This work examines the physiological and transcriptomic responses to soil drying in 17 diverse accessions of Arabidopsis thaliana, finding that acclimation to drought stress involved increased investment in photosynthesis, carbohydrate turnover, and root growth, as well as activation of abscisic acid signaling pathways and identifying functional variants of key stress response genes.
Abstract
Arabidopsis thaliana, like many species, is characterized by abundant genetic variation. This variation is rapidly being cataloged at the sequence level, but careful dissection of genetic variation in whole-organism responses to stresses encountered in the natural environment are lacking; this functional variation can be exploited as a natural mutant screen to determine gene function. Here, we document physiological and transcriptomic response to soil drying in 17 natural accessions of Arabidopsis. By imposing ecologically realistic stress conditions, we found that acclimation in Arabidopsis involved a strong signature of increased investment in photosynthesis, carbohydrate turnover, and root growth. Our results extend previous work in the Columbia accession suggesting that abscisic acid signaling pathways play an important role in drought stress response. We also identified several mechanisms, including an increase in leaf nitrogen concentration and upregulation of two-component signaling relays, that were common to most natural accessions but had not been identified in studies using only the Columbia accession. Principal component analysis reveals strong correlations between suites of genes and specific physiological responses to stress. The functional variants we identified may represent adaptive mutations in natural habitats and useful variants for agronomic improvement of crop species.
INTRODUCTION
Plants are repeatedly challenged by the abiotic environment and have evolved diverse strategies to cope with many types of environmental stress. Among these, temperature and soil water availability are factors that most strongly limit the natural distribution of plant species (Stebbins, 1952; Walter, 1964, 1968; Whittaker, 1975). Detailed genetic analysis in a small number of genotypes in a few crop and model species have begun to elucidate the molecular genetic basis of plant physiological responses to abiotic stress (Bohnert et al., 1995; Bray, 1997; Bartels and Sunkar, 2005; Seki et al., 2007; Bressan et al., 2009). In Arabidopsis thaliana, drought-responsive molecular pathways and networks have been described largely using single-gene knockouts and under- and overexpression lines. Additionally, several studies have characterized whole-genome transcriptional response of the Columbia (Col) accession to water deficit and osmotic stress (Seki et al., 2001, 2002; Kreps et al., 2002; Bray, 2004; Kilian et al., 2007; Huang et al., 2008). We have also recently reported transcriptome responses in two natural accessions of Arabidopsis (Juenger et al., 2010). These studies, and a long legacy of physiological studies (Hsiao, 1973; Ludlow, 1989; Kramer and Boyer, 1995; Chaves et al., 2003), reveal that plant water deficit stress response can involve coordinated changes in RNA transcription, developmental timing, growth allocation, sugar metabolism, cell wall composition, cytosolic chemistry, and photosynthetic activity, to name just a few identified responses found in different degrees and combinations in dehydration avoiding (i.e., Arabidopsis) or tolerant species.
Despite considerable progress in understanding the variety of mechanisms plants employ in stress responses, we remain largely unaware how these components vary or covary in nature or the identity of the genes and transcripts that underlie that variation. Inference based on a single genotype of Arabidopsis and comparison of Arabidopsis with highly divergent species, such as rice (Oryza sativa), make distinguishing between variable and conserved components challenging and has hindered progress in extrapolating the experimental findings derived from model systems. Identifying the magnitude, nature, and distribution of naturally segregating variation in drought stress response should be a priority for plant breeders and evolutionary biologists because such variation is the substrate for natural and artificial selection (Araus et al., 2002; Des Marais and Juenger, 2010). Of particular relevance to this endeavor are those traits showing genotype-dependent response to the environment, so-called genotype by environment interactors. Elucidating the genes and pathways underlying genotype by environment interactors is essential to the study of adaptive phenotypic plasticity and adaptation to local environments. Such an evolutionary approach will also identify response components that do not vary among genotypes because they may represent conserved core components of stress responses.
Arabidopsis provides a unique opportunity to explore variation in adaptive evolutionary responses to water stress as it has an extensive geographical distribution and has experienced a wide range of climatic selective regimes for thousands of generations (McKay et al., 2003; Alonso-Blanco et al., 2009). Common garden, quantitative genetic, and quantitative trait loci mapping experiments have identified abundant naturally segregating variation in plant-water relations in Arabidopsis (McKay et al., 2003, 2008; Hausmann et al., 2005; Juenger et al., 2005a, 2005b, 2010; Aguirrezabal et al., 2006; Bouchabke et al., 2008; Christman et al., 2008; Monda et al., 2011). Differences during water stress in the degree of expression of dehydration avoidance characteristics, such as early stomatal closure and reduced leaf growth, constitutive differences in integrated water use efficiency for biomass production, and differences in ultimate dehydration tolerance characteristics, have all been documented between or among different natural accessions of Arabidopsis.
Despite decades of research focused on plant abiotic stress responses, including drought, there are still many unresolved questions and contradictory patterns (Chaves et al., 2003). In particular, the extent to which growth, photosynthesis, and other aspects of primary metabolism are affected by drought stress remains an open question, as these responses seem to depend strongly on the severity of imposed stress and time scale under study. While some studies have found large decreases in primary metabolism, suggesting that carbon becomes limiting under stressful conditions (reviewed in Pinheiro and Chaves, 2011), other studies have found that moderate water stress has no effect or even a positive effect on primary metabolism (Hummel et al., 2010). We predict that these and other systemic responses to water stress may vary across the genetic diversity of Arabidopsis populations. A second major unresolved issue concerns the genetic architecture of abiotic stress responses: Does functional variation in natural populations arise from variation in core signaling components, such as transcription factors, kinases, or phosphatases, or is functional variation confined to downstream, effector genes, such as biosynthetic enzymes, redox regulators, and heat shock proteins?
In this study, we combine physiological data and whole-genome transcription profiling to argue that the traditional narrative of Arabidopsis water stress response, gained largely from a single ecotype under fairly severe stress (or stress shock), misses important details of the genetic basis of ecophysiological acclimation and adaptation. In particular, we find that, when subject to natural drying rates, dehydration avoidance responses in most Arabidopsis accessions are dominated by positive transcriptional control of photosynthesis and sugar metabolism resulting generally in a net increase in root biomass. We show that natural accessions differ in the extent to which these acclimation (plasticity) responses are expressed at the transcriptional and physiological levels. We also identify suites of genes whose expression under drought stress is strongly correlated with physiological parameters, such as specific leaf area (SLA), leaf nitrogen content, and water use efficiency. Our findings suggest that Arabidopsis harbors abundant natural variation in transcriptional responses to slowly imposed soil drying, possibly as a result of local adaptation.
RESULTS
We explored constitutive and soil moisture deficit-induced patterns of physiology and gene expression using factorial manipulation of genotype and environment. We exposed a diverse sampling of natural Arabidopsis accessions (Table 1; see Supplemental Figure 1 online) to a soil drying treatment designed to mimic drought in nature. This treatment resulted in roughly a 60% reduction in extractable soil moisture over a 7-d period, though this treatment was not so severe as to cause wilting (see details in Methods). This experimental design allowed us to test directly for genetic variability among accessions in the degree to which physiological parameters changed and transcripts responded to drying soil in terms of occurrence (responsive or not), magnitude (fold change), and pattern (up- or downregulated). Because both types of data were collected in a single experiment, we were able to identify statistical associations between transcriptional and physiological responses and thereby exploit the 17 studied accessions as a large screen for gene function. In addition, including the Col accession (represented here by Col-2) allowed us to ask if transcripts previously identified as responsive to water stress from genomic studies with Col are representative of general stress responses in Arabidopsis. Our sample included both spring and winter annual accessions (relatively quick-growing and not requiring vernalization treatment to induce flowering versus relatively slower growing and requiring vernalization, respectively; hereafter “Spring” and “Winter”). These two groups of accessions were grown in different blocks and analyzed in separate statistical analyses.
Table 1.
