This work identifies the maize cystatin CC9 as a compatibility factor in the biotrophic interaction with the fungal pathogen Ustilago maydis. The results show that CC9 functions as a suppressor of host defense responses by inhibiting defense-related apoplastic Cys proteases in the maize apoplast.
Abstract
Ustilago maydis is a biotrophic pathogen causing maize (Zea mays) smut disease. Transcriptome profiling of infected maize plants indicated that a gene encoding a putative cystatin (CC9) is induced upon penetration by U. maydis wild type. By contrast, cc9 is not induced after infection with the U. maydis effector mutant Δpep1, which elicits massive plant defenses. Silencing of cc9 resulted in a strongly induced maize defense gene expression and a hypersensitive response to U. maydis wild-type infection. Consequently, fungal colonization was strongly reduced in cc9-silenced plants, while recombinant CC9 prevented salicylic acid (SA)–induced defenses. Protease activity profiling revealed a strong induction of maize Cys proteases in SA-treated leaves, which could be inhibited by addition of CC9. Transgenic maize plants overexpressing cc9-mCherry showed an apoplastic localization of CC9. The transgenic plants showed a block in Cys protease activity and SA-dependent gene expression. Moreover, activated apoplastic Cys proteases induced SA-associated defense gene expression in naïve plants, which could be suppressed by CC9. We show that apoplastic Cys proteases play a pivotal role in maize defense signaling. Moreover, we identified cystatin CC9 as a novel compatibility factor that suppresses Cys protease activity to allow biotrophic interaction of maize with the fungal pathogen U. maydis.
INTRODUCTION
Plant pathogenic fungi cause significant yield losses in worldwide crop production (Sexton and Howlett, 2006). Necrotrophic pathogens kill the colonized tissue, which usually coincides with generation of reactive oxygen species, and then feed on the dead plant material (Glazebrook, 2005; van Kan, 2006). By contrast, biotrophic pathogens establish an intimate interaction with their hosts and colonize the living tissue, which they require to sequester nutrients from host cells (Glazebrook, 2005). Plant immune responses are induced by the perception of specific pathogen-associated molecular patterns (PAMPs), such as the fungal cell wall component chitin (Felix et al., 1993; Boller, 1995; Kaku et al., 2006). PAMP perception elicits basal defense signaling, including ion fluxes and (de)phosphorylation of proteins, generation of reactive oxygen species, as well as the production of signaling molecules, such as salicylic acid (SA) or jasmonic acid (JA) (Devoto and Turner, 2005; Berger et al., 2007). However, the activation of distinct hormone signaling pathways determines further defense programs, and these are defined by the lifestyle of the attacking pathogens (Glazebrook, 2005; Robert-Seilaniantz et al., 2011). Activation of JA signaling leads to the induction of defensins and chitinases as well as the accumulation of toxic secondary metabolites. This type of defense is effective against necrotrophic pathogens, and it has been shown that blockage of JA-dependent defenses confer enhanced susceptibility to necrotrophs, such as the gray mold fungus Botrytis cinerea (Rowe et al., 2010; El Oirdi et al., 2011). Activation of SA signaling results in the induction of pathogenesis-related (pr) genes, with the antifungal pr1 being the prime marker for induction of SA responses (Esfandiari et al., 2005; Seo et al., 2008). SA-dependent defense typically results in reactive oxygen species generation by peroxidase induction and, finally, the hypersensitive response (HR), including plant cell death (Torres et al., 2006; Vlot et al., 2009). It is widely accepted that this kind of defense is highly efficient against biotrophic pathogens, which need to prevent host cell death (Glazebrook, 2005). Pathogens overcome basal defense responses by secreting effectors that can target crucial components of distinct defense pathways (Chisholm et al., 2006; Jones and Dangl, 2006). In this context, there is increasing evidence that pathogens modulate the host’s signaling pathways to suppress plant responses and promote infection (Staskawicz et al., 2001; Robert-Seilaniantz et al., 2011).
The basidiomycetous smut fungus Ustilago maydis establishes a biotrophic interaction with its host plant maize (Zea mays), which leads to the formation of large tumors that can be formed in all aerial parts of the plant (Brefort et al., 2009; Skibbe et al., 2010). During the early stage of interaction, when the fungus differentiates on the host surface, PAMP-triggered immune responses can be observed (Doehlemann et al., 2008). However, this defense induction is attenuated after penetration of the plant epidermis, which coincides with the establishment of biotrophy (Doehlemann et al., 2008). At this stage, the fungus grows intracellularly and the colonized plant cells do not show obvious defense responses (Basse, 2005; Doehlemann et al., 2008; Doehlemann et al., 2009). Maize transcriptome profiling showed an induction of JA signaling during this stage, which is in line with the expected plant response to a compatible biotrophic pathogen (Doehlemann et al., 2008; Robert-Seilaniantz et al., 2011). It has recently been found that U. maydis actively reprograms hormone signaling in maize via a chorismate mutase that is transferred to the host cytoplasm. This fungal enzyme channels chorismate from SA synthesis toward prephenate and thereby contributes to the suppression of SA-associated host defenses (Djamei et al., 2011). Two additional effectors have already been identified in U. maydis that suppress defense responses by other means: Pep1 has been found to be an essential suppressor of early plant defense. Deletion mutants for pep1 induce various plant defenses, including elevated pr gene expression, accumulation of reactive oxygen species, and host cell death. These host defenses are activated immediately upon epidermal penetration. As a result, the pep1 deletion mutant fails to establish a compatible interaction and is blocked in its pathogenic development (Doehlemann et al., 2009). Another secreted effector, Pit2, was recently found to suppress plant defenses during later stages of host colonization (Doehlemann et al., 2011). In addition to fungal virulence factors, specific proteins of the host plant are also required to cause disease. These molecules are defined as compatibility or susceptibility factors (Vogel et al., 2002; Panstruga, 2003). For the biotrophic interaction of Blumeria graminis f. sp hordei with its host plant barley (Hordeum vulgare) as well as for barley nonhost resistance to the wheat (Triticum aestivum) pathogen B. graminis f. sp tritici, the conserved cell death suppressor Bax-Inhibitor-1 has been shown to enhance compatibility (Eichmann et al., 2004, 2010). An interesting group of plant genes with a potential role in pathogen interactions are the cystatins. These proteins are defined as inhibitors of papain C1A family Cys proteases and can be involved in various cellular processes (Martinez et al., 2009). Various reports describe defense functions of cystatins against insects, which results from inhibition of digestive proteases (Carrillo et al., 2011). Using stably transformed plants, the effects of cystatins against mites and pathogenic fungi were observed (Martínez et al., 2003; Carrillo et al., 2011). In wheat, it has recently been shown that induction of cystatin expression by exogenous application of JA increased resistance against Tilletia indica, a hemibiotrophic fungus that infects florets and damages grains in its destructive phase (Dutt et al., 2011). This suggests a JA-induced induction of resistance via cystatin activity against pathogens showing a necrotrophic phase. In tomato (Solanum lycopersicum), cystatins were found to be suppressed by SA, while their activity was associated with JA-induced gene expression (Doares et al., 1995). The Arabidopsis thaliana cystatin At CYS1, which is induced by wounding, nitric oxide, or avirulent pathogen attack, was shown to suppress hypersensitive cell death (Belenghi et al., 2003). The same authors speculated that At CYS1 inhibits the Cys proteases involved in the mitochondrial-dependent apoptosis pathway (Belenghi et al., 2003).
A transient virus-induced gene silencing (VIGS) system using the Brome mosaic virus (BMV) has recently been adapted to allow the functional study of maize genes during U. maydis infection (van der Linde et al., 2011a). Using this VIGS approach, we initiated a search for maize genes that contribute to compatibility in the U. maydis interaction. Here, we identified and functionally characterized maize Cystatin-9 (Corn Cystatin-9 [CC9]), which is a novel compatibility factor that suppresses maize immunity to U. maydis by the inhibition of apoplastic Cys proteases.
RESULTS
Identification of Cystatin-9 as a Compatibility Factor
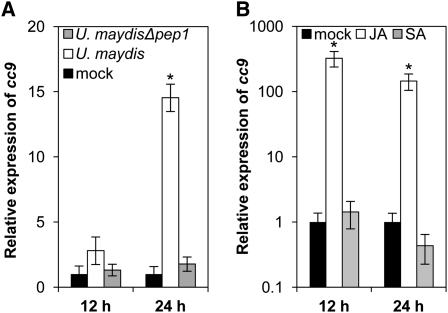
To identify novel compatibility factors in the maize–U. maydis interaction, transcriptome data of maize leaves after infection with the solopathogenic U. maydis strain SG200 and the effector mutant SG200Δpep1 were compared (Doehlemann et al., 2008, 2009). The maize cc9 gene appeared to be strongly induced early after infection with strain SG200, while there was only marginal induction after infection with the SG200Δpep1 mutant that elicits cell death (Doehlemann et al., 2009). This expression pattern of cc9 was verified by quantitative real-time PCR (qRT-PCR) (Figure 1A). As described previously, U. maydis wild-type infection is accompanied by the transcriptional activation of JA-associated genes, while no SA-dependent responses were activated upon establishment of biotrophy (Doehlemann et al., 2008). In this context, transcriptional regulation of cc9 in response to treatment with SA and methyl-JA was tested by qRT-PCR. JA treatment resulted in a strong induction of cc9 expression after 12 and 24 h, respectively, while SA treatment did not result in any detectable cc9 induction (Figure 1B). In Δpep1 infections, however, there is no indication that JA-responsive genes are activated, and cc9 expression is also not induced (Figure 1A; Doehlemann et al., 2009). This illustrates that cc9 expression in the compatible U. maydis wild-type infection correlates with the induction of JA signaling.
Figure 1.
Expression Profile of cc9.
(A) Expression of cc9 was analyzed by qRT-PCR 12 and 24 h after infection of maize seedlings with U. maydis strain SG200 (white bars) or SG200Δpep1 (gray bars) compared with mock controls (black bars).
(B) qRT-PCR analysis of cc9 expression in maize seedling leaves 12 and 24 h after treatment with 2 mM SA (gray bars) or 1 mM methyl-JA (white bars) compared with mock controls (black bars). Experiments were performed in three independent biological replicates. P values were calculated by an unpaired t test. Error bars represent the se; *P < 0.05.
