This work shows that degradation of oxalate via oxalyl-CoA synthetase is required for correct seed development and for pathogen defense in Arabidopsis. It identifies AAE3 as an oxalyl-CoA synthetase, providing support for an alternative pathway of oxalate degradation first proposed 50 years ago.
Abstract
Oxalate is produced by several catabolic pathways in plants. The best characterized pathway for subsequent oxalate degradation is via oxalate oxidase, but some species, such as Arabidopsis thaliana, have no oxalate oxidase activity. Previously, an alternative pathway was proposed in which oxalyl-CoA synthetase (EC 6.2.1.8) catalyzes the first step, but no gene encoding this function has been found. Here, we identify ACYL-ACTIVATING ENZYME3 (AAE3; At3g48990) from Arabidopsis as a gene encoding oxalyl-CoA synthetase. Recombinant AAE3 protein has high activity against oxalate, with Km = 149.0 ± 12.7 μM and Vmax = 11.4 ± 1.0 μmol/min/mg protein, but no detectable activity against other organic acids tested. Allelic aae3 mutants lacked oxalyl-CoA synthetase activity and were unable to degrade oxalate into CO2. Seeds of mutants accumulated oxalate to levels threefold higher than the wild type, resulting in the formation of oxalate crystals. Crystal formation was associated with seed coat defects and substantially reduced germination of mutant seeds. Leaves of mutants were damaged by exogenous oxalate and more susceptible than the wild type to infection by the fungus Sclerotinia sclerotiorum, which produces oxalate as a phytotoxin to aid infection. Our results demonstrate that, in Arabidopsis, oxalyl-CoA synthetase encoded by AAE3 is required for oxalate degradation, for normal seed development, and for defense against an oxalate-producing fungal pathogen.
INTRODUCTION
Oxalic acid, the smallest (two-carbon) dicarboxylic acid is thought to be present in most plant tissues. In many plant species, oxalate is known to be produced by the breakdown of ascorbate (vitamin C) via 4-0-oxalyl-threonate (Green and Fry, 2005; Franceschi and Nakata, 2005). Other demonstrated or proposed sources of oxalate in plants include oxidation of glycolate or glyoxylate by glycolate oxidase (Richardson and Tolbert, 1961; Yu et al., 2010) and oxidative degradation of oxaloacetate (Chang and Beevers, 1968; Franceschi and Nakata, 2005). In many plants, oxalate is not merely an intermediate in catabolic pathways. It has been observed to be synthesized in response to aluminum toxicity (Ma et al., 1997; Klug and Horst, 2010), as a carbon source for symbiotic nitrogen fixation (Trinchant et al., 1994), and, most broadly, for the production of calcium oxalate crystals in diverse plant species (Nakata, 2003; Franceschi and Nakata, 2005). In oxalate-accumulating plants, calcium oxalate crystal accumulation occurs in specialized idioblast cells. The crystals are thought to deter herbivore feeding (Korth et al., 2006) and may also serve a role in regulating free calcium levels (Franceschi and Nakata, 2005). Oxalic acid is toxic when applied to plant leaves (Dong et al., 2008), and some fungal pathogens, such as Sclerotinia sclerotiorum, secrete oxalate that acts as an important virulence factor during infection. Evidence indicates that oxalate may stimulate stomatal opening, interfere with cell wall structure, induce low pH–activated pectolytic enzymes and act as an elicitor of programmed cell death (Bateman and Beer, 1965; Lumsden, 1976; Guimarães and Stotz, 2004; Hegedus and Rimmer, 2005; Dong et al., 2008; Kim et al., 2008). S. sclerotiorum mutants deficient in oxalate synthesis have greatly reduced infectivity (Godoy et al., 1990; Guimarães and Stotz, 2004).
These considerations indicate that oxalate degradation is important for metabolism and plant health. Some plants contain oxalate oxidase enzymes that breakdown oxalate to CO2 and H2O2 (Srivastava and Krishnan, 1962; Suguira et al., 1979). Oxalate oxidase was shown to be an activity of some germin proteins that belong to a family of abundant cell wall proteins characterized in barley (Hordeum vulgare) and other monocot species (Lane et al., 1993; Druka et al., 2002). Importantly, expression of a wheat (Triticum aestivum) germin with oxalate oxidase activity in transgenic soybean (Glycine max) or rape (Brassica napus) plants led to enhanced resistance to S. sclerotiorum infection (Donaldson et al., 2001; Dong et al., 2008). Although Arabidopsis thaliana expresses a number of germin-like proteins, it does not contain detectable oxalate oxidase activity (Membré et al., 1997; 2000).
A less studied pathway of oxalate degradation was proposed in 1961 by Giovanelli and Tobin (Giovanelli and Tobin, 1961). Labeling experiments and enzyme assays in pea (Pisum sativum) seed extracts indicated a CoA- and ATP-dependent pathway leading to the production of formic acid and CO2 (Giovanelli and Tobin, 1964). Although some bacteria were known to synthesize oxalyl-CoA by a CoA transferase reaction, Giovanelli (1966) characterized an oxalyl-CoA synthetase activity in pea seed and other plant extracts that is proposed to be the first enzyme in this pathway. To date, however, no gene encoding oxalyl-CoA synthetase has been cloned from any organism. Here, we describe the identification of the Arabidopsis ACYL-ACTIVATING ENZYME3 (AAE3) gene (At3g48990) encoding a specific cytoplasmic oxalyl-CoA synthetase. Characterization of two aae3 mutants indicates that the AAE3 enzyme is required in Arabidopsis for oxalate degradation to CO2. Seeds of the mutants contain threefold higher levels of oxalate compared with the wild type and have reduced and delayed germination. In leaf tissue, increasing oxalyl-CoA synthetase activities are positively correlated with increased resistance to infection by S. sclerotiorum. These results indicate that oxalyl-CoA synthetase encoded by AAE3 has multiple potential roles in metabolism and plant health.
RESULTS
AAE3 Functions as an Oxalyl-CoA Synthetase
As one approach to identify the functions of AAE proteins encoded by the Arabidopsis genome, we surveyed genes whose patterns of expression are highly correlated with each AAE family member across the Arabidopsis Gene Expression database (Schmid et al., 2005) using the ATTEDII (www.atted.bio.titech.ac.jp) (Obayashi et al., 2009) and Gene Angler (http://bbc.botany.utoronot.ca) (Toufighi et al., 2005) search engines. These search engines calculate Pearson correlation coefficients for coexpression of a query gene with all of the genes represented on the Affymetrix ATH1 whole-genome array. Some genes coregulated with AAE3, such as those encoding formate dehydrogenase, putative malate dehydrogenase, formate-tetrahydrofolate ligase, isocitrate dehydrogenase, and glutamate dehydrogenase, indicated potential roles in organic acid metabolism (see Supplemental Table 1 online). Further investigation of the pathways associated with the genes coregulated with AAE3 was performed using the KEGG pathway database (Kanehisa and Goto, 2000). The pathway database indicated possible involvement of AAE3 within the glyoxylate pathway; therefore, potential substrates within this pathway were initially tested.
To discover whether any of the carboxylic acids involved in glyoxylate metabolism is a substrate for the AAE3 CoA synthetase, we set out to assay the activity of recombinant AAE protein with a range of potential substrates. A full-length cDNA of AAE3 (At3g48990) was obtained by RT-PCR from Arabidopsis mRNA and expressed as a His fusion protein in Escherichia coli using the pDEST17 vector. Expression of His-AAE3 was induced with arabinose and the recombinant protein purified by nickel-affinity chromatography. Upon electrophoresis in a SDS-polyacrylamide gel and visualization with Coomassie blue stain, the recombinant protein was estimated to be >90% pure (Figure 1A). A preparation was tested against 10 carboxylic acids at pH 7.5 and 300 μM substrate, using a spectrophotometric, coupled-enzyme assay (Ziegler et al., 1987; Chen et al., 2011). In these initial assays, His-AAE3 showed high activity only with oxalate, 5.8 μmol/min/mg protein. Activities against related compounds, including malonate succinate, glutarate, malate, glycolate, glyoxylate, acetate, lactate, and formate, were very low compared with that using oxalate as substrate, <0.1 μmol/min/mg AAE3 protein (Table 1). Additional assays indicated that AAE3 was inactive against a range of fatty acid substrates, including palmitic, stearic, oleic, and linolenic acids (data not shown). Although these assays are not completely exhaustive, they suggest that AAE3 is a highly specific oxalyl-CoA synthetase.
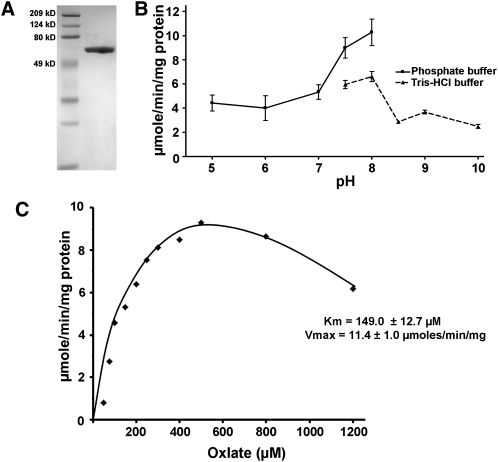
Figure 1.
Biochemical Analysis of Recombinant AAE3 Protein.
(A) SDS-PAGE gel of nickel affinity–purified His-AAE3 protein stained with Coomassie blue. Right lane, His-AAE3; left lane, molecular weight markers.
(B) Optimum pH for AAE3 was determined using 300 μM oxalate as substrate and two buffer systems: sodium phosphate for pH 5.0 to 8.0 and Tris-HCl for pH 7.5 to 10.0. Data are the mean ± se of two replicates.
(C) Kinetic analysis of AAE3 was performed using oxalate concentrations from 50 to 1200 μM oxalate. Km and Vmax were determined from nonlinear regression to the Michaelis-Menten equation for concentrations of oxalate up to 500 μM from five replicate experiments.
