Benzoxazinoids are constitutive protective compounds found in many grasses and in single dicot species. Glucosylation prevents autotoxicity, and upon attack by herbivores and pathogens, the reactive aglucon is produced. Isolation of the benzoxazinoid-specific UDP-glucosyltransferase and β-glucosidase from larkspur reveals convergent evolution of these functions in grasses and in the dicot species.
Abstract
Benzoxazinoids represent preformed protective and allelophatic compounds that are found in a multitude of species of the family Poaceae (Gramineae) and occur sporadically in single species of phylogenetically unrelated dicots. Stabilization by glucosylation and activation by hydrolysis is essential for the function of these plant defense compounds. We isolated and functionally characterized from the dicot larkspur (Consolida orientalis) the benzoxazinoid-specific UDP-glucosyltransferase and β-glucosidase that catalyze the enzymatic functions required to avoid autotoxicity and allow activation upon challenge by herbivore and pathogen attack. A phylogenetic comparison of these enzymes with their counterparts in the grasses indicates convergent evolution by repeated recruitment from homologous but not orthologous genes. The data reveal a great evolutionary flexibility in recruitment of these essential functions of secondary plant metabolism.
INTRODUCTION
Plants produce a vast array of secondary metabolites. Many of these compounds are toxic to plants and other organisms and are involved in defense against microbial attack or herbivore predation. Defense compounds may be synthesized de novo in response to pathogen attack (e.g., phytoalexins that are synthesized in plant–microbe interactions). Alternatively, protective compounds may be produced without challenge during normal development and are then termed constitutive or preformed defenses. To mitigate autotoxicity, these preformed compounds are frequently stored in special plant organs or organelles and are often inactivated by conjugation. Biosynthesis of preformed defenses can be distinguished by the following steps: (1) the branch point reaction committing the flux from primary to secondary metabolism, (2) biochemical modification of the initial intermediate leading to a biological active (toxic) secondary metabolite, (3) detoxification (stabilization) and storage of this metabolite, and (4) activation of the compound upon pathogen or herbivore attack. Individual classes of constitutive defenses occur with quite different frequencies within the plant kingdom. Cyanogenic glycosides are widespread and are present in more than 2650 plants from ferns to angiosperms (Bak et al., 2006). Contrarily, avenacins are restricted to the genus Avena (Hostettmann and Marston, 1995), and glucosinolates are found in the order Capparales and as an exception in the genus Drypetes (Rodman et al., 1998). Furthermore, the biosynthesis of specific preformed defense compounds can be characteristic for one plant family but additionally occur sporadically in single species of phylogenetically unrelated families. Such a distribution pattern is described for pyrrolizidine alkaloids (Hartmann and Witte, 1995) and benzoxazinoids (Sicker et al., 2000).
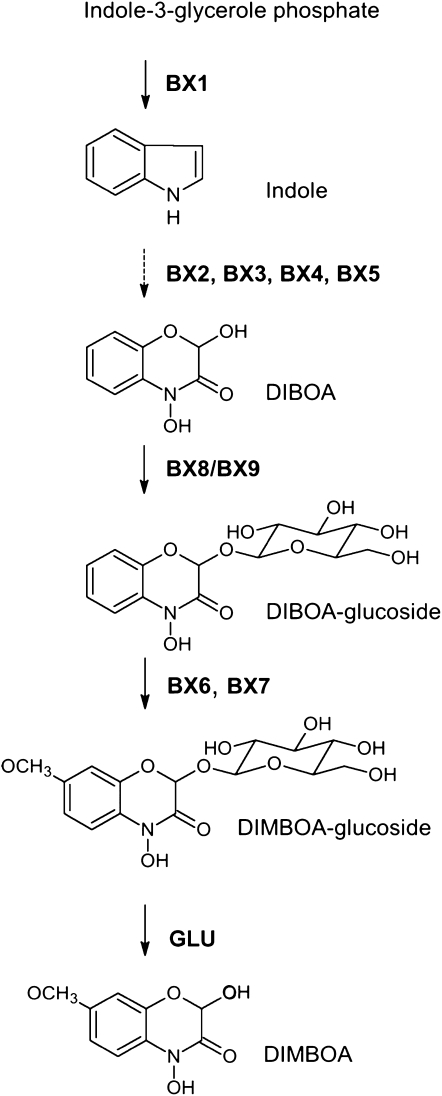
Benzoxazinoids represent protective and allelophatic defense compounds that are found in a multitude of species of the family Poaceae (Gramineae) of the monocot plants, including the major agricultural crops maize (Zea mays), wheat (Triticum aestivum), and rye (Secale cereale). Benzoxazinoid biosynthesis is fully elucidated in maize (Frey et al., 1997, 2003; von Rad et al., 2001; Jonczyk et al., 2008; Figure 1) and is characterized in part for wheat (Nomura et al., 2002, 2003, 2005; Sue et al., 2011), diploid Triticales (Nomura et al., 2007), and wild barley (Hordeum lechleri; Grün et al., 2005). DIBOA [2,4-dihydroxy-2H-1,4-benzoxazin-3(4H)-one] and its C-7-methoxy derivative DIMBOA are the predominant benzoxazinoids (Niemeyer, 1988). DIMBOA is the major benzoxazinoid in maize and wheat; DIBOA is prevalent in rye aboveground tissue, while DIMBOA is found in the root (Rice et al., 2005). Most of the benzoxazinoid-containing dicot species, including larkspur (Consolida orientalis), possess DIBOA.
Figure 1.
Benzoxazinoid Biosynthesis in Maize.
GDIMBOA is stored in the vacuole. Hydrolysis of the glucoside by Zm-GLU generates the reactive aglucone.
In the grasses, a series of five genes is sufficient to encode the enzymes for DIBOA biosynthesis (Frey et al., 1997; Nomura et al., 2002; Grün et al., 2005; Figure 1). The committing enzyme of benzoxazinoid biosynthesis, BX1, links primary to secondary metabolism. It evolved via gene duplication and neofunctionalization from the gene for the α-subunit of Trp synthase (TSA). Both enzymes are functionally indole-3-glycerol phosphate lyases that cleave indole-3-glycerol phosphate into indole and glyceraldehyde-3-phosphate. The following reactions in biosynthesis comprise the conversion of indole to DIBOA by the introduction of four oxygen atoms. These reactions are catalyzed by four cytochrome P450-dependent monooxygenases, termed BX2 to BX5, which are members of the CYP71C subfamily. Inhibitor studies indicate that in dicot benzoxazinoid biosynthesis, these reactions are also catalyzed by cytochrome P450 enzymes (Schullehner et al., 2008). Glucosylation by UDP-glucosyltransferases (UGTs) chemically stabilizes and increases the solubility of the constitutive defense compound rendering it suitable for storage in the vacuole. The two UGTs BX8 and BX9 catalyze the formation of the benzoxazinoid-glucoside. In maize, DIBOA-glucoside (GDIBOA) is converted into DIMBOA-glucoside (GDIMBOA) by the 2-oxoglutarate–dependent dioxygenase BX6 (Frey et al., 2003) and the O-methyltransferase BX7 (Jonczyk et al., 2008). In maize, as well as in other grasses, DIMBOA is exuded into the rhizosphere and has in addition to activity against nematodes and root-feeding insects (Xie et al., 1992; Zasada et al., 2005), allelopathic function (Sicker et al., 2000). Since DIMBOA is the preferred substrate for BX9, this UGT is rather involved in protecting root tissue from exogenous DIMBOA than in formation of GDIBOA during biosynthesis (von Rad et al., 2001). Hydrolysis of the glucosidic linkage by β-glucosidases (β-GLUs) activates the defense compound. In the intact maize plant, benzoxazinoid-glucosides and two specific-glucosidases (Zm-GLU1 and Zm-GLU2; Babcock and Esen, 1994) are kept in two different cellular compartments, the vacuole and the plastid, respectively. The toxic aglucone is produced upon disintegration of the cell due to pathogen or herbivore attack.
Outside the Poales, benzoxazinoids are detected in two distant orders of the eudicots, the Ranunculales and the Lamiales. Benzoxazinoid biosynthesis in these orders is restricted to single isolated species (Sicker et al., 2000). Two explanations exist for the parallel occurrence of the same metabolite in unrelated species: a monophyletic origin or convergent evolution. In the grasses, all DIBOA biosynthetic genes, including the UGT and the β-GLU, are of monophyletic origin (Nikus et al., 2003; Nomura et al., 2002, 2003; Frey et al., 2009; Sue et al., 2000, 2011). However, the branchpoint enzyme BX1 was independently recruited in three dicotyledonous species (with respect to each other and to grasses), although the orthologous enzyme of primary metabolism, TSA, was employed (Schullehner et al., 2008). TSA homologs in maize and rice (Oryza sativa) (indole-3-glycerol phosphate lyase; Frey et al., 2000; Zhuang et al., 2012), which catalyze the formation of indole as a volatile signal in tritrophic interaction, represent additional cases of independent recruitment and neofunctionalization of TSA.
Plant UGTs and β-GLUs form large gene families (e.g., 122 family 1 UGTs and 40 family 1 β-GLUs) are annotated in Arabidopsis thaliana. We were therefore interested in determining whether the functions that are responsible for the stabilization of benzoxazinoids and for the release of the toxic aglucone have a monophyletic origin in monocots and dicots or were established by recruiting different members from UGT and β-GLU gene pool. In the evolution of preformed defenses, the stabilization and activation of the reactive intermediate(s) can be considered as crucial points. To benefit from the biosynthesis of a toxin, the plant needs a tool to avoid autotoxicity and, complementarily, a function to release the toxic aglucone. Therefore, these functions may be required at the origin of an evolving pathway. Phylogenetic analysis of the glucosyltransferase and the glucosidase of sporadically occurring pathways can shed light on the specific constraints for these functions in constitutive defense compound biosynthesis.
We isolated and functionally characterized the DIBOA-specific glucosyltransferase and glucosidase from the dicot larkspur (C. orientalis; Ranunculaceae). These functions seem to have evolved independently from their counterparts in the Graminea. Specific biochemical features of the dicot enzymes corroborate the phylogenetic analysis based on amino acid sequences.