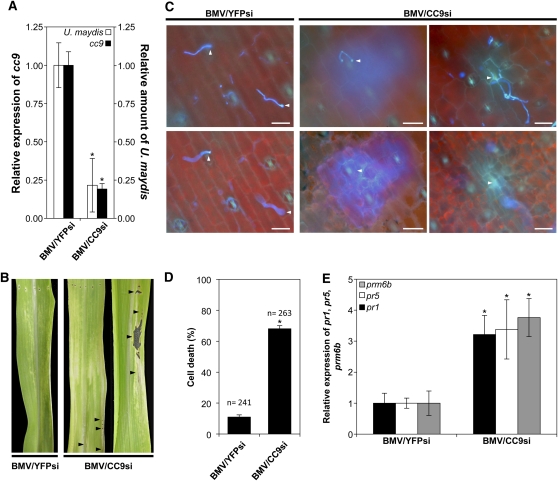
To test a potential role of cc9 for pathogenic development of U. maydis in maize, cc9 was silenced using a recently established VIGS method (van der Linde et al., 2011a; van der Linde and Doehlemann, 2012). Inoculation of maize seedlings with BMV containing a silencing construct for cc9 (BMV/CC9si) resulted in a significant decrease of cc9 transcript levels compared with control plants (inoculated with BMV/YFPsi; van der Linde et al., 2011a) (Figure 2A). Silencing of cc9 did not cause significant changes in expression of Cys protease genes (see Supplemental Table 1 online) or of pr-genes (for pr1 and prm6b). Furthermore, expression of pr5 was only weakly influenced by silencing of the cystatin (see Supplemental Table 2 online). Strikingly, in all cc9-silenced plants, U. maydis colonization was strongly reduced compared with control plants, as determined by quantitative PCR (Figure 2A). On average, silencing of cc9 by 81% (±4%) resulted in a 79% (±9%) reduction of U. maydis colonization (Figure 2A). Moreover, 3 d after U. maydis infection, extended chlorosis and necrotic lesions were observed at sites of fungal infection, indicating host cell death in the cc9-silenced plants (Figure 2B). Consequently, in cc9-silenced plants U. maydis–induced tumor formation was almost completely abolished (see Supplemental Figure 1 online), while BMV/YFPsi-inoculated control plants showed normal U. maydis infection (Figure 2A; see Supplemental Figure 1 online). These results indicate that expression of cc9 is essential for biotrophic development of U. maydis. This defines CC9 as a compatibility factor in the maize–U. maydis interaction.
Figure 2.
cc9-Silenced Maize Plants Show a Penetration Resistance to U. maydis.
(A) Fungal DNA and cc9 expression were quantified by RT-PCR using leaf samples from cc9-silenced plants and BMV/YFPsi-infected control plants 48 h after U. maydis infection. The relative amount of U. maydis DNA (white columns) and the relative expression of cc9 (black columns) of five BMV/YFPsi control plants were averaged. Column BMV/CC9si shows mean of fungal DNA and cc9 expression of five plants. For all qRT-PCRs, the relative expression or fungal amount in control plants was set to 1. Error bars represent the se; *P < 0.05.
(B) In BMV/CC9si-infected plants, U. maydis infection results in the formation of necrotic lesions along the leaf blade, as indicated by arrowheads. Representative photographs were taken 3 d after infection with U. maydis strain SG200.
(C) Microscopy analysis of BMV/YFPsi-inoculated control plants and BMV/CC9si-inoculated plants 2 d after infection with U. maydis strain SG200. U. maydis penetration attempts are indicated by arrowheads. Bars = 25 μm.
(D) Quantification of plant cells undergoing HR after U. maydis penetration in control plants (BMV/YFPsi) and cc9 silencing plants (BMV/CC9si) 2 d after infection; error bars represent the se; *P < 0.05. Experiments were performed in three independent biological replicates. P values were calculated by an unpaired t test.
(E) qRT-PCR analysis of marker gene pr1 (black bars), pr5 (white bars), and prm6b (gray bars) expression in control (BMV/YFPsi) and BMV/CC9si plants at 2 d after infection. n = 5; error bars represent se; *P < 0.05. P values were calculated by an unpaired t test.
To further characterize the role of CC9 during U. maydis infection and to understand why the fungal colonization is blocked in cc9-silenced plants, we followed U. maydis penetration by confocal microscopy. In BMV/YFPsi-infected control plants, only 11% of U. maydis appressoria induced plant cell wall autofluorescence, confirming previous observations from U. maydis wild-type infections (Figures 2C and 2D; Doehlemann et al., 2008). By contrast, in cc9-silenced maize leaves, ~68% of U. maydis appressoria induced strong autofluorescence, which is indicative of a HR of the penetrated host cell (Figure 2C; Koga et al., 1988). Furthermore, the U. maydis–induced cell wall autofluorescence spread from the site of infection to the surrounding epidermal cells as well as to mesophyll cells below (Figures 2C and 2D). In addition, transcriptional induction of the two SA-dependent genes pr1 and pr5 (van Loon et al., 2006) as well as the stress-induced marker prm6b (Nasser et al., 1990; Cordero et al., 1994; Didierjean et al., 1996) were monitored via qRT-PCR (Figure 2E). In the cc9-silenced plants, expression of pr1, pr5, and prm6b was significantly induced in response to U. maydis infection when compared with BMV/YFPsi control plants (Figure 2E). From these data, we conclude that silencing of cc9 results in an activation of maize defense after fungal penetration. As a consequence, U. maydis infection of cc9-silenced plants is blocked by a penetration-induced defense response.
Functional Characterization of Cystatin-9
cc9 encodes an 18.6-kD protein with an 18–amino acid N-terminal leader peptide that is predicted to be cleaved during secretion (Massonneau et al., 2005). CC9 contains a conserved plant cystatin consensus sequence in the central part of the protein (amino acids 104 to 113) and also the QxVxG motif (amino acids 131 to 135) that is predicted to be responsible for cystatin activity (see Supplemental Figure 2A online; Stubbs et al., 1990; Margis et al., 1998; Abraham et al., 2006). To functionally analyze CC9, the sequence encoding CC9 was amplified from cDNA of U. maydis–infected maize leaves and cloned into the pET16b vector for expression of a His-tagged fusion protein in Escherichia coli. Recombinant CC9 was purified via nickel-Sepharose affinity and subsequent gel filtration (see Supplemental Figure 2B online). Three point mutations were inserted into the putative active site (QxVxG into NxLxA) of CC9 to generate an inactive version of the protein (CC9ia) that also has been purified from E. coli (see Supplemental Figures 2A and 2B online). Because cystatins are defined as inhibitors of Cys proteases (Margis et al., 1998; Abraham et al., 2006), the functionality of recombinant CC9 compared with CC9ia was tested in an in vitro protease assay with the commercially available Cys protease papain. CC9 could inhibit papain activity in a concentration-dependent manner with a 1:1 stoichiometry (see Supplemental Figure 2C online). This shows that CC9 functions as a Cys protease inhibitor. By contrast, CC9ia did not inhibit papain activity, demonstrating loss of CC9 function after mutation of the QxVxG domain (see Supplemental Figure 2C online).
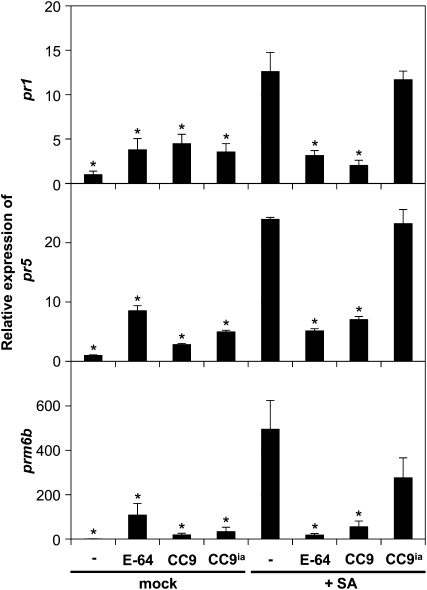
Since silencing of cc9 amplified the maize immune responses to U. maydis, we hypothesized that CC9 might act as a suppressor of plant defense. To test this, maize leaves were treated with 2 mM SA to trigger pr gene induction (Figure 3). Another set of plants was coinfiltrated with 0.5 mM recombinant CC9 and 2 mM SA. Remarkably, these plants did not show pr gene induction 24 h after the treatment, indicating suppression of the SA-induced signal by coinfiltration with CC9 (Figure 3). A comparable inhibition of pr gene induction was observed when SA was coinfiltrated with the specific Cys protease inhibitor E-64 (Barrett et al., 1982). By contrast, CC9ia did not suppress SA-triggered gene expression, demonstrating that suppression of pr gene expression by CC9 depends on its protease inhibitory activity (Figure 3).
Figure 3.
CC9 Suppresses SA-Induced pr Gene Expression.
pr gene expression in maize leaves after treatment with 2 mM SA (+SA) compared with buffer controls (mock). Recombinant CC9 (0.5 μM), CC9ia (0.5 μM), or E-64 (5 μM) was infiltrated at the same time as SA/mock treatment. qRT-PCR analysis shows expression of marker genes pr1, pr5, and prm6b 24 h after treatment. The experiment was performed in three biological replicates; error bars represent se; *P < 0.05. P values were calculated by an unpaired t test significant compared with pr gene expression of SA-treated plants.
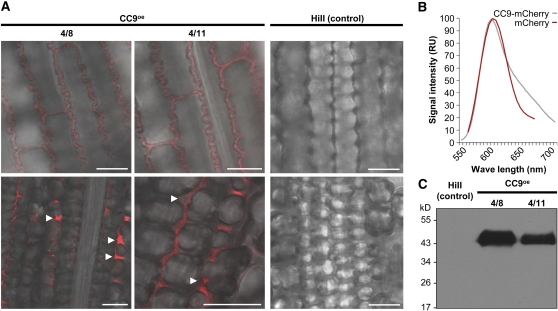
To confirm the predicted apoplastic localization and to facilitate functional analysis of CC9 in vivo, stable transgenic maize plants expressing a cc9:Human influenza hemagglutinin (HA):mCherry:HA fusion under control of the maize ubiquitin promoter were produced. These transgenic plants developed normally, did not show any phenotypic differences to nontransgenic controls, and were fully susceptible to U. maydis (see Supplemental Figure 3 online). The segregating T1 progeny of two independent cc9-overexpressing lines (CC9OE-4/8 and CC9OE-4/11) was used for further analysis. In these plants, CC9-mCherry was detected in the apoplastic space between epidermis cells as well as mesophyll cells (Figure 4A, left panel; see Supplemental Figure 4 online), which was absent in control plants (Figure 4A, right panel). Specificity of the fluorescence signal was confirmed by a spectral scan and comparison of the obtained spectrum to the expected spectrum of mCherry (Figure 4B). In addition, full-length CC9-HA-mCherry-HA was detected in apoplastic fluids of maize lines CC9OE-4/8 and CC9OE-4/11 by immunoblot analysis using anti-HA antibodies (Figure 4C; see Supplemental Figure 5 online). These results verify the in silico prediction of CC9 being secreted to the plant apoplast and show that CC9 is also secreted in the absence of U. maydis.
Figure 4.
Transgenic Maize Plants Overexpressing CC9-mCherry Secrete CC9-mCherry.
(A) Confocal images showing localization of CC9-mCherry (red) in two transgenic maize lines (4/8 and 4/11) that express cc9:HA:mCherry:HA (left two panels) and HiII control plants (right panel). Top panel: CC9-mCherry secreted by epidermal cells. Bottom panel: In mesophyll tissue, the CC9-mCherry accumulates in the apoplastic space between cells (arrowheads). Confocal images of HiII control plants taken with the same settings as the images of CC9oe plants exhibit no red fluorescence. Bars = 25 μm.
(B) Spectrum of mCherry fluorescence, comparing the signal observed in CC9oe maize lines (red) to the known spectrum of mCherry (gray) (n = 20). RU, relative units.
