The circadian clock is a timing device that allows plants to anticipate environmental changes rather than just respond to them. This work demonstrates that alternative splicing of clock gene transcripts is one of the mechanisms that regulate the clock, particularly in response to changes in temperature.
Abstract
Alternative splicing plays crucial roles by influencing the diversity of the transcriptome and proteome and regulating protein structure/function and gene expression. It is widespread in plants, and alteration of the levels of splicing factors leads to a wide variety of growth and developmental phenotypes. The circadian clock is a complex piece of cellular machinery that can regulate physiology and behavior to anticipate predictable environmental changes on a revolving planet. We have performed a system-wide analysis of alternative splicing in clock components in Arabidopsis thaliana plants acclimated to different steady state temperatures or undergoing temperature transitions. This revealed extensive alternative splicing in clock genes and dynamic changes in alternatively spliced transcripts. Several of these changes, notably those affecting the circadian clock genes LATE ELONGATED HYPOCOTYL (LHY) and PSEUDO RESPONSE REGULATOR7, are temperature-dependent and contribute markedly to functionally important changes in clock gene expression in temperature transitions by producing nonfunctional transcripts and/or inducing nonsense-mediated decay. Temperature effects on alternative splicing contribute to a decline in LHY transcript abundance on cooling, but LHY promoter strength is not affected. We propose that temperature-associated alternative splicing is an additional mechanism involved in the operation and regulation of the plant circadian clock.
INTRODUCTION
Alternative splicing (AS) is a well-established mechanism in eukaryotic cell function that regulates both transcriptome and proteome diversity and regulates protein structure/function and gene expression (Graveley, 2001; Black, 2003; Stamm et al., 2005; Chen and Manley, 2009). Following transcription, precursor mRNAs undergo splicing to remove introns and join exons to generate mRNAs for translation into proteins. Splicing is performed by the spliceosome, a large RNA-protein complex consisting of five small ribonucleoprotein particles and ~180 proteins (Wahl et al., 2009). Through the selection of alternative splice sites, different mRNA variant transcripts are produced from pre-mRNAs from the same gene. Alternatively, spliced mRNA variants can give rise to functionally different protein variants with altered amino acid sequences and protein domains resulting in, for example, changes in activity, localization, interaction partners, or posttranslational modifications (Black, 2003; Stamm et al., 2005). AS can also generate nonfunctional mRNAs containing premature termination codons (PTCs), which are targeted for degradation by nonsense-mediated decay (NMD), affecting transcript levels and expression, or giving rise to truncated proteins (Nicholson et al., 2010). Selection of alternative splice sites is determined by splice site sequences and sequences in exons and introns called splicing enhancer or silencer elements, which are binding sites for splicing factors such as Ser/Arg-rich and heterogeneous nuclear ribonucleoprotein particle protein families, and other cell-, stage-, or tissue-specific proteins (Wang et al., 2008; Barash et al., 2010). Regulation of AS depends on the relative levels and activities of these splicing factors that determine the use of different splice sites to produce more than one spliced mRNA from one gene.
In plants, AS is widespread with a current estimate of 42% of intron-containing genes in Arabidopsis thaliana being alternatively spliced (Filichkin et al., 2010). AS occurs in genes involved in growth and development, responses to biotic and abiotic stresses, and wounding (Dinesh-Kumar and Baker, 2000; Iida et al., 2004; Egawa et al., 2006; Palusa et al., 2007; Reddy, 2007; Barbazuk et al., 2008; Bove et al., 2008). The importance of splicing/AS is demonstrated by the variety of growth and developmental phenotypes of overexpression and knockout lines of plant Ser/Arg-rich proteins, including altered leaf and whole plant morphology, plant size, delayed flowering, and sensitivity to UV (Barta et al., 2008). AS has also been reported in the Arabidopsis circadian clock and its outputs. For example, an intron retention (IR) event in CIRCADIAN CLOCK ASSOCIATED1 (CCA1) is conserved in at least four plant species, and in Arabidopsis, two clock-regulated genes encoding RNA binding proteins, GLYCINE-RICH RNA BINDING PROTEIN7 (GRP7; also known as COLD CIRCADIAN RHYTHM AND RNA BINDING2 [CCR2]) and GRP8, are regulated by AS and NMD (Schöning et al., 2008; Filichkin et al., 2010). In Arabidopsis, the level of the intron-containing CCA1 transcript, which is nonfunctional owing to the presence of PTCs, was increased in high light and reduced in the cold (Filichkin et al., 2010). Recent work has provided further evidence of links between the circadian clock and AS. In Arabidopsis, PROTEIN ARGININE METHYL TRANSFERASE5 (PRMT5) is required for correct pre-mRNA splicing (Deng et al., 2010). PRMT5 methylates a wide variety of substrates, including histones and spliceosomal proteins (Sanchez et al., 2010), and prmt5 mutants show a long circadian period and dramatic changes in splicing of PSEUDO RESPONSE REGULATOR9 (PRR9) transcripts (Hong et al., 2010; Sanchez et al., 2010), which encode one component of the central circadian clock (McClung, 2006; Harmer, 2009). Temperature-dependent AS of the circadian clock genes Per and frq has been observed in Drosophila melanogaster and Neurospora crassa and is thought to play roles in determining phase and period (Majercak et al., 1999; Diernfellner et al., 2007; Akman et al., 2008). These observations raise the question of whether AS might be involved in the operation of the plant circadian clock or its response to altered conditions.
Organisms have evolved circadian clocks that drive rhythms at the molecular and cellular levels and thus temporally regulate their physiology and behavior to anticipate changes in the environment. Circadian rhythms are generated endogenously, persist in constant conditions, have a period close to 24 h, and can be entrained by light/dark or warm/cold cycles. In plants, many processes exhibit circadian rhythms, including photosynthesis, leaf movement, hormone responses, stem extension, and stomatal opening (McClung, 2006; Harmer, 2009; Pruneda-Paz and Kay, 2010). The expression of a large number of genes, estimated by different methods as about one-third of the genome (Michael and McClung, 2003; Covington et al., 2008), is under circadian regulation, and there is compelling evidence that possession of a circadian clock can confer a fitness benefit (Green et al., 2002; Dodd et al., 2005).
The framework of the plant circadian clock comprises three (or four) interlocking gene expression feedback loops (Locke et al., 2006; Zeilinger et al., 2006; Pokhilko et al., 2010), similar in concept to the feedback loops in other eukaryotic clocks (Bell-Pedersen et al., 2005); these transcriptional loops may be linked indirectly to nontranscriptional clocks (O’Neill et al., 2011). In the central loop of the plant transcriptional clock, two dawn-expressed, closely related, and partially redundant Myb transcription factors, CCA1 and LATE ELONGATED HYPOCOTYL (LHY), inhibit expression of TIMING OF CAB EXPRESSION1 (TOC1; also known as PRR1) by direct binding to the TOC1 promoter (Alabadí et al., 2001; Perales and Más, 2007). In turn, accumulation of TOC1 in the evening leads to expression of CCA1 and LHY; TOC1 functions at least partly by antagonizing CCA1 HIKING EXPEDITION (CHE), which is a transcriptional repressor of CCA1 but not of LHY (Pruneda-Paz et al., 2009). In the evening loop, TOC1 and CCAI/LHY repress expression of a component Y, which induces expression of TOC1; the model proposed by Locke et al. (2006) suggested that Y contains GIGANTEA (GI), but that of Pokhilko et al. (2010) separated the functions of Y and GI. In the morning loop, CCA1/LHY induce expression of PRR7 and PRR9 (Farré et al., 2005; Locke et al., 2006; Zeilinger et al., 2006); these function as transcriptional repressors to inhibit expression of CCA1 and LHY, as does PRR5 (Nakamichi et al., 2010). Other genes are also involved, including ARRYTHMO (LUX) and EARLY FLOWERING3 (ELF3), whose products repress PRR9 expression (Dixon et al., 2011; Helfer et al., 2011) and ELF4 (Doyle et al., 2002); ELF4, ELF3, and LUX form an evening complex that links the clock to diurnal regulation of growth (Nusinow et al., 2011). Operation of the plant transcriptional clock also involves functions at levels other than direct control of gene expression. For example, phosphorylation affects the DNA binding activity of CCA1 (Sugano et al., 1998; Portoles and Más, 2010). The proteasomal turnover of TOC1 and PRR5 is mediated by the F-box protein ZEITLUPE (ZTL) and is itself affected by phosphorylation (Fujiwara et al., 2008), and other components are also subject to turnover by the proteasome (Pruneda-Paz and Kay, 2010). Induction of TOC1 expression involves formation of transcriptionally permissive chromatin mediated by clock-regulated histone acetylation at TOC1 (Perales and Más, 2007). Recent work shows that mutations in jumonji domain histone demethylases affect the period of the clock (Jones et al., 2010; Lu et al., 2011).
The relationships between the circadian clock and temperature changes have been of great interest for several reasons (McClung and Davis, 2010). The term temperature compensation describes the observation that the period of circadian outputs varies little over a range of physiologically relevant temperatures. Identification of the mechanisms underlying temperature compensation has proved challenging. Genetic evidence has implicated CCA1, LHY, GI, and FLOWERING LOCUS C (Edwards et al., 2005, 2006; Gould et al., 2006), but a recent study suggested that the regulation of CCA1 and LHY expression by PRR7 and PRR9 plays a major role (Salomé et al., 2010). Entrainment refers to the synchronization of the circadian clock with the external environment. The Arabidopsis circadian clock can be entrained by low-amplitude temperature cycles; this seems to require PRR7 and PRR9 (Salomé and McClung, 2005). The clock gates the low-temperature induction of both C-REPEAT/DRE BINDING FACTOR gene expression and Ca2+ signals (Fowler et al., 2005; Dodd et al., 2006). Evidence from poplar (Populus spp), chestnut (Castanea sativa), and Arabidopsis shows that the clock can stall at low temperature, with the transcript levels of many clock genes becoming high and losing rhythmicity (Ramos et al., 2005; Bieniawska et al., 2008; Ibañez et al., 2008; Ibañez et al., 2010). However, the mechanisms underlying these relationships are not known.
In their natural environment, plants are subject to frequent and extensive temperature fluctuations. However, most research on the interaction of temperature with the circadian clock has been performed by comparison of plants at different fixed temperatures. It is often the case that analyzing transients can provide novel information not available from examination of the steady states themselves. Here, we investigated the expression of Arabidopsis circadian clock genes in response to temperature changes at the level of both transcription and AS. We show the occurrence of numerous AS events in the core clock genes, many of which are temperature sensitive and some of which have a major role in the effects of temperature on gene expression. Furthermore, we show that changes in AS, rather than changes in promoter strength, contribute to a decline in LHY transcript abundance on cooling. Collectively, the data implicate AS as a further mechanism involved in operation and regulation of the circadian clock.