RESULTS
Isolation of the C. orientalis UDPG:DIBOA-Glucosyltransferase Co-Bx8 and the GDIBOA-Glucosidase Co-Glu
To identify the functions for stabilization (BX8) and activation (termed BxGLU in the following) of benzoxazinoid biosynthesis in a dicot species, we isolated the specific glucosyltransferase and β-GLU from C. orientalis. The isolation was based on transcriptome data of 7- to 10-d-old seedling shoots. This plant material was chosen since it is available in large quantities and has high benzoxazinoid content and DIBOA biosynthesis activity (Schullehner et al., 2008). A total of 250,000 reads of the normalized cDNA were assembled using the Phred software (Ewing et al., 1998; Ewing and Green, 1998), which delivered 6.8 Mb of singlet sequences and 15.5 Mb of contigs.
Co-BX8 was identified by comparison of the transcriptome with peptide sequences of the DIBOA:UDP-glucosyltransferase (UDPG) activity that was purified almost to homogeneity from crude soluble protein extracts by ammonium sulfate precipitation and four chromatographic separations similar to the purification of the corresponding maize UGTs (von Rad et al., 2001; see Supplemental Table 1 online). Analysis of individual fractions on SDS-PAGE gels revealed a band with a molecular mass of ~55 kD that correlated with UDPG:DIBOA-glucosyltransferase activity in the enzyme assay (see Supplemental Figure 1 online). This band was excised and digested following the protocol of Schäfer et al. (2001). The sequence of several fragments of the trypsin digest was determined by mass spectroscopy.
In the C. orientalis transcriptome, 25 singlets and 37 contigs were assigned to UGT domain proteins due to the presence of the PFAM signature PF00201 (Finn et al., 2010; see Supplemental Table 2 online). Five contig sequences were assembled from at least 10 sequence reads each (see Supplemental Table 2 online) and thus represent abundant transcripts. The deduced amino acid sequences of these contigs were compared with the data from mass spectroscopy of the purified CoBX8. One contig with a size of 1174 bp included seven peptides of CoBX8 (see Supplemental Figure 2 online); no other significant similarities were detected in the trancriptome. The full-size cDNA corresponding to the candidate contig was isolated by screening a C. orientalis seedling cDNA library (Schullehner et al., 2008). The open reading frame of Co-Bx8 comprises 1428 bp; the deduced amino acid sequence includes the highly conserved plant secondary product glucosyltransferase box consensus sequence that characterizes the family 1 UGTs involved in plant secondary metabolism (Hughes and Hughes, 1994). The closest UGT homologs with defined biochemical functions are the cytokinin-O-glucosyltransferase At-COGT2 from Arabidopsis and the hydroxymandelonitril UGT Sb-HMNGT from Sorghum bicolor (49 and 43% identity on the amino acid level, respectively). For heterologous protein expression, the cDNA was cloned in the Escherichia coli expression vector pET28a as an N-terminal His-tag fusion protein. The recombinant enzyme had UDPG-DIBOA-glucosyltransferase activity and hence BX8 function (see below for detailed analysis).
The rationale for the isolation of the β-GLU enzyme was based on our experience that Bx genes are generally highly expressed in tissues with high benzoxazinoid content (Frey et al., 1995; von Rad et al., 2001; Jonczyk et al., 2008). The β-GLU genes present in 454 transcriptome data from C. orientalis shoot were identified by a Blast search using Zm-GLU1 (Esen, 1992) and the Arabidopsis myrosinase 1 At-TGG1 (Barth and Jander, 2006), two quite diverge members of the β-GLU family 1 (42% identity and 60% similarity), as query sequences (see Supplemental Table 2 online). Two abundant homologs were represented in full length in the seedling phage cDNA library. These genes have open reading frames of 1536 and 1527 bp and encode enzymes with 65% amino acid identity to each other. Both enzymes contain the sequence motifs ITENG and TFNEP that are characteristically conserved in glycoside hydrolases of family 1 (Davies and Henrissat, 1995). A signal peptide for targeting the proteins into the secretory pathway is predicted for both enzymes, and transit peptide sequences for localization to the plastid are not detected (TargetP 1.1, Emanuelsson et al., 2000; iSPORT, Bannai et al., 2002). The enzymes have ~45% amino acid identity to the query sequences Zm-GLU1 and At-TGG1 and ~60% amino acid identities to the closest homologous enzymes with defined function, the exoglucanase Os-4BGLU12 (Opassiri et al., 2006) and the hydroxynitril-glucoside hydrolyzing enzyme Lj-BGD2 (Morant et al., 2008a; Takos et al., 2010). Expression of the β-GLUs in E. coli failed probably due to instability caused by lack of plant-specific protein modification (Zhou et al., 2002; Morant et al., 2008b). Alternatively, transient expression of the proteins by magnifection (Marillonnet et al., 2005) with a Tobacco mosaic virus–based vector system was used. The vector system (Engler et al., 2008) was transferred via Agrobacterium tumefaciens into Nicotiana benthamiana leaves. Both enzymes were the major proteins in leaves 4 to 10 d after infection (see Supplemental Figure 3 online). The functionality of the heterologously expressed β-GLUs was verified by the detected hydrolysis of the general artificial substrate p-nitrophenyl-β-glucopyranoside (pNPG). One of the enzymes hydrolyzed GDIBOA and hence is functionally analogous to Zm-GLU1 (see below for detailed analysis). This enzyme is termed Co-GLU in the following. The second enzyme had no activity toward the benzoxazinoid substrate and thus was not further analyzed. The expression pattern of the other highly expressed β-GLUs differs substantially from the distribution of DIBOA in C. orientalis (see Supplemental Figure 4 online); therefore, it is unlikely that they encode benzoxazinoid-specific enzymes.
Expression Profiles of Co-Bx8 and Co-Glu Imply a Function in Benzoxazinoid Metabolism.
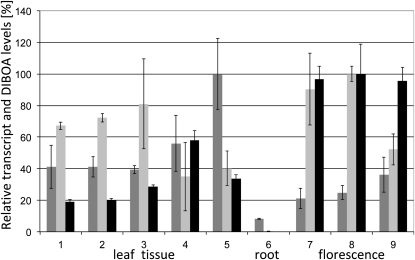
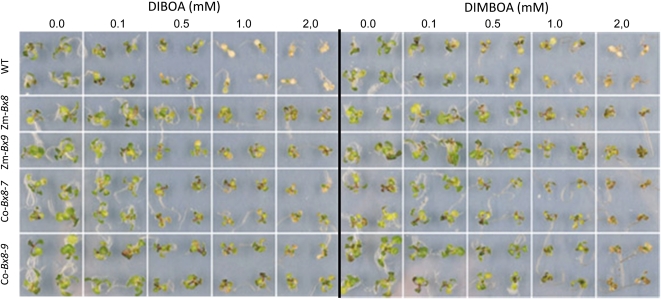
The expression pattern of hitherto characterized benzoxazinoid pathway genes in grasses and dicots (Frey et al., 2009) is largely connected to the accumulation of the metabolite in the plant. In C. orientalis, DIBOA is synthesized and present in significant concentrations in leaves of seedlings and mature plants and in flowers but has concentrations below the detection limit in the roots (Schullehner et al., 2008). The transcript levels of Co-Glu and Co-Bx8 were determined by quantitative RT-PCR for these tissues (Figure 2). Co-Glu transcript levels close to the values of the housekeeping gene GAPDH were found in leaves, flower buds, and open flowers; the level is below the detection limit in the root. Co-Glu displays essentially the same expression pattern as Co-Bx1, the first gene in the pathway (Schullehner et al., 2008). By contrast, the UGT Co-Bx8 is not only expressed in the shoot, leaves, and flower, but also in the root (Figure 2). This implies an additional function of the UGT, since no benzoxazinoid biosynthesis takes place and benzoxazinoids are not detectable within the root. Co-BX8 may protect the root tissue against exogenous benzoxazinoids that can originate from plant debris (Blum, 1999). In addition to the role in biosynthesis, Co-BX8 could function in protection against exogenous benzoxazinoids such as proposed for Zm-BX9. Zm-Bx9 is also expressed in older root tissue where benzoxazinoid biosynthesis is nearly absent (Schmälzlin, 2002). To determine the potential of Co-BX8 to restrict allelopathic effects of exogenous benzoxazinoids, transgenic Arabidopsis plants expressing Co-Bx8 were generated. BX8 activity was assayed in crude extracts of six independent transgenic plant lines. All transgenic but no wild-type control plants glucosylated DIBOA. Homozygous progeny of two transgenic lines with the highest Co-BX8 activity were tested for tolerance to DIBOA and DIMBOA (Figure 3). The benzoxazinoids were applied in concentrations of 0.1 to 2.0 mM to the two transgenic lines, as well as to wild-type control plants and Zm-Bx8 or Zm-Bx9 expressing homozygous transgenic Arabidopsis plants (von Rad et al., 2001). Even the lowest benzoxazinoid concentration impaired the growth of wild-type plant slightly, while 1.0 and 2.0 mM concentrations resulted in growth arrest and bleaching or browning of these plants. By contrast, all transgenic plants survived the treatment, although the Co-BX8 enzyme seems to be less effective in detoxification of DIMBOA compared with Zm-BX8 and Zm-BX9. This difference may result from the relatively high KmDIMBOA of Co-BX8 (see below). However, both C. orientalis and maize UDPG:DIBOA-glucosyltransferases were sufficient to overcome DIBOA and DIMBOA phytotoxicity.
Figure 2.
Pattern of Co-Bx8 and Co-Glu Expression and DIBOA Distribution.
Transcript levels were determined for 2-week-old seedlings (lane 1), hypocotyl and cotyledon (lane 2), and rosette leaves (lane 3) of 4-week-old plants, mature leaves (lane 4), and senescent leaves (lane 5) of 8- to 10-week-old plants, root (lane 6), green flower buds (lane 7), flower buds with violet petals (lane 8), and flowers (lane 9). The transcript levels were normalized with GAPDH expression, and DIBOA content was determined per milligram of fresh weight. Two biological replicates were analyzed in two technical replicates each. Co-Bx8, dark-gray bars; Co-Glu, light-gray bars; DIBOA, black bars. The sd is indicated.
Figure 3.
Reduction of Phytotoxicity of the Benzoxazinoids DIBOA and DIMBOA by Expression of the UGTs Zm-Bx8, Zm-Bx9, and Co-Bx8 (Transgenic Lines 7 and 9).