(C) Immunoblot of apoplastic fluids isolated from transgenic maize plants expressing CC9:HA:mCherry:HA and HiII control plants. Anti-HA antibody was used to detect full-length CC9-HA-mCherry-HA fusion protein (47 kD) in the apoplastic fluid extracts.
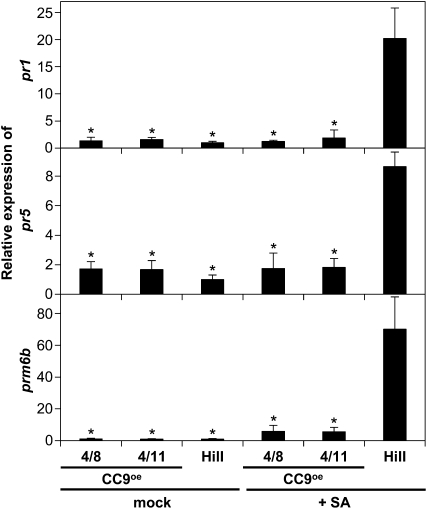
To test the influence of constitutive cc9 expression on maize defense signaling, CC9OE leaves were treated with 2 mM SA, and 24 h after treatment, pr gene expression was determined by qRT-PCR. Remarkably, the transgenic lines did not show any induction of pr gene expression in response to SA (Figure 5). These data provide further evidence that CC9 acts as a potent inhibitor of SA-induced defense signaling in maize.
Figure 5.
SA-Induced pr Gene Expression Is Blocked in CC9-mCherry–Overexpressing Maize Plants.
qRT-PCR analysis of marker genes pr1, pr5, and prm6b expression in wild-type HiII plants and cc9-overexpressing transgenic lines (4/8 and 4/11) 24 h after treatment with 2 mM SA or mock. For all qRT-PCRs, the relative expression in control plants (HiII mock) was set to 1. n = 3; error bars represent se; P values were calculated by an unpaired t test; *P < 0.05 compared with HiII treated with SA.
Inhibition of SA-Induced Apoplastic Cys Proteases by CC9
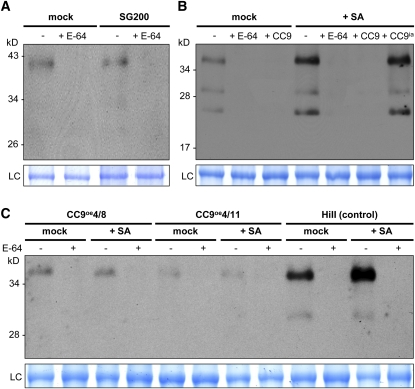
To investigate how CC9 mediates suppression of pr gene expression, we analyzed the activity of Cys proteases, which are the potential targets of CC9 in the maize plant. To this end, total protein extracts of maize leaves were isolated 48 h after mock infection, infection with the virulent U. maydis strain SG200, and treatment with SA. Active Cys proteases in the individual samples were visualized by protease activity profiling using DCG-04, which is a biotinylated form of E-64 that reacts with the catalytic Cys residue of papain-like Cys proteases (Greenbaum et al., 2000; van der Hoorn et al., 2004). Immunoblot detection of DCG-04–labeled proteases showed a weak signal at a size of ~40 kD in mock-treated controls as well as in U. maydis–infected plants (Figure 6A), indicating only weak activity of papain-like Cys proteases. By contrast, SA treatment resulted in a strong induction of protease activity, which was reflected by a markedly increased band at ~40 kD as well as additional bands at 30 and 25 kD (Figure 6B). However, these signals were absent when 0.5 μM recombinant CC9 was added to the protein extract before the DCG-04 labeling (Figure 6B), demonstrating the ability of CC9 to completely inhibit SA-induced protease activity. Furthermore, CC9ia did not inhibit DCG-04 labeling, indicating specificity of CC9 activity (Figure 6B). Preincubation with nonbiotinylated E-64 blocked labeling of the proteases, which confirmed that the detected signals were specific for papain-like Cys proteases (Figures 6A and 6B). Extracts of transgenic cc9-overexpressing plants exhibited only marginal protease levels, both in SA-treated and in mock-infiltrated control plants (Figure 6C). These low levels of Cys proteases in cc9-overexpressing plants correlate with the absence in pr gene expression and also demonstrate a substantial block in defense induction due to the presence of CC9.
Figure 6.
Activity-Based Protein Profiling Shows Activation of Maize Cys Proteases by SA.
(A) Immunoblot detection of DCG-04-labeled protein extracts from maize leaf extracts taken 2 d after infection with U. maydis strain SG200 compared with control samples taken 2 d after buffer (mock) treatment. Extracts were either untreated or pretreated with 5 μM E-64. LC, loading control.
(B) DCG-04 labeling of leaf extract from seedlings 2 d after treatment with 2 mM SA or buffer (mock). Extracts were pretreated with 5 μM E-64, 0.5 μM CC9, 0.5 μM CC9ia, or untreated, and subsequently labeled with DCG-04.
(C) DCG-04 labeling of protein extract from maize seedling leaves of two cc9-overexpressing lines (CC9oe4/8 and CC9oe4/11) and similar leaves of HiII wild-type plants as control. Leaves were taken 2 d after treatment with 2 mM SA or buffer (mock). Extracts were pretreated with 5 μM E-64 (+) or untreated (−), and subsequently labeled with DCG-04. Biotinylated proteases were detected using strep-HRP.
[See online article for color version of this figure.]
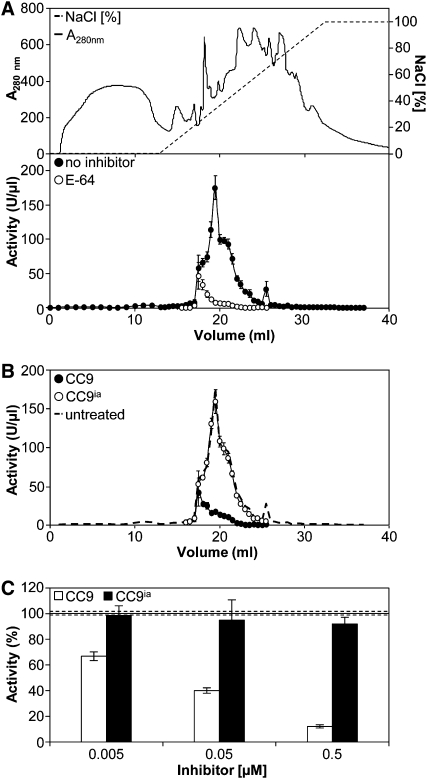
Because CC9 is secreted to the maize apoplast (Figure 4), we aimed to obtain more detailed insight into protease activity in the apoplastic fluid of maize leaves. To this end, apoplastic fluids of untreated plants, U. maydis–infected samples, and SA-treated leaves were collected at 48 h after treatment. The absence of subcellular contaminants in the apoplastic extracts was confirmed by immunoblot analysis and SDS-PAGE (see Supplemental Figures 5 and 6 online). Subsequently, the extracts were fractionated by anion-exchange chromatography, and protease activity was determined for all individual fractions (Figure 7A; see Supplemental Figure 7 online). In accordance with the DCG-04 assays, apoplastic fractions from untreated and U. maydis–infected leaves contained only low levels of protease activity (see Supplemental Figure 7 online). By contrast, apoplastic fractions of SA-treated maize leaves showed strongly induced protease activity, which was largely inhibited by E-64 (Figure 7A). The remaining E-64–insensitive protease activity, however, was found reproducibly in three biological replicates and can most likely be attributed to proteases other than Cys proteases.
Figure 7.
CC9 Inhibits Apoplastic Cys Proteases.
(A) Apoplastic fluid from maize seedling leaves 2 d after treatment with 2 mM SA, separated by anion-exchange chromatography. Top panel shows elution conditions and a typical profile of eluted proteins. Activity of proteases was tested in the presence (open symbols) and absence (closed symbols) of 5 μM E-64 using Z-Phe-Arg-AMC as substrate. Error bars represent se.
(B) Protease activity was determined after incubation with 0.5 μM CC9ia (open symbols) or 0.5 μM CC9 (closed symbols) using Z-Phe-Arg-AMC as substrate. The dashed line shows the protease activity without inhibitor (see [A]).
(C) Apoplastic fluid (peak fraction 19.5 mL of [A]) was preincubated with different concentrations of CC9 (white bars) or CC9ia (black bars). The protease activity of the untreated apoplastic fluid fraction was set to 100%. The dotted lines represent se of the untreated sample. Error bars represent se. Experiments were done in three biological replicates.
Next, apoplastic fluid fractions of SA-treated plants were incubated with recombinant CC9. This treatment resulted in a similar inhibition pattern as observed for E-64 (Figure 7B). Furthermore, protease inhibition by CC9 was concentration dependent (Figure 7C). By contrast, CC9ia did not inhibit apoplastic protease activity, indicating the specificity of this reaction (Figures 7B and 7C).
To elucidate on which level Cys proteases were activated upon SA treatment, apoplastic fluids of control- and SA-treated plants were incubated with 0.1 mM SDS, which is known to activate maize Cys proteases (Yamada et al., 2000). Interestingly, SDS treatment resulted in a strongly increased DCG-04 signal both in control and SA-treated fluids (see Supplemental Figure 8 online), suggesting that inactive proteases are constitutively present in the apoplast and are activated upon the SA stimulus. However, qRT-PCR analysis also showed a slight induction of Cys protease gene expression in response to SA (see Supplemental Table 3 online). A similar effect was also observed after infection of the U. maydis Δpep1 mutant (Doehlemann et al., 2009; see Supplemental Table 4 online). In addition, this effector mutant was found to induce an elevated Cys protease activity in maize leaves compared with wild-type infections (see Supplemental Figure 9 online).
SA-Induced Apoplastic Cys Proteases and Their Function in Defense Induction
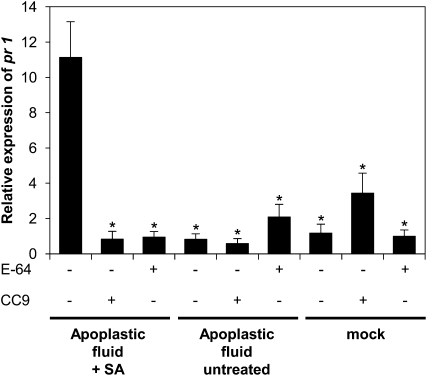
Based on the findings presented so far, we hypothesized that the SA-induced apoplastic Cys proteases function as a trigger for pr gene expression; in turn, their inhibition by CC9 would result in a block of defense activation. To test this assertion more directly, apoplastic protease fractions of controls as well as SA-treated plants were isolated and infiltrated into naïve maize plants (Figure 8; see Supplemental Figure 10 online). In addition, the apoplastic protease fractions were coinfiltrated with recombinant CC9 or E-64, respectively. As an additional control, plants were treated with CC9, E-64, or buffer alone. At 24 h after infiltration, RNA was extracted from the treated plants for subsequent qRT-PCR. As expected, neither infiltration of CC9 nor E-64 alone led to an induction of pr-1 gene expression (Figure 8). Similarly, the infiltration of apoplastic protease fractions that were isolated from untreated plants did not induce pr gene expression (Figure 8). By contrast, the protease fraction from SA-treated plants caused a significant induction of pr gene expression. Remarkably, this defense gene activation was completely abolished when either CC9 or E-64 had been coinfiltrated with the apoplastic protease fractions (Figure 8). This finding demonstrates that SA-induced apoplastic Cys proteases are sufficient to induce pr gene expression in a naïve plant. Moreover, the block of this induction by CC9 and E-64 showed that enzymatic activity of the Cys proteases is sufficient to cause defense activation.