Table 1.
Substrate Specificity of AAE3
| Substrate | Structure | Activity μmoles/min/mg Protein |
| Oxalate | HOOC-COOH | 5.8 |
| Malonate | HOOC-CH2-COOH | <0.1 |
| Succinate | HOOC-(CH2)2-COOH | <0.1 |
| Malate | HOOC-CH2 –CHOH-COOH | <0.1 |
| Acetate | CH3-COOH | <0.1 |
| Formate | H-COOH | <0.1 |
| Lactate | CH3-CHOH-COOH | <0.1 |
| Glycolate | CHOH-COOH | <0.1 |
| Glyoxylate | CHO-COOH | <0.1 |
| Glutarate | HOOC(CH2)3-COOH | <0.1 |
CoA synthetase activities were determined at pH 7.5, with a substrate concentration of 300 μM.
The His-AAE3 protein had activity against oxalate over a wide range of pH, with an optimum at pH 8.0 (Figure 1B). At pH 8.0 and with saturating concentrations of CoA (0.5 mM) and ATP (5 mM), the enzyme showed Michaelis-Menten kinetics with respect to oxalate concentration up to 500 μM, with substrate inhibition evident at 800 and 1200 μM oxalate that could be due to oxalate interfering with other enzymes or components present in the assay (Figure 1C). Using the data up to 500 μM provided a calculated Vmax of 11.4 ± 1.0 μmol/min/mg protein and a Km of 149.0 ± 12.7 μM. In comparison, the apparent Km values of oxalyl-CoA synthetases from partially purified protein extracts of pea and Lathyrus sativus were 2 and 1.33 mM, respectively, using a hydroxamate assay (Giovanelli, 1966; Malathi et al., 1970).
AAE3 Is Localized to the Cytosol
The omnibus database for bioinformatics-based predictions on the targeting of Arabidopsis proteins, SUBA (Heazlewood et al., 2005), contains conflicting information for the AAE3 protein. The TargetP program indicated a nonorganellar localization, while WoLFPSORT favored plastid localization, but both predictions came with low reliability scores. Peptides corresponding to AAE3 have been identified at low frequency in two proteomics studies of Arabidopsis chloroplasts (Joyard et al., 2009; Sun et al., 2009). To further investigate the subcellular localization of the AAE3 protein, we expressed green fluorescent protein (GFP) fusions in plant cells. Plasmid and binary vectors containing cDNAs encoding both AAE3-GFP and GFP-AAE3 fusion proteins, under control of the strong, constitutive 35S promoter were constructed. Bombardment of onion epidermal cells with gold particles coated with 35S:AAE3-GFP plasmid resulted in strong GFP signal in the cytoplasm of cells similar to that seen in the 35S:GFP control (Figure 2A). The 35S:GFP-AAE3 construct also localized to the cytoplasm (see Supplemental Figure 1 online).
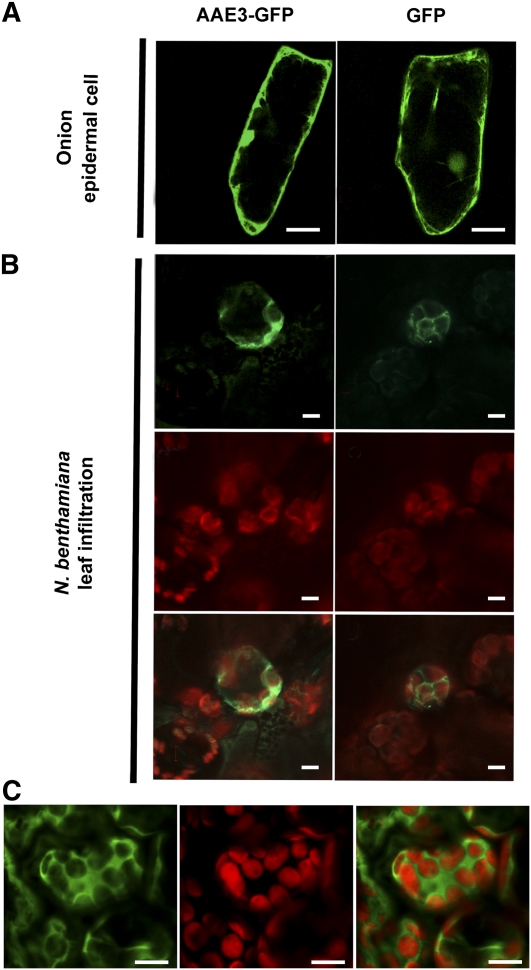
Figure 2.
Subcellular Localization of AAE3.
(A) GFP fluorescence images of onion epidermal cells transiently expressing AAE3-GFP (left panel) and free GFP (right panel). Bars = 50 μM.
(B) Transient expression of an AAE3-GFP fusion (left panels) and free GFP (right panels) in N. benthamiana leaf parenchyma cells introduced by Agrobacterium infiltration. GFP fluorescence images (top panels), chloroplast autofluorescence (middle panels), and merge of GFP and autofluorescence (bottom panels). Bars = 5 μM.
(C) Confocal image displaying the upper focal plane of mesophyll cells in Arabidopsis plants expressing a AAE3p:AAE3-GFP transgene. GFP fluorescence image (left panel), chloroplast autofluorescence (middle panel), and merge of GFP and autofluorescence (right panel). Bars = 10 μM.
The binary constructs were transfected into Agrobacterium tumefaciens and used for transient expression through infection of Nicotiana benthamiana leaves. Confocal imaging of mesophyll cells of N. benthamiana expressing the AAE3-GFP fusion protein showed GFP signal in the cytoplasm of cells and not coincident with the red chlorophyll autofluorescence that identifies the chloroplasts (Figure 2B). A similar pattern was observed for the GFP-AAE3 fusion protein (data not shown) and the GFP control (Figure 2B). Finally, we placed the AAE3-GFP coding sequence under control of the Arabidopsis AAE3 promoter (construct AAE3p:AAE3-GFP) and transformed this into the aae3 mutant. The transgene complemented the aae3 phenotype (see below), indicating that the AAE3-GFP fusion protein is functional. Confocal imaging of leaf cells from these transgenic plants again indicated cytoplasmic localization (Figure 2C). Taken together, our results indicate that the Arabidopsis AAE3 gene encodes a cytoplasmic oxalyl-CoA synthetase that may represent the activity first described by Giovanelli (1966) and possibly involved in a pathway of oxalate degradation (Giovanelli and Tobin, 1961, 1964).
Isolation of aae3 Mutants and Initial Phenotypic Analysis
To investigate the biological roles of oxalyl-CoA synthetase, we identified aae3 mutants by reverse genetics. Two T-DNA insertion lines are cataloged as having T-DNA inserted in the first (SALK_109915) or second (SALK_024432) exon of the AAE3 open reading frame, as shown in Figure 3A. Seed of both lines were obtained from the ABRC at Ohio State University. Individual plants of each line were genotyped by PCR using oligonucleotide primers P1/P2 and P3/P4, flanking the insertion sites, and the LBa1 primer designed to the left border of the T-DNA. DNA from homozygous mutant plants provided PCR products corresponding to the size expected for LBa1-P2 or LBa1-P4 amplification, respectively, while wild-type DNA yielded larger molecular weight products expected from amplification between the AAE3 specific primers (Figure 3B). Seeds from these homozygous plants were collected and designated aae3-1 (SALK_109915) and aae3-2 (SALK_024432).
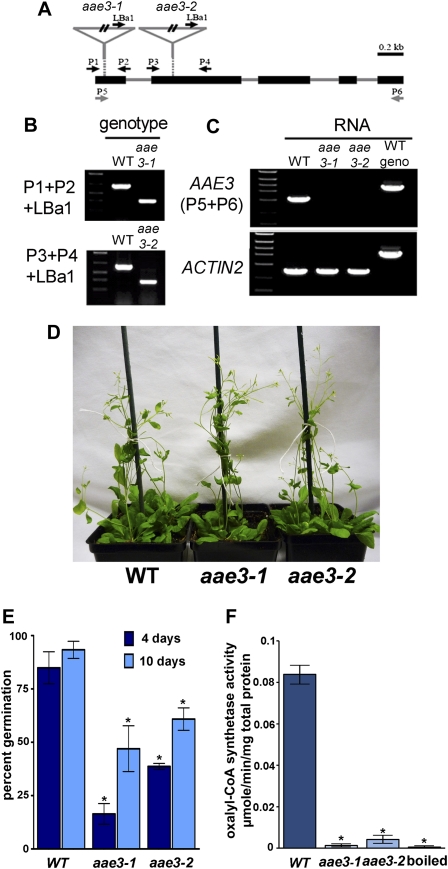
Figure 3.
Characterization of aae3 Mutants.
(A) Diagram of the aae3 locus, showing locations of the T-DNA inserts and the sites of primers used for analysis. Exons are represented by black rectangles and introns by solid lines. The T-DNA insertion sites for aae3-1 and aae3-2 are indicated by inverted triangles.
(B) Identification of the mutants by genomic PCR using gene-specific primers P1 and P2 for aae3-1 or P3 and P4 for aae3-2 and the T-DNA left border primer LBa1 using DNA from the wild type (WT), aae3-1, and aae3-2.
(C) RT-PCR analysis of AAE3 transcript levels in the wild type, aae3-1, and aae3-2 using primers P5 and P6. Arabidopsis ACT2 was amplified as a control for equal RNA levels.
(D) Image showing growth and development of 24-d-old wild-type, aae3-1, and aae3-2 plants grown in soil at 140 μmol photons/m2/s.
(E) Seed germination rates of the wild type, aae3-1, and aae3-2 on soil. Seeds were imbibed in water for 2 d at 4°C before planting. Germinated seedlings were counted 4 and 10 d after planting. Data are mean ± se of three independent sets of 100 seeds each. Asterisks denote significant difference from the wild type (*P < 0.05; Student’s t test).