Collection Information for the Accessions Used in This Study
| Accession | ABRC No. | Locale | Collection Coordinates | Life History | Mean Annual Precipitation (mm) |
| Bur-0 | CS6643 | Burren, Ireland | 54.1°N 6.2°W | Spring | 942 |
| CIBC-17 | CS22603 | Ascot, UK | 51.41°N 0.64°W | Spring | 701 |
| Col-2 | CS907 | Gorzow, Poland (est.) | 52.73°N 12.25°E | Spring | 1007 |
| HR-5 | CS22596 | Ascot, UK | 51.41°N 0.64°W | Spring | 701 |
| Knox-18 | CS22567 | Knox, IN, US | 41.28°N 86.62°W | Spring | 975 |
| Ler-1 | CS1686 | Gorzow, Poland | 52.73°N 12.25°E | Spring | 962 |
| NFA-10 | CS22599 | Ascot, UK | 51.41°N 0.64°W | Spring | 701 |
| SQ-8 | CS22601 | Ascot, UK | 51.41°N 0.64°W | Spring | 701 |
| Ts-1 | CS6868 | Tossa del Mar, Spain | 41.72°N 2.93°E | Spring | 644 |
| Ws-2 | CS2360 | Vasil’yevka, Belarus | 52.3°N 30°E | Spring | 624 |
| Ag-0 | CS6601 | Argentat, France | 45°N 1.3°E | Winter | 887 |
| Bil-5 | CS22578 | Billaberget, Sweden | 63.32°N 18.48°E | Winter | 615 |
| Eden-1 | CS22572 | Eden, Sweden | 62.88°N 18.18°E | Winter | 655 |
| Got-22 | CS22609 | Goettingen, Germany | 51.53°N 9.94°E | Winter | 652 |
| Omo2-3 | CS22585 | Ostra Mocklo, Sweden | 56.15°N 15.77°E | Winter | 546 |
| Tamm-2 | CS22604 | Tammisaari, Finland | 60°N 23.5°E | Winter | 609 |
| Ull2-5 | CS22586 | Ullstorp, Sweden | 56.06°N 13.97°E | Winter | 640 |
Data for precipitation are from Hijmans et al. (2005), and life history refers to the requirement of vernalization for flowering (Winter) versus no requirement (Spring) as determined by a common garden greenhouse experiment.
Soil Moisture Deficit Treatments
The main goal of the dry-down experiments was to subject all accessions to uniform soil moisture deficit with slow development of the stress so that acclimation could occur. Slow drying over 7 d in both experiments provided a simulation of the kind of soil drying that occurs often in agricultural and natural situations. The experiments achieved this goal with very uniform treatments across accessions and for both experiments (see Supplemental Figure 2A online). At harvest, the percentage of soil water remaining averaged 98.1% ± 0.6% and 39.0% ± 0.6% (least-square means ± 1 se for all values below) in the control and soil moisture deficit treatments, respectively, for the Spring accession experiment and 94.9% ± 0.7% and 37.7% ± 0.6% in the control and soil moisture deficit treatments, respectively, for the Winter accession experiment (soil moisture values indicate percentage of field capacity of the soil). Corresponding soil water potentials (Ψsoil), which measure the difficulty of extracting water from the soil, averaged for the entire soil volume for each pot were −0.02 ± 0.01 MPa for control treatments in both experiments and −1.12 ± 0.01 MPa and −1.27 ± 0.06 MPa for the soil moisture deficit treatments in the Spring and Winter accession experiments, respectively.
Treatment Effects on Water Relations
We quantified the impact of soil drying on plant water status with detailed data on leaf relative water content (RWC), leaf water content (WC), leaf water potential (Ψtot), leaf turgor (Ψp), and bulk leaf osmotic potentials (Ψsol) from experimental plants. RWC is a useful indicator of plant water balance as it expresses the WC of tissue relative to the WC at full hydration. Ψtot is an estimate of the overall potential of water in the leaf, while Ψp is a measure of the hydrostatic pressure (i.e., turgor) in plant cells and Ψsol indicates the effects of the presence of dissolved materials on the potential of the bulk leaf water. Soil drying consistently and significantly reduced leaf RWC, Ψtot, and Ψp in comparison to controls in both Spring and Winter accession experiments (all P < 0.0002; Figure 1; see Supplemental Figure 2B online). In the Winter accession experiment, the accessions responded differently to the treatments only for leaf RWC (significant interaction P = 0.03); for the other parameters, there was no significant accession treatment interaction (all P > 0.52). Only Ψtot differed among accessions and only in the Spring accession experiment (P = 0.04). In that experiment, Ψsol was slightly but significantly lower in the dry than the control treatment (−0.91 ± 0.03 versus −0.73 ± 0.03 MPa, respectively; P < 0.0001), but in the Winter accession experiment, there was no significant difference in Ψsol between the treatments or accessions (P > 0.36; overall average Ψsol = −1.50 ± 0.06 MPa).
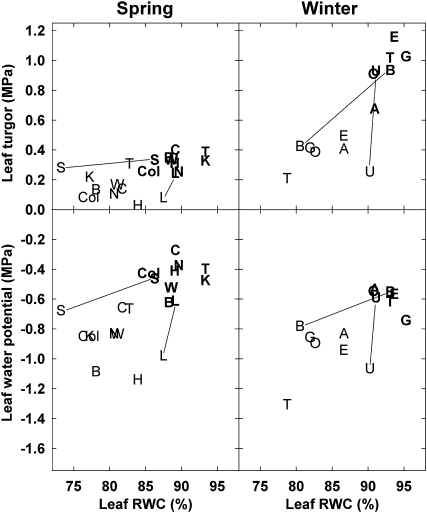
Figure 1.
Relationships of Leaf Water Potential (Ψtot) and Turgor (Ψp) to Leaf RWC for Each Accession.
Data are least-square means plotted as codes to indicate accession. Spring: Bur-0 = B, CIBC17 = C, Col-2 = Col, HR5 = H, Knox-18 = K, Ler-1 = L, NFA-10 = N, Sq-8 = S, Ts-1 = T, and Ws-2 = W. Winter: Ag-0 = A, Bil-5 = B, Eden-1 = E, Got-22 = G, Omo2-3 = O, Tamm-2 = T, and Ull2-5 = U. Controls are plotted in bold and the soil moisture deficit treatments in normal type. On each panel, lines link control and soil moisture deficit treatments of two example accessions. For clarity, not all lines are shown. n = 3 for each accession in each treatment.
Despite consistent effects of the soil moisture deficit treatments on plant water relations parameters, there was substantial diversity among accessions in the relationships among these parameters during drying. Differences in pressure-volume relationships shown in Figure 1 illustrate the wide spectrum of responses and likely reflect natural variation in leaf structural characteristics, including properties of cell wall elasticity. The leaf structural differences suggested by these relationships may be reflected by the large constitutive differences in leaf WC among both Spring and Winter accessions (both P < 0.0001; Figure 2). Among Spring accessions, Col-2 had by far the highest average leaf WC of 997% and Bur-0 was lowest at 490%, while among Winter accessions, Omo2-3 had the highest average leaf WC of 907% and Got-22 was lowest at 439%. We did not detect significant accession treatment interactions in either the Spring or Winter accession experiments (P > 0.09).
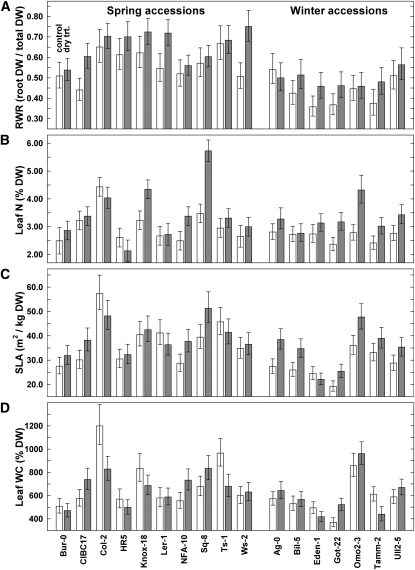
Figure 2.
Values for Developmental and Physiological Traits under Control and Soil Water Deficit Treatments in the Spring and Winter Accession Experiments.
Least-square means ± se (values are backtransformed from ln values used in the analyses) are shown for root to total biomass ratio (i.e., RWR) (A), leaf N concentration (B), SLA (C), and leaf WC (D). Bars show values for control (white) and soil water deficit treatment (dry trt.; gray). All values are relative to dry weight biomass (DW). n = 3 for each accession in each treatment. See Supplemental Figure 2 online for trait values not given here.
Developmental and Physiological Responses to Soil Moisture Deficit
We next addressed the impact of soil drying on components of whole-plant resource acquisition and partitioning. Several important developmental and physiological changes were evident (Figure 2; see Supplemental Figures 2C and 2D online). First, 16 of the 17 lines in both the Spring and Winter accession experiments increased root versus shoot biomass partitioning during the 7-d soil moisture dry-down; only the Winter accession Ag-0 failed to show this pattern (Figure 2A). Averaged across lines, root dry weight as a proportion of total plant dry weight, the root weight ratio (RWR), was 17% greater in the dry treatment than in controls for the Spring accession experiment (P = 0.003). The average increase was lower in the Winter accession experiment (7%) and was not significant overall (P = 0.08). There were no significant differences among accessions within each experiment and the interactions were also nonsignificant.
The second major change among nearly all lines in both experiments was a large and significant increase in leaf N concentration (N% dry weight) in response to the treatment (Figure 2B). Averaged across lines, leaf N% was 24% greater in the dry treatment than in controls for the Winter accession experiment (P = 0.0005). For the Spring accession experiment, the difference was 15% (P = 0.005). For the Spring accessions, but not the Winter accessions, there was also a significant difference among lines (P < 0.0001) and a significant interaction (P = 0.02). This interaction was caused by the decrease in leaf N% of 9% in Col-2 and 18% in HR5, while the other eight Spring lines had increased N% in the soil moisture deficit treatment. All Winter accessions had an increase in leaf N%; Bil-5 increased only 1% but Omo2-3 increased 55%.