Figure 8.
pr Gene Expression in Untreated Plants in Response to Infiltration of Apoplastic Cys Proteases.
qRT-PCR analysis of pr1 after infiltration of protease-containing apoplastic fluid fractions (17.5 to 22.5 mL) from SA-treated plants, mock-treated plants, or untreated controls. n = 3; error bars represent se; P values were calculated by an unpaired t test compared with pr1 gene expression of plants treated with protease-containing apoplastic fluid fractions from SA-treated plants. *P < 0.05. The relative expression in mock-treated plants was set to 1.
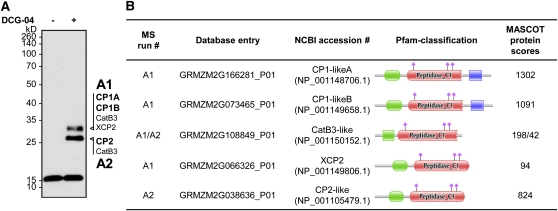
To elucidate which Cys proteases are present in the SA-induced apoplastic fluid, mass spectrometry (MS) was performed. Protease-active apoplastic fractions were treated with DCG-04 and labeled proteins were affinity purified on avidin beads. The DCG-04–labeled proteases were eluted and separated by SDS-PAGE (Figure 9A). Since protein concentrations after affinity purification were expected to be low, we visualized labeled proteins by immunoblot detection using streptavidin–horseradish peroxidase (strep-HRP). Since granulin-containing proteases are precipitated during the biotin-pulldown (Kaschani et al., 2010), only mature proteases were detected in two bands, at 28 and 32 kD. Gel regions containing labeled proteins were excised and subjected to in-gel digestion with trypsin. The obtained tryptic peptides were analyzed by nano-liquid chromatography–tandem MS (MS/MS) (Kaschani et al., 2009b). Peptide spectra were searched and assigned to proteins using MASCOT. In total, five proteins with a MASCOT score of >40 were identified (see Supplemental Data Set 1). PFAM analysis identified five papain-like Cys proteases, which all possess an N-terminal secretion signal (Figure 9). In addition, peptides were identified that correspond to a papain-like Cys protease (Cys protease1 [CP1]) but could not be assigned due to incomplete annotation of the maize genome sequence (see Supplemental Data Set 1). Most spectral counts were observed for the maize CP1, which appeared in the two highly similar isoforms CP1A and CP1B (Figure 9B) (Alexandrov et al., 2009). In addition, CP2, xylem Cys protease2 (XCP2), and a cathepsin B–like protease were identified (Figure 9). BLAST analysis showed that the five proteases are well conserved in other plant species and can be assigned to previously described homologs in Arabidopsis (Table 1). In summary, apoplastic fluids that trigger immune responses in naïve plants contain at least five Cys proteases.
Figure 9.
Identification of Biotinylated Cys Proteases in Maize Apoplastic Fluid.
Apoplastic fluid from maize seedling leaves treated for 2 d with 2 mM SA was separated by anion-exchange chromatography, and fractions containing activity (17.5 to 22.5 mL) were pooled and labeled with or without DCG-04.
(A) Small-scale samples were taken and analyzed with strep-HRP.
(B) A large portion of the labeling samples was subjected to SDS-PAGE, and the corresponding bands were analyzed by MS. Identified proteases for each band are indicated in (A). Protein domain structures of the identified proteases are given in (B). All proteases contain a signal peptide, a prodomain (green), and a peptidase C1 domain (red). CP1-likeA and CP1-likeB also carry a granulin domain (blue). The active site amino acids are indicated by pink diamonds. NCBI, National Center for Biotechnology Information.
Table 1.
Active Cys Proteases in the Maize Apoplast and Their Orthologs in Arabidopsis
| Identified Maize Cys Protease | Arabidopsis Ortholog | Arabidopsis Accession | Identity to Maize | E-Value |
| CP1-likeA | RD21A-like | At1g47128 | 67% | 0.0 |
| CP1-likeB | RD21A-like | At1g47128 | 69% | 0.0 |
| CathepsinB3-like | Cathepsin B-like | At1g02305 | 72% | 1e−175 |
| CP2-like | Cys proteinase AALP | At5g60360 | 73% | 0.0 |
| XCP2-like | XCP1-like | At4g35350 | 69% | 2e−176 |
DISCUSSION
Cystatin-9 Is a Novel Compatibility Factor in Maize
This study presents the functional characterization of the maize cystatin CC9. We show that CC9 represents a novel type of compatibility factor in the maize–U. maydis pathosystem. Plant cystatins have been defined as inhibitors of papain C1A family Cys proteases (Martinez et al., 2009) and were isolated in complexes with Cys proteases from different plant species (Yamada et al., 2001; Tajima et al., 2011). The maize genome encodes 12 putative cystatins, of which 11 contain an N-terminal secretion signal (Massonneau et al., 2005; Schnable et al., 2009). Transcriptome analysis showed that of the maize cystatins, only cc9 is induced during the early stage of U. maydis infection (Doehlemann et al., 2008). One remarkable feature of CC9 is its ability to efficiently inhibit all E-64–sensitive Cys proteases in maize. Both in whole-plant extract as well as in apoplastic fluid extracts, Cys protease activity was completely blocked by CC9, and commercial papain was inhibited in a 1:1 molecular stoichiometry. In the course of these studies, we identified SA-induced, CC9-sensitive apoplastic Cys proteases as potent inducers of immune responses in maize.
Silencing of cc9 resulted in a strong reduction of U. maydis colonization, which is associated with plant cell death and pr gene expression. Induction of HR-like responses in cc9-silenced plants is triggered during penetration of the maize epidermis by U. maydis. This indicates that CC9-mediated defense suppression is required at the early phase of the interaction, when biotrophy is established by the fungus, which is in line with the transient transcriptional induction of cc9 at this early stage of U. maydis infection. A remarkable observation was the complete block of SA-induced pr gene expression by recombinant CC9 that was infiltrated to the apoplast of maize leaves. This immune suppression requires the conserved QxVxG motif of CC9. In transgenic CC9-mcherry–overexpressing plants, the protein was found to localize to the apoplast, and these plants showed the same block in SA-induced pr gene expression that has been observed upon CC9 infiltration to wild-type plants. Therefore, we conclude that CC9 suppresses SA-induced immunity by inhibiting apoplastic proteases.
cc9 expression is induced in the compatible U. maydis wild-type infection that is, typically for biotrophic interactions, associated with an induction of JA signaling (Doehlemann et al., 2008; Robert-Seilaniantz et al., 2011), and this correlates with cc9 induction by direct JA treatment to maize leaves. In line with this, only marginal activity of papain-like Cys proteases was detected after U. maydis wild-type infection. By contrast, SA treatment strongly induced Cys proteases, and this defense-triggered activity could be inhibited by CC9. In line with this, the U. maydis Δpep1 mutant, which induces various defense responses including pr1 induction (Doehlemann et al., 2009), triggered increased Cys protease activity. Moreover, isolation of apoplastic fluids showed a strong induction of Cys proteases in this compartment, and this activity was inhibited by CC9 as well. SDS induction of apoplastic proteases suggests that inactive proteases are already present prior to an SA stimulus. Such posttranslational activation allows a quick induction of defense signaling. However, SA treatment as well as infection by the Δpep1 mutant also lead to a moderate induction of Cys protease genes on the transcriptional level, which may cause an additional de novo synthesis of proteases during the defense response. By contrast, U. maydis wild-type infection had a marginal effect only on one of the proteases. SA-induced activation of Cys proteases has also been reported in tomato, where treatment with the SA analog benzothiadiazole induced the activity of apoplastic Cys proteases (Shabab et al., 2008). Intriguingly, our studies revealed that the SA-induced protease activity was sufficient to induce pr gene expression in maize plants that did not experience prior treatment. This effect could be inhibited either by E-64 or by CC9, which demonstrated that no other molecules present in the fractionated apoplastic fluid were responsible for this activity. In addition, this result provides direct evidence for the defense-activating effect of SA-induced apoplastic Cys proteases. It furthermore provides an explanation for the ability of CC9 to suppress SA-induced pr gene expression, although at present, one cannot assess whether the proteases act directly as inducers of defense gene expression or trigger activation of a subsequent signaling cascade. In a simplified model, SA signaling, which could be triggered by an avirulent pathogen, activates Cys proteases, and these mediate a signal leading to defense activation. In the case of infection by a compatible biotroph, such as U. maydis, the induced JA signaling leads to cc9 expression, which in turn blocks Cys protease activity and thereby downstream defense responses (see Supplemental Figure 11 online). In addition, apoplastic Cys proteases turn out to be important factors in the maize immune system. On the one hand, they act upstream of SA-dependent defenses (i.e., they trigger pr gene expression). On the other hand, the apoplastic Cys proteases are activated themselves by SA as well.
SA-Induced Apoplastic Cys Proteases in Maize
MS analysis identified a set of five apoplastic Cys proteases of the C1A peptidase family (Rawlings et al., 2008; http://merops.sanger.ac.uk/). A previous study describes the isolation of CCPIP, a maize protease with high similarity to CP1A, in a complex with the two cystatins CC1 and CC2 (Yamada et al., 2000), but no functional analysis for this protease or these cystatins has been conducted so far. In the dicot plants Arabidopsis and tomato, several C1A proteases have been identified. The homologous tomato C14 protease was found to be targeted by Phytophthora infestans cystatin-like effector proteins (EPICs), and silencing of this protease resulted in an increased susceptibility to this oomycete pathogen (Tian et al., 2007; Kaschani et al., 2010). Both maize CP1A and tomato C14 contain, in addition to the N-terminal autoinhibitory domain, a C-terminal granulin domain that may negatively regulate protease activity (Bateman and Bennett, 1998). This domain is also found in the conserved Arabidopsis protease RD21. In both RD21 and C14, intermediate isoforms carry the granulin domain, whereas mature isoforms lack the granulin domain. However, the trigger for removal of the granulin domain is not known so far (Yamada et al., 2000; van der Hoorn et al., 2004). The identified protease CP2 shows only 30% identity to CP1A, although belonging to the C1 protease family as well (Rawlings and Morton, 2008). CP2 has been cloned from maize seeds and has been identified as an aleurain-like protease that is active during seed germination (Domoto et al., 1995; Rawlings and Morton, 2008). However, a putative function in pathogen interaction has not been described for CP2 so far. This also holds true for the XCP2-like proteases. XCPs are involved in autolysis of xyleme trachea elements during xylogenesis (Avci et al., 2008). Another protease identified in the maize apoplast was similar to Cathepsin B. In animal models, this type of protease has been implicated in various processes, including programmed cell death (Zeiss, 2003). In Nicotiana benthamiana, a Cathepsin B is required for the full HR and resistance to different bacterial pathogens, such as Erwinia amylovora and Pseudomonas syringae (Gilroy et al., 2007). Generation of triple mutants for all three Cathepsin B genes in Arabidopsis showed that cathepsins were functionally redundant in promoting basal resistance to P. syringae (McLellan et al., 2009). In addition, the pathogen-triggered activation of N. benthamiana Cathepsin B is associated with allocation of the protein to the apoplast, which is in line with the finding made in this study (Gilroy et al., 2007).