(F) Oxalyl-CoA synthetase activities in crude extracts from the wild type, aae3-1, and aae3-2, measured using 300 μM oxalate as substrate. The wild-type crude protein extract was boiled and used as control for background. Data are mean ± se (n = 3). Asterisks denote significant difference from the wild type (*P < 0.05; Student’s t test).
We prepared RNA from aae3-1, aae3-2, and wild-type plants and performed RT-PCR using primers designed to amplify the entire AAE3 open reading frame. An RT-PCR product of 1.6 kb, corresponding to the expected size of AAE3, was obtained from wild-type RNA, but no product was obtained using RNA from aae3-1 or aae3-2 plants (Figure 3C). Amplification of the ACT2 transcript indicates that RNA from the mutants was not degraded.
Under our normal culture conditions (22°C; 150 μmol/m2/s illumination) and in the absence of pathogen challenge, growth and development of aae3 mutant plants was substantially similar to the wild type (Figure 3D), except for apparently decreased and delayed seed germination. To quantify the germination, fully imbibed seeds of the wild type, aae3-1, and aae3-2 were allowed to germinate over 10 d. As shown in Figure 3E, wild-type seed averaged 85% germination after 4 d and >90% germination after 10 d. By contrast, seed of both mutant lines showed delayed and decreased germination; after 10 d, germination averaged 47 and 61% for seed of aae3-1 and aae3-2, respectively.
We assayed oxalyl-CoA synthetase in homogenates from wild-type and mutant seedlings. The enzyme was readily detected in extracts from the wild type (Figure 3F), but the extracts from both mutants contained very low activities, which were comparable to that measured in the wild-type extract after heat treatment at 100°C for 5 min. These results indicate that aae3-1 and aae3-2 are null mutants that are deficient in oxalyl-CoA synthetase. As described below, the characteristics of aae3-1 plants and aae3-2 plants were entirely similar; this strongly indicates that all the phenotypes we observed are caused by the defect in oxalyl-CoA synthetase.
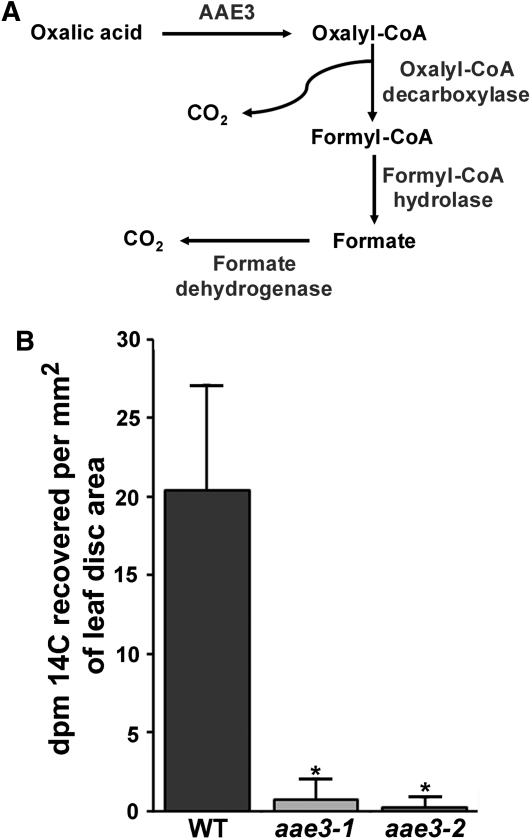
AAE3 Is Required for the Degradation Oxalate to CO2
It was proposed by Giovanelli and Tobin (1961) that an alternative to oxalate oxidase is found in some species to breakdown oxalate. Their results indicate that the catabolism pathway proceeds from oxalate to oxalyl-CoA to formyl-CoA to formate and eventually to CO2 by the pathway shown in Figure 4A (Giovanelli and Tobin, 1961, 1964). The initial step involves the ligation of oxalate and CoA to form oxalyl-CoA. To determine if AAE3 is required for oxalate catabolism in Arabidopsis, the aae3 mutants and wild-type plants were used for radiolabeled oxalate feeding experiments. Leaf disks cut from leaves of aae3-1, aae3-2, or the wild type were floated on 5 mL of 0.5 mM oxalate containing 5 μCi of 14C-oxalate in a sealed flask that contained a CO2 trap. After 5 h of incubation, the wild-type leaf disks had produced 14CO2 from the 14C-oxalate substrate, while flasks containing leaf disks from aae3-1 or aae3-2 plants yielded little or no 14CO2 (Figure 4B). These results indicate that the aae3 mutants are unable to convert oxalate into CO2 and support the proposed role of AAE3 in oxalate catabolism. Furthermore, these results imply that Arabidopsis does not have an alternative pathway for oxalate degradation to CO2. This is in agreement with previous studies indicating that Arabidopsis lacks oxalate oxidase activity (Membré et al., 1997, 2000).
Figure 4.
Measurements of Oxalate Degradation to CO2.
(A) Schematic of proposed oxalate degradation pathway in Arabidopsis.
(B) Radiolabeled CO2 measurements. Leaf discs of the wild type (WT), aae3-1, and aae3-2 were fed with 5 μCi of [14C] oxalate along with 500 μM nonlabeled oxalate for 5 h. The CO2 evolved was captured using 1 M KOH and the radioactivity measured by scintillation counting. Data are mean ± se (n = 4). Asterisks denote significant difference from the wild type (*P < 0.05; Student’s t test).
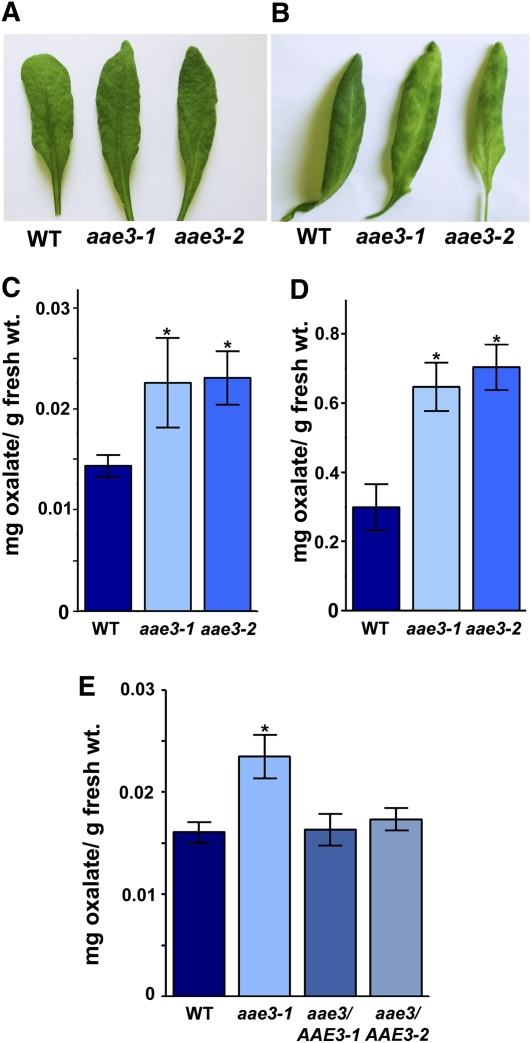
To further analyze the role of AAE3 in oxalate degradation, rosette leaves from the aae3 mutants and the wild type were removed and the petioles placed in a solution of 10 mM oxalate. After 48 h incubation in 10 mM oxalate the aae3 mutant leaves were chlorotic, whereas wild-type leaves were similar to leaves incubated in water as controls (Figures 5A and 5B). Furthermore, when oxalate concentrations were measured in leaves after 24 h of incubation in 10 mM oxalate, the results showed that both the aae3 mutants had more than twofold more oxalate than the wild type (Figure 5D). The results indicate that the mutants are sensitive to exogenous oxalate because of a deficiency in oxalyl-CoA synthetase activity. The water controls had much lower oxalate levels compared with the leaves incubated in 10 mM oxalate but also exhibited a 50% increase of oxalate in both aae3 mutants compared with the wild-type (Figures 5C and 5D). We repeated the experiment shown in Figure 5C and included leaves of two independent lines expressing an AAE3 transgene in the aae3-1 mutant background. In both lines, the AAE3 transgene restored oxalate levels to the wild type (Figure 5E). These results support a role for AAE3 in oxalate degradation.
Figure 5.
Analysis of Leaves Exposed to High Levels of Oxalate.
(A) and (B) Rosette leaves of 3-week-old wild type (WT), aae3-1, and aae3-2 were placed in either water for 48 h as control (A) or 10 mM oxalate for 48 h (B). The experiment was repeated three times with similar results.
(C) Samples of leaves from the experiment in (A) were removed after 24 h and oxalate content determined by HPLC. Data are mean ± se (n = 3). Asterisks denote significant difference from the wild type (*P < 0.05; Student’s t test).
(D) Samples of leaves from the experiment in (B) were removed after 24 h and oxalate content determined by HPLC. Data are mean ± se (n = 3); note different scale from (C). Asterisks denote significant difference from the wild type (*P < 0.05; Student’s t test).
(E) Oxalate content in rosette leaves of 3-week-old wild type, aae3-1, and aae3 complemented with AAE3-GFP under control of the AAE3 promoter (aae3/AAE3-1 and aae3/AAE3-2). Data are mean ± se (n = 4). Asterisks denote significant difference from the wild type (*P < 0.05; Student’s t test).