Wild-type (WT; first row) and transgenic Arabidopsis seedlings were incubated in different concentrations of the benzoxazinoids for 4 d, and the benzoxazinoid solution was exchanged every 24 h.
Detoxification and Activation Functions of the Benzoxazinoid Pathway in C. orientalis and in the Poaceae Are in Phylogenetic Groups That Separated before the Diversification of Monocots and Dicots
The expression patterns and enzymatic properties strongly suggest functionality of Co-BX8 and Co-GLU in benzoxazinoid metabolism. The phylogenetic relationship of the dicot and the respective monocot genes was predicted to reveal the evolutionary origin of these two functions.
Family 1 UGTs and family 1 β-GLUs comprise large gene families in plants (Paquette et al., 2003; Xu et al., 2004). Experimental evidence for enzyme function is only available for a subset of the enzymes. Family 1 UGTs involved in secondary metabolism are subdivided into 14 groups (Ross et al., 2001). UGTs involved in flavonoid metabolism are characterized across a wide phylogenetic range of plant species including maize. The monophyletic origin of the enzymes functioning in this common plant secondary metabolic pathway including UGTs is well established (Rauscher, 2006). The analysis of UGTs for flavonoid metabolism provides a measure for the phylogenetic relationship of the DIBOA-specific UGTs. To make equivalent data sets for both pathways available, we isolated the flavonoid-O-glucosyltransferases from C. orientalis and Lamium galeobdolon. In addition, the BX8/BX9 ortholog of wheat was isolated; this gene may be an allelic variant of the wheat Bx8 genes isolated recently (Sue et al., 2011) or represent a closely related duplicate thereof (98 to 96% identity on amino acid level). The genes were identified by screening the C. orientalis and L. galeobdolon transcriptome and the GenBank wheat EST collection for homologous sequences. The functions of the isolated genes were verified by heterologous expression and enzyme assays (see Supplemental Figure 5 online).
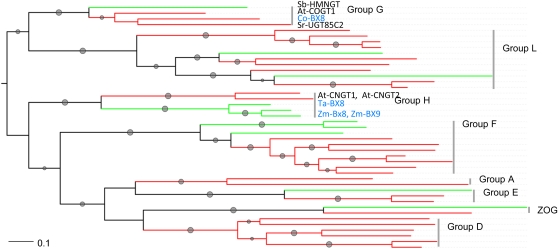
A phylogenetic tree (Figure 4; see Supplemental Data Set 1 online) was constructed that comprises plant secondary product glucosyltransferase box UGTs with defined biochemical functions and close homologs thereof. Family 1 UGTs for steroid metabolism were not included. Sequences from monocot and dicot plants were represented in almost all groups, demonstrating that diversification of the UGTs predates the divergence of monocots and dicots 180 million years ago (Brown et al., 2011). The flavonoid-3-O-glucosyltransferases enzymes (F3GTs) are found in group F; the analysis predicts a common precursor of F3GTs that existed before the separation of monocots and dicots. By contrast with F3GTs, the dicot Co-BX8 and the BX8 enzymes from grasses are found in distinct groups (G and H), and both groups separated before the diversification of dicots as predicted by the presence of dicot and monocot sequences in both groups. According to the standardized nomenclature system (Mackenzie et al., 1997), Co-BX8 is named UGT85N1, while the UGTs of grasses belong to family UGT710E. Except for BX8, group G enzymes are otherwise involved in N-glucosylation of cytokinins (At-COGT1) and biosynthesis of cyanogenic glucosides (Sb-HMNGT) and sweet glucosides (Sr-UGT85C2), while the group H enzymes At-CNGT1/At-CNGT2 are involved with N-glucosylate cytokinins.
Figure 4.
Phylogenetic Tree of Plant Family 1 UGTs.
Representative enzymes with biochemically defined functions were used in the analysis. One thousand bootstrap samples were generated; branches with bootstrap values higher than 50% are marked with gray dots, and the size is proportional to the value. The trees are colored by organism: monocots in green and dicots in red. The enzymes of benzoxazinoid metabolism are written in blue letters. The affiliation to UGT groups is indicated. UGTs and accession numbers from top to bottom: Group G: HMNGT_SORBI S. bicolor, COGT2_ARATH Arabidopsis, Co-BX8 C. orientalis, Sr-UGT85C2 (UGT85C2) Stevia rebaudiana. Group L: LGT_CITUN Citrus unshiu, UGT84A10 Brassica napus, AAF98390 B. napus. IABG_MAIZE Z. mays, UGT74B1 Arabidopsis, UGT74G1 St. rebaudiana. BAA36421 Perilla frutescens, BAD06874 Iris hollandia, UGT75B2 Arabidopsis, UGT75B1 Arabidopsis, NP_188793 Arabidopsis. Group H: At-CNGT1 (CNGT1_ARATH) Arabidopsis, At-CNGT2 (CNGT2_ARATH) Arabidopsis, Ta-BX8 Triticum aestivum, Zm-BX8, Zm-BX9 Z. mays. Group F: BZ1 Z. mays, UFOG_HORVU H. vulgare, BAD83701 I. hollandica, Co-F3GT C. orientalis, Lg-F3GT L. galeobdolon, UFOG_Gentr Gentiana triflora, NP_197207 Arabidopsis, NP_564357 Arabidopsis, AAU09442 Fragaria × ananassa, UFOG_VITVI Vitis vinifera. Group A: BAD95882 Ipomoea tricolor, Bp-UGAT Bellis perennis. Group E: NM0011539 Z. mays, NP_201470 Arabidopsis, NP_566938 Arabidopsis. Family 1 UGTs of zeatin metabolism: NP_001105017 Z. mays, ZOG_PHALU Phaseolus lunatus. Group D: UGT73K1 Medicago truncatula, BAD89042 Sa-GT4 Solanum aculeatissimum, BAD29722 Q6F4D5_CATRO C. roseus, CAB56231 Dorotheanthus bellidiformis, NP_181213 Arabidopsis, NP_181218 Arabidopsis.
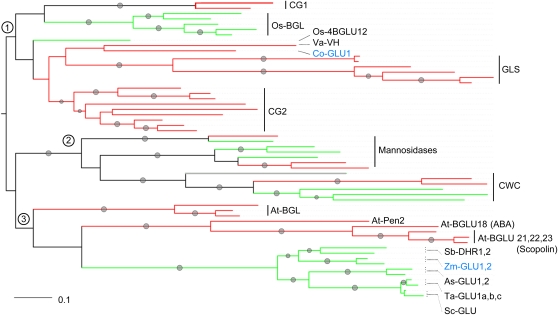
The phylogenetic analysis of the BxGLUs was done analogously. The data set included enzymes that display (additional) mannosidase activity and enzymes hydrolyzing cell wall compounds (common substrates), β-GLUs of defense pathways (benzoxazinoids, cyanogenic glucosides, glucosinolates, and saponins; summarized in Morant et al., 2008b), manually reannotated β-GLU sequences from Arabidopsis and rice (Xu et al., 2004; Opassiri et al., 2006), and β-GLUs annotated in the Uniprot database. The tree (Figure 5; see Supplemental Data Set 2 online) can be divided into three clades; one of these comprises common substrate glucosidases (clade 2). Monocot and dicot enzymes with mannosidase activity or cell wall compound-cleaving enzymes are present in separate branches of clade 2, indicating diversification of the respective precursor glucosidases before the separation of monocots and dicots. The two other clades each include enzymes cleaving benzoxazinoid glucosides (Co-GLU in clade 1 and Bx-GLUs of the grasses in clade 3). Interestingly, the BxGLUs are in subbranches with dicot and monocot enzymes that have cyanogenic glucoside as substrates (Co-GLU with β-vicianoside β-glucosidase; BxGLUs of the grasses with dhurrin β-GLU).
Figure 5.
Phylogenetic Tree of Plant β-GLUs.
Parameters and presentation analogous to Figure 4. Representative enzymes with biochemically defined functions and annotated close homologs were used in the analysis. The functions of β-GLUs in individual branches are indicated. The branch with the Pinus contorta sequence is marked in gray. β-GLUs and accession numbers from top to bottom: CG1, cyanogenic glucosidases; Q41172, Manihot esculenta; Q7Y073, Hevea brasiliensis. Os-BGL: BGL28_ORYSJ, BGL27_ORYSJ, BGL29_ORYSJ, BGL35_ORYSJ, BGL30_ORYSJ, O. sativa. Os4BGLU12: BGL12_ORYSJ exoglucanase, O. sativa. Va-VH: A2SY66, cyanogenic glucosidase, Vicia angustifolia. CoGLU1 C. orientalis. GLS: myrosinases, At-Myr5 (TGG5, Q3ECS3), At-Myr4 (TGG4 Q8GRX1), At-Myr2 (TGG2 Q9C5C2), At-Myr1 (TGG1 P37702), Sa-MYR (MB3 P29092), Bn-MYR1 (Q00326). CG2: cyanogenic glucosidases, Ps-AH1 (Q945G7), Ps-PH4 (Q945I4), Ps-PH3 (Q945N9) Prunus serotina; Dc-BGLU1(Q9SPK3) Dalbergia cochinchinensis; Tr-BGLU78 (P26205.1), Tr-BGLU78 (P26205.1) Trifolium repens; Lj-BGD7 (B2ZUU2), Lj-BGD2 (B2ZUU1), Lj-BGD4(B2ZUU0) L. japonicus. Mannosidases: At-BGLU40 (Q9FZE0), Arabidopsis; Os-BGL06 (Q8L7J2), Os-BGL08 (Q75I94), Os-3BGLU7 (Q42975) O. sativa; Hv-MAN (B5A496) H. vulgare; At-BGLU44 (Q9LV33) Arabidopsis; Sl-MAN (Q8VWL8) Solanum lycopersicum. CWC: cell wall compound glucosidases: Q9ZT64, P. contorta; At-BGLU46 (O80690), At-BGLU45 (O80689), Arabidopsis; Os-BGLU14 (Q7XPY7), Os-BGLU16 (Q7XSK2), Os-BGLU18 (Q7XSK0) O. sativa. At-BGL: At-BGLU16 (Q9M1D0), At-BGL12 (Q9FH03), At-BGLU15 (O64879), Arabidopsis. At-Pen2 (O64883), Arabidopsis. At-BGLU18 (ABA) (Q9SE50), Arabidopsis. At-BGLU 21,22,23 (Scopolin): At-BGLU23 (NM_111760), At-BGLU21 (Q9C525), At-BGLU22 (Q9C8Y9); Arabidopsis. Sb-DHR1,2: Q41290, Q93XR2, S. bicolor. Zm-GLU1,2: P49235, Q41761, Z. mays. As-GLU1,2: Q38786, Q9ZP27, A. sativa.Ta-GLU14,b,c: Q1XIR9, Q1XH05, Q1XH04, T. aestivum. Sc-GLU: Q9FYS3, S. cereale.