Plant Proteases in Pathogen Interactions
Papain-like Cys proteases are involved in various cellular processes (van der Hoorn, 2008). In animal systems, Cys proteases were identified as key regulators of apoptosis (Saraste and Pulkki, 2000; Fan et al., 2005). Also, in plants, a role in programmed cell death has been described for several enzymes of this class (van der Hoorn, 2008). Recent literature also provides increasing evidence that Cys proteases are involved in pathogen interaction (Rooney et al., 2005; Gilroy et al., 2007; van Esse et al., 2008). In soybean (Glycine max), Cys proteases were found to be involved in programmed cell death that was induced by the bacterial pathogen P. syringae (Solomon et al., 1999). This cell death–stimulating activity of Cys proteases was inhibited by ectopic expression of a cystatin. However, the mechanism suggested by the authors implies an intracellular activity of Cys proteases, which might be attributed to caspase-like proteases. In plant cells, these enzymes are considered to play a central role in mitochondrial-dependent programmed cell death, similar to the caspases in animal systems (Heath, 2000). By contrast, the mechanism described in this study relies on the regulation of apoplastic protease activity. Apoplastic Cys proteases were found to be targets of pathogen effectors. Examples are the P. infestans effectors EPIC1 and EPIC2B, which confer cystatin-like activity (Tian et al., 2007). The Cladosporium fulvum effector Avr2 was found to directly target the tomato proteases RCR3 and PIP1, which are both defense-induced Cys proteases localizing to the apoplast of tomato (Rooney et al., 2005; Shabab et al., 2008). Remarkably, both PIP1 and RCR3 are inhibited by P. infestans EPIC effectors as well, indicating that these proteases are an important component of the tomato immune system that is targeted by unrelated pathogens (Song et al., 2009). Interestingly, RCR3 belongs to the same protease family (C1A) as the most abundant proteases we identified in the maize apoplast, CP1 and CP2. Although sequence identity between these maize proteases and RCR3 is only 31% for CP1A and 33% for CP2, one might speculate that CP1A family proteases in different plant species have a conserved function in apoplastic defense signaling.
At present, it remains unclear how the apoplastic Cys proteases activate SA-induced defense signaling. Based on recent reports from other plant systems, one could consider different scenarios for the function of apoplastic Cys proteases in plant defense signaling. In Arabidopsis, tobacco, and soybean, it was shown that in addition to PAMP signaling, damage-associated molecular pattern (DAMP) signaling also occurs during pathogen attack (Pearce et al., 2001, 2010; Huffaker et al., 2006b). The released signals result in amplification of innate immune responses through the JA/ethylene and SA signaling pathways (Pearce et al., 2010). Besides cutin monomers and cell wall fragments, particularly small peptides that are released from larger propeptides can serve as DAMPs (Boller and Felix, 2009). One such peptide is Arabidopsis PEP1, which is released from the propeptide PROPEP1 by a yet unknown mechanism (Huffaker et al., 2006a). Recently, the maize homolog (Zm PEP1) has been identified and was shown to function as a DAMP signal in association with JA signaling to confer resistance against the necrotrophic pathogen Cochliobolus heterostrophus (Huffaker et al., 2011). Another possible mechanism of protease-mediated signaling is the proteolytic shedding of extracellular receptor domains by matrix metalloproteases. In animal systems, this mechanism has been identified as a common principle in various diseases, including cancer (Peschon et al., 1998; Choi et al., 2010), and also during invasion of fungal pathogens (Gazi et al., 2011). As another example, subtilisin-like proteases, termed phytaspases (plant Asp-specific protease), were identified in rice (Oryza sativa) and tobacco. Phytaspases have caspase-like activity and are synthesized as a pro-form and then processed by autocatalysis into the mature enzyme, which then localizes to the plant apoplast (Chichkova et al., 2010). During abiotic and biotic stress, the mature enzyme is recruited into the cell, where it causes reactive oxygen species accumulation and cytochrome c release (Chichkova et al., 2010). In oat (Avena sativa), a protease-mediated signaling mechanism was proposed for two subtilisin-like proteases (saspases), which show caspase-like activity (Coffeen and Wolpert, 2004). Induction of programmed cell death by the Cochliobolus victoria toxin victorin was found to coincide with relocalization of the saspases to the oat apoplast (Coffeen and Wolpert, 2004).
The variety of protease-mediated mechanisms in plant cell death regulation associated with the pathogen response demonstrates the importance of fine-tuning of protease activity by the interplay of activating triggers and inhibitors. The mechanism of host endogenous cystatin activity and defense-triggered apoplastic proteases presented in this study adds a new perspective on extracellular defense signaling in the monocot crop maize. Therefore, future research will aim to elucidate the roles of the individual proteases in defense signaling. A major challenge will be to identify the signal(s) that are released or removed by extracellular protease activity into the maize apoplast and to understand how such signals are perceived and transduced into downstream defense cascades.
METHODS
Growth Conditions and Plant Infections
For VIGS experiments, maize (Zea mays cv Va35) plants were grown as previously described (van der Linde et al., 2011a; van der Linde and Doehlemann, 2012). For all other experiments, Z. mays cv Early Golden Bantam or cv HiII (AxB) as well as respective transgenic lines were grown in a greenhouse at 28°C during the light period (26,000 lux, 14.5 h) and 22°C during the dark period (9.5 h). Nicotiana benthamiana plants were grown at 22°C during the light period (26,000 lux, 14.5 h) and 20°C during the dark period (9.5 h). To generate T1 progeny from primary transgenic HiII(AxB)x(AxB)/CC9oe lines, plants were self-pollinated. Brome mosaic virus (BMV) inoculations of maize and tobacco plants were done as previously described (van der Linde et al., 2011a; van der Linde and Doehlemann, 2012). For infections with Ustilago maydis, liquid cultures of strain SG200 (Kämper et al., 2006) and SG200Δpep1 (Doehlemann et al., 2009) were grown in YEPSL (0.4% yeast extract, 0.4% peptone, and 2% Suc), shaking at 200 rounds per minute at 28°C, to an OD600 of 0.6 to 0.8. Cells were centrifuged at 2500 rpm for 5 min and resuspended in water to an OD600 of 3.0 for infection of maize seedlings 11 d after BMV inoculation or 7 d after sowing of CC9oe T1 HiII plants.
Strain and Plasmid Constructions
Standard molecular biology methods were applied according to Sambrook et al. (1989). Oligonucleotides used for PCR are summarized in Supplemental Table 5 online.
pB3-3/CC9
For VIGS of cc9, a 248-bp PCR fragment of the cc9 coding region was amplified from U. maydis–infected maize cDNA using primers containing a HindIII restriction site (see Supplemental Table 5 online) and cloned in reverse orientation to RNA3 in pB3-3 as described (van der Linde et al., 2011a; van der Linde and Doehlemann, 2012). In silico analysis concerning small interfering RNA formation and silencing specificity of cc9 sequences was performed according to van der Linde et al. 2011a) and van der Linde and Doehlemann (2012). Plasmids pF1-11 and pF2-2 were provided by X. Ding and R. Nelson (Ding et al., 2006).
pET15bCC9
For overexpression of cc9 in Escherichia coli, the open reading frame was amplified from cDNA of U. maydis–infected maize with the forward primer containing a XhoI restriction site and binding downstream of the predicted signal peptide and the reverse primer containing a BamHI site (see Supplemental Table 5 online). Afterwards, PCR fragments and plasmid pET15b (Merck) were digested with XhoI and BamHI and ligated using T4 Ligase (NEB). The obtained plasmid was sequenced to check for errors and transformed into Tuner(DE3)pLysS cells for expression (Merck).
pET15bCC9ia
For the inactive version of cc9 in E. coli, the pET15bCC9 construct was used as PCR template with primers encoding for Asn instead of Gln at amino acid position 102, for Leu instead of Val at amino acid position 104, and Ala instead of Gly at amino acid position 106 (see Supplemental Table 5 online). The PCR product was digested with DpnI and transformed into E. coli. After sequencing, the plasmid was transformed into BL21(DE3)pLysS cells (Merck) for overexpression.
p6UCC9HAmcherryHA
A full-length PCR product of cc9 was amplified from cDNA of U. maydis–infected maize plants (see Supplemental Table 2 online) and PCR fragments of mCherry-HA by amplification using p123-pep1-mcherry-HA (Doehlemann et al., 2009) as template. Next, the cc9 PCR product was digested with BamHI and HindIII. mCherry-HA was digested with HindIII and EcoRI, and pUbi-ABM (DNA Cloning Service) was cut with BamHI and EcoRI. All three restriction fragments were ligated to the obtained pUbi-AB, containing cc9 fused at its C terminus to a HA tag, mCherry, and a second HA tag. This plasmid was restricted with SfiI to release the Ubiquitin promoter, cc9-HA-mCherry-HA, and the nos terminator. The resulting fragment was ligated into p6U (DNA Cloning Service) using SfiI.
Expression and Protein Purification
Expression and purification of CC9 and CC9ia was performed according to van der Linde et al. (2011b), while expression of cc9 was induced with 5 mM isopropyl β-d-1-thiogalactopyranoside and cc9ia with 1 mM isopropyl β-d-1-thiogalactopyranoside, followed by 2 h of incubation at 37°C. After purification by nickel-chelate chromatography, gel filtration chromatography was performed on an S-75 column (GE), and fractions containing CC9 were pooled and if necessary concentrated by Amicon Ultra centrifugation filters 3K (Millipore).
Microscopy
Fungal hyphae were stained with Calcofluor White (0.1 μg/mL in water). Samples were incubated in the staining solution for 1 min and washed in water prior to microscopy detection. Plasmolysis was induced by the addition of 1 M NaCl to the sample. Confocal images were taken on a TCS-SP5 confocal microscope (Leica) as described previously (Doehlemann et al., 2008). mCherry fluorescence of CC9-mCherry was excited at 561 nm and detected at 580 to 630 nm. For detection of Calcofluor White and HR by UV autofluorescence, an excitation of 405 nm and detection at 435 to 480 nm were used. Spectral measurement of mCherry fluorescence was performed using the LAS-AF software (version 2.5.1; Leica); the spectrum was calculated from raw data with the software Origin 6.1 (Origin Lab), using five-point fast Fourier transform smoothing. The obtained spectrum was compared with the published spectrum of mCherry (Shaner et al., 2004).
