Model for the orientational ordering of the plant microtubule cortical array. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 82: 011911. [DOI] [PubMed] [Google Scholar]
- Horn V., Thélu J., Garcia A., Albigès-Rizo C., Block M.R., Viallet J. (2007). Functional interaction of Aurora-A and PP2A during mitosis. Mol. Biol. Cell 18: 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johmura Y., Soung N.K., Park J.E., Yu L.R., Zhou M., Bang J.K., Kim B.Y., Veenstra T.D., Erikson R.L., Lee K.S. (2011). Regulation of microtubule-based microtubule nucleation by mammalian polo-like kinase 1. Proc. Natl. Acad. Sci. USA 108: 11446–11451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura E., Himmelspach R., Rashbrooke M.C., Whittington A.T., Gale K.R., Collings D.A., Wasteneys G.O. (2006). MICROTUBULE ORGANIZATION 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiol. 140: 102–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura E., Wasteneys G.O. (2008). MOR1, the Arabidopsis thaliana homologue of Xenopus MAP215, promotes rapid growth and shrinkage, and suppresses the pausing of microtubules in vivo. J. Cell Sci. 121: 4114–4123 [DOI] [PubMed] [Google Scholar]
- Kim S., Lee K., Rhee K. (2007). NEK7 is a centrosomal kinase critical for microtubule nucleation. Biochem. Biophys. Res. Commun. 360: 56–62 [DOI] [PubMed] [Google Scholar]
- Kirik V., Grini P.E., Mathur J., Klinkhammer I., Adler K., Bechtold N., Herzog M., Bonneville J.M., Hülskamp M. (2002a). The Arabidopsis TUBULIN-FOLDING COFACTOR A gene is involved in the control of the alpha/beta-tubulin monomer balance. Plant Cell 14: 2265–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Mathur J., Grini P.E., Klinkhammer I., Adler K., Bechtold N., Herzog M., Bonneville J.M., Hulskamp M. (2002b). Functional analysis of the tubulin-folding cofactor C in Arabidopsis thaliana. Current Biol. 12: 1519–1523 [DOI] [PubMed] [Google Scholar]
- Kong Z., Hotta T., Lee Y.R., Horio T., Liu B. (2010). The gamma-tubulin complex protein GCP4 is required for organizing functional microtubule arrays in Arabidopsis thaliana. Plant Cell 22: 191–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Patel U.K., Stearns T. (2006). GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8: 137–147 [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Gillmor C.S., Scheible W.R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineyuki Y., Wick S.M., Gunning B.E.S. (1988). Preprophase bands of microtubules and the cell cycle: Kinetics and experimental uncoupling of their formation from the nuclear cycle in onion root tip cells. Planta 174: 518–526 [DOI] [PubMed] [Google Scholar]
- Murata T., Sonobe S., Baskin T.I., Hyodo S., Hasezawa S., Nagata T., Horio T., Hasebe M. (2005). Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 7: 961–968 [DOI] [PubMed] [Google Scholar]
- Myers C., Romanowsky S.M., Barron Y.D., Garg S., Azuse C.L., Curran A., Davis R.M., Hatton J., Harmon A.C., Harper J.F. (2009). Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 59: 528–539 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Ehrhardt D.W., Hashimoto T. (2010). Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12: 1064–1070 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Hashimoto T. (2009). A mutation in the Arabidopsis gamma-tubulin-containing complex causes helical growth and abnormal microtubule branching. J. Cell Sci. 122: 2208–2217 [DOI] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Pastuglia M., Azimzadeh J., Goussot M., Camilleri C., Belcram K., Evrard J.L., Schmit A.C., Guerche P., Bouchez D. (2006). Gamma-tubulin is essential for microtubule organization and development in Arabidopsis. Plant Cell 18: 1412–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps J.D. (1969). The evolution of the mitotic apparatus: An attempt at comparative ultrastructural cytology in dividing plant cells. Cytobios 3: 2302–2313 [Google Scholar]
- Sampathkumar A., Lindeboom J.J., Debolt S., Gutierrez R., Ehrhardt D.W., Ketelaar T., Persson S. (2011). Live cell imaging reveals structural associations between the actin and microtubule cytoskeleton in Arabidopsis. Plant Cell 23: 2302–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardon T., Peset I., Petrova B., Vernos I. (2008). Dissecting the role of Aurora A during spindle assembly. EMBO J. 27: 2567–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N.G., Palmer A.E., Tsien R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Shaw S.L., Kamyar R., Ehrhardt D.W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Shi X.Q., Ma Y.Q. (2010). Understanding phase behavior of plant cell cortex microtubule organization. Proc. Natl. Acad. Sci. USA 107: 11709–11714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N., Nakagami H., Mochida K., Daudi A., Tomita M., Shirasu K., Ishihama Y. (2008). Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Yamagiwa A., Nishimura T., Mukai H., Ono Y. (2002). Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol. Biol. Cell 13: 3235–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixidó-Travesa N., Villén J., Lacasa C., Bertran M.T., Archinti M., Gygi S.P., Caelles C., Roig J., Lüders J. (2010). The gammaTuRC revisited: A comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell 21: 3963–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindemans S.H., Hawkins R.J., Mulder B.M. (2010). Survival of the aligned: Ordering of the plant cortical microtubule array. Phys. Rev. Lett. 104: 058103. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz R.A., Jürgens G. (1994). Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120: 2967–2978 [DOI] [PubMed] [Google Scholar]
- Traas J., Bellini C., Nacry P., Kronenberger J., Bouchez D., Caboche M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375: 676–677 [Google Scholar]
- Ueda K., Matsuyama T. (2000). Rearrangement of cortical microtubules from transverse to oblique or longitudinal in living cells of transgenic Arabidopsis thaliana. Protoplasma 213: 28–38 [Google Scholar]
- Whittington A.T., Vugrek O., Wei K.J., Hasenbein N.G., Sugimoto K., Rashbrooke M.C., Wasteneys G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613 [DOI] [PubMed] [Google Scholar]
- Wright A.J., Gallagher K., Smith L.G. (2009). discordia1 and alternative discordia1 function redundantly at the cortical division site to promote preprophase band formation and orient division planes in maize. Plant Cell 21: 234–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C.J.T., Lee Y.R.J., Liu B. (2009). The WD40 repeat protein NEDD1 functions in microtubule organization during cell division in Arabidopsis thaliana. Plant Cell 21: 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.Y., Chen Q., Feng J., Hou J.J., Yang F.Q., Liu J.J., Jiang Q., Zhang C.M. (2009). Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gammaTuRC to the centrosome. J. Cell Sci. 122: 2240–2251 [DOI] [PubMed] [Google Scholar]
- Zimmerman W.C., Sillibourne J., Rosa J., Doxsey S.J. (2004). Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15: 3642–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]