RESULTS
Extensive AS in Arabidopsis Core Clock Genes
In light of the recent reports of AS in CCA1 and PRR9 (Filichkin et al., 2010; Sanchez et al., 2010), we analyzed systematically the set of core clock genes (CCA1, LHY, TOC1 [PRR1], PRR3, 5, 7, and 9, ZTL, GI, and CHE) to identify AS events, which would generate mRNA splice variants. We employed a high-resolution RT-PCR system (HR RT-PCR) based on fluorescently labeled primers covering the length of each gene and separation of products, to single base pair resolution, on a capillary ABI3730 sequencer (Simpson et al., 2008; Raczynska et al., 2010; Kalyna et al., 2011). RT-PCR was performed on total RNA from a mixture of plant material grown at different temperatures and harvested at different time points across the diurnal cycle (see Methods). The primer strategies and representative HR RT-PCR panel electropherograms produced by the ABI 3730 genotyping software for a subset of the genes analyzed are shown in Supplemental Figures 1 to 3 online. In all, 63 different AS events (73 total transcripts, including normally spliced transcripts) were detected for the set of 10 clock genes (see Supplemental Data Set 1 online). The majority of AS events were novel with only eight AS events being currently annotated in The Arabidopsis Information Resource (TAIR) for LHY, CCA1, PRR9, PRR3, and ZTL. Additional splice variants reported for CCA1 (retention of intron 4) and PRR9 (retention of intron 3) (Filichkin et al., 2010; Sanchez et al., 2010) were confirmed here.
The identity of the majority of the novel events were determined by sequencing (either Sanger sequencing of RT-PCR products or by RNA-seq) or could be predicted (especially for IR events) based on precise knowledge of the size of the product, the sequence of introns, and potential alternative splice sites in the amplified regions (see Supplemental Data Set 1 online). Of these events, 37 had low expression (four were almost undetectable, 32 represented <1% of the total transcripts, and one was between 1 and 2%). Many of the AS events were IR events (27/63), of which 16 were found in low abundance (<1% of total transcripts) and probably represent partially spliced transcripts in the RNA samples. CHE had no AS events. ZTL had an AS event annotated in TAIR for which we found no evidence, and GI had three AS events that were all of low abundance (see Supplemental Data Set 1 online), but it should be noted that these AS events may be abundant only under environmental conditions not tested in this work.
Temperature-Dependent AS of Clock Genes
Having defined the AS events in the core clock genes of Arabidopsis, we investigated whether temperature influenced AS of these genes because temperature regulation of AS occurs in clock genes of Drosophila and N. crassa (Kojima et al., 2011) and the levels of the Arabidopsis CCA1 intron 4 retention are modulated by light and temperature (Filichkin et al., 2010). In addition, initial results showed that transitions to, and acclimation to, lower temperatures had profound and opposing effects on the levels of AS of CCA1 and LHY (see below). We used the HR RT-PCR system to examine the relative levels of fully spliced transcripts and transcripts containing the different AS events identified above for all of the core clock genes. Samples were collected at least every 3 h for 24 h at 20°C, for 24 h following transfer of plants to 4°C, and for 24 h after acclimation (day 4 after transfer to 4°C) (see Supplemental Figure 4 online). Fifteen important AS events in the core clock genes were observed, of which 13 were identified (Figure 1, Table 1). These either increased to between 10 and 50% of total transcripts in at least one phase of the transition and acclimation to 4°C or were virtually absent at 20°C and only visible in the cold treatment.
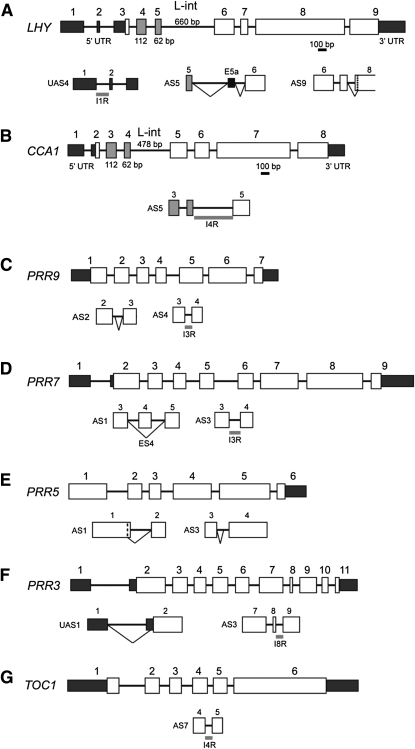
Figure 1.
Significant Temperature-Associated AS Events.
LHY (A), CCA1 (B), PRR9 (C), PRR7 (D), PRR5 (E), PRR3 (F), and TOC1 (G). Gene structures are shown with exons numbered; exons in the 5′ and 3′UTRs are smaller, dark-shaded boxes; coding sequences are larger open boxes, except the Myb-encoding exons of LHY and CCA1, which are shaded gray. AS events that comprise more than 10% of total transcripts or are cold specific are shown below the gene structures. Diagonal lines, splicing event; ES, exon skip (details in Supplemental Data Set 1 online). The vertical dashed line in LHY indicates the start of exon 8 in AS9.
Table 1.
Significant AS Events in Core Clock Genes
| Gene | AS Event | Sequencing | TAIR | AS Event | NMD |
| LHY | UAS4 | Sanger/NGS | TAIR.2 | IR1 (5′UTR) | NMD (P < 0.05)a |
| AS5 | Sanger/NGS | TAIR.5 | Alt 3′ss and alt 5′ss adds alternative exon of 82 nucleotides | NMD (P < 0.01)a | |
| AS9 | Sanger/NGS | Alt 3′ss in exon 8 removes 3 nucleotides (TAG/CAG) | In frame, no NMD observed | ||
| CCA1 | AS5 | Sanger/NGS/Filichkin et al. (2010) | IR4 | No NMD observed | |
| AS14 | Unknown | Unknown | Not determined | ||
| PRR9 | AS2 | Sanger | TAIR.2 | Alt 5′ss in IVS2 adds 8 nucleotides | NMD possible (P < 0.1) |
| AS4 | Sanger/NGS | IR3 | No NMD observed | ||
| PRR7 | AS1 | Sanger | ES Ex4 | NMD (P < 0.01) | |
| AS3 | Sanger | IR3 | No NMD observed | ||
| PRR5 | AS1 | Predictedb | Alt 5′ss in exon 1 or alt 3′ss in exon 2 each could remove nine nucleotides | No NMD observed | |
| AS3 | Sanger | Alt 3′ss in IVS3 adds 47 nucleotides | NMD (P < 0.01) | ||
| PRR3 | UAS1 | NGS | Alt 3′ss removes seven nucleotides from exon 2; uORF increased from nine to 29 amino acids | No NMD observed | |
| UAS2 | Unknown | Unknown | Not determined | ||
| AS3 | Sanger/NGS | IR8 | No NMD observed | ||
| TOC1 | AS7 | NGS | IR4 | NMD (P < 0.05) |
Features of the 15 AS events that change significantly after transfer of plants to 4°C. ASn, AS variant where n is the number to identify the product; UASn, unspliced transcript; ES, exon skip; ss, splice sites. The P values are for differences between Col-0, upf1-5, and upf3-1 by analysis of variance; n = 3.
NMD only observed in some conditions.
Predicted from size of RT-PCR product, sizes of introns, skipped exons, and positions of putative alternative 5′ or 3′splice sites.
Temperature-Dependent AS of LHY and CCA1
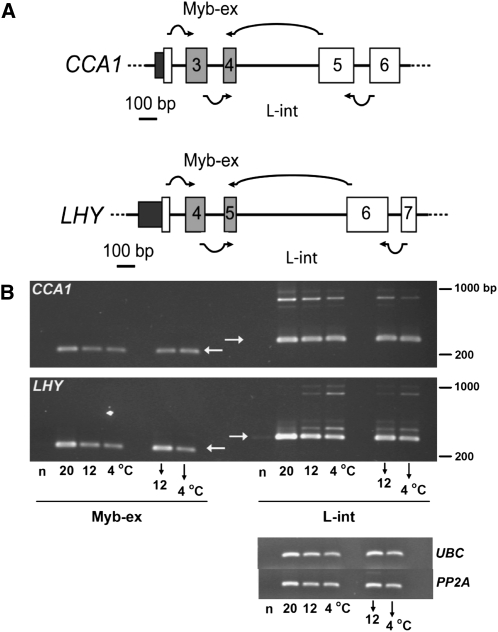
CCA1 and LHY are generally regarded as partially redundant components of the clock (McClung, 2006). Comparison of Figures 1A and 1B illustrates the similarities between the structures of the LHY and CCA1 genes. LHY contains an additional 26-bp exon (exon 2) in the 5′-untranslated region (UTR) compared with CCA1. In both genes, the Myb domain is encoded by two small exons (Myb-ex, exons 4 and 5 of LHY and 3 and 4 of CCA1), which are followed by a long intron (intron 4 in CCA1 and intron 5 in LHY), hereafter referred to as the long intron (L-int in figures and legends). The effects of transient or longer term temperature changes on CCA1 and LHY splicing patterns were tested by harvesting leaves at dawn from plants adapted to 20, 12, or 4°C, or grown at 20°C and transferred to 12 or 4°C for 12 h (Figure 2). Only a single product representing the fully or canonically spliced transcript (FS) was observed using RT-PCR primers spanning the Myb-ex. By contrast, primers spanning the long intron of each gene gave several products larger than the predicted size, which must retain either the full intron or parts of it (Figure 2) and were consistent with the products identified by HR RT-PCR (see Supplemental Data Set 1 online). Moreover, the AS products in the long intron of LHY were appreciably more abundant in plants adapting or already adapted to 12 or 4°C than in those adapted to 20°C, whereas for CCA1 the reverse was the case. This latter observation is consistent with Filichkin et al. (2010) in which the level of an intron-containing CCA1 transcript was reduced in the cold.
Figure 2.
Temperature Modulates AS of CCA1 and LHY.
(A) Primers targeting the Myb-ex and L-int regions of CCA1 spanned the splice junctions of exons 2/3 and 4/5 (CCA1 Myb-ex) and exons 3/4 and 5/6 (CCA1 L-int), respectively, as shown by the arrows; the corresponding primers for LHY spanned exons 3/4 and 5/6 (LHY Myb-ex) and exons 4/5 and 6/7 (LHY L-int).
(B) Temperature-dependent AS events detected by RT-PCR. Fully or canonically spliced transcript (FS) products are denoted by horizontal arrows in blank lanes. Samples were harvested at dawn in the denoted temperature conditions. Downward arrows denote plants 12 h after temperature had been reduced from 20 to 12°C or 4°C. n, negative control (no primer control). Amplification of UBC (UBC21; At5g25760) and PP2A (PP2AA3; At1g13320) served as transcript level reference genes. The figure is representative of three independent experiments.
We extended this analysis by examining samples across one light:dark cycle in plants adapted to temperatures either above or below 20°C, from both Columbia-0 (Col-0) plants and prr7-3 prr9-1 double mutant plants, which are unable to entrain to temperature cycles (Salomé and McClung, 2005) (see Supplemental Figure 5 online). The data confirm that temperature affects AS of CCA1 and LHY transcripts in opposite directions and show that AS events occur in both shoots and roots. The data for prr7-3 prr9-1, which expresses CCA1 and LHY at higher levels than does Col-0, show that the presence of AS products is clearly related to temperature rather than simply to the abundance of the transcript, implying that the capacity of the splicing machinery to produce the canonical transcript is not limiting.