Preparation of Total Plant Extract
For preparation of total leaf extract, seedling leaves were frozen in liquid N2, ground to powder, and mixed with one volume of 2× Laemmli loading buffer (Laemmli, 1970) and glass beads. Samples were boiled for 5 min and vortexed for 15 min, twice. Protein concentration in the sample supernatant was determined by the Bradford assay (Bradford, 1976).
Small-Scale Activity-Based Profiling with DCG-04 and Immunoblotting
Total soluble leaf protein of maize seedlings of cv Early golden Bantam or CC9oe T1 lines after treatment with SA, U. maydis infection, or the corresponding mock treatments was extracted by grinding the tissue in water containing 1 mM DTT. Protein concentration was determined by the Bradford assay (Bradford, 1976), and 0.2 mg/mL total protein was incubated with 5 μM E-64 and 0.5 μM CC9/CC9ia for 30 min at room temperature. Then, 15 mM sodium phosphate buffer, pH 6.0, 0.2 mM DTT, and 2 μM DCG-04 were added. After incubation for 5 h at room temperature, proteins were precipitated in acetone and resolved in 2× Laemmli loading buffer (Laemmli, 1970).
Immunoblotting and Immunodetection
For immunoblotting, 20 μg of protein per lane was separated on an SDS-PAGE gel (Laemmli, 1970) followed by immunoblotting. Biotinylated proteins were detected by strep-HRP (1:3000) (Sigma-Aldrich) (van der Hoorn et al., 2004). For detection of HA tag proteins Histone H3 and ribulose-1,5-bis-phosphate carboxylase/oxygenase, the blotting membrane was incubated for 1 h with anti-HA antibody (Sigma-Aldrich) (1:7500), anti-H3 antibody (Agrisera) (1:5000), or anti-ribulose-1,5-bis-phosphate carboxylase/oxygenase antibody (Sigma-Aldrich) (1:7500). After washing, the blots were incubated for 1 h with HRP-conjugated secondary antibodies (anti-mouse, 1:5000; anti-rabbit, 1:3000; both from Santa Cruz Biotechnology; or anti-chicken, 1:20,000; Sigma-Aldrich).
Large-Scale Activity-Based Profiling with DCG-04 and Target Identification by MS Using the Blind-Cut Method
Apoplastic fluid from maize seedling leaves 2 d after treatment with 2 mM SA was separated by anion-exchange chromatography. Protease activity containing fractions (17.5 to 22.5 mL) was pooled (final protein concentration 0.21 mg/mL) and incubated with DCG-04 according to Kaschani et al. (2010). Incubation with avidin beads and preparation for SDS-PAGE of the samples was performed as described by Kaschani et al. (2010) (see Supplemental Figure 12 online). The samples were divided in a smaller (2 μL) fraction and a bigger (30 μL) fraction and separated on the same SDS-PAGE (Laemmli, 1970) as indicated in Supplemental Figure 12 online. The part of the gel containing the smaller samples was then used for immunoblot procedure to localize biotinylated proteins (see lanes 2 and 3 in Supplemental Figure 5 online). Using the position of the biotinylated proteins from the strep-HRP blot (van der Hoorn et al., 2004) as guidelines, corresponding regions in the large sample containing gel were excised from lanes 7 and 9 (see Supplemental Figure 12 online). Next, the gel slabs were subjected to the in-gel digestion procedure, and obtained peptide mixes were used for the subsequent nano-liquid chromatography–electrospray ionization–MS/MS analysis (Kaschani et al., 2009a). The mass spectrometer was essentially set up as described by Nickel et al. (2012). Briefly, experiments were performed on a Thermo LTQ Velos mass spectrometer coupled to a Proxeon EASY-nLC. The LTQ Velos mass spectrometer was operated using Xcalibur software (version 2.1). The mass spectrometer was set in the positive ion mode, survey scanning between mass-to-charge ratio of 400 and 1600, with an ionization potential of 3.61 kV. Peptides were separated on a single reverse phase C18 column (inner diameter 75 μm, packed with 12-cm ReproSil- Pur C18-AQ [3 μm]) using an acetonitrile gradient (5 to 80% over 90 min), at a flow rate of 300 nL/min. Peptides were fragmented by collision-induced decay in a data-directed fashion, fragmenting the 20 most intense multiply charged precursors in each MS scan. Collision-induced decay energy was set to 35% for the generation of MS2 spectra. Following fragmentation, precursors were excluded from further MS/MS acquisition for 60 s with a list of excluded ions consisting of 500 members maximum.
MS2 spectra data were searched using the MASCOT algorithm (version 2.3.02) first against a database of known contaminants (as incorporated in MASCOT) followed by searching against the maize sequences from the database ZmB73_5b_FGS_translations_20110205.fasta (www.maizesequence.org/index.html) (Schaeffer et al., 2011). MASCOT searches allowed for oxidation of Met residues (16 D), static modification of Cys residues (57 D; due to alkylation with iodoacetamide), tryptic peptides with two missed cleavages allowed, and a mass tolerance set to ±0.25 D for precursor mass and ±0.35 D for product ion masses. The Decoy Database Search was turned on. The resulting MS2 spectra matches were assembled and filtered according to MASCOT protein and peptide scores. Proteins with a score above 40 based on an ion score of >35 were retained.
Transformation of Maize
Transgenic maize plants were produced by Agrobacterium tumefaciens–mediated gene transfer to F2 hybrid immature embryos of HiII A × HiII B largely following the protocol of Hensel et al. (2009). The full-length cDNA of cc9 was cloned into the maize transformation vector p6U, resulting in a cc9:HA:mcherry:HA fusion under control of the maize ubiquitin promoter. The construct was stably integrated into the genome of maize HiII (AxB)x(AxB) hybrid plants via Agrobacterium-mediated gene transfer to immature embryos. In this study, Agrobacterium strain AGL-1 (Lazo et al., 1991), containing p6UCC9HAmcherryHA, was used. The coculture medium contained 500 μM acetosyringone, and the concentration of the plant-selective agent Hygromycin B (Roche Diagnostics) was 200 μM.
Infiltration of Maize Leaves
Maize seedling leaves were infiltrated with 2 mM SA (Sigma-Aldrich) dissolved in 1% ethanol or 1 mM methyl-JA (Sigma-Aldrich) in 1% ethanol. For infiltration of apoplastic fluid fractions or SA with recombinant purified CC9, 2 mM SA, 0.5 μM CC9, 10% pooled fractions of MonoQ-separated apoplastic fluid, 1% ethanol, and 50 mM sodium phosphate buffer, pH 6,0 were used.
Nucleic Acid Preparation from Maize Tissue, cDNA Synthesis, and qRT-PCR
To determine the fungal biomass and gene expression of cc9, pr1 prmb6, pr5, cp1, cp2, cathB, and xcp2 in VIGS-targeted maize leaves, 2-cm sections of the infected leaves were excised 1 cm below the U. maydis or mock injection site 48 h after infection (13 d after BMV inoculation). To remove fungal cells from the leaf surface, the sections were washed three times with 0.1% Tween 20 in water. For gene expression analysis of cc9, cp1, cp2, cathB, and xcp2 after SA, mock, or JA treatment, infiltration spots were cut from leaves 12 and 24 h after infiltration. At these time points, leaf material of U. maydis–, mock-, and U. maydisΔpep1–infected plants was also collected as described above. To test the influence of SA, apoplastic fluid and CC9 infiltration into cv Early golden Bantam or CCoe T1 lines on pr1, prmb6, and pr5 expression, infiltrated spots were excised from plants 24 h after infiltration. For subsequent genomic DNA and RNA extraction, leaf material was frozen in liquid N2, ground to powder, and extracted using the MasterPure Complete DNA and RNA purification kit (Biozym). After extraction, the first-strand cDNA synthesis kit (Fermentas) was used to reverse transcribe 1 μg of total RNA with oligo(dT) Primers. The qRT-PCR analysis was performed using an iCycler (Bio-Rad) in combination with the iQ SYBR Green Supermix (Bio-Rad). For virus titer detection in tobacco, extracts were prepared and analyzed by quantitative PCR as described by van der Linde et al. (2011a). Primers for quantification of U. maydis biomass, maize gene transcription levels, and virus titer detection are summarized in Supplemental Table 5 online. Maize glyceraldehyde dehydrogenase (Doehlemann et al., 2009) or N. benthamiana actin (van der Linde et al., 2011a) were used as reference genes for normalization. For cycling conditions, see van der Linde et al. (2011a). Gene expression levels and relative amount of fungal DNA were then calculated relative to actin or gapdh expression levels or amount of gapdh DNA using the cycle threshold (Ct) 2−ΔCt method (Livak and Schmittgen, 2001). In the following, the average expression and the se of three independent experiments were calculated, and expression of the controls was set to 1. For analysis of VIGS, average expression and fungal amount of five silencing plants as well as se were calculated, and mean expression and fungal amount of five BMV/YFPsi plants were set to 1. P values were estimated using an unpaired t test. P values <0.05 were marked with an asterisk.
Protease Activity Tests
Protease activity was determined using the fluorimetric substrate Z-Phe-Arg 7-amido-4-methylcoumarin (Z-Phe-Arg-AMC) (Sigma-Aldrich), which leads to release of fluorescence at 460 nm when cleaved by protease activity (Zimmerman et al., 1976). Briefly, 3 mg/mL papain (2× crystallized; Sigma-Aldrich) was activated by incubation for 1.5 h in 50 mM sodium phosphate buffer, pH 6.0, and 10 mM DTT. After desalting in 100 mM sodium phosphate buffer, pH 6.0, the activity of 10 μM papain was measured using 10 μM substrate in 10 mM sodium phosphate buffer, pH 6.0, 150 mM NaCl, 1 mM EDTA, and 0.5 mM DTT in a total volume of 100 μL on a fluorometer (Tecan). The influence of CC9, CC9ia, or E-64 on protease activity was determined by incubating papain with various concentrations of CC9 or 5 μM E-64 (Sigma-Aldrich) for 10 min in the activity assay prior to the addition of substrate. For measurements of protease activity in apoplastic fluid fractions, 10 μL of the fraction was used in the assay as described above. For the calibration curve, different concentrations of Coumarin120 (Sigma-Aldrich) were used. One unit of activity was defined as the release of 0.01 μmol 7-amino-4-methylcoumarin per min.
Apoplastic Fluid Isolation
For the preparation of apoplastic fluid, seedling leaves were infiltrated with 2 mM SA, inoculated with SG200 culture, or left untreated as control. Two days after treatment, the leaves were harvested and cut into pieces of 2-cm length, followed by evacuation under water in a vacuum chamber for 3 × 15 min at 400 mbar. The evacuated leaf sections were then stacked into packs of 20 to 30 and squeezed into the barrel of a 50-mL syringe so that the cut edges of the leaves faced the ends of the barrel. The barrel was then put into a 50-mL falcon tube with the needle hub facing downwards and spun for 20 min at 2000g and 4°C. Afterwards, the extracted apoplastic fluid was collected from the falcon tube and stored at −20°C.