The phylogeny indicates independent evolution of UGTs and β-GLU for benzoxazinoid biosynthesis in monocots and dicots. This pattern suggests that these enzymes originated independently from enzymes that did not necessarily share functions in plant metabolism.
Monocot and Dicot BX8 and BxGLU Enzymes Display Distinct Differences in Enzymatic Function
To test the idea that the monocot and dicot BX8 and BxGLU functions arose independently, we investigated to what extent the C. orientalis and maize UGTs and β-GLUs exhibit distinct enzymatic specificities that may reflect different phylogenetic origins.
The steady state kinetic constants of E. coli–expressed Co-BX8 were determined for the substrates DIBOA, DIMBOA, and UDPG (Table 1). The reaction followed Michaelis-Menten kinetics for the three substrates (see Supplemental Figure 6A online). The Km values for DIBOA and UDPG are in the same range. Compared with Zm-BX8, the values are increased, but kcat values are similar (Table 1). Since the DIBOA concentrations in C. orientalis tissues (17 to 36 μmol/g fresh weight) exceed this concentration by at least a factor of 40, this enzymatic property is not contradictory to BX8 function of the enzyme. In addition, analysis of the DIBOA-glucosyltransferase activity in protein crude extract revealed a KmDIBOA similar to the heterologously expressed gene (see Supplemental Figure 7 online). The KmDIMBOA determined for Co-BX8 exceeds the KmDIBOA by a factor of ~8. Similarly, a difference of the Km value for DIBOA and DIMBOA is detected for Zm-BX9; however, in contrast with Co-BX8, the maize enzyme has a reduced affinity to DIBOA (Table 1). Further differences between Co-BX8 and the maize UGTs were found for the acceptance of other benzoxazinoid substrates (see Supplemental Figure 8 and Supplemental Table 3 online). N-hydroxylation influences only marginally Co-BX8 substrate specificity since HBOA and HMBOA values do not differ significantly from the data for DIBOA. By contrast, Zm-BX8 is sensitive to the lack of the N-OH group and has significantly reduced substrate specificity for HBOA and HMBOA. The presence of the 7-O-methyl group seems to be required for increased substrate specificity by Zm-BX9 (see Supplemental Table 3 online). Outside of the benzoxazinoids, auxins (indole acetic acid) and cytokinins (trans-zeatin) may be suspected as substrates since benzoxazinoid biosynthesis is connected with auxin biosynthesis via the Trp biosynthetic pathway. The phylogenetic analysis for the dicot and monocot BX8 enzymes displays a close homology to cytokinin-N-glucosylating enzymes. However, BX8 enzymes do not glucosylate the hormones (see Supplemental Table 3 online); likewise, flavonoids are not substrates. Hence, the dicot and the monocot BX8 enzymes are specific for benzoxazinoids but require different modifications of the core benzoxazinoid structure for highest activity.
Table 1.
Steady State Constants of the UDP-Glucosyltransferases Co-BX8, Zm-BX8, and Zm-BX9
| Enzyme | UDPG | DIBOA | DIMBOA | ||||||
| Km(μM) | kcat(s−1) | kcat/Km(mM−1 s−1) | Km(μM) | kcat(s−1) | kcat/Km(mM−1 s−1) | Km(μM) | kcat(s−1) | kcat/Km(mM−1 s−1) | |
| Co-BX8 | 743 ± 77 | 8.5 ± 0.2 | 11.4 | 441 ± 47 | 9.5 ± 0.2 | 21.5 | 3168 ± 549 | 17.6 ± 1.6 | 5.6 |
| Zm-BX8a | 81 | 22.6 | 280 | 61 | 12.5 | 205 | 81 | 22.7 | 280.0 |
| Zm-BX9a | 96 | 22.6 | 117 | 1300 | 12.5 | 6 | 71 | 11.6 | 163.0 |
The data for Zm-BX8 and Zm-BX9 are from von Rad et al. (2001).
To characterize the biochemical activity of Co-GLU, a range of β-glycosides was tested as substrates (see Supplemental Figure 9 and Supplemental Table 4 online). BxGLUs are phylogenetically related to cyanogenic glucoside-cleaving enzymes, and the cyanogenic glucoside dhurrin has been shown to be a competitive nonmetabolized inhibitor of Zm-GLU (Babcock and Esen, 1994). Furthermore, Zm-GLU (isoform Zm-p60.1) activity toward cytokinin glucosides has been demonstrated (Brzobohatý et al., 1993). These nonbenzoxazinoid substrates were included in our analysis of Co-GLU. Our results show that Co-GLU accepts the benzoxazinoids GDIBOA and GDIMBOA, the cyanogenic glucoside dhurrin, the cytokinin glucoside trans-zeatin-O-glucoside (tZOG), and pNPG as substrates. Hydrolysis of tZOG can be considered as a side reaction being 10 times lower in specific activity compared with the artificial glucoside pNPG (specific activity 61 ± 2 nmol·min−1·mg−1 compared with values of 550 ± 14 nmol·min−1·mg−1 for pNPG, at substrate concentrations of 500 μM). Km values and kcat values were determined for GDIBOA, GDIMBOA, and dhurrin (Table 2; see Supplemental Figure 6B online). A clear preference for the substrates GDIBOA and dhurrin was found. GDIBOA and dhurrin have similar catalytic values, while GDIMBOA is significantly less efficiently cleaved having a Km value ~6 times higher and a kcat 8 times lower compared with GDIBOA (Table 2). The Km values for GDIBOA (5.2 mM) and dhurrin (4.8 mM) hydrolysis were determined with C. orientalis crude extract and fit with the data from the heterologously expressed enzymes (see Supplemental Figure 7 online). In the grasses, GDIMBOA is the most efficiently hydrolyzed benzoxazinoid (Babcock and Esen, 1994; Sue et al., 2006); only in rye are the catalytic data for GDIBOA similar to the values for GDIMBOA (Nikus et al., 2003; Sue et al., 2006). Hence, the benzoxazinoid β-GLU functions of Co-GLU and monocot BxGLUs are distinct. Further substantial differences were displayed with respect to the enzyme activity toward dhurrin. While the maize enzyme does not hydrolyze dhurrin but GDIMBOA hydrolysis is inhibited by the cyanogenic glucoside, Co-GLU hydrolyzes dhurrin efficiently (Table 2), and the presence of dhurrin inhibits GDIBOA hydrolysis in the assay with heterologously expressed Co-GLU and in the crude extract (see Supplemental Figure 10 online).
Table 2.
Steady State Constants of Heterologously Expressed Co-GLU with the Substrates GDIBOA, GDIMBOA, and Dhurrin
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (s−1·mM−1) |
| GDIBOA | 4.6 ± 0.4 | 32.0 ± 1.55 | 7.00 |
| GDIMBOA | 28.4 ± 4.4 | 4.2 ± 0.5 | 0.15 |
| Dhurrin | 4.8 ± 1.0 | 52 ± 6.4 | 10.80 |
DISCUSSION
Plants use chemical defense as a major barrier to protect against herbivores and microbial pathogens. The large chemical and structural diversity of the reactive natural products within the plant kingdom assures that the development of generalists is restricted. On a genomic level, the biosynthetic potential of plants is reflected by the expansion of genes encoding enzymes functioning in secondary metabolic pathways, such as oxygenases, methyltransferases, UGTs, etc., into large gene families. The proliferation of the P450 monooxygenase gene family in plants dates to ~450 million years ago, when the first vascular plants, mosses, colonized land (Gonzalez and Nebert, 1990; Nelson, 2006). In the case of UGTs, vascular plant speciation was accompanied by an increase in the number of ancestral genes and a remarkable expansion occurring in a lineage-specific manner (Yonekura-Sakakibara and Hanada, 2011). Here, we were interested in the evolution of the UGTs and β-GLUs that are required for stabilization and activation of the benzoxazinoids.
The UGT Co-BX8 and the β-GLU Co-GLU catalyze the ultimate steps of benzoxazinoid metabolism in C. orientalis. The data from protein purification and transcriptome analysis identified CoBX8 as the UDPG:DIBOA-glucosyltransferase present in shoots. The analysis of Co-BX8 in transgenic Arabidopsis demonstrated in planta the potential of the enzyme to glucosylate benzoxazinoids. Evidence for Co-GLU representing a BxGLU function is given by its ability to hydrolyze GDIBOA in vitro and its expression in GDIBOA synthesizing tissue. For C. orientalis, no mutant lines or special ecotypes are available. Hence, we cannot exclude by genetic analysis that additional genes with DIBOA-glucosylation or GDIBOA-hydrolyzing function exist in C. orientalis. However, since the Km values determined for the protein extracts and heterologously expressed proteins correspond and the genes are highly expressed in benzoxazinoid-rich tissue, Co-BX8 and Co-GLU are concluded to be UGT and β-GLU enzymes catalyzing in planta the respective steps in benzoxazinoid metabolism.
Catalytic data for BxGLUs have been determined for maize (Cicek et al., 2000), wheat (Sue et al., 2006), and rye (Nikus et al., 2003; Sue et al., 2006). The grass enzymes display a range of Km values from 0.1 to 1.4 mM for the substrates. The variation in this value might not be of great relevance for the in vivo function of the enzymes due to the high concentration of the substrate present in the tissues; likewise, the KmGDIBOA value of Co-GLU is in the millimolar range (4.6 mM) and fits well with the substrate concentration. BxGLUs in the grasses are often present as set of paralogous genes (Figure 5), and each gene may have a distinct pattern of expression. Homodimeric Zm-GLU complexes and multimers thereof are catalytically active (Cicek and Esen, 1998). Heterohexamers are described for wheat (Sue et al., 2006). Our transcriptome analysis did not reveal a functional homolog of Co-GLU that is expressed in green seedling tissue. A major difference between Co-GLU and the grasses BxGLUs is the subcellular localization. Plastidic localization has been shown for BxGLUs of the grasses (Nikus et al., 2001) and is predicted by the presence of an analogous signal peptide for the closely related β-GLUs of sorghum and oat (Avena sativa; Morant et al., 2008b). Such a signal peptide is not detected for CoGLU; rather, transfer into the secretory pathway is strongly predicted by the programs TargetP1.1 (Emanuelsson et al., 2000) and iPSORT (Bannai et al., 2002).