The changes in leaf N% caused similarly large proportional changes in C:N ratios (−12% in Spring, P = 0.02; −20% in Winter, P = 0.0005; see Supplemental Figure 2D online). Although there was a significant treatment effect on leaf C concentration (C% dry weight) for the Spring accessions (P = 0.03), the relative change from controls (38.5% ± 0.2%) to treatment (38.0% ± 0.2%) was only −1%. Thus, changes in leaf C:N were almost entirely due to changes in leaf N% and not changes in C%. There were significant differences among Spring accessions in leaf C% (P = 0.03) and leaf C:N (P < 0.0001; see Supplemental Figure 2D online). No treatment by accession interactions were found for either the Spring or Winter accession experiments (all P > 0.24).
Two other developmental and physiological changes should be noted. In the Winter accession experiment, SLA, which measures the amount of leaf area produced per unit biomass invested in leaves and impacts leaf surface area display for photosynthetic light capture and gas exchange, was on average 23% greater in the soil drying treatment plants than in the controls (Figure 2C; P = 0.0009). There was a 7% increase in SLA in response to the treatment in the Spring experiment, but this was not significant (P = 0.23). In both experiments, accessions differed constitutively in SLA (both P < 0.0002), and these were strongly correlated with their constitutive differences in leaf WC (both r > 0.72, P < 0.002; Figure 2D). Variation in SLA is often positively correlated with variation in relative growth rate and many other whole-plant physiological and developmental parameters (Lambers et al., 2008). Finally, in both experiments, there were small shifts in leaf carbon isotope composition (δ13C) consistent with a small increase in water use efficiency in the soil moisture deficit treatment compared with controls, but these were not significant (Winter P = 0.12, Spring P = 0.28; see Supplemental Figure 2D online). However, as found previously (McKay et al., 2003), the Winter accessions had much greater apparent water use efficiency (less negative δ13C) than the Spring accessions.
Genetic Diversity of Transcriptional Response
We used a statistical approach to detect differentially expressed genes on the ATH1 array, which allowed us to identify genes with consistent, though possibly small magnitude, expression differences among accessions and between treatments. At a positive false discovery rate (pFDR) of 0.05, we found 8671 (41.8% of those in the analysis) and 6465 (30.5%) genes exhibited constitutively differing levels of expression in the Spring and Winter accession analyses, respectively. Our data therefore suggest that a large fraction of the genome is differentially expressed in rosette leaves among natural accessions irrespective of environmental conditions, as shown previously in Arabidopsis (van Leeuwen et al., 2007; Gan et al., 2011). Because we removed probes containing hybridization polymorphisms, genes showing constitutive differences are not artifacts of sequence divergence in probe sites (see details of probe filtering for hybridization polymorphism in Methods).
Across the ten Spring accessions, 6734 genes (32.4%) showed a significant response to soil drying. Roughly equal numbers of these reflect upregulation (3527 genes) and downregulation (3207 genes) of expression in response to soil drying. Similarly, across the seven Winter accessions, 5270 genes (24.9%) showed a significant treatment effect with 2690 genes upregulated and 2580 downregulated.
We next asked whether individual genes showed different treatment effects among the accessions using two criteria. First, we identified genes with a significant accession by treatment interaction based on the initial factorial analysis of variance (ANOVA) performed on robust multichip average (RMA) (log2)-transformed fold change in expression. Second, we identified genes with both significant treatment and accession effects but a nonsignificant interaction effect; this pattern suggests an additive relationship on the log2 scale but an interaction (multiplicative relationship) on the raw scale (for a discussion of scale and interaction effects, see Juenger et al., 2010). Genes in this latter group may also be considered to respond differentially to the stress, although they are unlikely to exhibit strong rank changing patterns (Berrington de Gonzalez and Cox, 2007). There was considerable heterogeneity among accessions in the response to the soil moisture deficit treatment with 4488 (of which 1016 show significant interaction in log2 transformed expression values) and 2697 (500 in log2 values) genes exhibiting significant accession treatment interactions for the Spring and Winter analyses, respectively.
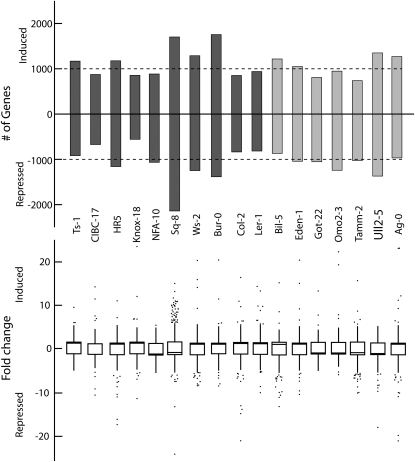
To further explore the heterogeneity in drought-responsive gene expression, we also used a series of independent one-way ANOVAs testing for treatment effects within each accession. These analyses found that the number of genes with significant treatment effects varied considerably among accessions and that many genes were significant in only one or a small number of accessions. For example, at a pFDR of 0.05, 1689 genes had a significant treatment effect in the Col-2 accession, whereas the expression of 3842 genes was significantly affected by the treatment in the Sq-8 accession. Most accessions were intermediate to these two extremes (Figure 3A). The accessions also varied considerably in the magnitude of expression changes of individual genes (Figure 3B). For example, no transcript in the NFA-10 accession showed greater than a fivefold change in expression, whereas transcript abundance for individual genes from several other accessions increased >25-fold.
Figure 3.
Diversity of Transcriptional Response.
(A) Number of significant soil drying treatment effect genes at pFDR = 0.05 for each accession identified from one-way ANOVAs. Black boxes are Spring annual accessions; gray boxes are Winter-annual accessions. Dashed lines indicate 1000 genes on the y axis.
(B) Box plots of fold change for significant soil moisture deficit treatment effect genes in each accession. Boxes enclose 50% quantile, and dotted whiskers indicate 97.5% quantile ranges. Some extreme outliers (fold change > 25) have been omitted. n = 3 for each accession in each treatment.
Only two genes showed significant treatment effect in all ten Spring accessions: the protein phosphatase PP2CA (At3G11410), which was upregulated, and the heat stress transcription factor AT-Heat Shock Transcription FactorA7A (At3G51910), which was downregulated. Eight genes were significantly affected in nine of the 10 Spring accessions, including Histone H1-3 (His1-3) (At2g18050) and Responsive to ABA18 (RAB18) (At5g66400), which are commonly found to be upregulated by abiotic stress in the Col accession (Ascenzi and Gantt, 1997; Seki et al., 2002; Urano et al., 2009), and a DREB family protein (At1g77640). Several other genes frequently found to be stress responsive in Col were also found to be strongly responsive in this study (Table 2). It is interesting to note that several genes were up- or downregulated in multiple Spring accessions but in neither Col-2 nor the other popular lab strain Landsberg erecta (Ler). Among these are the two-component response regulators Arabidopsis response regulator4 (ARR4) (At1g10470) and ARR7 (At1g19050), which were upregulated in the eight other Spring accessions.
Table 2.