Fractionation of Apoplastic Fluid by Anion-Exchange Chromatography
For separation of apoplastic fluid by anion-exchange chromatography, 5 mg of apoplastic fluid was loaded on a 1-mL MonoQ resin column (GE) equilibrated with 20 mM sodium phosphate buffer, pH 6.0. The column was washed with 2 mL 20 mM sodium phosphate buffer, pH 6.0. The flow-through and the wash were collected in 1-mL fractions. Then, proteins were eluted in 500-μL fractions with a 20-mL linear NaCl gradient from 0 to 1 M NaCl in 20 mM sodium phosphate buffer, pH 6.0. Afterwards, the column was washed with 5 mL of 1 M NaCl in 20 mM sodium phosphate buffer, pH 6.0, and collected in 1-mL fractions.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: maize cc9, BN000513; maize gapdh, NM001111943; N. benthamiana actin, AY594294; maize pr1, BM351351; maize pr5, BM075306; maize pr6mb, AY111675; maize xcp2, NP_001149806.1; maize cp2, NP_001105479.1; maize cathepsinB, NP_001150152.1; maize cp1-likeA, NP_001148706.1; maize cp1-likeB, NP_001149658.1; and U. maydis ppi, XM_754780.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. U. maydis Symptoms in BMV/YFPsi- and BMV/CC9si-Inoculated Plants after Infection with the Solopathogenic U. maydis Strain SG200.
Supplemental Figure 2. CC9 Structure, Purification, and Inhibitory Activity on Papain.
Supplemental Figure 3. CC9oe Plants Show Normal Growth and Are Fully Susceptible to U. maydis.
Supplemental Figure 4. Secretion of CC9 in Leaves of Transgenic Maize Line (4/8) Expressing cc9:HA:mCherry:HA.
Supplemental Figure 5. Immunoblot Analysis of CC9-Overexpressing Maize Plants.
Supplemental Figure 6. Test for Subcellular Contaminations in the Apoplastic Fluid Extracts.
Supplemental Figure 7. Protease Activity in Apoplastic Fluids of Untreated and U. maydis–Infected Maize Leaves.
Supplemental Figure 8. Activation of Cys Proteases by SDS.
Supplemental Figure 9. Maize Cys Protease Activity after U. maydis Infection.
Supplemental Figure 10. Experimental Setup to Test the Biological Activity of SA-Induced Cys Proteases in Apoplastic Fluid by qRT-PCR.
Supplemental Figure 11. Model Describing the Interplay of Maize Defense Induction by Apoplastic Cys Proteases and Their Inhibition by CC9.
Supplemental Figure 12. Illustration of the Blind Cut Method.
Supplemental Table 1. Expression of Cys Proteases in BMV/CC9si and BMV/YFPsi Plants after Infection with U. maydis.
Supplemental Table 2. Expression of pr1, pr5, and prm6b in BMV/CC9si and BMV/YFPsi Plants.
Supplemental Table 3. Expression of Cys Proteases in SA-Treated Plants.
Supplemental Table 4. Expression of Cys Proteases in U. maydis Wild-Type and U. maydis Δpep1 Infected Plants.
Supplemental Table 5. PCR Primers Used in This Study.
Supplemental Data Set 1. Proteins Identified by Mass Spectrometry.
Acknowledgments
We thank Daniela Aßmann for expert technical assistance and Stefan Schmidt for diligent greenhouse support. We also thank Heike Büchner and Sandra Wolf for excellent technical assistance in maize transformation. We thank Christian Herrberger and André Mueller for the extraction of apoplastic fluids. We also thank Regine Kahmann and Alga Zuccaro for helpful comments on the manuscript. This project was financially supported by the Max Planck Society and the Deutsche Forschungsgemeinschaft via Projects DO 1421/1-1 (Deutsche Forschungsgemeinschaft Research Group FOR666) and HO 3983/4-1/7-1.
AUTHOR CONTRIBUTIONS
K.v.d.L., C.H., C.K., and F.K. performed the research. K.v.d.L. and G.D. designed the research. R.A.L.v.d.H. and J.K. contributed analytical and experimental tools. K.v.d.L., F.K., and G.D. analyzed data. K.v.d.L. and G.D. wrote the article.
References
- Abraham Z., Martinez M., Carbonero P., Diaz I. (2006). Structural and functional diversity within the cystatin gene family of Hordeum vulgare. J. Exp. Bot. 57: 4245–4255 [DOI] [PubMed] [Google Scholar]
- Alexandrov N.N., Brover V.V., Freidin S., Troukhan M.E., Tatarinova T.V., Zhang H., Swaller T.J., Lu Y.P., Bouck J., Flavell R.B., Feldmann K.A. (2009). Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol. Biol. 69: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avci U., Petzold H.E., Ismail I.O., Beers E.P., Haigler C.H. (2008). Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J. 56: 303–315 [DOI] [PubMed] [Google Scholar]
- Barrett A.J., Kembhavi A.A., Brown M.A., Kirschke H., Knight C.G., Tamai M., Hanada K. (1982). L-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem. J. 201: 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basse C.W. (2005). Dissecting defense-related and developmental transcriptional responses of maize during Ustilago maydis infection and subsequent tumor formation. Plant Physiol. 138: 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Bennett H.P. (1998). Granulins: The structure and function of an emerging family of growth factors. J. Endocrinol. 158: 145–151 [DOI] [PubMed] [Google Scholar]
- Belenghi B., Acconcia F., Trovato M., Perazzolli M., Bocedi A., Polticelli F., Ascenzi P., Delledonne M. (2003). AtCYS1, a cystatin from Arabidopsis thaliana, suppresses hypersensitive cell death. Eur. J. Biochem. 270: 2593–2604 [DOI] [PubMed] [Google Scholar]
- Berger S., Benediktyová Z., Matouš K., Bonfig K., Mueller M.J., Nedbal L., Roitsch T. (2007). Visualization of dynamics of plant-pathogen interaction by novel combination of chlorophyll fluorescence imaging and statistical analysis: differential effects of virulent and avirulent strains of P. syringae and of oxylipins on A. thaliana. J. Exp. Bot. 58: 797–806 [DOI] [PubMed] [Google Scholar]
- Boller T. (1995). Chemoperception of microbial signals in plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 189–214 [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brefort T., Doehlemann G., Mendoza-Mendoza A., Reissmann S., Djamei A., Kahmann R. (2009). Ustilago maydis as a pathogen. Annu. Rev. Phytopathol. 47: 423–445 [DOI] [PubMed] [Google Scholar]
- Carrillo L., Martinez M., Álvarez-Alfageme F., Castañera P., Smagghe G., Diaz I., Ortego F. (2011). A barley cysteine-proteinase inhibitor reduces the performance of two aphid species in artificial diets and transgenic Arabidopsis plants. Transgenic Res. 20: 305–319 [DOI] [PubMed] [Google Scholar]
- Chichkova N.V., Shaw J., Galiullina R.A., Drury G.E., Tuzhikov A.I., Kim S.H., Kalkum M., Hong T.B., Gorshkova E.N., Torrance L., Vartapetian A.B., Taliansky M. (2010). Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO J. 29: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Choi S., Lee H., Choi J.R., Oh E.S. (2010). Shedding; towards a new paradigm of syndecan function in cancer. BMB Rep. 43: 305–310 [DOI] [PubMed] [Google Scholar]
- Coffeen W.C., Wolpert T.J. (2004). Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell 16: 857–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero M.J., Raventos D., Sansegundo B. (1994). Differential expression and induction of chitinases and beta-1,3-glucanases in response to fungal infection during germination of maize seeds. Mol. Plant Microbe Interact. 7: 23–31 [Google Scholar]
- Devoto A., Turner J.G. (2005). Jasmonate-regulated Arabidopsis stress signalling network. Physiol. Plant. 123: 161–172 [Google Scholar]
- Didierjean L., Frendo P., Nasser W., Genot G., Marivet J., Burkard G. (1996). Heavy-metal-responsive genes in maize: Identification and comparison of their expression upon various forms of abiotic stress. Planta 199: 1–8 [DOI] [PubMed] [Google Scholar]
- Ding X.S., Schneider W.L., Chaluvadi S.R., Mian M.A., Nelson R.S. (2006). Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Mol. Plant Microbe Interact. 19: 1229–1239 [DOI] [PubMed] [Google Scholar]
- Djamei A., et al. (2011). Metabolic priming by a secreted fungal effector. Nature 478: 395–398 [DOI] [PubMed] [Google Scholar]
- Doares S.H., Narváez-Vasquez J., Conconi A., Ryan C.A. (1995). Salicylic acid inhibits synthesis of proteinase-inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol. 108: 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Reissmann S., Assmann D., Fleckenstein M., Kahmann R. (2011). Two linked genes encoding a secreted effector and a membrane protein are essential for Ustilago maydis-induced tumour formation. Mol. Microbiol. 81: 751–766 [DOI] [PubMed] [Google Scholar]
- Doehlemann G., van der Linde K., Assmann D., Schwammbach D., Hof A., Mohanty A., Jackson D., Kahmann R. (2009). Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 5: e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Wahl R., Horst R.J., Voll L.M., Usadel B., Poree F., Stitt M., Pons-Kühnemann J., Sonnewald U., Kahmann R., Kämper J. (2008). Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 56: 181–195 [DOI] [PubMed] [Google Scholar]
- Domoto C., Watanabe H., Abe M., Abe K., Arai S. (1995). Isolation and characterization of two distinct cDNA clones encoding corn seed cysteine proteinases. Biochim. Biophys. Acta 1263: 241–244 [DOI] [PubMed] [Google Scholar]
- Dutt S., Pandey D., Kumar A. (2011). Jasmonate signal induced expression of cystatin genes for providing resistance against Karnal bunt in wheat. Plant Signal. Behav. 6: 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann R., Bischof M., Weis C., Shaw J., Lacomme C., Schweizer P., Duchkov D., Hensel G., Kumlehn J., Hückelhoven R. (2010). BAX INHIBITOR-1 is required for full susceptibility of barley to powdery mildew. Mol. Plant Microbe Interact. 23: 1217–1227 [DOI] [PubMed] [Google Scholar]
- Eichmann R., Schultheiss H., Kogel K.H., Hückelhoven R. (2004). The barley apoptosis suppressor homologue BAX inhibitor-1 compromises nonhost penetration resistance of barley to the inappropriate pathogen Blumeria graminis f. sp. tritici. Mol. Plant Microbe Interact. 17: 484–490 [DOI] [PubMed] [Google Scholar]
- El Oirdi M., El Rahman T.A., Rigano L., El Hadrami A., Rodriguez M.C., Daayf F., Vojnov A., Bouarab K. (2011). Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23: 2405–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfandiari N., Kohi Habibi M., Mosahebi G.H., Mozafari J. (2005). Detection of Alfalfa mosaic virus (AMV) in pea field in Iran. Commun. Agric. Appl. Biol. Sci. 70: 407–410 [PubMed] [Google Scholar]
- Fan T.-J., Han L.-H., Cong R.-S., Liang J. (2005). Caspase family proteases and apoptosis. Acta Biochim. Biophys. Sin. (Shanghai) 37: 719–727 [DOI] [PubMed] [Google Scholar]
- Felix G., Regenass M., Boller T. (1993). Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein-phosphorylation, and establishment of a refractory state. Plant J. 4: 307–316 [Google Scholar]
- Gazi U., Rosas M., Singh S., Heinsbroek S., Haq I., Johnson S., Brown G.D., Williams D.L., Taylor P.R., Martinez-Pomares L. (2011). Fungal recognition enhances mannose receptor shedding through dectin-1 engagement. J. Biol. Chem. 286: 7822–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy E.M., et al. (2007). Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J. 52: 1–13 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Greenbaum D., Medzihradszky K.F., Burlingame A., Bogyo M. (2000). Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem. Biol. 7: 569–581 [DOI] [PubMed] [Google Scholar]