Independent Evolution of the Benzoxazinoid-Specific UGTs and β-GLUs by Repeated Recruitment of Nonorthologous Members of the Gene Families
Benzoxazinoid biosynthesis is monophyletic in the grasses (Nomura et al., 2003; Grün et al., 2005; Frey et al., 2009; Sue et al., 2011; this article), but the analysis of the first gene of the pathway, Bx1, in grasses and benzoxazinoid-producing dicots revealed an evolution by repeated independent duplications of the TSA gene (Schullehner et al., 2008). For the evolution of sporadically occurring biosynthetic pathways, further data are available for pyrrolizidine alkaloids (Reimann et al., 2004). Like in benzoxazinoid biosynthesis, the branchpoint enzyme homospermidine synthase evolved by repeated independent recruitment of a primary function, deoxyhypusine synthase. The analysis of widespread plant secondary pathways, such as flavonoid and cyanogenic glucoside biosynthesis, demonstrated recruitment of genes from the same gene family for the respective enzymatic steps in the pathways, such as the CYP75 flavonoid hydroxylases in angiosperm flavonoid biosynthesis (Nelson and Werck-Reichhart, 2011), CYP79 and CYP71E monooxygenases (Jørgensen et al., 2011), and UGT85 glucosyltransferases in cyanogenic glucoside biosynthesis (Jones et al., 1999; Franks et al., 2008; Kannangara et al., 2011). However, the recruited genes are not necessarily orthologous as shown recently for cyanogenic glucoside biosynthesis by Takos et al. (2011). It has been suggested that repeated recruitment might represent the majority of convergent evolution events in plant secondary metabolism (Cseke et al., 1998; Pichersky and Lewinsohn, 2011; Takos et al., 2011), and the elucidation of cyanogenesis in insects revealed convergent evolution even across the plant and animal kingdoms by recruitment of members of the same gene functions (Jensen et al., 2011). Although the encoded enzymes, two multifunctional P450 monooxygenases, and one UGT in plant and insect use identical substrates to synthesize the same pathway intermediates, all three genes evolved independently. To investigate the evolutionary origin of the UGTs and β-GLUs of benzoxazinoid biosynthesis, we reconstructed phylogenies using representative amino acid sequences and rigorous statistical evaluation of the results (Figures 4 and 5). In principle, the number of characters in the alignment limits the resolution of any gene tree. In our trees, this is reflected by high bootstrap support for most of the nodes but poor bootstrap values for the most ancient groupings. However, comparisons of these topologies with trees obtained by an alternative tree reconstruction method (Bayesian approach) resulted in good overall agreement. The C. orientalis F3GT is found together with the respective monocot and dicot enzymes in the group F branch of UGT in a position that reflects the phylogeny of the Ranunculaceae (Figure 4). Like UGTs of flavonoid biosynthesis, UGTs of cyanogenic glucoside pathways have been recruited from the same UGT group. By contrast, benzoxazinoid-specific UGTs belong to different groups, UGT85 and UGT710. The analysis locates the BxGLUs in two distinct clades of the β-GLU tree; therefore, our phylogenetic data indicate independent recruitment of stabilization and activation function from homologous but clearly not orthologous genes in the dicot C. orientalis compared with the monocot lineage.
Substrate specificity of UGTs can be relaxed, and enzymatic versatility of UGT family members enables glucosylation of broad spectra of endogenous and nonendogenous substrates (Jones et al., 1999; Schwab 2003). For example, linalool produced by heterologous expression of S-linalool synthase in petunia (Petunia hybrida) was readily glucosylated by an endogenous glucosyltransferase (Lücker et al., 2001). The sensitivity of Arabidopsis to exogenous benzoxazinoids indicates that there is no universal efficient protection by plant UGTs against this nonendogenous substrate. Similarly, no Arabidopsis UGT was capable to convert p-hydroxy-mandelonitrile into dhurrin (Tattersall et al., 2001). Hence, distinct secondary metabolites might require specific UGTs to be glucosylated. Our results for UGTs involved in benzoxazinoid biosynthesis support the notion that these UGTs independently acquired their ability to recognize the specific substrates after the expansion that occurred in vascular plant lineages (Yonekura-Sakakibara and Hanada, 2011). The BX8 UGTs of maize and C. orientalis have narrow substrate spectra; activity is restricted to benzoxazinoids, a rather unique class of metabolites (see Supplemental Table 3 online). However, within the benzoxazinoids, maize and C. orientalis BX8 functions discriminate differentially with respect to modifications of the benzoxazinoid basic structure (see Supplemental Table 3 and Supplemental Figure 8 online). C. orientalis BX8 has optimal catalytic properties for DIBOA, the principal benzoxazinoid-aglucone in this plant. Zm-BX8, on the other hand, does not differentiate between DIBOA and DIMBOA, although DIBOA is glucosylated prior to the modifications at C-7 to yield DIMBOA (Figure 1). The difference in specificity may reflect intrinsic enzymatic properties of the distinct progenitor UGTs or may have evolved in maize as a consequence of the demand to detoxify exogenous DIMBOA exuded by neighboring plants. The closest characterized relatives of the monocot and dicot BX8 functions have substrate specificities ranging from cytokinins to steviol glycosides (Figure 4). It can be speculated that the progenitor enzymes of benzoxazinoid biosynthesis and cytokinin homeostasis had activity toward both classes of substrates. However, the fraction of UGT sequences with biochemically defined function is relatively small and thus biases the phylogenetic analysis. There is no experimental evidence for an overlap of substrates of UGTs of benzoxazinoid biosynthesis and cytokinin metabolism.
As discussed above, the phylogenetic analysis demonstrates that respective progenitors of the BxGLUs are not orthologous (Figure 5). Similar to the finding for the UGTs of the pathway, the specificity of the BxGLUs reflects the predominant benzoxazinoid present in a particular plant species, and specificity probably has been selected during evolution of the genes. GDIMBOA is the main benzoxazinoid and the preferred substrate for the maize and wheat enzymes (Babcock and Esen, 1994; Sue et al., 2006). In S. cereale, both GDIBOA and GDIMBOA are prevalent, and Sc-GLU uses both these substrates with similar efficiencies (Nikus et al., 2003; Sue et al., 2006), while for Co-GLU, only GDIBOA is a suitable substrate.
Promiscuity of Monocot and Dicot BxGLUs
The substrate specificity of plant β-GLUs ranges from highly selective to broadly general substrate spectra (Morant et al., 2008b). BxGLUs are promiscuous enzymes. According to catalytic data, the maize, wheat, and C. orientalis β-GLUs each cleave a second substrate as efficiently as the benzoxazinoid, the cytokinin tZOG (Campos et al., 1992; Brzobohatý et al., 1993), the isoflavonoid genistein glucoside (Sue et al., 2000), and dhurrin (this work), respectively. The relevance of these activities on dhurrin and genistein in C. orientalis and wheat is unclear since the compounds have not been detected in these species. A function in cytokinin homeostasis, as proposed for the maize enzyme, can be excluded in C. orientalis since Co-GLU has only minor activity on tZOG. Remarkably, the cyanogenic glucoside dhurrin represents an inhibitor for Zm-GLU but functions as a substrate for Co-GLU. Similarily, β-GLUs of cyanogenesis also show notable second activities. The enzyme mixture of the purified β-GLUs Lj-BGD2 and Lj-BGD4 that are involved in activation of cyanogenic and noncyanogenic hydroxynitril glucosides in Lotus japonicas (Takos et al., 2010) hydrolyze the isoflavonoid-glucoside daidzin (Morant et al., 2008a), although isoflavonoids are not natural substrates for these enzymes. Based on phylogenetic analysis and the promiscuity of the β-GLUs, it has been hypothesized that isoflavonoid glucoside-cleaving enzymes have evolved from cyanogenic glucoside activating β-GLUs (Chuankhayan et al., 2007; Morant et al., 2008a). Parallels exist for benzoxazinoids: Cyanogenic glucosides and benzoxazinoids are found in grasses and in Ranunculaceae (Sharples et al., 1972; Vetter, 2000). Our phylogeny (Figure 5) suggests that the BxGLUs and an enzyme involved in cyanogenic glucoside cleavage (Sb-DHR or Va-VH) have a close common ancestor. However, since repeated independent recruitment has been shown for cyanogenic glucoside biosynthesis (Takos et al., 2011), this may reflect promiscuity of β-GLUs rather than a direct origin of secondary metabolite β-GLUs from functions of cyanogenic glucoside biosynthesis. The fact that the phylogeny of grass β-GLUs in clade 3 (Figure 5) mirrors the plant phylogeny independent of specific enzyme function underlines the notion that an enzyme with broad substrate specificity might have been recruited and evolved toward β-GLUs with specificities for such diverse substance classes as benzoxazinoids, cyanogenic glucosides, and saponins.
Required Settings at the Advent of Evolution of Constitutive Defense Compounds
The evolution of a pathway leading to the accumulation of an autotoxic metabolite seems only realizable in the presence of a suitable stabilization function, as it would be otherwise deleterious for the organism. This idea is reminiscent of the original concept of retrograde evolution of a biosynthetic pathway that was developed by Horowitz (1945). Stabilization by glucosylation can be considered the endpoint in the biosynthesis of a preformed defense compound. In order to become fixed in evolution, expression of a pathway should confer a selective advantage to the plant. In this respect, the β-GLU function that activates the stabilized compound in response to pathogens and herbivore attack is equally important. Such a selective advantage has been shown by the transgenic expression of Sb-DHR from sorghum in barley. Barley normally synthesizes the Leu-derived cyanogenic glucoside epiheterodendrin but lacks an activating β-GLU (Nielsen et al., 2002). Transgenic expression of Sb-DHR established cyanogenesis and increased the resistance of the plant against the pathogen Blumeria graminis (Nielsen et al., 2006).