Twenty Highest Fold Changes for Induced and Repressed Genes in Response to Soil Drying Treatment Compared to Control in Spring Accessions
| Spring Rank | AGI | Gene | Spring Fold-Change | Winter Rank | Winter Fold Change | Significant in Col-2? | Significant in ≥Half of Spring Accessions? |
| Induced transcripts | |||||||
| 1 | AT5G59310 | LTP4; lipid transfer protein 4 | 9.646 | 2 | 8.211 | Yes | Yes |
| 2 | AT5G59320 | LTP3; lipid transfer protein 3 | 7.548 | 4 | 6.048 | Yes | No |
| 3 | AT2G18050 | HIS1-3; histone H1-3 | 6.437 | 48 | 2.047 | Yes | Yes |
| 4 | AT1G52400 | BGL1/BGLU18/ATBG1; β-glucosidase 18 | 6.354 | 8 | 2.798 | No | No |
| 5 | AT3G53980 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily | 6.157 | 1 | 9.948 | Yes | Yes |
| 6 | AT3G28220 | TRAF-like family protein | 5.420 | 7 | 3.315 | No | Yes |
| 7 | AT2G37870 | Bifunctional inhibitor/lipid transfer protein/seed storage 2S albumin superfamily 3 | 4.956 | 3 | 7.584 | Yes | Yes |
| 8 | AT1G73480 | α/β-Hydrolase superfamily protein | 4.046 | NS | NS | No | No |
| 9 | AT2G42530 | COR15B; cold-regulated 15b | 3.681 | NS | NS | Yes | No |
| 10 | AT5G66400 | RAB18/ATDI8; dehydrin family protein | 3.464 | 50 | 2.032 | Yes | Yes |
| 11 | AT4G04330 | Chaperonin-like RbcX protein | 3.342 | 112 | 1.808 | No | No |
| 12 | AT2G07698 | ATPase, F1 complex, α-subunit protein | 3.168 | NS | NS | No | Yes |
| 13 | ATMG01190 | ATP1; ATP synthase subunit 1 | 3.161 | NS | NS | No | Yes |
| 14 | AT4G16000 | Unknown protein | 3.138 | 263 (down) | −1.672 | No | Yes |
| 15 | AT1G52000 | Mannose binding lectin superfamily protein | 3.134 | NS | NS | No | Yes |
| 16 | AT2G39030 | Acyl-CoA N-acyltransferases (NAT) superfamily | 3.038 | NS | NS | No | No |
| 17 | AT5G02020 | SIS; encodes a protein involved in salt tolerance | 2.982 | NS | NS | No | No |
| 18 | AT2G46680 | ATHB-7/ATHB7/HB-7; homeobox 7 | 2.982 | 251 | 1.607 | Yes | Yes |
| 19 | AT2G42540 | COR15A; cold-regulated 15a | 2.871 | 44 | 2.078 | Yes | Yes |
| 20 | AT2G29450 | ATGSTU5/ATGSTU1/AT103-1A/GSTU5; glutathione S-transferase τ 5 | 2.855 | 83 | 1.882 | Yes | Yes |
| Repressed transcripts | |||||||
| 1 | AT1G75750 | GASA1 | −6.989 | NS | NS | Yes | Yes |
| 2 | AT4G12470 | AZI1; azelaic acid induced 1 | −5.700 | 178 | −1.827 | Yes | Yes |
| 3 | AT3G30775 | ERD5/PRODH/ATPOX/PRO1; methylenetetrahydrofolate reductase family protein | −5.473 | NS | NS | Yes | Yes |
| 4 | AT1G22690 | Gibberellin-regulated family protein | −4.615 | NS | NS | Yes | Yes |
| 5 | AT3G16670 | Pollen Ole e 1 allergen and extensin family protein | −4.538 | 2 | −4.654 | Yes | No |
| 6 | AT5G16570 | GLN1;4 | Gln synthetase 1;4 | −4.427 | 100 | −2.052 | No | No |
| 7 | AT5G64120 | Peroxidase superfamily protein | −4.270 | 16 | −3.216 | Yes | Yes |
| 8 | AT4G37800 | XTH7; xyloglucan endotransglucosylase/hydrolase 7 | −4.228 | 225 | −1.726 | No | No |
| 9 | AT1G62480 | Vacuolar calcium binding protein-related | −4.157 | 227 | −1.725 | Yes | Yes |
| 10 | AT5G44420 | PDF1.2, PDF1.2A, LCR77 | plant defensin 1.2 | −3.980 | 1 | −7.590 | No | Yes |
| 11 | AT2G43620 | Chitinase family protein | −3.924 | 7 | −4.095 | Yes | Yes |
| 12 | AT4G08300 | Nodulin MtN21 /EamA-like transporter family protein | −3.697 | 147 | −1.915 | No | Yes |
| 13 | AT1G77640 | Integrase-type DNA binding superfamily protein | −3.414 | NS | NS | Yes | Yes |
| 14 | AT3G16530 | Legume lectin family protein | −3.340 | 24 | −2.937 | No | No |
| 15 | AT2G22170 | Lipase/lipooxygenase, PLAT/LH2 family protein | −3.181 | 609 | −1.423 | No | Yes |
| 16 | AT4G36410 | UBC17; ubiquitin-conjugating enzyme 17 | −3.118 | NS | NS | Yes | Yes |
| 17 | AT2G44790 | UCC2; uclacyanin 2 | −3.102 | 20 | −3.066 | Yes | Yes |
| 18 | AT1G23090 | AST91, SULTR3; sulfate transporter 91 | −3.063 | 38 | −2.558 | Yes | Yes |
| 19 | AT2G28190 | CSD2, CZSOD2; Cu/Zn superoxide dismutase 2 | −2.935 | NS | NS | Yes | No |
| 20 | AT1G06830 | Glutaredoxin family protein | −2.893 | 187 | −1.803 | Yes | No |
AGI, Arabidopsis Genome Initiative. NS, not significant.
The most striking finding of our analysis of the Winter accession experiment was the paucity of genes previously identified by studies of drought stress in the Col accession. For example, six genes showed significant treatment effect in all seven Winter accessions. An unknown protein (At1g21520) was the lone gene to be downregulated among all Winter accessions. A putative chaperonin (At1g26230), the SIGF sigma-factor gene (At2g36990), another unknown protein (At3g48200), a major facilitator protein (At3g21670), and Methionine over-accumulator2 (At4g29840) were upregulated in all seven Winter accessions. Only two of these transcripts were significant in half of the Spring accessions. The difference in specific genes responding to stress in Spring and Winter accessions can also be seen in the genes showing the highest fold change (Table 3).
Table 3.
Twenty Highest Fold Changes for Induced and Repressed Genes in Response to Soil Drying Treatment Compared to Control in Winter Accessions
| Winter Rank | AGI | Gene | Winter Fold Change | Spring Rank | Spring Fold Change | Significant in Col-2? | Significant in ≥Half of Winter Accessions? |
| Induced transcripts | |||||||
| 1 | AT3G53980 | Bifunctional inhibitor/lipid transfer protein/seed storage 2S albumin superfamily | 9.948 | 5 | 6.157 | Yes | Yes |
| 2 | AT5G59310 | LTP4; lipid transfer protein 4 | 8.211 | 1 | 9.646 | Yes | Yes |
| 3 | AT2G37870 | Bifunctional inhibitor/lipid transfer protein/seed storage 2S albumin superfamily | 7.584 | 7 | 4.956 | Yes | Yes |
| 4 | AT5G59320 | LTP3; lipid transfer protein 3 | 6.048 | 2 | 7.548 | Yes | Yes |
| 5 | AT4G33550 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily | 5.811 | 45 | 2.442 | No | Yes |
| 6 | AT5G18840 | Major facilitator superfamily protein | 4.123 | 331 (down) | −1.553 | No | No |
| 7 | AT3G28220 | TRAF-like family protein | 3.315 | 6 | 5.420 | No | No |
| 8 | AT1G52400 | BGL1/BGLU18; β-glucosidase 18 | 2.798 | 4 | 6.354 | No | No |
| 9 | AT5G62520 | SRO5; similar to RCD one 5 | 2.748 | 113 | 2.002 | No | Yes |
| 10 | AT1G64110 | DAA1; DUO1-activated ATPase | 2.714 | 146 | 1.891 | Yes | No |
| 11 | AT4G16146 | cAMP-regulated phosphoprotein 19-related protein | 2.696 | NS | NS | No | No |
| 12 | AT2G34510 | Protein of unknown function, DUF642 | 2.638 | 114 | 1.999 | No | Yes |
| 13 | AT3G16250 | NDF4; NDH-dependent cyclic electron flow 1 | 2.600 | 102 | 2.039 | No | Yes |
| 14 | AT4G23670 | Polyketide cyclase/dehydrase and lipid transport superfamily protein | 2.597 | NS | NS | No | No |
| 15 | AT2G36145 | Unknown protein | 2.565 | NS | NS | No | No |
| 16 | AT4G15910 | ATDI21; drought-induced 21 | 2.533 | 30 | 2.660 | Yes | No |
| 17 | AT1G61800 | GPT2; glucose-6-phosphate/phosphate translocator 2 | 2.503 | NS | NS | No | No |
| 18 | AT1G48750 | Bifunctional inhibitor/lipid transfer protein/seed storage 2S albumin superfamily | 2.502 | 147 | 1.883 | No | No |
| 19 | AT5G23060 | CaS; calcium sensing receptor | 2.421 | NS | NS | No | yes |
| 20 | AT4G34160 | CYCD3; CYCLIN D3;1 | 2.398 | 57 | 2.326 | No | No |
| Repressed transcripts | |||||||
| 1 | AT5G44420 | PDF1.2; plant defensin 1.2 | −7.590 | 10 | −3.980 | No | No |
| 2 | AT3G16670 | Pollen Ole e 1 allergen and extensin family protein | −4.654 | 5 | −4.538 | Yes | No |
| 3 | AT2G21650 | MEE3; Homeodomain-like superfamily protein | −4.607 | 68 | −2.124 | Yes | Yes |
| 4 | AT1G21520 | Unknown protein | −4.536 | 59 | −2.195 | No | Yes |
| 5 | AT1G72060 | Ser-type endopeptidase inhibitors | −4.315 | 153 | −1.809 | No | No |
| 6 | AT5G56870 | BGAL4; β-galactosidase 4 | −4.153 | NS | NS | No | No |
| 7 | AT2G43620 | Chitinase family protein | −4.095 | 11 | −3.924 | Yes | Yes |
| 8 | AT3G13790 | ATBFRUCT1; glycosyl hydrolase family 32 protein | −3.876 | 62 | −2.183 | Yes | Yes |
| 9 | AT3G16360 | AHP4; HPT phosphotransmitter 4 | −3.869 | NS | NS | Yes | Yes |
| 10 | AT3G22060 | Unknown protein | −3.805 | 79 | −2.056 | No | No |
| 11 | AT1G17700 | PRA1; prenylated RAB acceptor 1.F1 | −3.542 | 110 | −1.923 | No | Yes |
| 12 | AT5G61160 | AACT1; anthocyanin 5-aromatic acyltransferase 1 | −3.505 | 34 | −2.553 | No | No |
| 13 | AT3G26830 | PAD3/CYP71B15; cytochrome P450 superfamily protein | −3.378 | NS | NS | No | No |
| 14 | AT2G28630 | KCS12; 3-ketoacyl-CoA synthase 12 | −3.369 | 85 | −2.016 | No | No |
| 15 | AT1G06160 | ORA59; octadecanoid-responsive Arabidopsis AP2/ERF 59 | −3.364 | 261 | −1.628 | No | No |
| 16 | AT5G64120 | Peroxidase superfamily protein | −3.216 | 7 | −4.270 | Yes | No |
| 17 | AT2G26690 | Major facilitator superfamily protein | −3.198 | NS | NS | No | No |
| 18 | AT4G11890 | Protein kinase superfamily protein | −3.131 | 302 | −1.584 | No | No |
| 19 | AT1G26380 | FAD binding berberine family protein | −3.131 | 246 | −1.646 | No | No |
| 20 | AT2G44790 | UCC2; uclacyanin 2 | −3.066 | 17 | −3.102 | Yes | No |
AGI, Arabidopsis Genome Initiative. NS, not significant.