We confirmed that alternatively spliced products in the L-int region of LHY were not simply artifacts of an amplification-based method using a ribonuclease protection assay to detect transcripts containing the 3′ end of the LHY long intron (see Supplemental Figure 6A online). The probe contained 219 bases from the long intron, 62 bases from exon 6, and 46 bases of linker sequence derived from the cloning construct. Protected fragments corresponding to the 62 bases of exon 6 and the fully retained intron (281 bases) were observed (see Supplemental Figure 6B online). In addition, two fragments between 62 and 100 bases were detected; these are consistent with protection of a novel exon E5a (82 bases, AS5; Figure 1A) and either AS4 or AS7 (see Supplemental Data Set 1 online). As with the RT-PCR experiments, the abundances of the AS products of LHY increased at lower temperatures (see Supplemental Figures 6B and 6C online).
We also used RNA-seq data to confirm the extent of temperature-dependent AS of LHY (see Supplemental Figure 7 online). Analysis of read data ab initio (see Methods) provided independent confirmation of the existence of the 82-bp E5a. In plants acclimated at 20°C, the vast majority of reads in the vicinity of the long intron (194 of 204) came from the canonical spliced product and spanned exon 5 to exon 6 with only a small number of reads from intronic sequence or E5a (see Supplemental Figures 7A and 7C online). However, in plants harvested after transfer to 4°C, a significant proportion of reads came from within the long intron. These included reads within E5a, some of which spanned exon 5 to E5a and some E5a to exon 6 and thus defined the boundaries of E5a (see Supplemental Figures 7B and 7C online). In addition, an increased number of reads were obtained within the long intron upstream of E5a, consistent with increased retention of this region as seen in AS7 (see Supplemental Figure 7D online). Collectively, the data indicate extensive AS of CCA1 and LHY transcripts, the extent of which changes both immediately on transfer to and following acclimation to lower temperatures. Thus, temperature has a major effect on the AS of these transcripts.
AS Plays a Significant Role in Regulating the Expression of LHY in Response to Temperature Changes
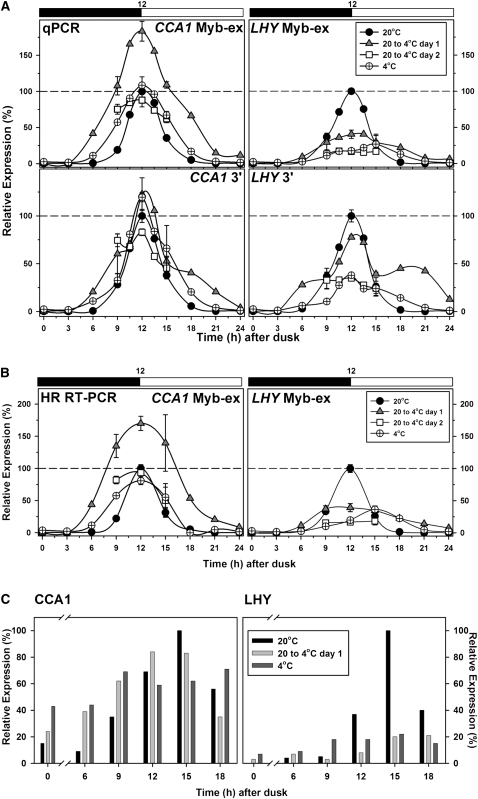
We next asked how temperature affects the expression of CCA1 and LHY and whether the changes in splicing could influence accumulation of functional protein. Figure 3A shows that the genes respond to temperature in different ways. When temperature was reduced from 20 to 4°C at dusk, CCA1 transcript abundance measured by quantitative real-time RT-PCR (qPCR) with two different sets of primers rose earlier, reached a higher value, and formed a broader peak. After adaptation at 4°C, the peak height was similar to that obtained at 20°C but the peak remained broader. By contrast, temperature reduction caused a marked reduction in the peak height of LHY transcripts with a substantial broadening of the peak (Figure 3A).
Figure 3.
Temperature Alters Expression of CCA1 and LHY.
(A) and (B) Expression was measured across a 12:12 h diurnal cycle (black and white bar; dark and light, respectively) for the denoted temperature conditions and normalized relative to the dawn peak level (100%) for plants acclimated to 20°C. Data at time points 9, 12, and 15 h after dusk show the mean and sd (n = 3).
(A) Expression of CCA1 and LHY measured by qPCR using primers for the Myb-ex region and a 3′ region as shown.
(B) Expression measured by HR RT-PCR using primer sets #398 and #397 for CCA1 and LHY, respectively (see Supplemental Figure 1 online).
(C) Immunoblot analysis of CCA1 (left) and LHY (right) protein levels across a diurnal cycle at the denoted temperatures. Protein abundance is expressed relative to the highest value at 20°C.
The abundance of differently spliced transcripts was also assessed by HR RT-PCR. First, the abundance of several different transcripts was measured after 20 to 26 cycles of amplification. The data (see Supplemental Figure 8 online) show that, over this range, RT-PCR products increase exponentially with cycle number and the relative levels of different products from the same gene do not vary, thus validating quantitation by this method. The response of transcript abundance to temperature changes as assessed by HR RT-PCR (Figure 3B) was in good agreement with the data from qPCR (Figure 3A). Measurement of protein abundance by immunoblot analysis provided a different picture. Profiles across one light:dark cycle showed that a reduction in temperature brought about a sharp decline in the peak height for LHY protein, while CCA1 protein levels were much less affected (Figure 3C). As a result, the relative abundance of CCA1 protein at its peak (3 h after dawn) 15 h after a reduction in temperature from 20 to 4°C was significantly higher, 77% ± 21% of the 20°C control (mean ± sd of triplicate values), than that of LHY protein, 25% ± 2% of the 20°C value.
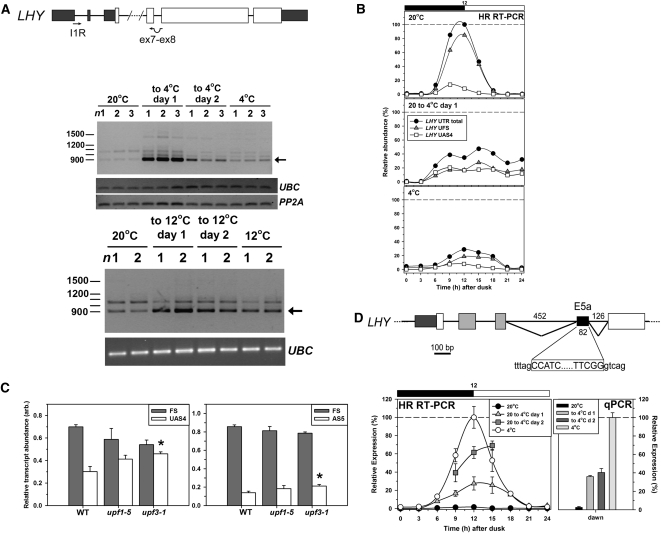
Thus, the effects of temperature on protein abundance and transcript abundance are not identical. Given that temperature differentially affects AS of these genes, we asked whether the proportion of transcripts that are nonfunctional (unable to encode full-length protein) could depend on temperature. Figure 4A illustrates a dramatic but transient retention of LHY intron 1 (LHY-UAS4; Figure 1A, Table 1) during the transition from 20°C to either 12 or 4°C. Quantitative analysis by HR RT-PCR, in which total LHY transcript was assessed as the sum of all the isoforms detected with primer set 355 (see Supplemental Figure 1 online), confirmed the decline seen using qPCR (Figure 3A). It showed that UAS4 increased both in absolute terms and as a proportion of total LHY transcripts, changing from some 10% of total transcripts at 20°C to ~50% during the first day after the reduction in temperature and ~30% after acclimation to 4°C (Figure 4B). The additional 5′UTR sequence due to retention of intron 1 in UAS4 increases the number of upstream open reading frames (uORFs) in the transcript. Although it is not possible to predict the effects of the uORFs in UAS4 on translatability of the normal LHY ORF, it is clear that UAS4 transcripts can undergo NMD, as in some conditions their relative abundance increases significantly in the NMD-impaired mutants upf1-5 and upf3-1 (Figure 4C) (see Methods for the assessment of NMD).
Figure 4.
Retention of LHY Intron 1 and Splicing of an Alternative Exon in the LHY Long Intron on Cooling.
(A) RT-PCR amplification of LHY transcripts retaining intron 1 using the indicated primers. Top gel: cooling from 20 to 4°C. Boxes are shaded as in Figure 1. The arrow shows the size of the fragment expected for retention of intron 1 but otherwise undergoing canonical splicing. n1,2,3, triplicate samples harvested at dawn under the conditions indicated. UBC and PP2A served as reference genes. Bottom gel: cooling from 20 to 12°C; n1,2, duplicate samples harvested at dawn under the conditions indicated with UBC as reference gene.
(B) HR RT-PCR was used to measure the abundance of canonically spliced transcripts (UFS) and those retaining intron 1 (UAS4) in the 5′UTR of LHY across a diurnal cycle at the denoted temperature conditions. Both transcripts were amplified from a single primer set (primer set #355; see Supplemental Figure 1A online). UFS and UAS4 transcripts are expressed relative to total transcript (UAS1, UAS2, UAS3, UAS4, and UFS) at dawn at 20°C. Data at time points 9, 12, and 15 h after dusk represent the mean and sd (n = 3). Black and white bars represent dark and light, respectively.
(C) Relative abundances of FS and AS LHY transcripts in Col-0 and upf mutant seedlings assessed using HR RT-PCR. Left, FS and UAS4, harvested at dawn from plants in a 16:8 photoperiod. Right, FS and AS5, harvested 3 h after dawn from plants in a 12:12 photoperiod. Data are means ± sd; n = 3. Asterisks indicate a significant increase relative to the wild type; P < 0.01 by Student’s t test. arb., arbitrary units.
(D) Top: AS within the L-int of LHY generating E5a (splicing event AS5; Figure 1A, Table 1). The alternative 3′ and 5′ splice sites are shown; exon sequence, capitals; intron sequence, lowercase. Bottom: Abundance of transcripts containing E5a across a diurnal cycle (black and white bar; dark and light, respectively) for the denoted temperature condition as determined (left) by HR RT-PCR using primer set #292 (see Supplemental Figure 1A online) and (right) by qPCR using primers spanning the novel E5a splice junctions. Data at time points 9, 12, and 15 h after dusk are means ± sd (n = 3). Increases on cooling measured at dawn by qPCR are all significant; P < 0.001.