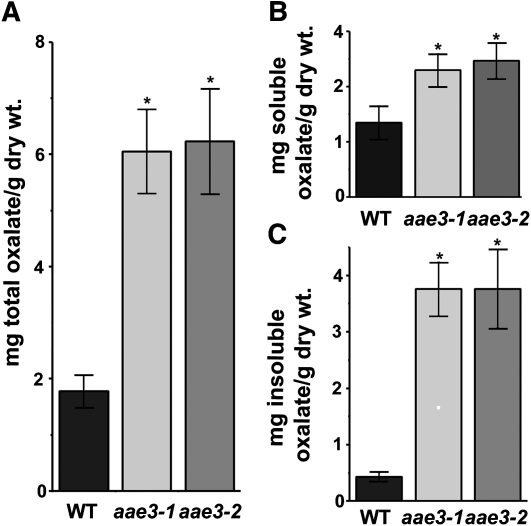
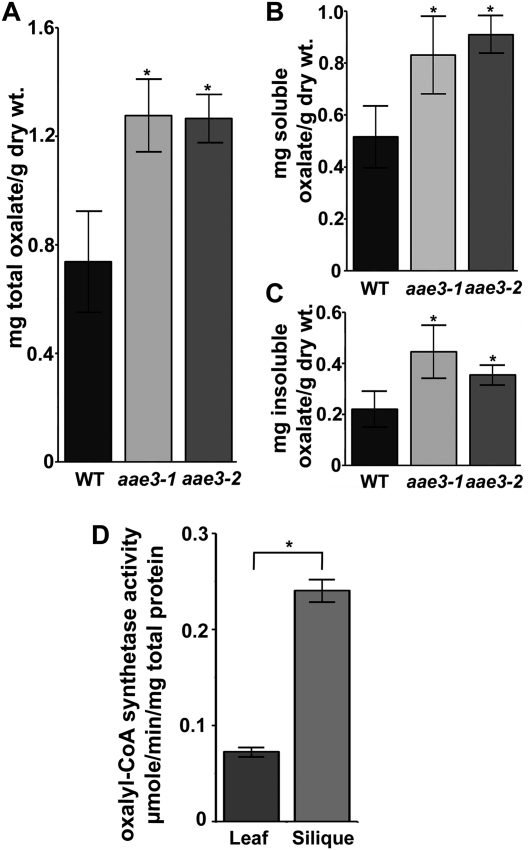
Due to the incapacity of aae3 mutants to degrade oxalate to CO2 in leaves, we inferred that soluble and insoluble oxalate concentrations might be increased within the mutant plants in the absence of exogenous supplementation. To test this, oxalate levels were measured in seeds and leaves of the wild type and the aae3 mutants. Total oxalate levels were shown to be more than threefold higher in aae3 mutant seeds (6.2 mg oxalate/g dry seeds) compared with the wild type (1.8 mg oxalate/g dry seeds) (Figure 6A). Further analyses of the soluble and insoluble fractions of seeds revealed the largest increase was within the insoluble fraction (Figures 6B and 6C), where the aae3 mutants were found to contain 3.8 mg oxalate/g dry seeds, ninefold higher than the wild type (0.42 mg oxalate/g dry seeds). The effects of aae3 mutations were less pronounced in leaf tissue. As shown in Figure 7, total oxalate in aae3 mutant and wild-type leaves was 1.27 and 0.74 mg oxalate/g dry weight, respectively. These values are lower than those found in plants that accumulate calcium oxalate crystals in leaf tissue, such as spinach (Spinacia oleracea; 65 to 157mg oxalate/dry weight) (Libert and Franceschi, 1987) and Medicago truncatula (22.8 mg oxalate/g dry weight) (Nakata and McConn, 2003). In wild-type plants, the relative enzyme activity of AAE3 was more than threefold higher in mature siliques than in rosette leaves (Figure 7D), which underscores the importance of AAE3 during seed maturation. These results provide additional support for the involvement of AAE3 in oxalate degradation.
Figure 6.
Oxalate Measurements in Dry Seeds.
Oxalate contents were determined by HPLC for seeds of the wild type (WT), aae3-1, and aae3-2. The total (A), soluble (B), and insoluble (C) oxalate contents are shown. Soluble oxalate was extracted with water from 200 ground seeds, while the remaining insoluble fraction was solubilized by incubation at pH 2.8 and 65°C for 1 h. The data are mean ± se (n = 7). Asterisks denote significant difference from the wild type (*P < 0.05; Student’s t test).
Figure 7.
Oxalate Measurements in Rosette Leaves.
(A) to (C) Oxalate contents were determined by HPLC for leaves of the wild type (WT), aae3-1, and aae3-2. The total (A), soluble (B), and insoluble (C) oxalate contents are shown. Soluble oxalate was extracted with water from 20 mg lyophilized leaf tissue, while the remaining insoluble fraction was solubilized by incubation at pH 2.8 and 65°C for 1 h. The data are mean ± se (n = 8).
(D) Oxalyl-CoA synthetase activities in crude extracts from wild-type leaves and mature siliques measured using 300 μM oxalate as substrate. The data are mean ± se (n = 3).
Asterisks denote significant difference from the wild type (*P < 0.05; Student’s t test).
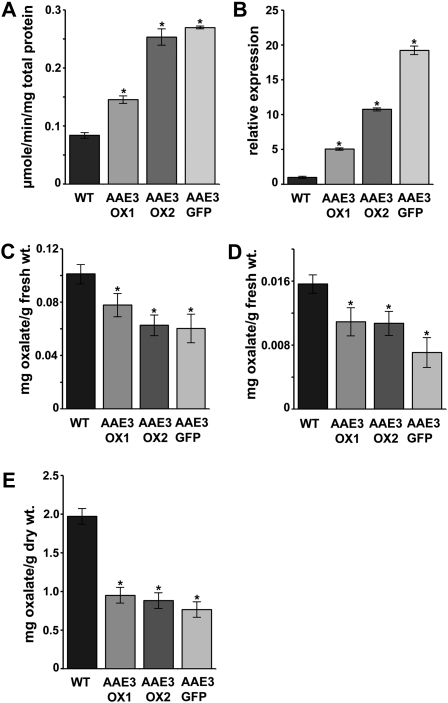
Since the loss of AAE3 decreases oxalate catabolism, we wanted to determine if elevated AAE3 would increase oxalate degradation. To do this, we generated transgenic Arabidopsis in which an AAE3 cDNA, or an AAE3-GFP fusion, was expressed under control of the 35S promoter. We screened transgenic lines by assaying tissue homogenates for oxalyl-CoA synthetase activity and chose three lines, AAE3-OX1, AAE3-OX2, and AAE3-GFP, that showed modest increases (1.7-, 3-, and 3.2-fold, respectively) in enzyme activity, relative to the wild type (Figure 8A). The AAE3 mRNA levels in these three lines were increased 5-, 11-, and 19-fold, respectively, relative to the wild type (Figure 8B). To determine the consequence of elevated oxalyl-CoA synthetase activity, leaves of the AAE3 overexpressing lines and the wild type were placed in 10 mM oxalate for 5 h before measurement of oxalate concentrations. Leaf tissue of all three overexpression lines contained substantially less oxalate than the wild type (Figure 8C). In the absence of exogenous oxalate, both leaves and seeds of the overexpression lines exhibited decreased levels of oxalate compared with the wild-type (Figures 8D and 8E).
Figure 8.
Analysis of Transgenic Plants Overexpressing AAE3.
(A) Oxalyl-CoA synthetase activities in crude extracts from wild-type (WT) and AAE3 overexpression lines using 300 μM oxalate as substrate. Data are mean ± se (n = 3).
(B) Relative expression of AAE3 in rosette leaf tissue from 3-week-old wild-type and AAE3 overexpression lines. Data are mean ± se (n = 3).
(C) and (D) Oxalate concentrations in rosette leaves from 3-week-old wild-type and AAE3 overexpression lines that were placed in either 10 mM oxalate for 5 h (C) or water for 5 h as control (D). Data are mean ± se (n = 8).
(E) Oxalate contents of mature seeds from wild-type and AAE3 overexpression lines. Data are mean ± se (n = 3).
Asterisks denote significant difference from the wild type (*P < 0.05; Student’s t test).
Loss of AAE3 Interferes with Seed Development
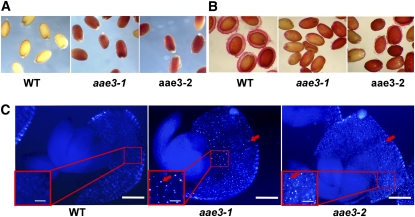
Our initial experience with the aae3 mutant lines indicated they both have decreased germination on agar plates and in soil, compared with wild type, and this was confirmed by our quantitative analysis of germination (Figure 3E). Visual observation of mutant seed revealed no readily discernable difference in size or appearance relative to the wild type. However, when seeds were incubated in 2,3,5-triphenyltetrazolium chloride (Beisson et al., 2007), the aae3-1 and aae3-2, but not wild-type, seeds became intensely stained indicating increased permeability of the seed coat in both mutants (Figure 9A). When wild-type seeds were stained with ruthenium red dye during imbibition, a mucilage layer surrounding the seeds was visualized, but this layer was absent or greatly reduced in aae3-1 and aae3-2 seed (Figure 9B). Taken together, these staining tests indicate that oxalate accumulation in aae3-1 and aae3-2 seed affects the integrity and function of the seed coat.
Figure 9.
Phenotypes of aae3 Mutant Seeds.
(A) Seeds stained with 2,3,5-triphenyltetrazolium chloride for 24 h at 30°C; the red color indicates permeability of the seed coat. WT, wild type.
(B) Seeds stained with ruthenium red for 15 min at room temperature to visualize mucilage formation on the seed coats.
(C) Under polarized light, calcium oxalate crystals are visible (red arrows) in aae3-1 and aae3-2 but not in the wild type. Bars = 37.5 μM and 150 μM (inset).
To find out if the high levels of insoluble oxalate measured in aae3-1 and aae3-2 seeds (Figure 6C) resulted in deposition of calcium oxalate crystals, seeds were cleared overnight in 5% bleach and then gently squashed to extrude the embryo from the seed coat. Under polarized light, numerous crystals were visible in seed coats of aae3-1 and aae3-2 but not the wild type (Figure 9C). Crystals were not observed in embryo tissue from any of the lines.