Functional Annotation
We used the program Athena (O’Connor et al., 2005) to ask whether the promoters of genes showing a significant treatment effect were enriched for known cis-regulatory elements. Because the complete genome sequences of our 17 accessions are not yet available, this analysis was constrained to querying predicted promoter elements found in the upstream regions of genes annotated in the Col accession. Looking first at elements common to upregulated genes in both Spring and Winter accessions, we found that most of the overrepresented elements were variations on the abscisic acid (ABA)–responsive element (ABRE) core motif ACGTG (Table 4). ABREs have been found in many studies to play a key role in water or osmotic stress signaling (reviewed in Yamaguchi-Shinozaki and Shinozaki, 2005). We also found an overrepresentation among the Spring accessions of several variations on the G-box motif, which also contains the core ABRE motif and is often found in the promoters of light-regulated genes (Giuliano et al., 1988). The evening element motif, which is believed to play a role in genes controlled by circadian rhythm (Michael et al., 2008), was also enriched in both Spring and Winter accessions. Notably, we did not find an enrichment of the Drought-Responsive/Cold Responsive Element Elements, which have been the focus of several past studies of drought stress in the Col accession (Stockinger et al., 1997; Liu et al., 1998; Yamaguchi-Shinozaki and Shinozaki, 2005). Overrepresented promoter motifs in downregulated genes included the TATA-box and W-box motifs, which are very abundant in the genome as a whole. ABREs were also statistically overrepresented among the downregulated genes, though not to the extent found in the upregulated genes. In general, there were many more significantly overrepresented motifs among the loci downregulated by the treatment than those upregulated in the Winter accessions. In particular, there were many more downregulated genes with ABRE core motifs in their promoters (Table 4).
Table 4.
Enrichment of Annotated Promoter Motifs in Soil Drying Treatment Effect Genes as Estimated in Athena
| Element | No. of Genes with Element | P Value | Element | No. of Genes with Element | P Value |
| Spring | |||||
| Up (3527 Genes) | Down (3207 Genes) | ||||
| ABRE-like* | 954 | 10E-10 | TATA | 2626 | 10E-6 |
| CACGTG motif* | 792 | 10E-10 | ATHB5ATCORE | 167 | 10E-6 |
| ACGTABREMOTIF20SEM* | 739 | 10E-10 | UPRMOTIFIAT | 156 | 10E-6 |
| GADOWNAT* | 438 | 10E-10 | TGA1* | 156 | 10E-6 |
| ABRE* | 269 | 10E-10 | ATHB1 | 124 | 10E-6 |
| ABF* | 183 | 10E-8 | ACGTABREMOTIF20SEM* | 581 | 10E-5 |
| Evening element | 362 | 10E-7 | CARGCW8GAT | 2056 | 10E-5 |
| GBOXLERBCS* | 145 | 10E-6 | W-box | 2183 | 10E-5 |
| ABREATRD22* | 135 | 10E-5 | ABRE-like* | 793 | 10E-4 |
| I-box | 1474 | 10E-4 | L1-box | 632 | 10E-4 |
| Winter | |||||
| Up (2690 Genes) | Down (2580 Genes) | ||||
| ACGTABREMOTIF20SEM* | 496 | 10E-5 | ABRE-like* | 802 | 10E-10 |
| ABREATRD22* | 109 | 10E-5 | CACGTGMotif* | 609 | 10E-10 |
| Evening element | 275 | 10E-4 | ACGTABREMOTIF20SEM* | 578 | 10E-10 |
| ABRE* | 179 | 10E-4 | GADOWNAT* | 360 | 10E-9 |
| W-box | 1886 | 10E-9 | |||
| ABRE* | 195 | 10E-8 | |||
| MYCATERD1 | 1077 | 10E-8 | |||
| AtMYC2 BS in RD22 | 1077 | 10E-8 | |||
| GBF1/2/3 in ADH1* | 78 | 10E-7 | |||
| CARGCW8GAT | 1713 | 10E-7 | |||
| TATA | 2139 | 10E-7 | |||
| TGA1* | 131 | 10E-6 | |||
| UPRMOTIFIAT | 131 | 10E-6 | |||
| GBOXLERBCS* | 114 | 10E-6 | |||
| T-box | 1498 | 10E-6 | |||
| ABF* | 138 | 10E-6 | |||
| UPRE2AT | 40 | 10E-6 | |||
| I-box | 1129 | 10E-5 | |||
| RY repeat | 143 | 10E-4 | |||
Asterisk indicates that the listed motif contains an ABRE core motif.
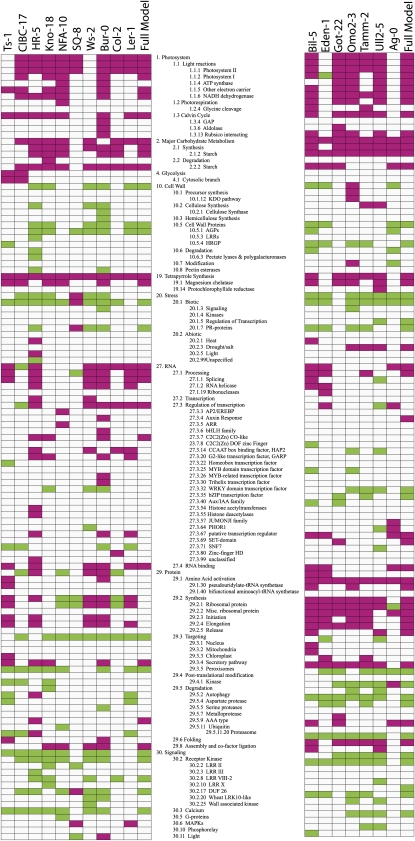
We next asked if any functional classes of genes were overrepresented in our lists of significant treatment effect genes. We performed Wilcoxon rank sum tests of curated functional bins as implemented in the program MapMan (Usadel et al., 2006). The PageMan module of MapMan tests, for a given experiment, if a particular functional bin has a higher median expression value than other bins in the genome. We found that most accessions, including both Winter and Spring accessions, showed strong upregulation of bins involved in photosynthesis (Figure 4; see Supplemental Figure 3 online). This response included synthesis of the proteins and pigments of photosystem I and II, including tetrapyrrole synthesis; for many accessions, we also saw enrichment of bins involved in photorespiration and the Calvin cycle. We also found significant upregulation of carbohydrate synthesis and degradation, particularly those processes involving starch. Protein synthesis bins, especially those containing prokaryotic ribosomal proteins as well as chloroplast protein targeting bins, were also enriched for upregulation in many accessions. Several accessions, including Ws-2, Ts-1, and Ag-0, did not show a statistical enrichment for photosynthetic response, and Ws-2 and Ts-1 also lacked a significant upregulation of carbohydrate degradation genes. Ws-2 and Eden-1 showed the typical upregulation of carbohydrate synthesis. The Eden-1 accession had a significant enrichment of downregulated genes involved in photosystem I and showed a nonsignificant response in bins associated with photosystem II, the Calvin cycle, photorespiration, and the synthesis of tetrapyrroles and carotenoids.
Figure 4.
Functional Enrichments of Genes with Significant Treatment Effect.