- Heath M.C. (2000). Hypersensitive response-related death. Plant Mol. Biol. 44: 321–334 [DOI] [PubMed] [Google Scholar]
- Hensel G., Kastner C., Oleszczuk S., Riechen J., Kumlehn J. (2009). Agrobacterium-mediated gene transfer to cereal crop plants: current protocols for barley, wheat, triticale, and maize. Int. J. Plant Genomics 2009: 835608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Dafoe N.J., Schmelz E.A. (2011). ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiol. 155: 1325–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Ryan C.A. (2006a). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 103: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Ryan C.A. (2006b). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 103: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper J., et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101 [DOI] [PubMed] [Google Scholar]
- Kaschani F., Gu C., Niessen S., Hoover H., Cravatt B.F., van der Hoorn R.A. (2009a). Diversity of serine hydrolase activities of unchallenged and botrytis-infected Arabidopsis thaliana. Mol. Cell. Proteomics 8: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F., Shabab M., Bozkurt T., Shindo T., Schornack S., Gu C., Ilyas M., Win J., Kamoun S., van der Hoorn R.A. (2010). An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154: 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F., Verhelst S.H., van Swieten P.F., Verdoes M., Wong C.S., Wang Z., Kaiser M., Overkleeft H.S., Bogyo M., van der Hoorn R.A. (2009b). Minitags for small molecules: detecting targets of reactive small molecules in living plant tissues using ‘click chemistry’. Plant J. 57: 373–385 [DOI] [PubMed] [Google Scholar]
- Koga H., Zeyen R.J., Bushnell W.R., Ahlstrand G.G. (1988). Hypersensitive cell death, autofluorescence, and insoluble silicon accumulation in barley leaf epidermal cells under attack by Erysiphe graminis f. sp. hordei. Physiol. Mol. Plant Pathol. 32: 395–409 [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lazo G.R., Stein P.A., Ludwig R.A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (NY) 9: 963–967 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Margis R., Reis E.M., Villeret V. (1998). Structural and phylogenetic relationships among plant and animal cystatins. Arch. Biochem. Biophys. 359: 24–30 [DOI] [PubMed] [Google Scholar]
- Martinez M., Cambra I., Carrillo L., Diaz-Mendoza M., Diaz I. (2009). Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiol. 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez M., López-Solanilla E., Rodríguez-Palenzuela P., Carbonero P., Díaz I. (2003). Inhibition of plant-pathogenic fungi by the barley cystatin Hv-CPI (gene Icy) is not associated with its cysteine-proteinase inhibitory properties. Mol. Plant Microbe Interact. 16: 876–883 [DOI] [PubMed] [Google Scholar]
- Massonneau A., Condamine P., Wisniewski J.P., Zivy M., Rogowsky P.M. (2005). Maize cystatins respond to developmental cues, cold stress and drought. Biochim. Biophys. Acta 1729: 186–199 [DOI] [PubMed] [Google Scholar]
- McLellan H., Gilroy E.M., Yun B.W., Birch P.R., Loake G.J. (2009). Functional redundancy in the Arabidopsis Cathepsin B gene family contributes to basal defence, the hypersensitive response and senescence. New Phytol. 183: 408–418 [DOI] [PubMed] [Google Scholar]
- Nasser W., deTapia M., Burkard G. (1990). Maize pathogenesis-related proteins: Characterization and cellular distribution of 1,3-β-glucanases and chitinases induced by Brome mosaic virus infection or mercuric-chloride treatment. Physiol. Mol. Plant Pathol. 36: 1–14 [Google Scholar]
- Nickel S., Kaschani F., Colby T., van der Hoorn R.A., Kaiser M. (2012). A para-nitrophenol phosphonate probe labels distinct serine hydrolases of Arabidopsis. Bioorg. Med. Chem. 20: 601–606 [DOI] [PubMed] [Google Scholar]
- Panstruga R. (2003). Establishing compatibility between plants and obligate biotrophic pathogens. Curr. Opin. Plant Biol. 6: 320–326 [DOI] [PubMed] [Google Scholar]
- Pearce G., Moura D.S., Stratmann J., Ryan C.A., Jr (2001). RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 98: 12843–12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G., Yamaguchi Y., Barona G., Ryan C.A. (2010). A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc. Natl. Acad. Sci. USA 107: 14921–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon J.J., et al. (1998). An essential role for ectodomain shedding in mammalian development. Science 282: 1281–1284 [DOI] [PubMed] [Google Scholar]
- Rawlings N.D., Morton F.R. (2008). The MEROPS batch BLAST: A tool to detect peptidases and their non-peptidase homologues in a genome. Biochimie 90: 243–259 [DOI] [PubMed] [Google Scholar]
- Rawlings N.D., Morton F.R., Kok C.Y., Kong J., Barrett A.J. (2008). MEROPS: The peptidase database. Nucleic Acids Res. 36(Database issue): D320–D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D.G. (2011). Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Rooney H.C., Van’t Klooster J.W., van der Hoorn R.A., Joosten M.H., Jones J.D., de Wit P.J. (2005). Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308: 1783–1786 [DOI] [PubMed] [Google Scholar]
- Rowe H.C., Walley J.W., Corwin J., Chan E.K., Dehesh K., Kliebenstein D.J. (2010). Deficiencies in jasmonate-mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog. 6: e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Saraste A., Pulkki K. (2000). Morphologic and biochemical hallmarks of apoptosis. Cardiovasc. Res. 45: 528–537 [DOI] [PubMed] [Google Scholar]
- Schaeffer M.L., Harper L.C., Gardiner J.M., Andorf C.M., Campbell D.A., Cannon E.K.S., Sen T.Z., Lawrence C.J. (2011). MaizeGDB: Curation and outreach go hand-in-hand. Database (Oxford) 2011: bar022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Seo P.J., Lee A.-K., Xiang F., Park C.-M. (2008). Molecular and functional profiling of Arabidopsis pathogenesis-related genes: Insights into their roles in salt response of seed germination. Plant Cell Physiol. 49: 334–344 [DOI] [PubMed] [Google Scholar]
- Sexton A.C., Howlett B.J. (2006). Parallels in fungal pathogenesis on plant and animal hosts. Eukaryot. Cell 5: 1941–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabab M., Shindo T., Gu C., Kaschani F., Pansuriya T., Chintha R., Harzen A., Colby T., Kamoun S., van der Hoorn R.A.L. (2008). Fungal effector protein AVR2 targets diversifying defense-related cys proteases of tomato. Plant Cell 20: 1169–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Skibbe D.S., Doehlemann G., Fernandes J., Walbot V. (2010). Maize tumors caused by Ustilago maydis require organ-specific genes in host and pathogen. Science 328: 89–92 [DOI] [PubMed] [Google Scholar]
- Solomon M., Belenghi B., Delledonne M., Menachem E., Levine A. (1999). The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11: 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Win J., Tian M., Schornack S., Kaschani F., Ilyas M., van der Hoorn R.A., Kamoun S. (2009). Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. USA 106: 1654–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B.J., Mudgett M.B., Dangl J.L., Galan J.E. (2001). Common and contrasting themes of plant and animal diseases. Science 292: 2285–2289 [DOI] [PubMed] [Google Scholar]
- Stubbs M.T., Laber B., Bode W., Huber R., Jerala R., Lenarčič B., Turk V. (1990). The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: A novel type of proteinase inhibitor interaction. EMBO J. 9: 1939–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima T., Yamaguchi A., Matsushima S., Satoh M., Hayasaka S., Yoshimatsu K., Shioi Y. (2011). Biochemical and molecular characterization of senescence-related cysteine protease-cystatin complex from spinach leaf. Physiol. Plant. 141: 97–116 [DOI] [PubMed] [Google Scholar]
- Tian M., Win J., Song J., van der Hoorn R., van der Knaap E., Kamoun S. (2007). A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 143: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A., Jones J.D., Dangl J.L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn R.A.L. (2008). Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 59: 191–223 [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A.L., Leeuwenburgh M.A., Bogyo M., Joosten M.H.A.J., Peck S.C. (2004). Activity profiling of papain-like cysteine proteases in plants. Plant Physiol. 135: 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linde K., Doehlemann G. (2012). Utilizing virus induced gene silencing for the functional characterization of maize genes during infection with the fungal pathogen Ustilago maydis. In Methods in Molecular Biology, Becker A., (New York: Humana Press/Springer Science and Business Media; ), in press [DOI] [PubMed] [Google Scholar]
- van der Linde K., Gutsche N., Leffers H.M., Lindermayr C., Müller B., Holtgrefe S., Scheibe R. (2011b). Regulation of plant cytosolic aldolase functions by redox-modifications. Plant Physiol. Biochem. 49: 946–957 [DOI] [PubMed] [Google Scholar]
- van der Linde K., Kastner C., Kumlehn J., Kahmann R., Doehlemann G. (2011a). Systemic virus-induced gene silencing allows functional characterization of maize genes during biotrophic interaction with Ustilago maydis. New Phytol. 189: 471–483 [DOI] [PubMed] [Google Scholar]
- van Esse H.P., Van’t Klooster J.W., Bolton M.D., Yadeta K.A., van Baarlen P., Boeren S., Vervoort J., de Wit P.J.G.M., Thomma B.P.H.J. (2008). The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 20: 1948–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kan J.A. (2006). Licensed to kill: The lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11: 247–253 [DOI] [PubMed] [Google Scholar]
- van Loon L.C., Rep M., Pieterse C.M.J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.A., Klessig D.F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Vogel J.P., Raab T.K., Schiff C., Somerville S.C. (2002). PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Kondo A., Ohta H., Masuda T., Shimada H., Takamiya K. (2001). Isolation of the protease component of maize cysteine protease-cystatin complex: Release of cystatin is not crucial for the activation of the cysteine protease. Plant Cell Physiol. 42: 710–716 [DOI] [PubMed] [Google Scholar]
- Yamada T., Ohta H., Shinohara A., Iwamatsu A., Shimada H., Tsuchiya T., Masuda T., Takamiya K. (2000). A cysteine protease from maize isolated in a complex with cystatin. Plant Cell Physiol. 41: 185–191 [DOI] [PubMed] [Google Scholar]
- Zeiss C.J. (2003). The apoptosis-necrosis continuum: insights from genetically altered mice. Vet. Pathol. 40: 481–495 [DOI] [PubMed] [Google Scholar]
- Zimmerman M., Yurewicz E., Patel G. (1976). A new fluorogenic substrate for chymotrypsin. Anal. Biochem. 70: 258–262 [DOI] [PubMed] [Google Scholar]