AAE3 Affects Susceptibility of Arabidopsis to an Oxalate-Producing Fungal Pathogen
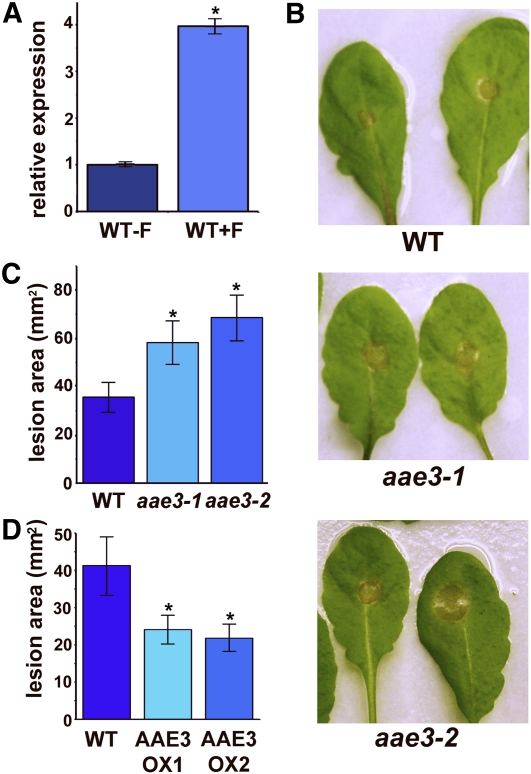
Oxalate is an important virulence determinant for some pathogenic fungi. The role of oxalate during infection has been studied extensively in S. sclerotiorum (Bateman and Beer, 1965; Guimarães and Stotz, 2004; Hegedus and Rimmer, 2005). Excretion of oxalate by these fungi enables infection of the plant, and fungal mutants that have lost the ability to produce oxalate exhibit greatly reduced virulence. Interestingly, microarray data available from the AtGenExpress database (Schmid et al., 2005) and the Gene Expression Omnibus (accession number GSE5684) (Edgar et al., 2002) indicate that AAE3 is induced following inoculation of plants with the oxalate-producing fungus Botrytis cinerea. To determine if S. sclerotiorum also induces AAE3 expression, wild-type Arabidopsis leaves were inoculated with S. sclerotiorum. After 36 h, the fungal lesion area was cut out and the remaining tissue was collected for quantitative PCR analysis. The results showed a fourfold increase in AAE3 levels compared with noninoculated leaf tissue (Figure 10A). Because the oxalyl-CoA synthetase encoded by AAE3 mediates oxalate degradation in Arabidopsis, the induction of AAE3 by S. sclerotiorum and the importance of oxalate for fungal virulence led us to hypothesize that AAE3 may be involved in plant defense against oxalate-producing fungi in a similar fashion to the oxalate-degrading enzyme, oxalate oxidase, in other plant species (Donaldson et al., 2001; Dong et al., 2008). To test if AAE3 is important for plant defense against oxalate-producing fungi, leaves of the wild type and both aae3 mutants were inoculated with S. sclerotiorum. After 48 h of incubation, leaves were photographed (Figure 10B) and lesion areas measured. Both aae3-1 and aae3-2 displayed significantly more leaf area infected by the fungus than did the wild type (Figure 10C). Thus, absence of oxalyl-CoA synthetase in the aae3 mutants is associated with increased susceptibility to S. sclerotiorum.
Figure 10.
Sclerotinia sclerotiorum Inoculation of Detached Rosette Leaves.
(A) Quantitative RT-PCR of AAE3 expression in detached wild-type (WT) leaves inoculated with S. sclerotiorum (+F) for 36 h and control leaves (−F). Data are mean ± se (n = 3). For +F, asterisk denotes significant difference from −F (*P < 0.05; Student’s t test).
(B) Images of wild-type, aae3-1, and aae3-2 leaves 48 h after inoculation with S. sclerotiorum.
(C) Measurements of fungal spread for the experiment shown in (B). Data are mean ± se (n = 15). Asterisks denote significant difference from the wild type (*P < 0.05; Student’s t test).
(D) Measurements of fungal spread in leaves of wild-type and AAE3 overexpression lines inoculated with S. sclerotiorum. Lesion areas were measured 48 h after inoculation. Data are mean ± se (n = 18).
Given the increased susceptibility of the aae3 mutants to S. sclerotiorum, we wanted to know whether increased oxalyl-CoA synthetase activity would assist in plant defense against this oxalate-producing fungus. Leaves of AAE3-OX1 and AAE3-OX2 plants along with wild-type controls were inoculated with S. sclerotiorum. After 48 h, the lesions on leaves from both overexpression lines were ~40% smaller than those on wild-type leaves (Figure 10D). These results indicate that the pathway of oxalate degradation initiated by oxalyl-CoA synthetase is important in Arabidopsis for defense against S. sclerotiorum infection and that modest increases in oxalyl-CoA synthetase activity can further decrease susceptibility to this pathogen.
DISCUSSION
AAE3 Is an Oxalyl-CoA Synthetase and Is Required for Oxalate Degradation in Arabidopsis
AAE3 is a member of the large superfamily of AAEs in Arabidopsis that comprises 63 proteins (Shockey et al., 2003; Shockey and Browse, 2011), but the AAE3 amino acid sequence is not closely related to any other member. The most closely related proteins in Arabidopsis are two 4-coumaroyl-CoA ligase-like proteins that exhibit only 30% sequence identity and 50% similarity to AAE3. Our screen of potential substrates for AAE3, based on coexpression data, led to the discovery that it functions as an oxalyl-CoA synthetase. Screening of other potential substrates indicated that AAE3 is very specific for oxalate, since other molecules with structures similar to oxalate were unable to function as substrates (Table 1). Some CoA synthetase enzymes activate a wide range of substrates in vitro; examples include several long-chain acyl-CoA synthetases that can use multiple fatty acids as substrates (Shockey et al., 2002) and some 4-coumaroyl-CoA ligase-like proteins that were found to use both fatty acids and jasmonate precursors as substrates in vitro (Kienow et al., 2008). By contrast, other AAEs are very specific for their substrate, such as the malonyl-CoA synthetase encoded by AAE13 (Chen et al., 2011). The high specificity of the His-AAE3 recombinant enzyme for oxalate identifies AAE3 as the first gene cloned from any organism to encode oxalyl-CoA synthetase activity. Earlier studies have described possible oxalyl-CoA synthetase activity from plant protein extracts, but no gene has previously been isolated that encodes this function (Giovanelli and Tobin, 1961, 1964; Malathi et al., 1970; Adsule and Barat, 1977).
Leaf tissue from both aae3-1 and aae3-2 mutant plants was unable to degrade radiolabeled oxalate (Figure 4), indicating that AAE3 is required for the catabolism of oxalate to CO2. Oxalate oxidase is predominantly described in the literature to perform oxalate degradation, in a reaction that directly converts oxalate to CO2 and H2O2 (Franke et al., 1943; Finkle and Arnon, 1954; Srivastava and Krishnan, 1962; Suguira et al., 1979; Lane et al., 1993; Davidson et al., 2009). An alternative multistep pathway was proposed for oxalate catabolism by Giovanelli and Tobin (1961). In this pathway, oxalate is also eventually broken down to CO2, but it is achieved through the coordination of four enzymes (Figure 4A). The first step in this pathway involves the ligation of oxalate to CoA by oxalyl-CoA synthetase. The subsequent steps hypothesized in this pathway include oxalyl-CoA decarboxylase, formyl-CoA hydrolase, and formate dehydrogenase. In support of this pathway, Giovanelli and Tobin (1964) demonstrated that radiolabeled oxalate, via a CoA- and ATP-dependent mechanism, is converted to CO2 with formic acid as an intermediate. Our results indicate that AAE3 encodes an oxalyl-CoA synthetase similar to that characterized from pea seeds (Giovanelli, 1966). In Arabidopsis, formate dehydrogenase is encoded by At5g14780 (Li et al., 2002), and this gene is coexpressed with AAE3 during pathogen infection (Pearson correlation coefficient of 0.77; see Supplemental Table 1 online). The protein encoded by At5g17380 is a homolog of the well-characterized oxalyl-CoA decarboxylase of Oxalobacter formigenes (Berthold et al., 2005; Svedruzić et al., 2005). The sequences of these two proteins share 41% identity and 63% similarity. The expression of At5g17380 is correlated with that of AAE3 in the database of Nottingham Arabidopsis Stock Centre arrays (Toufighi et al., 2005) (Pearson correlation coefficient of 0.53; see Supplemental Table 1 online). Additional mutational and molecular-genetic studies will be required to test the possible roles of these two genes in encoding enzymes of oxalate degradation.
Activity of oxalayl-CoA synthetase has been described in protein extracts from several plant species. Enzyme activity was shown to be highest in legumes such as pea, soybean, and L. sativus but was also observed in pumpkin (Cucurbita pepo) and wheat germ (Giovanelli and Tobin, 1964; Giovanelli, 1966; Malathi et al., 1970; Adsule and Barat, 1977). Our results indicate that Arabidopsis uses this alternative oxalate catabolism pathway and is not able to degrade oxalate to CO2 in leaf discs without oxalyl-CoA synthetase activity. These data also support previous investigations that failed to detect oxalate oxidase activity in Arabidopsis (Membré et al., 1997, 2000). The pathway initiated by oxalyl-CoA synthetase may be an alternative to oxalate oxidase in some species, such as Arabidopsis, but may also exist in species alongside the oxalate oxidase pathway. For example, oxalyl-CoA synthetase activity was reported in wheat germ (Giovanelli, 1966), which is also known to contain oxalate oxidase.
Loss of AAE3 Increases Oxalate Levels and Renders Arabidopsis More Susceptible to an Oxalate-Producing Fungal Pathogen
The inability of the aae3 mutants to degrade oxalate results in increased oxalate concentrations within tissues of the plants, relative to concentrations in the wild type (Figures 6 and 7). In seeds of the aae3 mutants, total oxalate is ~6 mg/g dry weight compared with <2 mg/g dry weight in wild-type seeds (Figure 6A). The high concentrations of oxalate in the seeds could be due to the degradation of ascorbic acid (Green and Fry, 2005). During seed development, ascorbate increases during early development and is then broken down as the seed matures (Arrigoni et al., 1992). Interestingly, the period of ascorbate breakdown coincides with high AAE3 expression during seed development (Toufighi et al., 2005). The high levels of oxalate result in the formation of calcium oxalate crystals in the seed coat (Figure 9).