The proportion of LHY transcripts specifically containing E5a increased dramatically on cooling to 4°C (Figure 4D), and the proportion of AS transcripts retaining some or all of intron 5 (e.g., AS5 and AS7) reached 20% of the total LHY transcripts at dawn in plants acclimated to 4°C as judged by HR RT-PCR. These transcripts are nonfunctional because of the presence of PTCs, and AS5 undergoes NMD (Figure 4C). In contrast with LHY, the proportion of CCA1 transcripts that are incorrectly spliced and, therefore, nonfunctional falls at low temperature (Figure 2; see also Filichkin et al., 2010), consistent with the smaller relative decrease in CCA1 than LHY.
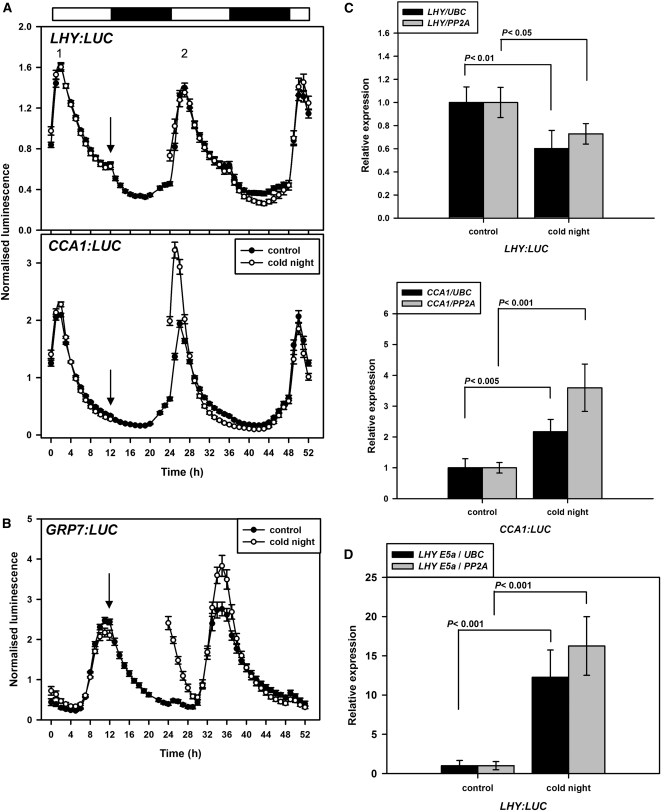
LHY Promoter Strength Is Not Affected by a Cold Night
To assess whether the reduction in LHY transcripts on cooling is caused by transcriptional or posttranscriptional events, we monitored the effect of temperature on promoter strength. Duplicate sets of plants expressing LHY:LUC+, CCA1:LUC+, or GRP7:LUC+ were imaged over one night/day cycle at 20°C. One set of plants was cooled to 4°C for the next 12-h dark period and then returned to the imaging chamber at dawn to allow comparison of LUC activity at 20°C with the second set, which had remained at 20°C throughout. This allowed us to assess the effect of a cold night on promoter strength by calculating for individual plants the ratio of the height of the second peak (after a cold night or a control night) to that of the first, labeled 2 and 1, respectively in Figure 5A. As a positive control, we used GRP7, well known to be induced by cold (Carpenter et al., 1994); the cold night significantly increased luminescence from GRP7:LUC+ (Figure 5B). The plants used for imaging were in the Wassilewskija background. We therefore used qPCR and RT-PCR to confirm that a cold night affected both LHY and CCA1 transcript abundance (Figure 5C) and LHY AS (Figure 5D) in these plants as in Col-0. The imaging data show that cooling does not affect LHY promoter strength but significantly increases CCA1 promoter strength (Figure 5A). Overall, the data suggest that the transient formation of LHY UAS4 transcripts and the progressive formation of LHY AS5 transcripts on cooling, and the resulting NMD, contribute significantly to the large decline in functional LHY transcripts and LHY protein in the cold.
Figure 5.
LHY Promoter Strength Is Not Affected by a Cold Night.
(A) Imaging started at dusk but the first 12 h are not shown; time 0 represents dawn. Control plants (closed symbols) were kept at 20°C. Plants exposed to a cold night (open symbols) were removed from the imaging chamber at the point indicated by the arrow, placed in the dark at 4°C for 12 h, and then returned to the imaging chamber at 20°C. Luminescence data were not collected during the cold night. Plants expressed either LHY:LUC+ or CCA1:LUC+. Data are means ± se; n = 15 for LHY:LUC+ control plants, and n = 20 for all other conditions.
(B) Imaging as in (A); plants expressed GRP7:LUC+. Data are means ± se; n = 5 at 20°C, and n = 7 at 4°C.
(C) Plants were harvested at dawn after a night at 20 or 4°C. LHY and CCA1 transcripts were assessed relative to UBC and PP2A by qPCR for ProLHY:LUC+ plants and ProCCA1:LUC+ plants, respectively. Data are means ± se, n = 4, and significance was assessed by Student’s t test.
(D) As for (C); LHY E5a transcripts were assessed relative to UBC and PP2A by qPCR for ProLHY:LUC+ plants. Data are means ± se, n = 4, and significance was assessed by Student’s t test.
AS Modulates the Response of PRR Expression to Temperature Changes
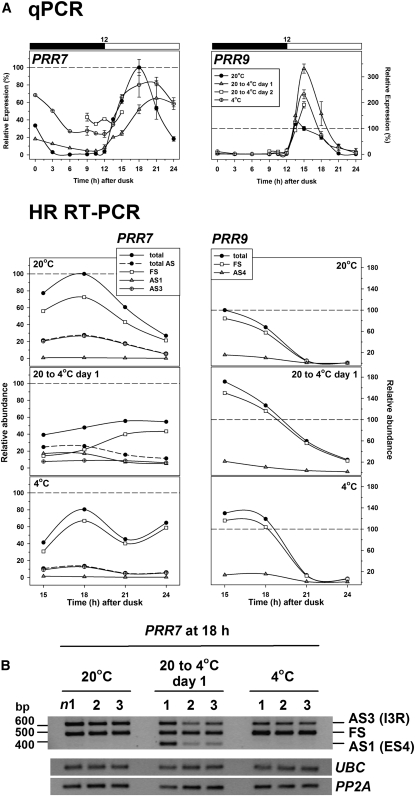
Since both PRR7 and PRR9 have been implicated in responses to temperature (Salomé and McClung, 2005; Salomé et al., 2010), and splicing of PRR9 transcripts is markedly affected by mutation of PRMT5 (Sanchez et al., 2010), we evaluated the effect of temperature on splicing of all of the PRR clock genes. PRR9 and PRR7 are at least partly redundant and were initially modeled as a single component (Locke et al., 2006). However, Figure 6A shows that they respond in opposite ways to the reduction in temperature from 20 to 4°C. PRR7 transcripts respond to the temperature change with a reduction in peak height, but recover on acclimation with a flattened peak. Remarkably, the abundance of an AS PRR7 transcript in which exon 4 is skipped (PRR7-AS1; Figure 1D, Table 1; see Supplemental Figure 2A online) is substantially but transiently increased (Figures 6A and 6B), reaching more than 30% of the total transcript at 15/18 h after transfer. The events AS3 (retention of intron 3) and AS1 are mutually exclusive (see Discussion) and together comprise >50% of total transcript at 18 h after transfer. Their products are predicted to be nonfunctional because they introduce PTCs and AS1 transcripts where exon 4 is skipped are targets of NMD (see Supplemental Figure 2C online). Hence, these events, which add to the reduced total expression of PRR7, would be expected to have a major effect in reducing the abundance of functional PRR7 in the first day after the transition to low temperature. By contrast, PRR9 transcripts reach a substantially higher peak value in the first cycle after a reduction in temperature (Figure 6A). The increased peak height of PRR9 is partly retained after acclimation to 4°C. PRR9 transcripts in which intron 3 is retained (PRR9-AS4) or an additional 8 nucleotides of intron 2 is included due to the use of an alternative 5′ splice site in the intron (PRR9-AS2) (Figure 1C, Table 1; see Supplemental Figure 2B online) would both give rise to nonfunctional products as the ORF is disrupted. Although both of these events are relatively abundant (when combined they can make up nearly 30% of transcripts), they are little affected by the temperature drop (Figure 6A) and do not seem to modulate the expression of PRR9 in response to low temperature.
Figure 6.
Temperature Alters Expression and AS of PRR7 and PRR9.
(A) Top: transcript abundance measured by qPCR across a 12:12-h diurnal cycle (black and white bar; dark and light, respectively) for the denoted temperature condition. Data for 9, 12, 15, 18, 21, and 24 h after dusk represent mean ± sd values (n = 3). Expression levels were normalized relative to the peak values in plants acclimated to 20°C (at 15 and 18 h after dusk for PRR9 and PRR7, respectively). Bottom: relative levels of total and AS transcripts determined by HR RT-PCR over the period 15 to 24 h after dusk in the conditions indicated. Transcripts were amplified using primer set #327 (PRR7) and #170 (PRR9) to identify significant AS events (see Supplemental Figure 2 online). Totals were the sum of the FS and AS transcripts. Expression levels were normalized relative to the peak values of total transcript in plants acclimated to 20°C as above.
(B) RT-PCR amplification of PRR7 using primers (see Supplemental Data Set 2 online) to detect AS1 and AS3 during cooling from 20 to 4°C. n1, 2, and 3 refer to triplicate samples taken 18 h after dusk under the conditions indicated, harvested and processed independently. UBC and PP2A served as reference genes.
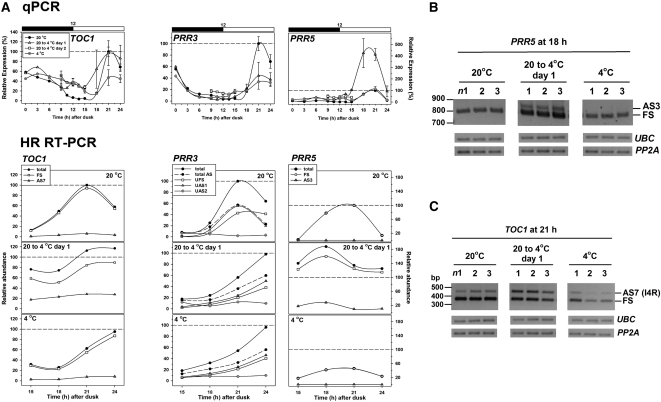
PRR5 and PRR3 also show opposite effects on cooling (Figure 7A). The first peak of PRR5 expression following transfer to 4°C is substantially higher and somewhat broader than the peak at 20°C, but this is reversed on acclimation to 4°C. By contrast, the peak PRR3 level is reduced both during the temperature transition and after acclimation to 4°C. Strikingly, two AS events in PRR5 seem to be transition specific. Event AS3 in PRR5 (Figure 1E, Table 1; see Supplemental Figure 3 online) is virtually absent at 20°C but increases to around 10% of transcripts during the transition (Figures 7A and 7B) and AS1 responds similarly (see Supplemental Figure 3 online). AS1 is an in frame removal of 9 nucleotides (Figure 1E, Table 1; see Supplemental Figure 3B online) whose functional effects cannot be predicted, while the retention of part of intron 3 in AS3 would disrupt the ORF and introduce PTCs; event AS3 induces NMD (Table 1). Three AS events (UAS1, UAS2 [unknown], and AS3) are detectable for PRR3 (Figure 7A). PRR3-UAS1 (Figure 1F, Table 1; see Supplemental Figure 3A online) comprised close to 50% of total transcripts in all conditions studied. This AS event only removes 7 nucleotides from the 5′UTR but leads to an increase in the length of a uORF from 9 to 29 amino acids. However, unlike the intron 1 retention in LHY, this does not trigger NMD (see Supplemental Figure 3D online). PRR3-AS3 (Figure 1F, Table 1; see Supplemental Figure 3A online) transcripts have intron 8 retained, which disrupts the ORF. This event increased from some 6 to 16% of total transcript following acclimation to 4°C, so would have a modest effect on PRR3 protein level.