Mechanisms for the evolution of metabolic pathways have been a matter of debate for a long time. Jensen (1976) proposed proliferation of pathways based on the substrate ambiguity of enzymes. Promiscuous functions are latent and not under selection pressure. Experimental data (e.g., Aharoni et al., 2005) demonstrated the possibility for generation of a new enzyme activity based on promiscuity. Interestingly, additional enzyme function is gained without loss of the original activity, demonstrating a plasticity that can serve as a starting point in evolution. Substrate ambiguity of UGTs and β-GLUs may provide an easily accessible starting point for the establishment of suitable stabilization and activation functions and hence enable repeated evolution of benzoxazinoid biosynthesis. Subsequently, gene duplication and functionalization can deliver more specific enzymes.
Is the finding that the enzymes for stabilization and activation of benzoxazinoids have been recruited independently in the grasses and in C. orientalis an indication of convergent evolution of the pathway? Two scenarios are conceivable: convergent evolution of the whole pathway in these taxa or monophyletic establishment followed by loss of individual functions and consecutive secondary recruitment of suitable functions from their respective gene pools. Repeated recruitment of the branchpoint enzyme BX1 of the pathway has been shown (Schullehner et al., 2008). Hence, the module that consists of the modifying P450 enzymes remains the only part of the pathway that might be of monophyletic origin. At present, members of the P450 Cyp71C subfamiliy to which Bx2 to Bx5 belong have not been detected in the genomes of dicot plants (cytochrome p450 homepage D.R. Nelson, http://drnelson.uthsc.edu/CytochromeP450.html). This finding suggests that the P450 enzymes required in DIBOA biosynthesis and, thus, the complete pathway evolved by repeated evolution in the dicot and grass lineages. Final proof, however, requires isolation of the respective P450 genes from C. orientalis. In any case, the independent origin of the enzymes for the branchpoint reaction, for stabilization, and for activation in the grasses and in C. orientalis indicates a great evolutionary flexibility in recruitment of these essential functions for secondary plant metabolism.
METHODS
Standards and Reference Chemicals
Benzoxazinoids were gifts from Dieter Sicker (University of Leipzig, Germany) or prepared as described by von Rad et al. (2001). Trans-zeatin and zeatin-riboside were purchased from Duchefa Biochemie, trans-zeatin-O-glucoside from OlChemim, and pNPG from Sigma-Aldrich.
Plant Materials and Growth Conditions
Plants were grown at 19°C at a 12-h-light/12-h-dark day cycle. Larkspur (Consolida orientalis) seed was provided by Margot Schulz (Institut für Molekulare Physiologie und Biotechnologie der Pflanzen, University Bonn, Germany). Seeds were vernalized at 4°C in soil. Arabidopsis thaliana seed, ecotype Columbia-0, was vernalized at 4°C and grown on half-strength Murashige and Skoog solidified medium or on soil. For selection, the medium was supplemented with 25 mg/L dl-phosphinotricine (Duchefa Biochemie). Nicotiana benthamiana was grown on soil and used for infiltration 5 to 6 weeks after germination.
Generation of Transgenic Plants
Arabidopsis was transformed by floral dip and selected as described by von Rad et al. (2001). Transient transgenic expression of β-GLU in N. benthamiana was as described by Marillonnet et al. (2005) with vectors described by Engler et al. (2008).
Molecular Biology Methods
DNA and RNA isolation, cDNA synthesis, cloning, and PCR amplification were as described by Schullehner et al. (2008). To determine the C. orientalis transcriptome, RNA of seedling shoots was prepared and first-strand cDNA was synthesized using degenerate primers. After normalization and ligation of adaptor sequences (Vertis Biotechnologie), the data were collected by 454 sequencing (Eurofins MWG Operon). The sequence reads were assembled using Phred software (Ewing et al., 1998; Ewing and Green, 1998). The transcriptome average sequence size was 340 bp. A C. orientalis cDNA library (young shoot; Schullehner et al., 2008) was screened for full-size transcripts of the respective gene. Probes for screening were partial gene sequences amplified by PCR using the sequence information from the transcriptome.
Isolation of His-tagged proteins and enzyme assays were as described by von Rad et al. (2001).
A list of primers used for cloning and expression analysis is given in Supplemental Table 5 online.
Enzyme Purification
Co-BX8 purification was essentially as described by von Rad et al. (2001) using 7- to 14-d-old leaves (see Supplemental Table 1 online). After ammonium sulfate precipitation (45 to 65% saturation), the protein was dissolved in 20 mM HEPES, 14 mM 2-mercaptoethanol, 0,5 mM EDTA, and 20% glycerol, pH 7.5 (buffer R), and the ammonium sulfate concentration was adjusted to 1 M. The protein solution was loaded to a HiTrap ButylFF column (2.5 × 1.6 cm, 5 mL; GE Healthcare). Elution was with a linear gradient of 25 column volumes at 0.5 mL/min to reach 100% buffer R. Fractions with Co-BX8 activity were pooled, and KCl was added to a final concentration of 0.1 M. The protein solution was applied to a sizing column (HiLoad 16/60 Superdex 200 prep grade, 60 × 1.6 cm, 120 mL, flow rate 0.5 mL/min; GE Healthcare,). Co-BX8 activity eluted with proteins of masses between 45 and 60 kD, the size range expected for UGT monomers of plant secondary metabolism (Vogt and Jones, 2000). The fractions with Co-BX8 activity were concentrated by centrifugation with Vivaspin (Sartorius) vials and transferred to a buffer containing 50 mM MES, 14 mM 2-mercaptoethanol, 0.5 mM EDTA, and 20% glycerol, pH 6.0, by application on NAP columns (GE Healthcare). The protein solution was applied to affinity chromatography (HiTrap Blue HP column; GE Healthcare). Elution was with a KCl gradient. The activity eluted at 200 mM KCl, and the fractions were immediately neutralized by addition of HEPES buffer, pH 8.0, concentrated, and transferred into buffer R as described above. Anion exchange chromatography (Mono Q, 5.0 cm × 0.5 cm, 1 mL; GE Healthcare) was employed as a final polishing step, and elution was with a gradient in buffer B to 1 M KCl. Analysis of individual fractions on SDS-PAGE gels revealed a band with a molecular mass of ~55 kD that correlated with DIBOA:UGT activity in the enzyme assay (see Supplemental Figure 1 online).
The enzyme preparation was used for the determination of peptide sequences as described by Jonczyk et al. (2008). Co-BX8 was identified by comparison of the transcriptome data with the peptide sequences
Determination of Catalytic Properties
Glucosylation of benzoxazinoids was assayed as described by von Rad et al. (2001). UDPG was used as glucose donor with either benzoxazinoid as substrate. For determination of enzymatic parameters with DIBOA and DIMBOA as acceptor substrates, 2 mM UDPG was used and the aglucone concentration varied from 0.175 to 21 mM and 0.175 to 10.5 mM, respectively. Determination was in three replicates. Enzyme parameters were determined using GraphPad Prism Version 4.03 with nonlinear regression (see Supplemental Figure 6A online).
Glucosylation of kaempferol, quercetin, myricetin, pelargonidin, and delphinidin was assayed as described by Griesser et al. (2008). For analysis of indole-3-acetic acid glucosylation, the protocol of Jackson et al. (2001) was used, and trans-zeatin glucosylation was tested according to Hou et al. (2004).
β-GLU activity with the artificial substrate pNPG was assayed as described by Esen and Cokmus (1990). The assays for GDIBOA, GDIMBOA, and dhurrin hydrolysis by β-GLU are described by Cicek et al. (2000), and trans-zeatin glucosidase activity was tested according to Brzobohatý et al. (1993). For the determination of catalytic parameters, the substrate GDIBOA was used at concentrations between 0.175 and 10.5 mM, GDIMBOA varied between 0.175 and 17.5 mM, and dhurrin was used in concentrations between 1 and 5 mM. Determination was in three replicates. Enzyme parameters were determined using GraphPad Prism Version 4.03 with nonlinear regression (see Supplemental Figure 6B online). Plant protein extracts were prepared as described by Marillonnet et al. (2005). Protein crude extracts used for the determination of Km values were passed through gel filtration columns (Illustra NAP-10/25 columns; GE Healthcare) to remove endogenous substrates and products. Boiled protein extracts were used as negative controls.
Phylogenetic Analysis
The initial files containing manually selected UGT and β-GLU protein sequences were aligned using the multiple alignment program MUSCLE 3.8.31 (Edgar, 2004). The alignments were manually trimmed to remove poorly conserved terminal regions and signal peptides (see Supplemental Data Sets 1 and 2 online). Phylogenetic trees were calculated using the program RAxML 7.0.4 (Stamatakis, 2006) using the PROTGAMMAJTT model, fast bootstrap mode (-f a option), and 1000 bootstrap samples. As the comparison of the derived topologies with trees generated by an alternative method (Bayesian approach) did not result in major differences, we used the RAxML trees for further analysis. The trees were visualized and colored by organism (monocots in green and dicots in red) using the iTOL software (Letunic and Bork, 2007). Internal nodes found in more than 500 of the bootstrap samples were visualized by circles, which were scaled according to their bootstrap values.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: HM559229 (Co-Bx8, UGT85N1), HM559224 (Co-F3GT, UGT78B3), HM559224 (Lg-F3GT, UGT78J1), HM559230 (Ta-Bx8, cv Kanzler UGT710E6), HM559225 (Co-Glu), and HM559226 (C. orientalis β-Glu with unknown function).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Purification of Co-BX8.
Supplemental Figure 2. Amino Acid Sequence of Co-BX8.
Supplemental Figure 3. Heterologous Expression of Proteins by Magnifection.
Supplemental Figure 4. Gene Expression Pattern of Candidate β-Glucosidases.
Supplemental Figure 5. Substrate Specificity of Co-F3GT and Lg-F3GT and Michaelis-Menten Kinetics of Ta-BX8.
Supplemental Figure 6. Michaelis Menten Kinetics of Co-BX8 and Co-GLU.
Supplemental Figure 7. Kinetics of DIBOA Glucosylation, GDIBOA, and Dhurrin Hydrolysis in C. orientalis Protein Crude Extract.
Supplemental Figure 8. Substrates Analyzed for Glucosylation by Co-BX8.
Supplemental Figure 9. Substrates Analyzed for Hydrolysis by Co-GLU.
Supplemental Figure 10. Kinetics of GDIBOA Cleavage in the Presence of Dhurrin.
Supplemental Table 1. Purification of Co-BX8 from C. orientalis.