Maroon boxes indicate significant upregulation as determined by Wilcoxon rank sum tests of curated functional bins as implemented in the PageMan module of the MapMan software package. Green boxes indicate bins that are significantly downregulated.
Other generalized responses shared by most accessions included upregulation of amino acid and protein synthesis and of the poorly understood pentatricopeptide repeat (PPR) proteins. Most lines had downregulation of histone bins as well as bins involved in protein secretion, the ubiquitin proteasome, vesicle transport, and most bins involved in signaling. We saw an enrichment of downregulated genes involved in biotic stress and a paucity of significant bins annotated as having a role in abiotic stress response.
We used Fisher’s exact test to ask if accession treatment interaction (AxT) genes had functional enrichments. Accounting for multiple testing with a pFDR of 0.05 made these analyses quite conservative: Only three bins were significantly enriched among the Spring accessions and none among the Winter accessions (see Supplemental Table 1online). One of these bins, the Domain of Unknown Function 26 (DUF26; also known as the Cys-Rich Receptor-like Kinases [CRKs]) was overrepresented in the Spring accessions with 18 of 42 genes significant. Most of these genes were downregulated in response to the soil moisture deficit treatment. The histone synthesis bin and protein targeting to the chloroplast bin were also strongly overrepresented among AxT interactors in the Spring accessions. There were no significant MapMan bins enriched among the genes showing AxT interaction in the Winter accessions.
While we did not find a statistical enrichment of annotated abiotic stress genes, we did observe moderate treatment effects in a number of previously studied drought-responsive members of the Drought Responsive Element Binding (DREB)/Dwarf and Delayed Flowering (DDF) family of transcription factors. These included DREB1e/DDF2 (At1g63030) and DREB1f/DDF1 (At1g12610), which were upregulated in both Spring and Winter accessions, and a number of putative DREB proteins containing AP2 domains (At1g12630, At1g21910, At1g33760, At2g23340, At3g60490, At4g32800, and At5g52020).
Integrating Gene Expression and Physiology
We performed principal components analysis on the expression and physiology treatment responses to ask whether major axes of variation in the two types of data were correlated. This approach is conservative because, with a sample size of 7 (as in the Winter accession experiment) or 10 (as in the Springs), only r values above 0.707 or 0.602, respectively, are significant at P < 0.05 (Rohlf and Sokal, 1981). The first principle component (PC1) of the Spring expression data, which explained 23.6% of the expression variance, was strongly negatively correlated (r2 = −0.604) with the first component of physiological variation, which explained 43.0% of the variance, though this correlation was marginally nonsignificant (P = 0.064). Spring expression PC2 was likewise strongly but nonsignificantly correlated with physiology PC2 (r2 = −0.569, P = 0.086).
We next asked whether expression PCs were significantly correlated with individual physiology traits. Spring expression PC1 was significantly correlated with leaf N% (r2 = −0.718, P = 0.019), while expression PC2 was significantly correlated with SLA (r2 = −0.675, P = 0.032) and there was a nonsignificant correlation with leaf RWC (r2 = −0.606, P = 0.063). However, the correlation between expression PC1 and leaf N% appears to be driven almost entirely by the Sq-8 accession, which showed particularly strong responses in leaf N% and SLA (Figures 2B and 2C). In the absence of Sq-8, there was no correlation between PC1 and leaf N% (r2 = −0.319, P = 0.403), but there was a strong correlation between expression PC1 and the total number of genes expressed in an accession (r2 = 0.408, P = 0.004). The correlation between expression PC2 and SLA was stronger after excluding Sq-8 (r2 = −0.797, P = 0.010).
Winter physiology PC1 was correlated with expression PC1, though this was not significant (r2 = −0.733, P = 0.061), while expression PC2 show a significant correlation with physiology PC3 (r2 = −0.820, P = 0.024). Winter expression PC1 was strongly and significantly correlated with leaf δ13C (r2 = −0.904, P = 0.005), and there was a trend toward a correlation with leaf Ψp (r2 = −0.730, P = 0.064). Winter expression PC2 was strongly and significantly correlated with shoot dry weight (r2 = −0.884, P = 0.008).
We next tested for functional enrichment (MapMan terms) in transcripts with the strongest loadings on the expression principle components that showed significant correlations with particular physiological traits. Spring expression PC1 (correlated with leaf N% or total genes expressed; see above) was strongly enriched for genes involving histones and light signaling. Spring expression PC2 (correlated with SLA and leaf WC) was enriched for several photosynthesis-related bins, thioredoxins, photosystems I and II subunits, as well as genes involved in autophagy (Table 5). Winter expression PC1 (correlated with leaf δ13C and leaf Ψp) was enriched for transcripts involved in arabinogalactan cell wall proteins, histones, CCAAT-box binding transcription factors, and tonoplast intrinsic proteins. No functional bins were enriched in genes with the highest loading on winter expression PC2 at a pFDR of 0.05. Specific genes found in the enriched bins are reported in Table 6.
Table 6.
Enrichment of MapMan Functional Bins for Genes with the Highest Loadings on Expression PC1 in the Winter Accessions
| Winter Expression PC1 (Significantly Correlated with Carbon Isotope Ratio) | ||||||
| MapMan Bin | P Value | pFDR P Value | Bin Genes in PC1 Tail | Bin Genes in Test | Total Genes in Test | |
| DNA, synthesis/chromatin structure, histone | 1.27E-09 | 1.29E-06 | 15 | 45 | 20,899 | |
| AT1G09200 | Histone superfamily protein | AT3G54560 | HTA11; histone H2A 11 | |||
| AT1G52740 | HTA9; histone H2A protein 9 | AT5G10390 | Histone superfamily protein | |||
| AT2G28740 | HIS4; histone H4 | AT5G22880 | H2B; histone B2 | |||
| AT2G37470 | Histone superfamily protein | AT5G54640 | HTA1; histone superfamily protein | |||
| AT2G38810 | HTA8; histone H2A 8 | AT5G59870 | HTA6; histone H2A 6 | |||
| AT3G20670 | HTA13; histone H2A 13 | AT5G59910 | HTB4; histone superfamily protein | |||
| AT3G45980 | H2B; Histone superfamily protein | AT5G65360 | Histone superfamily protein | |||
| AT3G46030 | HTB11; histone superfamily protein | |||||
| Cell wall proteins, AGPs | 1.83E-06 | 0.0009 | 11 | 40 | 20,899 | |
| AT1G55330 | AGP21; arabinogalactan protein 21 | AT4G37450 | AGP18; arabinogalactan protein 18 | |||
| AT2G14890 | AGP9; arabinogalactan protein 9 | AT5G40730 | AGP24; arabinogalactan protein 24 | |||
| AT2G47930 | AGP26; arabinogalactan protein 26 | AT5G53250 | AGP22; arabinogalactan protein 18 | |||
| AT3G06360 | AGP27; arabinogalactan protein 27 | AT5G55730 | FLA1; FASCICLIN-like arabinogalactan 1 | |||
| AT3G13520 | AGP12; arabinogalactan protein 12 | AT5G56540 | AGP14; arabinogalactan protein 14 | |||
| AT3G60900 | FLA10; FASCICLIN-like arabinogalactan-protein 10 | |||||
| CCAAT-box binding factor | 7.87E-05 | 0.0266 | 8 | 31 | 20,899 | |
| AT1G30500 | NF-YA7; nuclear factor Y, subunit A7 | AT3G12480 | NF-YC11; nuclear factor Y, subunit C11 | |||
| AT1G54160 | NF-YA5; nuclear factor Y, subunit A5 | AT3G14020 | NF-YA6; nuclear factor Y, subunit A6 | |||
| AT1G72830 | HAP2C; nuclear factor Y, subunit A3 | AT3G20910 | NF-YA9; nuclear factor Y, subunit A9 | |||
| AT3G05690 | UNE8; nuclear factor Y, subunit A2 | AT5G06510 | NF-YA10; nuclear factor Y, subunit A10 | |||
| Major intrinsic proteins, TIP | 0.00009 | 0.02772 | 5 | 11 | 20,899 | |
| AT1G52180 | Aquaporin-like superfamily protein | |||||
| AT2G25810 | TIP4; tonoplast intrinsic protein 4;1 | |||||
| AT2G36830 | GAMMA-TIP; γ-tonoplast intrinsic protein | |||||
| AT3G16240 | DELTA-TIP; δ-tonoplast integral protein | |||||
| AT3G47440 | TIP5; tonoplast intrinsic protein 5;1 | |||||
Winter expression PC2 (significantly correlated with shoot dry weight). No bins were significant at pFDR = 0.05.
Table 5.