Figure 7.
Temperature Alters Expression and AS of TOC1, PRR3, and PRR5.
(A) Top: Transcript abundance measured by qPCR across a 12:12-h diurnal cycle (black and white bar; dark and light, respectively) for the denoted temperature condition. Data for 9, 12, 15, 18, 21, and 24 h after dusk represent mean ± sd values (n = 3). Expression levels were normalized relative to the peak values in plants acclimated to 20°C (21 h for each gene). Bottom: Relative levels of total and AS transcripts determined by HR RT-PCR over the period 15 to 24 h after dusk in the conditions indicated. Transcripts were amplified using primer set #174 (TOC1), #374 (PRR3), and #457 (PRR5) to identify significant AS events (see Supplemental Figure 3 online). Totals were the sum of the FS and AS transcripts. Expression levels were normalized relative to the peak values of total transcript in plants acclimated to 20°C as above.
(B) and (C) RT-PCR amplification of PRR5 using primers to detect AS3 and of TOC1 using primers to detect AS7 (see Supplemental Data Set 2 online) during cooling from 20 to 4°C. n1, 2, and 3 refer to triplicate samples harvested at 18 h after dusk (PRR5) and 21 h after dusk (TOC1) under the conditions indicated. UBC and PP2A served as reference genes.
The first peak of TOC1 expression, 21 h after transfer to 4°C, is substantially broader than the corresponding peak at 20°C. This peak height is markedly reduced after acclimation to 4°C with the result that TOC1 expression is much flatter at 4°C than at 20°C (Figure 7A). One AS event (TOC1-AS7, retention of intron 4; Figure 1G, Table 1; see Supplemental Figure 3C online) increases transiently to form nearly 25% of total transcripts during the first day after transfer to 4°C (Figures 7A and 7C). This would reduce the level of full-length TOC1, as retention of intron 4 disrupts the ORF. This event has also been observed transiently during cooling from 20 to 12°C (see Supplemental Figure 3F online).
Several of the AS events that are observed transiently on cooling induce NMD (e.g., UAS4 in LHY, AS1 in PRR7, AS3 in PRR5, and AS7 in TOC1). This led us to ask whether there might be a transient decline in the activity of the NMD pathway on cooling. We therefore tested the effects of cooling on several AS events in other genes that are known to induce NMD (Kalyna et al., 2011). In four out of five cases, the relative abundances of the AS species were not affected by cooling (see Supplemental Figure 9 online). Hence, the transient appearance of some AS events on cooling is not caused by a nonspecific decline in NMD activity in these conditions.
The Responses of Clock and Output Genes to Temperature Reduction
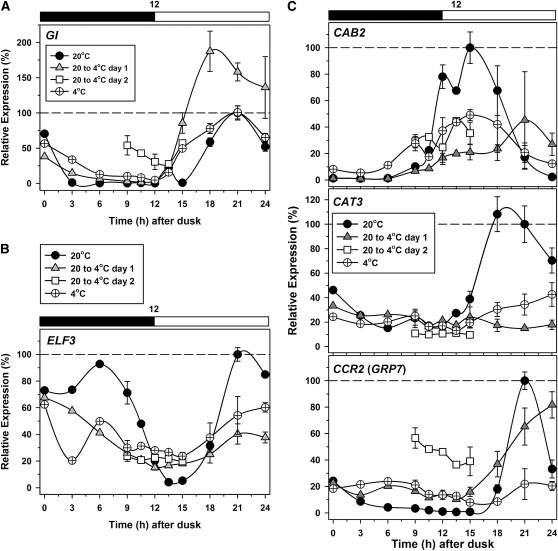
The expression profile of GI resembled that for CCA1 and PRR5, with a large increase in peak height on the first day after transfer to cold that was not retained in acclimated plants (Figure 8A). GI is a multiexon gene, and, as noted above, three AS events were identified for GI here (see Supplemental Data Set 1 online). However, all were found at low abundance relative to fully spliced transcripts and would not therefore affect protein levels. ZTL has two gene models in TAIR with one and two introns, respectively. We were unable to detect AS by HR RT-PCR. The response of ELF3 to the temperature shift (Figure 8B) resembled those of PRR7 and PRR3 (Figures 6A and 7A); we found no evidence for AS events. For both LUX and ELF4, which are intronless, acclimation to 4°C resulted in a reduction in transcript peak height (see Supplemental Figure 10 online). Finally, CHE is annotated as a single exon gene with no introns and we did not detect AS. Therefore, expression of GI, ZTL, ELF3, and CHE is unaffected by AS, unlike other core clock genes. We also assessed some well-established output genes (Figure 8C). The temperature reduction rapidly suppressed oscillation in CAT3 transcripts and suppressed that in GRP7 after a delay, so that its expression profile resembled that of TOC1. By contrast, oscillation in CAB2 transcripts was suppressed initially but partially recovered on adaptation to 4°C.
Figure 8.
Temperature Alters Expression and AS of Other Clock Components and Clock Outputs.
Transcript abundance was measured by qPCR across a 12:12-h diurnal cycle (black and white bar; dark and light, respectively) at the denoted temperature condition. GI (A), ELF3 (B), and CAB2, CAT3, and GRP7 (CCR2) (C). Data for 9, 12, 15, 18, 21, and 24 h after dusk represent mean and sd values (n = 3). Expression levels were normalized relative to values in plants acclimated to 20°C, at 15 h after dusk for CAB2, and 21 h after dusk for CAT3, GI, ELF3, and CCR2/GRP7.
The Effects of Temperature on AS Events and Clock Gene Expression Are Reversible
Clearly a reduction in temperature from 20 to 4°C elicits several posttranscriptional changes to clock gene expression both transiently and on acclimation. We next asked whether an increase in temperature from 4 to 20°C reversed these adaptations. Plants were grown at 20°C in 12 h light:dark diurnal cycles, acclimated to 4°C for 5 d, and then shifted to 20°C. Unlike the temperature reduction regime, which was initiated at dusk, we started the temperature increase at dawn to avoid any conflict between clock-entraining light:dark and warm:cool cycles. Using this experimental design, dawn-phased clock genes, such as LHY and CCA1, were assayed a full diurnal cycle (24 h) after the temperature increase, whereas for the temperature reduction, these genes were measured 12 h after transfer. Conversely, late afternoon and evening phased clock genes were assayed 6 to 12 h after increase or 18 to 24 h after decrease. To compare changes in gene expression between the temperature increase and decrease treatments, transcript levels were expressed relative to the levels determined for the starting acclimated conditions.
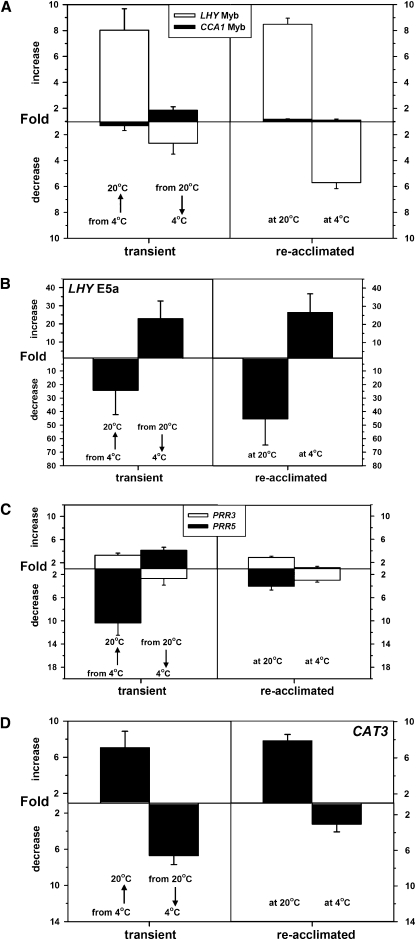
In the temperature increase regime, expression of LHY increased 8.0-fold transiently and 8.5-fold on acclimation to 20°C (Figure 9A), in comparison to decreases of 2.5- and 5.5-fold in the temperature decrease regime (Figures 3A and 9A). The exon inclusion event in the LHY long intron (E5a; Figure 1A) that was upregulated following the temperature decrease also showed the opposite behavior following the temperature increase (Figure 9B). Like the temperature decrease, the increase had little effect on CCA1 expression after acclimation (Figure 9A). The expression levels of PRR5 and PRR3 respond transiently to temperature decrease in opposite directions (up- and downregulated, respectively; Figure 7A) and similarly respond transiently to the temperature increase in opposite directions (Figure 9C). Thus, transient and longer term changes in clock gene expression and AS pattern can be detected irrespective of the direction of temperature change. This suggests that they are part of a temperature response mechanism rather than a cold stress response. The reduced expression of CAT3 in the cold is also reversed following a temperature increase (Figure 9D), confirming that the circadian temperature response can feed through to outputs.
Figure 9.
Temperature Increases and Decreases Have Opposite Effects on Both Gene Expression and Splicing.
Transcript abundances were measured by qPCR during the first day (left panels) or on the 5th day (right panels) after a temperature shift. For each gene, expression was measured at the time of maximum expression in the starting conditions, and data are expressed as a fold increase or fold decrease relative to the starting values at 4 or 20°C. Data represent mean and sd values (n = 3).
(A) Total LHY and CCA1 expression as measured by qPCR of the Myb-ex region.
(B) E5a of LHY by qPCR as in Figure 4D.
(C) PRR3 and PRR5 expression as in Figure 7A.
(D) CAT3 expression as in Figure 8C.
DISCUSSION
This work provides several important conclusions about AS in the operation and functions of the plant circadian clock. First, we established the extent of AS at a system-wide level. TAIR currently contains 18 annotated splice variants for the core clock genes (TOC1, LHY, CCA1, PRR3/5/7/9, ZTL, GI, and CHE), including the 10 fully or normally spliced transcripts. LHY has five splice variants; CCA1, PRR9, PRR3, and ZTL have two each; and PRR7, PRR5, TOC1, GI, and CHE have a single annotated transcript each. Additional splice variants have also been reported for CCA1 (retention of intron 4) and PRR9 (retention of intron 3) (Filichkin et al., 2010; Sanchez et al., 2010). We extend this by identifying 63 AS events, 15 of which are significantly modulated by temperature. AS is clearly much more widespread, complex, and quantitatively important than has previously been appreciated. Second, AS of clock genes is clearly sensitive to temperature in the normal ambient range, and it plays a significant role in the response to temperature transitions. This is shown in particular by the marked transient accumulation of nonproductive LHY, PRR7, and PRR5 transcripts following a reduction in temperature. Moreover, some AS events trigger NMD and thus contribute directly to transcript stability and changes in transcript abundance in response to temperature shifts. Indeed, for LHY, we provide evidence that temperature affects transcript abundance posttranscriptionally rather than by promoter strength. Third, the data reveal strong differences in the behavior of clock genes that are regarded as partially redundant.