Supplemental Table 2. Transcriptome Statistics.
Supplemental Table 3. Substrate Specificity of Co-BX8, Zm-BX8, and Zm-BX9.
Supplemental Table 4. Determination of Enzyme Specificity of Co-GLU.
Supplemental Table 5. Lists of Primers.
Supplemental Data Set 1. Amino Acid Alignment Used to Generate the Phylogenetic Tree of Plant Family 1 UDP-Glucosyltransferases Shown in Figure 4 (Phylip File).
Supplemental Data Set 2. Amino Acid Alignment Used to Generate the Phylogenetic Tree of Plant β-Glucosidases Shown in Figure 5 (Phylip File).
Acknowledgments
We thank Trang Pham Vu Thuy and Inge Göpfrich for the isolation and characterization of Co-Glu, Niklas Bechtel for work on C. orientalis and L. galebodolon F3GT, Agnes Luzak and Miriam Hillenmeyer for isolation and characterization of Ta-Bx8, and Linlin Zheng for expert quantitative RT-PCR analysis. We also thank Margot Schulz and Dieter Sicker for valuable suggestions and the provision of seed and standard compounds and Peter I. Mackenzie for the classification of the UGTs. We thank Michael G. Muszynski for critically reading the manuscript and for helpful comments. This work was supported by grant GI140/11-3 of the Deutsche Forschungsgemeinshaft Priority Programme 1152 “Evolution of Metabolic Diversity” to A.G.
AUTHOR CONTRIBUTIONS
R.D. performed the research. T.R. contributed new computational tools for phylogenetic analysis. M.H. provided the peptide sequence data for identification of Co-BX8. W.S. contributed tools for the analysis of substrates and products. A.G. and M.F. designed the research and wrote the article.
References
- Aharoni A., Gaidukov L., Khersonsky O., McQ Gould S., Roodveldt C., Tawfik D.S. (2005). The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 37: 73–76 [DOI] [PubMed] [Google Scholar]
- Babcock G.D., Esen A. (1994). Substrate specificity of maize β-glucosidase. Plant Sci. 101: 31–39 [Google Scholar]
- Bak S., et al. (2006). Cyanogenic glycosides: A case study for evolution and application of cytochromes P450. Phytochem. Rev. 5: 309–329 [Google Scholar]
- Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. (2002). Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18: 298–305 [DOI] [PubMed] [Google Scholar]
- Barth C., Jander G. (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46: 549–562 [DOI] [PubMed] [Google Scholar]
- Blum U. (1999). Designing laboratory plant debris–soil bioassays: some reflections. In Principles and Practices in Plant Ecology: Allelochemical Interactions. Inderjit, Dakshini K.M.M., Foy C.L., (Boca Raton, FL: CRC; ), pp.17–23 [Google Scholar]
- Brown N.J., Newell C.A., Stanley S., Chen J.E., Perrin A.J., Kajala K., Hibberd J.M. (2011). Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331: 1436–1439 [DOI] [PubMed] [Google Scholar]
- Brzobohatý B., Moore I., Kristoffersen P., Bako L., Campos N., Schell J., Palme K. (1993). Release of active cytokinin by a beta-glucosidase localized to the maize root meristem. Science 262: 1051–1054 [DOI] [PubMed] [Google Scholar]
- Campos N., Bako L., Feldwisch J., Schell J., Palme K. (1992). A protein from maize labeled with azido-IAA has novel beta-glucosidase activity. Plant J. 2: 675–684 [Google Scholar]
- Chuankhayan P., Rimlumduan T., Tantanuch W., Mothong N., Kongsaeree P.T., Metheenukul P., Svasti J., Jensen O.N., Cairns J.R.K. (2007). Functional and structural differences between isoflavonoid beta-glycosidases from Dalbergia sp. Arch. Biochem. Biophys. 468: 205–216 [DOI] [PubMed] [Google Scholar]
- Cicek M., Blanchard D., Bevan D.R., Esen A. (2000). The aglycone specificity-determining sites are different in 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA)-glucosidase (maize beta-glucosidase) and dhurrinase (sorghum beta-glucosidase). J. Biol. Chem. 275: 20002–20011 [DOI] [PubMed] [Google Scholar]
- Cicek M., Esen A. (1998). Structure and expression of a dhurrinase (beta-glucosidase) from sorghum. Plant Physiol. 116: 1469–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cseke L., Dudareva N., Pichersky E. (1998). Structure and evolution of linalool synthase. Mol. Biol. Evol. 15: 1491–1498 [DOI] [PubMed] [Google Scholar]
- Davies G., Henrissat B. (1995). Structures and mechanisms of glycosyl hydrolases. Structure 3: 853–859 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Engler C., Kandzia R., Marillonnet S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen A. (1992). Purification and partial characterization of maize (Zea mays L.) beta-glucosidase. Plant Physiol. 98: 174–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen A., Cokmus C. (1990). Maize genotypes classified as null at the glu locus have beta-glucosidase activity and immunoreactive protein. Biochem. Genet. 28: 319–336 [DOI] [PubMed] [Google Scholar]
- Ewing B., Green P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194 [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Finn R.D., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38(Database issue): D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T.K., Yadollahi A., Wirthensohn M.G., Guerin J.R., Kaiser B.N., Sedgley M., Ford C.M. (2008). A seed coat cyanohydrin glucosyltransferase is associated with bitterness in almond (Prunus dulcis) kernels. Funct. Plant Biol. 35: 236–246 [DOI] [PubMed] [Google Scholar]
- Frey M., Chomet P., Glawischnig E., Stettner C., Grün S., Winklmair A., Eisenreich W., Bacher A., Meeley R.B., Briggs S.P., Simcox K., Gierl A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699 [DOI] [PubMed] [Google Scholar]
- Frey M., Huber K., Park W.J., Sicker D., Lindberg P., Meeley R.B., Simmons C.R., Yalpani N., Gierl A. (2003). A 2-oxoglutarate-dependent dioxygenase is integrated in DIMBOA-biosynthesis. Phytochemistry 62: 371–376 [DOI] [PubMed] [Google Scholar]
- Frey M., Kliem R., Saedler H., Gierl A. (1995). Expression of a cytochrome P450 gene family in maize. Mol. Gen. Genet. 246: 100–109 [DOI] [PubMed] [Google Scholar]
- Frey M., Schullehner K., Dick R., Fiesselmann A., Gierl A. (2009). Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70: 1645–1651 [DOI] [PubMed] [Google Scholar]
- Frey M., Stettner C., Pare P.W., Schmelz E.A., Tumlinson J.H., Gierl A. (2000). An herbivore elicitor activates the gene for indole emission in maize. Proc. Natl. Acad. Sci. USA 97: 14801–14806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F.J., Nebert D.W. (1990). Evolution of the P450 gene superfamily: animal-plant ‘warfare’, molecular drive and human genetic differences in drug oxidation. Trends Genet. 6: 182–186 [DOI] [PubMed] [Google Scholar]
- Griesser M., Vitzthum F., Fink B., Bellido M.L., Raasch C., Munoz-Blanco J., Schwab W. (2008). Multi-substrate flavonol O-glucosyltransferases from strawberry (Fragaria x ananassa) achene and receptacle. J. Exp. Bot. 59: 2611–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün S., Frey M., Gierl A. (2005). Evolution of the indole alkaloid biosynthesis in the genus Hordeum: Distribution of gramine and DIBOA and isolation of the benzoxazinoid biosynthesis genes from Hordeum lechleri. Phytochemistry 66: 1264–1272 [DOI] [PubMed] [Google Scholar]
- Hartmann T., Witte L. (1995). Chemistry, biology and chemoecology of the pyrrolizidine alkaloids. In Alkaloids: Chemical and Biological Perspectives, Pelletier S.W., (Oxford, UK: Pergamon Press; ), pp. 155–233 [Google Scholar]
- Horowitz N.H. (1945). On the evolution of biochemical syntheses. Proc. Natl. Acad. Sci. USA 31: 153–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostettmann K., Marston A. (1995). Saponins. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Hou B.K., Lim E.K., Higgins G.S., Bowles D.J. (2004). N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 279: 47822–47832 [DOI] [PubMed] [Google Scholar]
- Hughes J., Hughes M.A. (1994). Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq. 5: 41–49 [DOI] [PubMed] [Google Scholar]
- Jackson R.G., Lim E.K., Li Y., Kowalczyk M., Sandberg G., Hoggett J., Ashford D.A., Bowles D.J. (2001). Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 276: 4350–4356 [DOI] [PubMed] [Google Scholar]
- Jensen N.B., Zagrobelny M., Hjernø K., Olsen C.E., Houghton-Larsen J., Borch J., Møller B.L., Bak S. (2011). Convergent evolution in biosynthesis of cyanogenic defence compounds in plants and insects. Nat. Commun. 2: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.A. (1976). Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30: 409–425 [DOI] [PubMed] [Google Scholar]
- Jonczyk R., Schmidt H., Osterrieder A., Fiesselmann A., Schullehner K., Haslbeck M., Sicker D., Hofmann D., Yalpani N., Simmons C., Frey M., Gierl A. (2008). Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: characterization of Bx6 and Bx7. Plant Physiol. 146: 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.R., Møller B.L., Hoj P.B. (1999). The UDP-glucose:p-hydroxymandelonitrile-O-glucosyltransferase that catalyzes the last step in synthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor. Isolation, cloning, heterologous expression, and substrate specificity. J. Biol. Chem. 274: 35483–35491 [DOI] [PubMed] [Google Scholar]
- Jørgensen K., Morant A.V., Morant M., Jensen N.B., Olsen C.E., Kannangara R., Motawia M.S., Møller B.L., Bak S. (2011). Biosynthesis of the cyanogenic glucosides linamarin and lotaustralin in cassava: Isolation, biochemical characterization, and expression pattern of CYP71E7, the oxime-metabolizing cytochrome P450 enzyme. Plant Physiol. 155: 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannangara R., Motawia M.S., Hansen N.K., Paquette S.M., Olsen C.E., Møller B.L., Jørgensen K. (2011). Characterization and expression profile of two UDP-glucosyltransferases, UGT85K4 and UGT85K5, catalyzing the last step in cyanogenic glucoside biosynthesis in cassava. Plant J. 68: 287–301 [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2007). Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128 [DOI] [PubMed] [Google Scholar]