In both the soluble and insoluble forms, oxalate strongly chelates calcium and other divalent cations. The high amounts of oxalate measured in aae3-1 and aae3-2 seeds suggest the possibility that calcium-signaling processes in the seed may be affected by calcium sequestration into crystals and a consequent reduction in free calcium levels. For example, calcium-dependent protein kinases have important roles during seed dormancy and germination (Luan, 2009; Nakashima et al., 2009). Calcium is also important for the production and cross-linking of cell wall components, so that the formation of the crystals and free oxalate could interfere with cell wall formation and suberin synthesis (Hirschi, 2004), rendering the seeds more permeable to 2,3,5-triphenyltetrazolium chloride and limiting production of mucilage (Figure 9B). The mucilage layer is composed mainly of hydrated pectin from the seed coat. Whereas it has been proposed to have roles in seed hydration and seed dispersal, it is not required for germination under well-hydrated conditions (Western et al., 2000). Although crystal formation in the seed coat is common in many species, Arabidopsis does not normally form crystals and may not be able to cope with the sequestration of calcium and the physical disturbance caused by the crystals (Franceschi and Nakata, 2005). In this respect, the aae3 mutants represent a good system to study calcium oxalate crystal formation in plants, and its potential consequences. In our experiments, the lack of oxalyl-CoA synthetase results in reduced and delayed germination (Figure 3E).
In leaf tissue, the difference in oxalate accumulation between the aae3 mutants and the wild type was substantially less than in seeds, with mutants exhibiting a 70% increase in oxalate relative to the wild type (Figure 6A). The relative importance of AAE3 for oxalate degradation in seeds compared with leaves was consistent with the enzyme activity in mature siliques being fourfold higher than leaves (Figure 7D). The rate of oxalate production in Arabidopsis leaves is not known, but we consider it conceivable that leaves on mutant plants are able to remove oxalate, possibly by transport to the roots or other parts of the plant. Secretion of oxalate from the roots may obviate the need for metabolism to prevent accumulation of this compound to toxic levels. Alternatively, there may be pathways to convert oxalate to other metabolites that are not converted to CO2 during the 5-h incubation of our radiotracer experiments (Figure 4). The aae3 mutants may serve as a good system to study oxalate metabolism and movement that is not feasible in other organisms that have an oxalate degradation pathway. In any case, mutant plants remained healthy under our growth conditions (Figure 3D), indicating that the observed increases in oxalate levels are not sufficient to cause the damage symptoms seen in leaves incubated with exogenous oxalate (Figure 5B).
Previous studies have shown that oxalate is an essential virulence factor in some fungal species (Bateman and Beer, 1965; Guimarães and Stotz, 2004; Hegedus and Rimmer, 2005). These fungi require the excretion of oxalate to penetrate and infect the host leaf. The failure of the aae3 mutants to degrade oxalate suggested that they might be more susceptible to fungal infection; indeed, our results demonstrate a role for plant AAE3 oxalyl-CoA synthetase in defense against the oxalate producing fungus, S. sclerotiorum (Figures 10B and 10C). This is in line with expression data that indicate AAE3 is induced during infection by the oxalate-producing pathogens B. cinerea (Toufighi et al., 2005) and S. sclerotiorum (Figure 10A). Although AAE3 is important for plant defense against oxalate producing fungi, wild-type Arabidopsis is still quite susceptible to these fungi, and modest overexpression of AAE3 oxalyl-CoA synthetase resulted in a measurable decrease in susceptibility to S. sclerotiorum infection (Figure 10D).
Most researchers consider the main source of oxalate in plant tissue to be from the degradation of ascorbate (Franceschi and Nakata, 2005; Green and Fry, 2005), but additional pathways from oxaloacetate (Chang and Beevers, 1968; Franceschi and Nakata, 2005), glycolate, and glyoxylate (Richardson and Tolbert, 1961; Yu et al., 2010) probably also contribute significantly to oxalate production. In some plant species, but not Arabidopsis, oxalate is degraded by oxalate oxidase, an enzyme activity of some germin proteins (Lane et al., 1993; Druka et al., 2002). Our identification of AAE3 as an oxalyl-CoA synthetase in Arabidopsis provides support for an alternative pathway of oxalate degradation first proposed 50 years ago by Giovanelli and Tobin (1961). It is likely that oxalyl-CoA synthetase functions as a step in this pathway mainly for the metabolism of endogenous oxalate. However, our characterization of two allelic aae3 mutants indicates that the enzyme is also important for defense against pathogens that synthesize oxalate to aid their infection of plants.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia seeds were imbibed at 4°C overnight and sown in soil consisting of peat (60%), pumice (20%), and sand (20%) (Sunshine Professional Growing Mix). Plants were grown in environmentally controlled growth chambers with 14, 16, or 24 h light at 150 to 200 μmol photons/m2/s, 50% humidity, and 22°C. The Nicotiana benthamiana and some sets of Arabidopsis were grown in greenhouse conditions with 16 h of light at 22°C. For in vitro cultures, seeds were sterilized by soaking in 30% bleach with 0.1% Triton X-100 and were rinsed five times with sterile water. In vitro cultures were performed on Murashige and Skoog (MS) medium, pH 5.7 (Murashige and Skoog, 1962), supplemented with 8 g/L agar, 1% Suc, and 0.5 g/L MES.
AAE3 Construct Preparation
AAE3 full-length cDNAs with and without a stop codon were obtained from first-strand cDNAs of rosette leaves from Arabidopsis ecotype Columbia using RT-PCR with the following primers: 5′-CACCATGGATAGCGATACTCTCTCA-3′ and 5′-TCAGGGCTTCTCAAGGAAATG-3′ or 5′-GGGCTTCTCAAGGAAATG-3′. The PCR products were cloned into the Gateway entry vector pENTR TOPO (Invitrogen) and transferred using LR clonase (Invitrogen) to the destination vectors p2GWF7 and p2FGW7 for onion cell bombardment, pB7WGF2 and pB7FWG2 for leaf infiltration and Arabidopsis transformation, pDEST17 for protein purification, and pB2GW7 for overexpression (Karimi et al., 2002). For complementation of aae3-1, the AAE3 promoter (1406 bp) was obtained via PCR using Arabidopsis genomic DNA with the following primers containing XbaI and BamHI restriction sites: 5′-GCTCTAGAATGAACCCACCAATATAAGTATAAG-3′ and 5′-CGGGATCCGGTTACGTCGGAGAGATAAACA-3′. The PCR product was cloned into pGEM-T Easy (Promega). The promoter was excised from pGEM-T Easy with XbaI and BamHI and ligated to the corresponding restriction sites of pCAMBIA1300 binary vector. The AAE3-GFP was excised from the p2GWF7 vector by the restriction sites SpeI and SacII, blunted, and transferred to the AAE3 promoter pCAMBIA1300 vector that was digested with SacI and BamHI and blunted.
Subcellular Localization of AAE3
Transient expression of AAE3-GFP, GFP-AAE3, and GFP fusion constructs was performed in onion (Allium cepa) epidermal cells using particle bombardment (PDS-1000; Bio-Rad) with 1.1 μm gold particles and 1150 rupture discs according to the manufacturer’s protocol. Transient expression in N. benthamiana was achieved by leaf infiltration with Agrobacterium tumefaciens containing 35S:AAE3-GFP or 35S:GFP-AAE3 constructs (Voinnet et al., 2003; Sparkes et al., 2006). Images were captured with a Zeiss LSM 510 META confocal microscope. For stable expression of AAE3-GFP, Arabidopsis plants were transformed with the pB7FWGF2 binary construct containing 35S-AAE3-GFP. Images were captured with a FV300 laser scanning confocal microscope (Olympus America).
Isolation of T-DNA Insertion Lines
The AAE3 (At3g48990) T-DNA insertions lines aae3-1 (Salk_109915) and aae3-2 (Salk_ 024432) were obtained from the ABRC at the Ohio State University. To identify plants homozygous for each T-DNA, right and left gene-specific genomic primers were designed using T-DNA primer design server athttp://signal.salk.edu/tdnaprimers.2 html. Three primers, right and left gene-specific primers located outside of the inserted T-DNA and a T-DNA left border sequence primer, were used to amplify PCR product template from extracted genomic DNA of rosette leaf tissue. The primers used to screen aae3-1 were 5′-TCCCATGTGATTCTTCAGCAC-3′ and 5′-TCATTCAAAGTCATTTTTACCA-3′; for aae3-2, they were 5′-ATATATCAGCGACTCCCACCG-3′ and 5′-TGGTTCACGTAGTGGGCCATCG-3′ and a T-DNA left border sequence primer (LBa1) 5′-TGGTTCACGTAGTGGGCCATCG-3′. To verify that these were null mutants for the AAE3 gene, PCR (30 cycles) was conducted on cDNA prepared from 200 ng of leaf total RNA using the AAE3-specific primers 5′-CACCATGGATAGCGATACTCTCTCA-3′ and 5′-TCAGGGCTTCTCAAGGAAATG-3′. The ACT2 cDNA, encoded by gene At5g09810, was used as an internal control and was amplified using the primers 5′-ACCATTTCTCTATCTTTCTCTCTCGCTG-3′ and 5′-CAAACTCACCACCACGAACCAGATAA-3′.
AAE3 His-Tagged Protein Purification
Escherichia coli strain BL21AI competent cells (Invitrogen) were transformed with the N-terminal His-tagged pDEST17 bacterial expression vector containing AAE3. A small culture was grown overnight at 37°C and used to inoculate 500 mL of Luria-Bertani medium supplemented with carbenicillin. The large culture was incubated at 37°C until it reached an OD600 of 0.4. To induce expression, Ara was added at 0.2% (w/v), and the culture was grown for an additional 4 h at 30°C. The cells were then collected by centrifugation and frozen until analysis.