We systematically assessed AS in the core clock genes of Arabidopsis. We confirmed important AS events that have already been noted, for example, the retention of intron 1 of LHY (At1g01060.2) and the retention of intron 4 in CCA1 (Filichkin et al., 2010), and identified many novel, unannotated AS events in LHY, CCA1, PRR3/5/7/9, and TOC1, such as the additional exon 5a in LHY, the exon 4 skip in PRR7, the retention of intron 4 in TOC1, and three and two AS events in PRR3 and PRR5, respectively. We then used our HR RT-PCR system to analyze dynamic changes in levels of fully spliced and alternatively spliced transcripts during transition and acclimation to low temperatures. This high-resolution system has allowed the contribution of the multiple different AS events to the total level of mRNAs to be followed over many different time points and conditions. Transcript level changes measured by this system are in good agreement with those of fully spliced and AS transcripts analyzed by qPCR. Several of the AS events are likely to be of physiological importance on the grounds that (1) the AS transcripts comprise at least 10 to 50% of the total transcripts under some conditions and/or (2) the extent of AS is gene specific and dependent on conditions. In addition, the majority of the AS events generate nonproductive mRNA transcripts through the introduction of PTCs. It is important to note that while these transcripts are distinguishable from productive normally spliced mRNA transcripts by HR RT-PCR, they would not be distinguished in microarrays.
The pattern of accumulation of AS transcripts varies. As noted above, some events become abundant only during temperature transitions but are barely detectable in the steady states. Some, such as the additional exon 5a in LHY, change in abundance progressively during the acclimation from one temperature to another, while others, such as those in PRR9, do not change in abundance in response to temperature. The quantitatively significant AS events involve IR, exon skipping, and the use of alternative 3′ and 5′ splice sites. The consequences of these events also vary. For example, the majority disrupt the ORF and introduce PTCs, leading to nonproductive mRNAs. PTC-containing transcripts can trigger NMD (LHY-UAS4, LHY-AS5, PRR9-AS2, and PRR7-AS1) or can be immune to NMD (e.g., the majority of the IR events). The effects of the upf mutants on the abundances of these PTC-containing transcripts are variable. This is consistent with the data of Kalyna et al. (2011), which showed substantial variation in the degree of increase of such transcripts in the upf mutants. In the absence of antibodies for most of the clock proteins, we are currently only able to extrapolate effects on protein levels from the relative abundance of the AS isoforms. It is also possible that truncated proteins could be produced from the PTC-containing transcripts. Finally, more than half of the AS events were not abundant (<1% of total transcripts) under the conditions used here. Low-abundance transcripts with IR events may simply represent partially spliced transcripts, but it is possible that some of these transcripts could increase under different conditions or may represent more abundant transcripts in particular cell types (our analysis is based on RNA extracted from whole shoots). Hence, a systematic examination of AS under a wide range of environmental conditions, stress related and otherwise, would prove valuable. Moreover, since nonproductive transcripts can represent up to 50% of the total for some genes, it is clear that conclusions based on measurement of total transcript by microarrays or qPCR must be treated with caution.
It is also necessary to consider the transcript diversity of the alternatively spliced clock genes in terms of how the multiple different AS events are distributed among different transcripts. The AS events in each gene can be mutually exclusive, independent, or interact such that one splicing event affects whether another occurs. For example, PRR7 has two significant mutually exclusive events, retention of intron 3 and skipping of exon 4 (which also removes introns 3 and 4), both of which give nonproductive mRNAs. These events cannot occur in the same transcript, so the abundance of nonproductive PRR7 transcripts is the sum of the individual occurrence of these events, which reaches over 50% during the transition to 4°C. AS3 and AS5 in PRR5 are also mutually exclusive. Events can be independent, particularly when in different regions of the pre-mRNA. For example, two of the three significant events in LHY (retention of intron 1 in the 5′UTR and the additional exon E5a from intron 5) are in different regions of the pre-mRNA, and RT-PCR covering both events showed that they occur independently. Thus, AS transcript isoforms have also been identified containing combinations of the individual AS events.
Our experiments were designed on the basis that the circadian clock is a homeostatic mechanism that has likely evolved to maintain its period relatively constant even though environmental conditions, such as light intensity and temperature, can fluctuate rapidly and frequently. Hence, monitoring the transitions between states can be at least as informative as monitoring steady states. The strength of this approach is illustrated by our detection of several AS events that become quantitatively significant only in plants adapting to a reduction in temperature but not in plants acclimated to the higher or lower temperature. This behavior is not caused by a transient reduction in general NMD pathway activity on cooling. Overall, the data provide evidence for the functional importance of AS because in most cases the effect of temperature on AS complements its effect on total transcript abundance. Thus, a reduction in temperature decreases expression of LHY, PRR7, TOC1, and PRR3 and also increases the proportion of nonfunctional transcripts transiently and/or after acclimation. The functional importance of AS is particularly clear for LHY. On cooling, the UAS4 event transiently reaches nearly 50% of the total transcript and triggers NMD. Since cooling does not affect the promoter strength of LHY (Figure 5), it is highly likely that this AS event contributes significantly to the loss of total transcript and LHY protein in these conditions. The same argument applies to AS1 in PRR7. The relationship between these effects and other rapid responses to temperature changes, such as in Ca2+ levels (Knight et al., 1991), remains to be seen. Transient AS events have been observed during shifts from 20 to 4°C and 20 to 12°C. However, the extent and duration of temperature changes required to trigger changes in AS need to be determined.
We detected specific changes in the splicing of CCA1 and LHY transcripts across the range of temperatures between 28 and 4°C. Clock gene transcripts oscillate in light:dark cycles across this range, though with reduced amplitude at 4°C. Moreover, in all of the cases that we have examined, where a change in splicing pattern is observed on temperature reduction, the opposite effect is seen in a temperature increase. By contrast, splicing of the majority of introns is not affected by temperature. Thus, the changes in splicing pattern that we detected are not simply a splicing response to a sudden cold stress (e.g., inactivation or lower efficiency of splicing) but represent subtle and specific modulation of functional expression levels. The changes in AS of specific introns may reflect features of the introns themselves (e.g., secondary structure) or temperature-dependent changes in levels and activity of splicing or other factors. For example, recent work showed that the prmt5 mutation massively increased the proportion of two AS events in PRR9 transcripts corresponding to those listed as AS2 and AS4 in Table 1 (Sanchez et al., 2010). These events were readily detectable in our conditions, but their relative abundance was not affected by temperature and was dramatically less than in the prmt5 mutant (for AS4, compare Figure 2A of Sanchez et al., 2010 with Figure 6A). Examination of all AS events in the prmt5 mutant will be needed to assess whether PRMT5 modulates any of the other AS events found here or whether it is involved in the effects of temperature on AS. Overall, our observations suggest strongly that AS provides one mechanism through which plants detect and/or respond to temperature cycles and possibly also to fluctuations in temperature throughout the day.
Previous work on the effect of cold on the plant circadian clock has shown that oscillations in most clock components in Arabidopsis, chestnut, and poplar become severely dampened or eliminated on acclimation to low temperature (Ramos et al., 2005; Bieniawska et al., 2008; Ibañez et al., 2008, 2010). For example, the data of Bieniawska et al. (2008) show that oscillations in Arabidopsis GI and PRR7 are undetectable after 2 d in a light:dark cycle at 4°C, while those in TOC1, LHY, and CCA1 are severely dampened. Surprisingly, oscillations in LUX were maintained, albeit with a small phase shift. In our experiments, oscillations were still detectable, though expression of TOC1 was markedly flattened at 4°C. However, there is one significant difference between the two sets of results. In our work, the peak expression levels of LHY, TOC1, PRR3, and ELF3 were all appreciably reduced at 4°C, whereas Bieniawska et al. (2008) showed that expression of these genes dampened at a high, rather than low, level. This may be due to differences in experimental conditions, as both the growth conditions (hydroponic versus soil) and the photoperiod (12:12 h versus 16:8 h) were different. However, given our evidence for the accumulation of some nonfunctional transcripts of clock genes at low or high temperatures, it is clear that further studies of the effect of temperature, and indeed other environmental changes, on the clock and associated processes need to consider the involvement of AS events rather than just the measurement of total transcript levels.
Our data cast additional light on the operation of the plant circadian clock. The clock components CCA1 and LHY have been regarded as partially redundant (McClung, 2006); indeed, in some studies, they have been modeled as a single component (Locke et al., 2005, 2006). However, some functional differences between them, particularly in the ways that they contribute to temperature compensation, have been noted previously (Gould et al., 2006). Our data emphasize differences between them; changes in temperature transiently affect expression of the two genes in opposite directions and also have opposite effects on AS of their transcripts. Likewise, PRR9 and PRR7 have been regarded as partially redundant and have been modeled as a single component (Locke et al., 2006; see also Zeilinger et al., 2006; Pokhilko et al., 2010), but our data stress that they respond in opposite ways to temperature changes. These opposite responses need to be considered in developing further models of the clock. CCA1 and LHY can form both homo- and heterodimers (Lu et al., 2009; Yakir et al., 2009), and the different combinations are likely to have different properties. Given our data on the effects of temperature on gene expression and protein abundance, CCA1-containing dimers might primarily target PRR9 and PRR5, whereas LHY-containing dimers might target PRR7. This and related hypotheses should be tested, for example, by chromatin immunoprecipitation studies. Our data also have implications for studies of output pathways, since it is noteworthy that there are differences in the responses of output genes to temperature changes. Previous work has shown that CAT3 and CAB2 are regulated by distinct clocks that differ in their temperature sensitivities (Michael et al., 2003a). Our work extends this by showing that expression of CAB2 in the cold recovers after a transient decline, while that of CAT3 does not. Further studies of these and other outputs may allow delineation of distinct output pathways by relating the responses of outputs with those of specific clock components.
In conclusion, we established that AS plays a significant role in the responses of the circadian clock to changes in temperature and is thus an additional mechanism involved in the operation and regulation of the clock.