- Lücker J., Bouwmeester H.J., Schwab W., Blaas J., van der Plas L.H.W., Verhoeven H.A. (2001). Expression of Clarkia S-linalool synthase in transgenic petunia plants results in the accumulation of S-linalyl-beta-D-glucopyranoside. Plant J. 27: 315–324 [DOI] [PubMed] [Google Scholar]
- Mackenzie P.I., et al. (1997). The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7: 255–269 [DOI] [PubMed] [Google Scholar]
- Marillonnet S., Thoeringer C., Kandzia R., Klimyuk V., Gleba Y. (2005). Systemic Agrobacterium tumefaciens-mediated transfection of viral replicons for efficient transient expression in plants. Nat. Biotechnol. 23: 718–723 [DOI] [PubMed] [Google Scholar]
- Morant A.V., Bjarnholt N., Kragh M.E., Kjaergaard C.H., Jørgensen K., Paquette S.M., Piotrowski M., Imberty A., Olsen C.E., Møller B.L., Bak S. (2008a). The beta-glucosidases responsible for bioactivation of hydroxynitrile glucosides in Lotus japonicus. Plant Physiol. 147: 1072–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant A.V., Jørgensen K., Jørgensen C., Paquette S.M., Sánchez-Pérez R., Møller B.L., Bak S. (2008b). Beta-glucosidases as detonators of plant chemical defense. Phytochemistry 69: 1795–1813 [DOI] [PubMed] [Google Scholar]
- Nelson D., Werck-Reichhart D. (2011). A P450-centric view of plant evolution. Plant J. 66: 194–211 [DOI] [PubMed] [Google Scholar]
- Nelson D.R. (2006). Plant cytochrome P450s from moss to poplar. Phytochem. Rev. 5: 193–204 [Google Scholar]
- Nielsen K.A., Hrmova M., Nielsen J.N., Forslund K., Ebert S., Olsen C.E., Fincher G.B., Møller B.L. (2006). Reconstitution of cyanogenesis in barley (Hordeum vulgare L.) and its implications for resistance against the barley powdery mildew fungus. Planta 223: 1010–1023 [DOI] [PubMed] [Google Scholar]
- Nielsen K.A., Olsen C.E., Pontoppidan K., Møller B.L. (2002). Leucine-derived cyano glucosides in barley. Plant Physiol. 129: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer H.M. (1988). Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones), defense chemicals in the Gramineae. Phytochemistry 27: 3349–3358 [Google Scholar]
- Nikus J., Daniel G., Jonsson L.M.V. (2001). Subcellular localization of beta-glucosidase in rye, maize and wheat seedlings. Physiol. Plant. 111: 466–472 [DOI] [PubMed] [Google Scholar]
- Nikus J., Esen A., Jonsson L.M.V. (2003). Cloning of a plastidic rye (Secale cereale) beta-glucosidase cDNA and its expression in Escherichia coli. Physiol. Plant. 118: 337–345 [Google Scholar]
- Nomura T., Ishihara A., Imaishi H., Endo T.R., Ohkawa H., Iwamura H. (2002). Molecular characterization and chromosomal localization of cytochrome P450 genes involved in the biosynthesis of cyclic hydroxamic acids in hexaploid wheat. Mol. Genet. Genomics 267: 210–217 [DOI] [PubMed] [Google Scholar]
- Nomura T., Ishihara A., Imaishi H., Ohkawa H., Endo T.R., Iwamura H. (2003). Rearrangement of the genes for the biosynthesis of benzoxazinones in the evolution of Triticeae species. Planta 217: 776–782 [DOI] [PubMed] [Google Scholar]
- Nomura T., Ishihara A., Iwamura H., Endo T.R. (2007). Molecular characterization of benzoxazinone-deficient mutation in diploid wheat. Phytochemistry 68: 1008–1016 [DOI] [PubMed] [Google Scholar]
- Nomura T., Ishihara A., Yanagita R.C., Endo T.R., Iwamura H. (2005). Three genomes differentially contribute to the biosynthesis of benzoxazinones in hexaploid wheat. Proc. Natl. Acad. Sci. USA 102: 16490–16495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opassiri R., Pomthong B., Onkoksoong T., Akiyama T., Esen A., Ketudat Cairns J.R. (2006). Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 beta-glucosidase. BMC Plant Biol. 6: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette S., Møller B.L., Bak S. (2003). On the origin of family 1 plant glycosyltransferases. Phytochemistry 62: 399–413 [DOI] [PubMed] [Google Scholar]
- Pichersky E., Lewinsohn E. (2011). Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 62: 549–566 [DOI] [PubMed] [Google Scholar]
- Rauscher M.D. (2006). The evolution of flavonoids and their genes. In The Science of Flavonoids, Grotewold E., (New York: Springer Science and Business Media; ), pp. 175–211 [Google Scholar]
- Reimann A., Nurhayati N., Backenköhler A., Ober D. (2004). Repeated evolution of the pyrrolizidine alkaloid-mediated defense system in separate angiosperm lineages. Plant Cell 16: 2772–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C.P., Park Y.B., Adam F., Abdul-Baki A.A., Teasdale J.R. (2005). Hydroxamic acid content and toxicity of rye at selected growth stages. J. Chem. Ecol. 31: 1887–1905 [DOI] [PubMed] [Google Scholar]
- Ross J., Li Y., Lim E.K., Bowles D.J. (2001). Higher plant glycosyltransferases. Genome Biol. 2: S3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodman J., Soltis P., Soltis D., Sytsma K., Karol K. (1998). Parallel evolution of glucosinolate biosynthesis inferred from congruent nuclear and plastid gene phylogenies. Am. J. Bot. 85: 997. [PubMed] [Google Scholar]
- Schäfer H., Nau K., Sickmann A., Erdmann R., Meyer H.E. (2001). Identification of peroxisomal membrane proteins of Saccharomyces cerevisiae by mass spectrometry. Electrophoresis 22: 2955–2968 [DOI] [PubMed] [Google Scholar]
- Schmälzlin K. (2002). Experimentelle Analyse der DIMBOA-Biosynthese in Zea mays. PhD Dissertation (Munich, Germany: Technische Universität München).
- Schullehner K., Dick R., Vitzthum F., Schwab W., Brandt W., Frey M., Gierl A. (2008). Benzoxazinoid biosynthesis in dicot plants. Phytochemistry 69: 2668–2677 [DOI] [PubMed] [Google Scholar]
- Schwab W. (2003). Metabolome diversity: Too few genes, too many metabolites? Phytochemistry 62: 837–849 [DOI] [PubMed] [Google Scholar]
- Sharples D., Spring M.S., Stoker J.R. (1972). Biosynthesis of the major cyanogenic glycoside of Thalictrum aquilegifolium. Phytochemistry 11: 2999–3002 [Google Scholar]
- Sicker D., Frey M., Schulz M., Gierl A. (2000). Role of natural benzoxazinones in the survival strategy of plants. Int. Rev. Cytol. 198: 319–346 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Sue M., Ishihara A., Iwamura H. (2000). Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings. Plant Sci. 155: 67–74 [DOI] [PubMed] [Google Scholar]
- Sue M., Nakamura C., Nomura T. (2011). Dispersed benzoxazinone gene cluster: Molecular characterization and chromosomal localization of glucosyltransferase and glucosidase genes in wheat and rye. Plant Physiol. 157: 985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sue M., Yamazaki K., Yajima S., Nomura T., Matsukawa T., Iwamura H., Miyamoto T. (2006). Molecular and structural characterization of hexameric beta-D-glucosidases in wheat and rye. Plant Physiol. 141: 1237–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos A., Lai D., Mikkelsen L., Abou Hachem M., Shelton D., Motawia M.S., Olsen C.E., Wang T.L., Martin C., Rook F. (2010). Genetic screening identifies cyanogenesis-deficient mutants of Lotus japonicus and reveals enzymatic specificity in hydroxynitrile glucoside metabolism. Plant Cell 22: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos A.M., Knudsen C., Lai D., Kannangara R., Mikkelsen L., Motawia M.S., Olsen C.E., Sato S., Tabata S., Jørgensen K., Møller B.L., Rook F. (2011). Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant J. 68: 273–286 [DOI] [PubMed] [Google Scholar]
- Tattersall D.B., Bak S., Jones P.R., Olsen C.E., Nielsen J.K., Hansen M.L., Høj P.B., Møller B.L. (2001). Resistance to an herbivore through engineered cyanogenic glucoside synthesis. Science 293: 1826–1828 [DOI] [PubMed] [Google Scholar]
- Vetter J. (2000). Plant cyanogenic glycosides. Toxicon 38: 11–36 [DOI] [PubMed] [Google Scholar]
- Vogt T., Jones P. (2000). Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 5: 380–386 [DOI] [PubMed] [Google Scholar]
- von Rad U., Hüttl R., Lottspeich F., Gierl A., Frey M. (2001). Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J. 28: 633–642 [DOI] [PubMed] [Google Scholar]
- Xie Y., Arnason T.J., Philogene B.J.R., Olechowski H.T., Hamilton R.I. (1992). Variation of hydroxamic acid content in maize roots in relation to geographic origin of maize germ plasm and resistance to Western corn rootworm (Coleoptera: Chrysomelidae). J. Econ. Entomol. 85: 2478–2485 [Google Scholar]
- Xu Z.W., Escamilla-Treviño L.L., Zeng L.H., Lalgondar M., Bevan D.R., Winkel B.S.J., Mohamed A., Cheng C.L., Shih M.C., Poulton J.E., Esen A. (2004). Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Biol. 55: 343–367 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Hanada K. (2011). An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 66: 182–193 [DOI] [PubMed] [Google Scholar]
- Zasada I.A., Meyer S.L., Halbrendt J.M., Rice C. (2005). Activity of hydroxamic acids from Secale cereale against the plant-parasitic nematodes Meloidogyne incognita and Xiphinema americanum. Phytopathology 95: 1116–1121 [DOI] [PubMed] [Google Scholar]
- Zhou J.M., Hartmann S., Shepherd B.K., Poulton J.E. (2002). Investigation of the microheterogeneity and aglycone specificity-conferring residues of black cherry prunasin hydrolases. Plant Physiol. 129: 1252–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Fiesselmann A., Zhao N., Chen H., Frey M., Chen F. (2012). Biosynthesis and emission of insect herbivory-induced volatile indole in rice. Phytochemistry 73: 15–22 [DOI] [PubMed] [Google Scholar]