The lysis and purification of the protein was performed according the protocol for native conditions described in the protein purification kit from Qiagen. Frozen cells were thawed on ice for 15 min and resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8) supplemented with lysozyme (1 mg/mL) and benzonase. After 30 min on ice, the cells were sonicated and centrifuged at 10,000g for 25 min at 4°C. The supernatant was collected and loaded onto a column packed with Ni-nickel-nitriloacetic acid agarose to bind the protein. The column was washed with wash buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole at pH8) and eluted using 1 mL of elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8). Salts were removed by passing the protein sample through a column packed with Sephadex G-25 (Amersham Biosciences) and equilibrated with 100 mM Tris-HCl, pH 7.5. The protein concentration of the eluate was determined by performing a Bradford assay (Bradford, 1976). The size and purity of the protein were assessed by running the sample on an SDS-polyacrylamide gel and staining with Coomassie Brilliant Blue R 250.
Determining AAE3 Enzyme Activity and Kinetics
AAE3 enzyme activity was determined by a coupled enzyme assay (Ziegler et al., 1987; Koo et al., 2006). The assay was initiated by the addition of 5 μg of purified protein to the buffered reaction mixture containing 0.1 M Tris-HCl, pH 8, or 0.1 M NaPO4, pH 8, 2 mM DTT, 5 mM ATP, 10 mM MgCl2, 0.5 mM CoA, 0.4 mM NADH, 1 mM phosphoenolpyruvate, and 10 units each of myokinase, pyruvate kinase, and lactate dehydrogenase and the carboxylic acid substrate, in a final volume of 800 μL. The reaction was then observed by measuring the oxidation of NADH at 340 nm on a Beckman DU530 spectrophotometer. To determine optimal pH and substrate specificity, 300 μM of each potential substrate was used, whereas a range of oxalate concentrations were employed to determine Km and Vmax. The substrates tested for CoA synthetase activity were oxalate, malonate, succinate, glutarate, malate, glycolate, glyoxylate, acetate, lactate, and formate.
To test the enzyme activity in plants, seeds were sterilized and placed in liquid MS medium, pH 5.7, supplemented with 8 g/L agar, 1% Suc, and 0.5 g/L MES. Plants were grown for 3 weeks in Erlenmeyer flasks on a rotary shaker at 25°C, 50 μmol photons/m2/s, and a 16-h photoperiod. The plants were then rinsed, frozen in liquid nitrogen, and ground. Rosette leaves and mature siliques were harvested from plants grown on soil, frozen in liquid nitrogen, and ground. A crude protein extraction method was used to extract the enzyme from the ground tissue as described by Chen et al. (2011). Briefly, the frozen ground tissue was added to an extraction buffer solution containing 100 mM Tris-HCl, pH 7.5, 1mM EDTA, 1% (v/v) 2-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride and centrifuged at 15,500 rcf for 10 min at 4°C. For the rosette leaves and siliques, the soluble protein in the supernatant was washed and concentrated using a 30-kD cellulose centrifugal filter (Millipore). The supernatants were then used for enzyme assays as described above.
Radiolabeled Oxalate Feeding
Leaf discs of wild-type Columbia, aae3-1, and aae3-2 were isolated using an 8.5-mm borer. The leaf discs were then placed in an Erlenmeyer flask containing 5 mL of MS medium, pH 5.7, supplemented with 0.5% Suc, 0.05% MES, 500 μM oxalate, and 5 μCi of [14C] oxalate (American Radiolabeled Chemicals). A glass vial containing 500 μL of 1 M KOH acted as a CO2 trap and the flask sealed with a neoprene stopper. The flasks were slowly shaken at room temperature for 5 h, and then the reaction was stopped by the addition of 1 mL of 0.25 M HCl through the stopper using a syringe. The leaf disks were shaken for 10 more minutes and then the apparatus was taken apart to retrieve the radiolabeled CO2 trapped in within the KOH. The radioactivity in the trap was measured by liquid scintillation counting and adjusted for radioactivity trapped in control flasks with no leaf disks.
Analysis of aae3 Seed Phenotypes
To visualize seed mucilage, seeds were incubated for 15 min in an aqueous solution of 0.03% (w/v) ruthenium red at room temperature and rinsed with water before observation under a stereomicroscope (Western et al., 2000). To determine seed permeability, seeds were incubated in 1% (w/v) 2,3,5-triphenyltetrazolium chloride at 30°C for 24 h (Debeaujon et al., 2000; Beisson et al., 2007). Oxalate crystals were observed by first imbibing seeds overnight in 5% bleach to clear the tissues. The seeds were then visualized using polarized light on an Olympus BH-2 light microscope and photographs taken with a ProgRes C12 Plus digital camera (Jenoptik).
Determining Oxalate Concentrations in the Plant
Oxalate concentrations in rosette leaves and dry seeds were measured by HPLC. Rosette leaves of 3-week-old Arabidopsis were harvested from three independently grown sets and ground in liquid nitrogen. Dry seeds were harvested from four independently grown sets, and 50 seeds from each plant were pooled and used for oxalate measurements. Oxalate extraction was performed as described by Nakata and McConn (2003). Soluble oxalate was obtained by grinding the sample in water followed by centrifugation and collection of the supernatant. Insoluble oxalate was extracted from the resulting pellet by incubation at pH 2.8 and 65°C for 1 h. The samples were filtered (0.2 μm) and analyzed for oxalate by HPLC (Waters model 2695) coupled to a photodiode array detector (Waters model 2996) with a Bio-Rad Aminex HPX-87H ion exclusion column (300 × 7.8 mm, 0.6 mL/min, 35°C). The data were integrated and measured using the Empower 2 software (Waters). External standards of oxalate were used to determine sample oxalate concentrations with a retention time of 6.0 min.
Oxalate Sensitivity Assay
Rosette leaves of aae3 mutants and the wild type were excised from 3-week-old plants and the petioles recut under distilled water using a razor blade. The petioles of the leaves were then placed in 500 μL of 10 mM oxalate or water in a 15-mL falcon tube. The falcon tubes were sealed with micropore tape and placed under 115 μmol photons/m2/s light for 5, 24, or 48 h. To measure oxalate concentrations, leaves were taken out after 5 h for the AAE3-overexpressing lines and 24 h for the aae3 mutants. Oxalate was extracted and measured with HPLC as described above.
Quantitative RT-PCR Analysis
Quantitative RT-PCR was performed with a Bio-Rad CFX-96 teal-time PCR detection system and CFX manager software using a SYBR Green master mix (Clontech). The PCR conditions used were as follows: 95°C for 2 min, 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. RNA was extracted using the SV total RNA isolation system (Promega) and reverse transcribed. The reverse transcription reaction was then diluted and used as template. AAE3 levels were determined using the AAE3-specfic primers 5′-TGGGGAAGAGATTAACTGTGC-3′ and 5′-GCGCTGAATCTTACCAGAGG-3′. The housekeeping gene TUB2 was used as a control to normalize the cycle threshold values, using the TUB2-specific primers 5′-ACTGTCTCCAAGGGTTCCAGGTTT-3′ and 5′-ACCGAGAAGGTAAGCATCATGCGA-3′, and relative transcript levels were then calculated.
Fungal Growth Assays
Fungal growth experiments were performed with Sclerotinia sclerotiorum (kindly provided by Weidong Chen, Washington State University) and fully expanded rosette leaves from 4-week-old plants grown under a 14-h light regime. Application of S. sclerotiorum to the leaf tissue was performed as described by Bessire et al. (2007). The fungus was grown on potato dextrose agar (Difco), and a 2-cm2 region of hyphae was cut and transferred to a liquid culture of 0.5 potato dextrose broth (Difco) and incubated for 2 d. The 5-mL culture was then diluted 1:10 with fresh 0.5 potato dextrose broth and homogenized using a polytron. Rosette leaves were then cut and placed on wet Whatman paper within a Petri dish, and 5 μL of fungal suspension was placed on each leaf. After 48 h, leaves were photographed and the lesion areas measured using the ImageJ software (Girish and Vijayalakshmi, 2004). For analysis of AAE3 expression during S. sclerotiorum infection, cut leaves of the wild type were placed on wet Whatman paper in a Petri dish and 5 μL of fungal suspension, or water as control, was placed on each leaf. After 36 h, the tissue containing the fungal lesion was cut out and the remaining tissue was collected for quantitative PCR analysis.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number AF503762 (AAE3, At3g48990).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Subcellular Localization of AAE3.
Supplemental Table 1. Genes Whose Expression Is Correlated with AAE3 (At3g48990).
Acknowledgments
We thank Mechthild Tegeder (Washington State University) for use of the Bio-Rad Aminex HPX-87H ion exclusion column and Qiumin Tan (Washington State University) for advice and help with the HPLC analyses. We also thank Weidong Chen (Washington State University) for providing the S. sclerotiorum. This work was supported by the National Science Foundation (Grant MCB-0420199) and by the Agricultural Research Center at Washington State University.
AUTHOR CONTRIBUTIONS
J.F., H.U.K., and J.B. designed the research. J.F. and H.U.K. performed the research. J.F., H.U.K., and P.A.N. analyzed the data. J.F. and J.B. wrote the article with input from the other authors.