METHODS
Plant Material and Growth Conditions
All plant material was the Col-0 ecotype unless stated otherwise. The prr7-3 prr9-1 T-DNA insertion mutant (Michael et al., 2003b; Salomé and McClung, 2005) was a gift of Rob McClung (Dartmouth College, Hanover, NH). Seeds were surface sterilized, stratified in the dark at 4°C for 4 d, and grown hydroponically to maturity (5 weeks) in Microclima environment-controlled cabinets (Snijders Scientific), maintaining 20°C (55% relative humidity) and 12 h light (130 to 150 μE m−2 s1):12 h dark as described previously (James et al., 2008). In the main, plants were harvested at 3-h intervals over 24 h at 20°C, and on days 1 and 4 after transfer to 4°C with additional samples around dawn on day 2 (see Supplemental Figure 4 online). Each temperature experiment was performed in a single growth cabinet to eliminate the potential effects of minor changes in light intensities and light quality on gene expression. The switch to 4°C from 20°C was initiated at dusk and the switch to 20°C from 4°C was initiated at dawn. Typically in a temperature reduction, the cabinet took 40 min to reach 12°C, 2 h to reach 6°C, and 6 h to reach 4°C air temperature. The plants acclimated to 28, 12, and 4°C were exposed to these temperatures for 4 d. Tissue was rapidly frozen over liquid N2 and stored at −80°C until isolation of RNA and preparation of cDNA. For NMD experiments, Col-0, upf1-5 (Arciga-Reyes et al., 2006), and upf3-1 (Hori and Watanabe, 2005) were grown on agar plates at 22°C in 16 h light:8 h dark or in 12 h light:12 h dark for 10 d, transferred to 4°C for 12 h, and harvested as noted in the figure legends. In some cases, the NMD pathway mutations only altered the steady state level of particular transcripts in some experimental conditions, probably because the extent of any change must depend on the relative rates of normal and NMD turnover. A significant increase (P < 0.05) in the relative abundance of a transcript in upf3-1or upf1-5 compared with Col-0 was taken to indicate NMD.
RNA Extraction, cDNA Synthesis, RT-PCR, and qPCR
Total RNA (1 μg) was extracted and DNase treated prior to cDNA synthesis for qPCR analysis as described by James et al. (2008). cDNAs were amplified using PerfeCTa SyBr Green FastMix (Quanta BioSciences) using a Mx3000P real-time PCR system (Agilent Technologies). Cycle threshold (Ct) values for the genes of interest were normalized by subtracting the mean Ct values for three reference genes, UBC, PP2A, and At4g32680. The stability of these three genes was confirmed using geNORM (Vandesompele et al., 2002), but ACT2 (At3g18780) was rejected as a reference gene. The delta-delta Ct algorithm (Livak and Schmittgen, 2001) was used to determine relative changes in gene expression. RT-PCR was performed using cDNAs and HotStarTaq DNA polymerase (Qiagen). Products were coelectrophoresed on 1.5% agarose and 0.5× TBE gels with 100-bp markers (Roche Diagnostics) and stained with ethidium bromide. Primer sequences employed for RT-PCR and qPCR are detailed in Supplemental Data Set 2 online.
Circadian Clock HR RT-PCR Panel
Total RNA was extracted from 162 samples representing three biological replicates of 54 different time points throughout the day-night cycle, before and after transfer to 4°C. Up to 100 mg of tissue was extracted with the RNeasy plant mini kit (Qiagen) following the manufacturer’s instructions. RNA preparations were DNase treated (on column) to remove DNA using Qiagen RNase-Free DNase Set. The RT-PCR reactions were essentially as described by Simpson et al. (2008). Briefly, 4 μg of total RNA was used in first-strand cDNA synthesis by reverse transcription with oligo(dT)18 and random hexamer primers using the Sprint RT Complete-Double PrePrimed kit (Clontech) in a final volume of 20 μL. For each gene, a number of overlapping primers were designed to cover the whole sequence. These gene-specific primer pairs (with one 6-carboxyfluorescein labeled primer) were designed to amplify between ~400 and 1200 bp and to capture different splicing events. cDNA reactions were diluted by adding 50 μL water to each reaction, and 1 μL of each cDNA was aliquoted into 96-well plates containing the gene-specific primers. RT-PCR reactions were performed for 24 cycles as described by Simpson et al. (2008). The resultant RT-PCR products representing AS transcripts were detected on an ABI3730 automatic DNA sequencer along with GeneScan 1200 LIZ size standard (Applied Biosystems). RT-PCR products were accurately sized and mean peak areas calculated using GeneMapper software. To measure changes in AS of particular isoforms, the peak areas were normalized relative to the peak area values for two reference genes, UBC21 and PP2AA3.
To determine the sequences of unknown alternatively spliced isoforms, RT-PCR products were also cloned into Promega pGEM-T vector and transformed using high-efficiency Promega JM109 cells. Following transformation, colony PCRs were performed on ~30 selected white colonies to identify clones with different sized insets using T7 and SP6 promoter primers flanking the cloning site in the pGEM-T vector. To detect transcripts with small size changes due to AS, ~50 random colonies were also sequenced. Thus, ~50 to 75 clones from each primer pair were analyzed.
Primer sequences used for HR RT-PCR are detailed in Supplemental Table 2 online.
RNA-seq
Total RNA for dawn samples across the temperature regime was converted to a library of short DNA fragments for sequencing on the Illumina platform. Briefly, RNA integrity was assessed (RNA integrity number >8) using a 2100 Bioanalyzer (Agilent Technologies). mRNA was purified from total RNA (8 μg) using oligo(dT) beads, fragmented, and converted to double-stranded cDNA according to the Illumina protocols. Adapters were added to the ends of the DNA fragments to permit hybridization to Illumina single-read flow cells. The size (~200 bp), purity, and concentration of the resultant DNA libraries were validated by assessing aliquots on a 2100 Bioanalyzer. DNA libraries were sequenced (Illumina) to generate 50-bp reads. Ab initio splice site discovery for LHY was performed by aligning reads, using Soap2 aligner (Li et al. 2009), to the TAIR9 Arabidopsis thaliana genomic sequence with the LHY genomic sequence omitted. Nonaligned reads were realigned against the LHY genomic sequences using the BLAST algorithm (Altschul et al., 1997). Subsequent alignments were filtered to enable the characterization of exonic reads by identifying alignments of at least 48 bp and reads covering splice junctions by identifying broken alignments, where two consecutive parts of a given read (covering together at least 48 bp) aligned to two separated genomic regions. The coordinates of the 82-bp region within the long intron defined as E5a were annotated for the LHY reference sequence and the original Illumina read files realigned using the RNA-seq tool of the CLC Genomics Workbench, version 3.7. Raw sequence data are available from the authors on request.
Ribonuclease Protection Assay
A 787-bp region of the LHY long intron region was amplified from Col-0 genomic DNA using the primers forward, 5′-CAGAACATATTGGGACAAAGACTGC-3′, and reverse, 5′-GGAATTTCTATGTCCAAAGCTTGGC-3′, and the resultant product cloned into pPCR-Script Amp SK(+) (Stratagene). Vector with cloned insert in the appropriate orientation was linearized with EcoRI to provide the template for the in vitro synthesis (MAXIscript kit; Ambion), by phage T3 RNA polymerase, of a 327-base antisense RNA probe incorporating [α-32P]UTP (Perkin-Elmer). Full-length synthesized RNA probe was gel purified in 8 M urea 5% acrylamide gels and subsequently used in RNase protection assays (RPAIII; Ambion). Briefly, ~200 pg of radiolabeled probe was hybridized at 42°C with 20 μg of shoot total RNA or, for control assays, 20 μg of yeast RNA. Nontarget RNA and nonhybridized probe were digested with a mixture of RNaseA/T1. Remaining radiolabeled RNA:RNA hybrids were precipitated and coelectrophoresed on 8 M urea 5% acrylamide gels with an aliquot of [α-32P]UTP-labeled RNA Century Plus markers (Ambion). Gels were dried and exposed to a phosphor screen for 5 d and the screen scanned with a FujiFilm FLA-5000 phosphor imager. Data were analyzed with Image Gauge software, version 4.0. Similarly, RNase protection assays employing an [α-32P]-labeled 460-b probe protecting a 407-b fragment of UBC in combination with 10 μg of shoot total RNA (or 10 μg of yeast RNA) were performed as references. The UBC fragment was synthesized from shoot cDNA using the primers forward, 5′-TTAGAGATGCAGGCATCAAGAGCGC-3′, and reverse, 5′-GATCTTAGAAGATTCCCTGAGT CGC-3′, and the resulting amplicon cloned into pCR4-TOPO (Invitrogen). Vector with appropriately orientated insert was NotI digested prior to in vitro transcription, using phage T3 RNA polymerase.
Immunoblotting
Immunoblots for LHY were performed as previously described (James et al., 2008). The anti-LHY antibody was raised in rabbits against His-tagged LHY (Kim et al., 2003). The antibody against CCA1 (Wang and Tobin, 1998) was a gift from E.M. Tobin (University of California, Los Angeles, CA). Blots were quantified using ImageJ software.
Imaging
Plants expressing promoter:LUC+ constructs in the Wassilewskija background (CCA1:LUC+, LHY:LUC+, and GRP7:LUC+) were used. Seeds were from A. Millar (University of Edinburgh, UK). Plants were grown hydroponically (James et al., 2008) for 3 weeks in 12-h-white-light (75 μmol m−2 s−1):12-h-dark cycles and then transferred to an imaging room under the same conditions, except that light (15 μmol m−2 s−1) was provided by blue and red light-emitting diodes. Plants were sprayed with 5 mM d-Luciferin (Promega) in 0.01% Triton for ~2 h before the start of imaging at dusk. They were imaged for 24 h at 20°C before some were removed at dusk, kept in the dark in a cold room (4°C) overnight, and then returned to 20°C just before dawn. Bioluminescence was detected using a Photek 225/18 intensified charge-coupled device camera with a 16-mm lens. For each time point, the images were integrated for 10 min in photon counting mode without any filter and analyzed using IFS32 software (Photek). Fifteen to 20 individual plants were imaged in each condition. Data are expressed relative to the average signal for each individual plant across the first light cycle, before cold treatment. The graphs show the means and se of these normalized data.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: CAT3, At1g20620; CCA1, At2g46830; ELF4, At2g40080; GI, At1g22770; GRP7, At2g21660; LHY, At1g01060; LUX, At3g46640; PP2AA3, At1g13320; PRR3, At5g60100; PRR5, At5g24470; PRR7, At5g02810; PRR9, At2g46790; TOC1, At5g61380; UBC21, At5g25760; and ZTL, At5g57360.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alternative Splicing in LHY and CCA1.
Supplemental Figure 2. Alternative Splicing in PRR7 and PRR9.
Supplemental Figure 3. Alternative Splicing in PRR3, PRR5, and TOC1.
Supplemental Figure 4. Graphical Overview of the Sampling Strategy.
Supplemental Figure 5. Temperature-Dependent Alternative Splicing of CCA1 and LHY Occurs in Both Shoots and Roots of Wild-Type and Morning Loop Mutant Plants.
Supplemental Figure 6. Temperature-Dependent Alternative Splicing of LHY L-int as Revealed by Ribonuclease Protection Assay.
Supplemental Figure 7. Temperature-Dependent Inclusion of an Alternative Exon (Exon 5a) in the LHY Long Intron.