Enrichment of MapMan Functional Bins for Genes with the Highest Loadings on Expression PC1 and 2 in the Spring Accessions
| Spring Expression PC1 (Significantly Correlated with Leaf N Content) | ||||||
| MapMan Bin | P Value | pFDR P Value | Bin Genes in PC1 Tail | Bin Genes in Test | Total Genes in Test | |
| DNA synthesis/chromatin structure, histone | 7.754E-6 | 0.0003 | 11 | 45 | 20,500 | |
| AT1G06760 | winged-helix transcription factor protein | AT4G27230 | HTA2; histone H2A 2 | |||
| AT1G07790 | HTB1; histone superfamily protein | AT4G40030 | Histone superfamily protein | |||
| AT1G08880 | HTA5; histone superfamily protein | AT4G40040 | Histone superfamily protein | |||
| AT1G54690 | HTA3; γ-histone variant H2AX | AT5G54640 | HTA1; histone superfamily protein | |||
| AT2G28720 | Histone superfamily protein | AT5G59910 | HTB4; histone superfamily protein | |||
| AT2G28740 | HIS4; histone H4 | |||||
| Signaling, light | 3.863E-5 | 0.0131 | 14 | 84 | 20,500 | |
| AT1G04400 | CRY2 | AT4G15090 | FAR1; FRS (FAR1 Related Sequences) transcription factor | |||
| AT1G30440 | Phototropic-responsive NPH3 family protein | AT4G18130 | PHYE; phytochrome E | |||
| AT1G76320 | FRS4; FAR1-related sequence 4 | AT4G31820 | ENP; phototropic-responsive NPH3 family protein | |||
| AT1G80010 | FRS8; FAR1-related sequence 8 | AT4G37590 | NPY5; phototropic-responsive NPH3 family protein | |||
| AT2G18790 | PHYB; phytochrome B | AT5G18960 | FRS12; FAR1-related sequence 12 | |||
| AT2G46340 | SPA1; SPA (suppressor of phyA-105) protein family | AT5G35840 | PHYC; phytochrome C | |||
| AT4G08920 | CRY1 | AT5G47800 | Phototropic-responsive NPH3 family protein | |||
| Spring Expression PC2 (Significantly Correlated with SLA) | ||||||
| MapMan Bin | P Value | pFDR P Value | Bin Genes in PC2 Tail | Bin Genes in Test | Total Genes in Test | |
| Photosystem I polypeptide subunits | 8.528E-7 | 0.0004 | 7 | 13 | 20,500 | |
| AT2G46820 | PTAC8; photosystem I P subunit | AT1G52230 | PSAH2; photosystem I subunit H2 | |||
| AT3G16140 | PSAH-1; photosystem I subunit H-1 | AT2G20260 | PSAE-2; photosystem I subunit E-2 | |||
| AT4G12800 | PSAL; photosystem I subunit l | AT5G64040 | PSAN; photosystem I reaction center subunit PSI-N, chloroplast, putative | |||
| AT4G28750 | PSAE-1; photosystem I reaction center subunit IV/PsaE protein | |||||
| Photosystem II polypeptide subunits | 8.970E-7 | 0.0004 | 10 | 30 | 20,500 | |
| AT1G14150 | PQL1; PsbQ-like 2 | AT3G50820 | PSBO2; photosystem II subunit O-2 | |||
| AT1G67740 | PSBY; photosystem II BY | AT4G05180 | PSBQ-2; photosystem II subunit Q-2 | |||
| AT1G76450 | Photosystem II reaction center PsbP family protein | AT4G15510 | Photosystem II reaction center PsbP family protein | |||
| AT2G30570 | PSBW; photosystem II reaction center W | AT4G21280 | PSBQ-A; photosystem II subunit QA | |||
| AT2G39470 | PPL2; PsbP-like protein 2 | AT5G02120 | OHP; one-helix protein | |||
| Protein degradation, autophagy | 1.456E-6 | 0.0005 | 8 | 19 | 20,500 | |
| AT1G54210 | APG12; ubiquitin-like superfamily protein | AT3G15580 | ATG8I; ubiquitin-like superfamily protein | |||
| AT1G62040 | ATG8C; ubiquitin-like superfamily protein | AT4G04620 | ATG8B; ubiquitin-like superfamily protein | |||
| AT2G45170 | ATATG8E; AUTOPHAGY 8E | AT4G16520 | ATG8F; ubiquitin-like superfamily protein | |||
| AT3G06420 | ATG8H; ubiquitin-like superfamily protein | AT4G21980 | APG8A; ubiquitin-like superfamily protein | |||
| Redox, thioredoxin | 7.30E-4 | 0.0145 | 12 | 66 | 20,500 | |
| AT1G03680 | ATHM1; thioredoxin M-type 1 | AT2G15570 | ATHM3; thioredoxin superfamily protein | |||
| AT1G11530 | ATCXXS; C-terminal Cys residue changed to Ser 1 | AT3G02730 | TRXF1; thioredoxin F-type 1 | |||
| AT1G43560 | Aty2; thioredoxin Y2 | AT3G15360 | ATHM4; thioredoxin M-type 4 | |||
| AT1G50320 | ATHX; thioredoxin X | AT5G16400 | TRXF2; thioredoxin F2 | |||
| AT5G08410 | FTRA2; ferredoxin/thioredoxin reductase subunit A (variable subunit) 2 | |||||
| AT5G23440 | FTRA1; ferredoxin/thioredoxin reductase subunit A (variable subunit) 1 | |||||
| AT1G76080 | ATCDSP32; chloroplastic drought-induced stress protein of 32 kD | |||||
| AT2G04700 | ferredoxin thioredoxin reductase catalytic beta chain protein | |||||
DISCUSSION
Our results are consistent with prior studies showing that the plant drought stress response is highly complex, involving many levels of biological organization, including changes in developmental, cellular, and physiological processes. Our study extends prior work by documenting natural variation in both physiology and gene expression responses to moderate soil drying within a single widely distributed species. Moreover, by collecting transcriptome and physiological measurements in a common experiment, we were able to identify statistical associations between whole-plant traits and the molecular mechanisms that may drive variation in these traits. We demonstrate that Arabidopsis, as a species, is characterized by considerable variation in the number of genes, the magnitude and direction of their response, and their functions, which are deployed in response to drought stress.
Global Response
We found strong correlations between Arabidopsis physiological and whole-genome measures of expression. Gene-level expression data, in particular, are inherently “noisy,” as they include experimental, environmental, and genetic variance. However, these components can be adequately separated with the experimental design and statistical analyses we employed. PC analysis allowed us to reduce many axes of expression variation into a small number of variables, revealing that >70% of the variance in expression response among the 10 Spring accessions in >20,000 genes was explained by just five PCs; only four PCs were needed to explain >70% of the variance in the Winter accessions. This dimensionality reduction allowed us to identify a strong correlation between major axes of expression variance and the major axes of physiology variance. While this connection should, perhaps, be expected, we are unaware of any prior attempts to link natural variation in transcriptome environmental response with natural variation in physiological response in plants.
More detailed analyses of these PCs allowed us to identify a strong, statistically significant, correlation between the first major axes of expression variance with variance in leaf N% in the Spring accessions and variance in leaf δ13C, an integrated proxy for apparent water use efficiency, in the Winter accessions. These correlations suggest a role for variation in nutrient allocation in differences among Arabidopsis accessions and indicate the transcriptional basis of this organism-level trait. We also find that variance in SLA was significantly correlated with expression PC2 in the Spring accessions, suggesting that expression variance along that axis correlates with variance in developmental response to stress. Our finding of statistical correlations between expression PCs and physiological traits suggests that variation in identifiable molecular components may drive variation in ecologically important functional traits.
Photosynthesis and Plant Carbon Status
Our results show that Arabidopsis whole-plant carbon status increased slightly under moderate soil water deficit over a short time scale. Several lines of evidence support this hypothesis. First, we found that total biomass was higher in the soil moisture deficit treatment in nine of 10 Spring accessions and five of seven Winter accessions. In most accessions, this change was driven primarily by increased root biomass in the water-limited plants relative to the control plants (Figure 2A), though several Spring accessions also added biomass in their aboveground tissues (see Supplemental Figure 2C online). Second, functional analysis of transcriptional activity shows that many photosynthesis-related transcripts were significantly upregulated in nearly all of the accessions studied here (Figure 4). We found overrepresented induced groups of transcripts involved in photosystem protein and pigment synthesis as well as protein regulation and transport to the chloroplast. Third, we found an increase in leaf N% and increased SLA (Figures 2B and 2C), perhaps reflecting the increased investment in photosynthetic machinery. Strong correlations between Spring expression PC2 (enriched for many genes involved in photosynthesis) and SLA and Winter expression PC1 and δ13C suggest candidates for the genetic basis of physiological variation in plant carbon status. Finally, the many upregulated major carbohydrate metabolism transcripts (Figure 4) may be a signature of increased photosynthesis.