References
- Adsule R.N., Barat G.K. (1977). Occurrence of oxalyl-CoA synthetase in Indian pulses. Experientia 33: 416–417 [DOI] [PubMed] [Google Scholar]
- Arrigoni O., De Gara L., Tommasi F., Liso R. (1992). Changes in the ascorbate system during seed development of Vicia faba L. Plant Physiol. 99: 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman D.F., Beer S.V. (1965). Simultaneous production and synergistic action of oxalic acid and polygalacturonase during pathogenesis by Sclerotium rolfsii. Phytopathology 55: 204–211 [PubMed] [Google Scholar]
- Beisson F., Li Y., Bonaventure G., Pollard M., Ohlrogge J.B. (2007). The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19: 351–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold C.L., Moussatche P., Richards N.G., Lindqvist Y. (2005). Structural basis for activation of the thiamin diphosphate-dependent enzyme oxalyl-CoA decarboxylase by adenosine diphosphate. J. Biol. Chem. 280: 41645–41654 [DOI] [PubMed] [Google Scholar]
- Bessire M., Chassot C., Jacquat A.-C., Humphry M., Borel S., Petétot J.-M., Métraux J.-P., Nawrath C. (2007). A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J. 26: 2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chang C.C., Beevers H. (1968). Biogenesis of oxalate in plant tissues. Plant Physiol. 43: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Kim H.U., Weng H., Browse J. (2011). Malonyl-CoA synthetase, encoded by ACYL ACTIVATING ENZYME13, is essential for growth and development of Arabidopsis. Plant Cell 23: 2247–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R.M., Reeves P.A., Manosalva P.M., Leach J.E. (2009). Germins: A diverse protein family important for crop improvement. Plant Sci. 177: 499–510 [Google Scholar]
- Debeaujon I., Léon-Kloosterziel K.M., Koornneef M. (2000). Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson P.A., Anderson T., Lane B.G., Davidson A.L., Simmonds D.H. (2001). Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotina sclerotiorum. Physiol. Mol. Plant Pathol. 59: 297–307 [Google Scholar]
- Dong X.B., Ji R.Q., Guo X.L., Foster S.J., Chen H., Dong C.H., Liu Y.Y., Hu Q., Liu S.Y. (2008). Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta 228: 331–340 [DOI] [PubMed] [Google Scholar]
- Druka A., Kudrna D., Kannangara C.G., von Wettstein D., Kleinhofs A. (2002). Physical and genetic mapping of barley (Hordeum vulgare) germin-like cDNAs. Proc. Natl. Acad. Sci. USA 99: 850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkle B.J., Arnon D.I. (1954). Metabolism of isolated cellular particles from photosynthetic tissues. Physiol. Plant. 7: 614–624 [Google Scholar]
- Franceschi V.R., Nakata P.A. (2005). Calcium oxalate in plants: Formation and function. Annu. Rev. Plant Biol. 56: 41–71 [DOI] [PubMed] [Google Scholar]
- Franke W., Schumann F., Banerjee B. (1943). Zur biologischen oxydation der oxalsaure. II. Hoppe Seylers Z. Physiol. Chem. 278: 24–42 [Google Scholar]
- Giovanelli J. (1966). Oxalyl-coenzyme A synthetase from pea seeds. Biochim. Biophys. Acta 118: 124–143 [DOI] [PubMed] [Google Scholar]
- Giovanelli J., Tobin N.F. (1961). Adenosine triphosphate- and coenzyme A-dependent decarboxylation of oxalate by extracts of peas. Nature 190: 1006–1007 [DOI] [PubMed] [Google Scholar]
- Giovanelli J., Tobin N.F. (1964). Enzymic decarboxylation of oxalate by extracts of plant tissue. Plant Physiol. 39: 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girish V., Vijayalakshmi A. (2004). Affordable image analysis using NIH Image/ImageJ. Indian J. Cancer 41: 47. [PubMed] [Google Scholar]
- Godoy G., Steadman J.R., Dickman M.B., Dam R. (1990). Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol. Mol. Plant Pathol. 37: 179–191 [Google Scholar]
- Green M.A., Fry S.C. (2005). Vitamin C degradation in plant cells via enzymatic hydrolysis of 4-O-oxalyl-L-threonate. Nature 433: 83–87 [DOI] [PubMed] [Google Scholar]
- Guimarães R.L., Stotz H.U. (2004). Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol. 136: 3703–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J.L., Tonti-Filippini J., Verboom R.E., Millar A.H. (2005). Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol. 139: 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus D.D., Rimmer S.R. (2005). Sclerotinia sclerotiorum: When “to be or not to be” a pathogen? FEMS Microbiol. Lett. 251: 177–184 [DOI] [PubMed] [Google Scholar]
- Hirschi K.D. (2004). The calcium conundrum. Both versatile nutrient and specific signal. Plant Physiol. 136: 2438–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2009). Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol. Plant 2: 1154–1180 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kienow L., Schneider K., Bartsch M., Stuible H.-P., Weng H., Miersch O., Wasternack C., Kombrink E. (2008). Jasmonates meet fatty acids: Functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana. J. Exp. Bot. 59: 403–419 [DOI] [PubMed] [Google Scholar]
- Kim K.S., Min J.-Y., Dickman M.B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microbe Interact. 21: 605–612 [DOI] [PubMed] [Google Scholar]
- Klug B., Horst W.J. (2010). Oxalate exudation into the root-tip water free space confers protection from aluminum toxicity and allows aluminum accumulation in the symplast in buckwheat (Fagopyrum esculentum). New Phytol. 187: 380–391 [DOI] [PubMed] [Google Scholar]
- Koo A.J.K., Chung H.S., Kobayashi Y., Howe G.A. (2006). Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 281: 33511–33520 [DOI] [PubMed] [Google Scholar]
- Korth K.L., Doege S.J., Park S.H., Goggin F.L., Wang Q., Gomez S.K., Liu G., Jia L., Nakata P.A. (2006). Medicago truncatula mutants demonstrate the role of plant calcium oxalate crystals as an effective defense against chewing insects. Plant Physiol. 141: 188–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane B.G., Dunwell J.M., Ray J.A., Schmitt M.R., Cuming A.C. (1993). Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 268: 12239–12242 [PubMed] [Google Scholar]
- Li R., Moore M., Bonham-Smith P.C., King J. (2002). Overexpression of formate dehydrogenase in Arabidopsis thaliana resulted in plants tolerant to high concentrations of formate. J. Plant Physiol. 159: 1069–1076 [Google Scholar]
- Libert B., Franceschi V.R. (1987). Oxalate in crop plants. J. Agric. Food Chem. 35: 926–938 [Google Scholar]
- Luan S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14: 37–42 [DOI] [PubMed] [Google Scholar]
- Lumsden R.D. (1976). Pectolytic enzymes of Scerlotinia sclerotiorum and their localization in infected bean. Can. J. Bot. 54: 2630–2641 [Google Scholar]
- Ma J.F., Zheng S.J., Matsumoto H., Hiradate S. (1997). Detoxifying aluminium with buckwheat. Nature 390: 569–5709403684 [Google Scholar]
- Malathi K., Padmanaban G., Sarma P.S. (1970). Biosynthesis of β-N-oxalyl-l-α,β-diaminopropionic acid, the Lathyrus sativus neurotoxin. Phytochemistry 9: 1603–1610 [Google Scholar]
- Membré N., Berna A., Neutelings G., David A., David H., Staiger D., Sáez Vásquez J., Raynal M., Delseny M., Bernier F. (1997). cDNA sequence, genomic organization and differential expression of three Arabidopsis genes for germin/oxalate oxidase-like proteins. Plant Mol. Biol. 35: 459–469 [DOI] [PubMed] [Google Scholar]
- Membré N., Bernier F., Staiger D., Berna A. (2000). Arabidopsis thaliana germin-like proteins: Common and specific features point to a variety of functions. Planta 211: 345–354 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Nakata P.A. (2003). Advances in our understanding of calcium oxalate crystal formation and function in plants. Plant Sci. 164: 901–909 [Google Scholar]
- Nakata P.A., McConn M.M. (2003). Calcium oxalate crystal formation is not essential for growth of Medicago truncatula. Plant Physiol. Biochem. 41: 325–329 [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37(Database issue): D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson K.E., Tolbert N.E. (1961). Oxidation of glyoxylic acid to oxalic acid by glycolic acid oxidase. J. Biol. Chem. 236: 1280–1284 [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Shockey J., Browse J. (2011). Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants. Plant J. 66: 143–160 [DOI] [PubMed] [Google Scholar]
- Shockey J.M., Fulda M.S., Browse J. (2003). Arabidopsis contains a large superfamily of acyl-activating enzymes. Phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme a synthetases. Plant Physiol. 132: 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey J.M., Fulda M.S., Browse J.A. (2002). Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 129: 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Srivastava S.K., Krishnan P.S. (1962). An oxalic acid oxidase in the leaves of Bougainvillea spectabilis. Biochem. J. 85: 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suguira M., Yamamura H., Hirano K., Sasaki M., Morikawa M., Tsuboi M. (1979). Purification and properties of oxalate metabolism: unexpected structures and mechanisms. Chem. Pharm. Bull. (Tokyo) 27: 2003–2007 [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P.D.B., van Wijk K.J. (2009). PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37(Database issue): D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedruzić D., Jónsson S., Toyota C.G., Reinhardt L.A., Ricagno S., Lindqvist Y., Richards N.G.J. (2005). The enzymes of oxalate metabolism: Unexpected structures and mechanisms. Arch. Biochem. Biophys. 433: 176–192 [DOI] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The Botany Array Resource: E-Northerns, expression angling, and promoter analyses. Plant J. 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Trinchant J.C., Guerin V., Rigaud J. (1994). Acetylene reduction by symbiosomes and free bacteroids from broad bean (Vicia faba L.) nodules (role of oxalate). Plant Physiol. 105: 555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Western T.L., Skinner D.J., Haughn G.W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Jiang J.Z., Zhang C., Jiang L.R., Ye N.H., Lu Y.S., Yang G.Z., Liu E., Peng C.L., He Z.H., Peng X.X. (2010). Glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis in rice. J. Exp. Bot. 61: 1625–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K., Braun K., Bockler A., Fuchs G. (1987). Studies on the anaerobic degradation of benzoic acid and 2-aminobenzoic acid by a denitrifying Pseudomonas strain. Arch. Microbiol. 149: 62–69 [Google Scholar]