Supplemental Figure 8. Validation of the Use of HR RT-PCR for Quantitation of Transcript Abundance.
Supplemental Figure 9. Cooling Does Not Transiently Reduce the Effectiveness of the NMD System.
Supplemental Figure 10. Effects of Cooling on the Expression of LUX and ELF4.
Supplemental Data Set 1. Alternative Splicing in Core Clock Genes Detected by High-Resolution RT-PCR.
Supplemental Data Set 2. Primer Sequences.
Acknowledgments
We thank Julie Russell and Julie Galbraith (University of Glasgow) for excellent technical assistance, Scott Ramsay (University of Glasgow) for help with RNA-seq, José Monreal (Universidad de Sevilla) for help with sample processing, Karen Halliday (University of Edinburgh) for RNA samples, Isabelle Carré (University of Warwick) for supply of the LHY-expressing construct, and Elaine Tobin (University of California, Los Angeles, CA) for the anti-CCA1 antibody. We thank Yamile Marquez, Maria Kalyna, and Andrea Barta (Max F. Perutz Laboratories, Medical University of Vienna) for RNA-Seq data on clock genes. The work was supported by Biotechnology and Biological Science Research Council Grant BB/G008735/1 to H.G.N. and G.I.J., Leverhulme Trust Fellowship RF/2/RFG/2008/0309 to H.G.N., and Biotechnology and Biological Science Research Council Grant (BB/G024979/1) and the European Union FP6 Programme Network of Excellence on AS (EURASNET) (LSHG-CT-2005-518238) to J.W.S.B.
AUTHOR CONTRIBUTIONS
This article is the result of an equal collaboration between the Nimmo and Brown labs. A.B.J., J.W.S.B., N.H.S., G.I.J., and H.G.N. conceived the study and designed experiments. A.B.J., N.H.S., S.B., J.M., and G.A.N. performed experiments. A.B.J., N.H.S., S.B., J.M., G.A.N., P.H., J.W.S.B., and H.G.N. analyzed data. A.B.J., J.W.S.B., and H.G.N. wrote the article.
References
- Akman O.E., Locke J.C., Tang S., Carré I., Millar A.J., Rand D.A. (2008). Isoform switching facilitates period control in the Neurospora crassa circadian clock. Mol. Syst. Biol. 4: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Arciga-Reyes L., Wootton L., Kieffer M., Davies B. (2006). UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47: 480–489 [DOI] [PubMed] [Google Scholar]
- Barash Y., Calarco J.A., Gao W., Pan Q., Wang X., Shai O., Blencowe B.J., Frey B.J. (2010). Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Barbazuk W.B., Fu Y., McGinnis K.M. (2008). Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 18: 1381–1392 [DOI] [PubMed] [Google Scholar]
- Barta A., Kalyna M., Lorković Z.J. (2008). Plant SR proteins and their functions. Curr. Top. Microbiol. Immunol. 326: 83–102 [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J. (2005). Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 6: 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniawska Z., Espinoza C., Schlereth A., Sulpice R., Hincha D.K., Hannah M.A. (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 147: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.L. (2003). Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Bove J., Kim C.Y., Gibson C.A., Assmann S.M. (2008). Characterization of wound-responsive RNA-binding proteins and their splice variants in Arabidopsis. Plant Mol. Biol. 67: 71–88 [DOI] [PubMed] [Google Scholar]
- Carpenter C.D., Kreps J.A., Simon A.E. (1994). Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 104: 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Manley J.L. (2009). Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S., Cao X. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 107: 19114–19119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diernfellner A., Colot H.V., Dintsis O., Loros J.J., Dunlap J.C., Brunner M. (2007). Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms. FEBS Lett. 581: 5759–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar S.P., Baker B.J. (2000). Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 97: 1908–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.E., Knox K., Kozma-Bognar L., Southern M.M., Pokhilko A., Millar A.J. (2011). Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 21: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Jakobsen M.K., Baker A.J., Telzerow A., Hou S.-W., Laplaze L., Barrot L., Poethig R.S., Haseloff J., Webb A.A.R. (2006). Time of day modulates low-temperature Ca2+ signals in Arabidopsis. Plant J. 48: 962–973 [DOI] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Doyle M.R., Davis S.J., Bastow R.M., McWatters H.G., Kozma-Bognár L., Nagy F., Millar A.J., Amasino R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Edwards K.D., Anderson P.E., Hall A., Salathia N.S., Locke J.C., Lynn J.R., Straume M., Smith J.Q., Millar A.J. (2006). FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K.D., Lynn J.R., Gyula P., Nagy F., Millar A.J. (2005). Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170: 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa C., Kobayashi F., Ishibashi M., Nakamura T., Nakamura C., Takumi S. (2006). Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes Genet. Syst. 81: 77–91 [DOI] [PubMed] [Google Scholar]
- Farré E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.K., Mockler T.C. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S.G., Cook D., Thomashow M.F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S., Wang L., Han L., Suh S.S., Salomé P.A., McClung C.R., Somers D.E. (2008). Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Gould P.D., Locke J.C., Larue C., Southern M.M., Davis S.J., Hanano S., Moyle R., Milich R., Putterill J., Millar A.J., Hall A. (2006). The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B.R. (2001). Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17: 100–107 [DOI] [PubMed] [Google Scholar]
- Green R.M., Tingay S., Wang Z.Y., Tobin E.M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129: 576–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Song H.R., Lutz K., Kerstetter R.A., Michael T.P., McClung C.R. (2010). Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21211–21216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., Watanabe Y. (2005). UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 43: 530–540 [DOI] [PubMed] [Google Scholar]
- Ibáñez C., Kozarewa I., Johansson M., Ogren E., Rohde A., Eriksson M.E. (2010). Circadian clock components regulate entry and affect exit of seasonal dormancy as well as winter hardiness in Populus trees. Plant Physiol. 153: 1823–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez C., Ramos A., Acebo P., Contreras A., Casado R., Allona I., Aragoncillo C. (2008). Overall alteration of circadian clock gene expression in the chestnut cold response. PLoS ONE 3: e3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K., Seki M., Sakurai T., Satou M., Akiyama K., Toyoda T., Konagaya A., Shinozaki K. (2004). Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 32: 5096–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A.B., Monreal J.A., Nimmo G.A., Kelly C.L., Herzyk P., Jenkins G.I., Nimmo H.G. (2008). The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 322: 1832–1835 [DOI] [PubMed] [Google Scholar]
- Jones M.A., Covington M.F., DiTacchio L., Vollmers C., Panda S., Harmer S.L. (2010). Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. USA 107: 21623–21628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., et al. (December 20, 2011) Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. http://dx.doi.org/10.1093/nar/gkr932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-Y., Song H.-R., Taylor B.L., Carré I.A. (2003). Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J. 22: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M.R., Campbell A.K., Smith S.M., Trewavas A.J. (1991). Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Kojima S., Shingle D.L., Green C.B. (2011). Post-transcriptional control of circadian rhythms. J. Cell Sci. 124: 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. (2009). SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Locke J.C., Kozma-Bognár L., Gould P.D., Fehér B., Kevei E., Nagy F., Turner M.S., Hall A., Millar A.J. (2006). Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J.C., Southern M.M., Kozma-Bognar L., Hibberd V., Brown P.E., Turner M.S., Millar A.J. (2005). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 1: 2005.0013 [DOI] [PMC free article] [PubMed]
- Lu S.X., Knowles S.M., Andronis C., Ong M.S., Tobin E.M. (2009). CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 150: 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Knowles S.M., Webb C.J., Celaya R.B., Cha C., Siu J.P., Tobin E.M. (2011). The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 155: 906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J., Sidote D., Hardin P.E., Edery I. (1999). How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24: 219–230 [DOI] [PubMed] [Google Scholar]
- McClung C.R. (2006). Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C.R., Davis S.J. (2010). Ambient thermometers in plants: From physiological outputs towards mechanisms of thermal sensing. Curr. Biol. 20: R1086–R1092 [DOI] [PubMed] [Google Scholar]
- Michael T.P., McClung C.R. (2003). Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 132: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., Salome P.A., McClung C.R. (2003a). Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc. Natl. Acad. Sci. USA 100: 6878–6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., Salomé P.A., Yu H.J., Spencer T.R., Sharp E.L., McPeek M.A., Alonso J.M., Ecker J.R., McClung C.R. (2003b). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P., Yepiskoposyan H., Metze S., Zamudio Orozco R., Kleinschmidt N., Mühlemann O. (2010). Nonsense-mediated mRNA decay in human cells: Mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 67: 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J.S., van Ooijen G., Dixon L.E., Troein C., Corellou F., Bouget F.-Y., Reddy A.B., Millar A.J. (2011). Circadian rhythms persist without transcription in a eukaryote. Nature 469: 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa S.G., Ali G.S., Reddy A.S. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 49: 1091–1107 [DOI] [PubMed] [Google Scholar]
- Perales M., Más P. (2007). A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19: 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A., Hodge S.K., Stratford K., Knox K., Edwards K.D., Thomson A.W., Mizuno T., Millar A.J. (2010). Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol. Syst. Biol. 6: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portolés S., Más P. (2010). The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 6: e1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Kay S.A. (2010). An expanding universe of circadian networks in higher plants. Trends Plant Sci. 15: 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska K.D., Simpson C.G., Ciesiolka A., Szewc L., Lewandowska D., McNicol J., Szweykowska-Kulinska Z., Brown J.W., Jarmolowski A. (2010). Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 38: 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A., Pérez-Solís E., Ibáñez C., Casado R., Collada C., Gómez L., Aragoncillo C., Allona I. (2005). Winter disruption of the circadian clock in chestnut. Proc. Natl. Acad. Sci. USA 102: 7037–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A.S. (2007). Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 58: 267–294 [DOI] [PubMed] [Google Scholar]
- Salomé P.A., McClung C.R. (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A., Weigel D., McClung C.R. (2010). The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22: 3650–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S.E., et al. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468: 112–116 [DOI] [PubMed] [Google Scholar]
- Schöning J.C., Streitner C., Meyer I.M., Gao Y., Staiger D. (2008). Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 36: 6977–6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C.G., Fuller J., Maronova M., Kalyna M., Davidson D., McNicol J., Barta A., Brown J.W. (2008). Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J. 53: 1035–1048 [DOI] [PubMed] [Google Scholar]
- Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H. (2005). Function of alternative splicing. Gene 344: 1–20 [DOI] [PubMed] [Google Scholar]
- Sugano S., Andronis C., Green R.M., Wang Z.-Y., Tobin E.M. (1998). Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc. Natl. Acad. Sci. USA 95: 11020–11025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accur\ate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034 [DOI] [PMC free article] [PubMed]
- Wahl M.C., Will C.L., Lührmann R. (2009). The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Tobin E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Yakir E., Hilman D., Kron I., Hassidim M., Melamed-Book N., Green R.M. (2009). Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol. 150: 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger M.N., Farré E.M., Taylor S.R., Kay S.A., Doyle F.J., III (2006). A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol. Syst. Biol. 2: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]