The prevalence and direction of altered growth during the dehydration stress response remains an unresolved issue in the literature (Pinheiro and Chaves, 2011). Our finding that sublethal soil drying led to increased photosynthetic activity contradicts the traditional view that carbon rapidly becomes limiting during drought stress. According to the traditional view, carbon limitation arises from the combined effects of restriction of photosynthesis due to stomatal closure and the reallocation of fixed carbon to nonphotosynthetic organs (e.g., roots) and to soluble sugars as osmoprotectants (Taiz and Zeiger, 2010). However, recent work by Hummel et al. (2010) on the Col accession using a stress treatment very similar to ours (though with lower light level) found enzymatic evidence for an increase in plant carbon status during mild soil water deficit. Moreover, they found increased allocation to root growth and only a small contribution of Suc to osmotic adjustment. We also identified an increase in RWR in response to soil drying in 16 of 17 accessions (Figure 2A). In many cases, the increased RWR was because of greater absolute root growth with smaller shoots. Increased photosynthetic capacity per unit leaf dry weight is a likely result of the changes we observed in leaf N%, SLA, and leaf WC at the physiological level as well as the consistent changes observed in photosynthesis-related gene expression. Taken together, the increase in photosynthetic activity and increase in root growth represents a drought escape strategy (see below), wherein plants increase water extraction capacity and carbon gain relative to carbon investment in leaves. This is consistent with increased plant carbon status under nonlethal soil drying and perhaps is important for accelerating the transition from vegetative growth to the reproductive phase in an effort to produce seed before soil dehydration becomes fatal. Further research is needed on the molecular and developmental aspects of this stress-induced transition in crops and model systems.
Our experimental design allowed us to discover transcripts that responded to drought. For example, the PPR proteins constitute a large (~440 predicted genes in the Col-0 genome) family of poorly understood genes. We found that these genes were generally upregulated in both Spring and Winter accessions, with nearly all Winter accessions showing enriched upregulation of the PPR MapMan bin. Several PPR genes were also among the transcripts with high loadings on expression PCs. Past work has suggested that most PPRs are constitutively expressed at a low level and that they may play a role in posttranscriptional control of mitochondrial and chloroplast RNAs (Lurin et al., 2004). In maize (Zea mays) and Arabidopsis, PPRs are essential components of RNA editing in chloroplasts where complex transcriptional units often require considerable posttranscriptional modification (Meierhoff et al., 2003; Kotera et al., 2005; Okuda et al., 2006). Our data suggest that these proteins also play a role in dehydration stress response, perhaps functioning in the apparent expansion of photosynthetic activity that we observed.
The Genetic Basis of Variation in Drought Escape
The enrichment of Spring expression PC1 for genes involved in light signaling and histones and the correlation between expression PC1 and the raw number of responsive genes may indicate that this PC describes major sources of variation in genome-wide transcription and signaling processes. Recent studies have suggested a role for chromatin remodeling at specific genetic loci in response to osmotic stress (Sokol et al., 2007; Kim et al., 2008). Remodeling also occurs at a genome scale in response to light signals and temperature changes (Kumar and Wigge, 2010; Tittel-Elmer et al., 2010; van Zanten et al., 2010) and during the transition to flowering (Tessadori et al., 2007). This final observation may suggest a mechanism for our observed correlation between light signaling, histones, and drought stress response. Cryptochrome2 (CRY2), which was among the genes in the tail of expression PC1 and was upregulated in all of our Spring accessions except Sq-8, is hypothesized to play a role in the reorganization of chromatin prior to the transition to flowering (Tessadori et al., 2007). Interestingly, the CRY2 gene and several others in the enriched light signaling bin have known osmotic stress–responsive motifs in their promoter regions, including the drought-responsive element (Yamaguchi-Shinozaki and Shinozaki, 1994), and a MYB binding motif (Abe et al., 1997, 2003). Spring expression PC1 may then, in part, describe variation in a drought escape strategy via early flowering and thereby reveal crosstalk between drought stress signaling and the flowering time pathway.
Comparison to Past Studies: How Representative Is the Col Genotype?
In addition to including a diverse panel of Arabidopsis accessions, our study differs from prior transcriptome analyses in two important ways that are worth considering. First, our stress treatment was quite gradual, consisting of a controlled ~10% reduction in soil water remaining each day over 7 d. Four other widely cited transcriptome analyses used much more rapid and severe drought treatments. Kreps et al. (2002) subjected liquid cultured plants to mannitol treatment and Seki et al. (2002) removed agar-grown seedlings to dry filter paper. Both of these approaches were used by Kilian et al. (2007), who applied mannitol as an osmotic stress and used benchtop drying as drought stress. Huang et al. (2008) withheld water from soil-grown plants until soil WC was ~5%. Of these, only the Kilian et al. study used the ATH1 array reported herein. It is worth noting that the results of the Kilian et al. study are the source for many of the Gene Ontology (GO) “response to water deprivation” and “response to osmotic stress” terms reported in The Arabidopsis Information Resource (TAIR) and other online resources. Because gene annotations in Arabidopsis are often the basis for annotation of other plant genomes, the results of one experiment can quickly propagate to many experimental systems.
A second major difference between our study and earlier studies is the criterion used to identify treatment responsive genes. Three of these prior studies applied a fold-change threshold for determining significance, reporting only genes with greater than 2 to 3 times induction or 0.5 to 3 times repression. To identify accession and accession by treatment interaction effects on gene expression and to avoid arbitrarily excluding genes that may have important functional effects despite low fold changes of expression, we relied on a statistical criterion to generate our lists of differentially expressed genes. Huang et al. (2008) also used a statistical approach and identified a raw number of drought-responsive genes in Col (1969 genes) that is comparable to those identified in Col-2 in our study (1689 genes).
Most published studies concerning Arabidopsis functional biology rely on the Col accession. Bray (2004) compared the Col drought-responsive gene lists of Kawaguchi et al. (2004), Kreps et al. (2002), and Seki et al. (2002) and found that these lists shared 30 genes in common (27 induced and three repressed). We found that 21 of these genes had a significant response in the Spring accession experiment and only seven were significant in the Winter accession experiment. This comparison suggests that studies in the Col accession may be somewhat representative of other Spring accessions, but reliance on Col alone will certainly miss many stress-responsive genes in Spring accessions and many more responsive genes in Winter accessions. It must be noted, however, that our Winter accessions were not subjected to cold treatment; therefore, some observed differences between Col and the Winter accessions may arise from the latter being in an extended vegetative phase. That transcriptional stress response in Col is not a strong predictor of the response of other accessions has been noted previously with respect to cold stress response (Hannah et al., 2006) and exogenous treatment with auxin (Delker et al., 2010) and salicylic acid (van Leeuwen et al., 2007).
Like many model systems, the Col genotype has been propagated for many decades as a laboratory strain. The precise collection locale for Col is not known nor is its breeding history prior to establishment as a laboratory standard. How representative of other accessions is the drought stress response in Col? Our results suggest that, qualitatively, the Col-2 genotype used here is quite similar to other Spring accessions in that its transcriptome shows similar enrichment of functional categories and deploys the conserved responses discussed above (Figure 4). However, Col-2 is among the most conservative in terms of the raw number of genes that responded to the soil moisture deficit treatment we used (Figure 3). Interestingly, we found that two signaling genes, ARR4 (At1g10470) and ARR7 (At1g19050), showed significant positive expression responses to soil moisture deficit in eight Spring accessions but not in Col-2 or in Ler. Both ARR4 and ARR7 were upregulated in Col-2, but neither significantly so at the pFDR of 0.05; these genes were nonresponsive to our imposed stress in Ler-1. Neither of these genes has GO annotations associated with drought or osmotic stress (although GO annotations for drought and osmotic stress were derived from experiments in Col; see above). ARR proteins comprise a small family that participates in two component signaling cascades via interaction with members of the Arabidopsis His kinase (AHK) receptors. AHK2 and AHK3 are believed to be negative regulators of osmotic stress and both ARR4 and ARR7 are downregulated in AHK2/AHK3 double mutants (Tran et al., 2007). AHK proteins are also involved in cytokinin signal transduction and thereby play a role in regulating development of both shoot and root growth (Nishimura et al., 2004). To our knowledge, ARR proteins have not been shown to directly interact with AHK1, which is hypothesized to be an osmotic sensor in Arabidopsis (Urao et al., 1999).
Diversity of Transcriptional Response
We observed a greater than twofold difference among accessions in the number of genes induced by the soil moisture deficit treatment (Figure 3A). Variation in the raw numbers of genes induced by our treatment could be due proximately to two types of genetic architecture. First, the transcriptional differences may be caused by polymorphisms in upstream regulators, which would change the topology of response networks or activate different downstream targets. This might take the form of a transcription factor exhibiting quantitatively different levels of DNA binding activity, thereby promoting the transcription of more (or fewer) targets, or of structural variation in the transcription factor that affects its own activation (e.g., via polymorphism in transcriptional [cis-regulatory] or protein [phosphorylation, etc.] activation domains). A small number of such changes could have large effects on the number of downstream genes that show transcriptional response. Alternatively, variation in the total number of genes activated/deactivated in a given accession may be due to variation in the target genes themselves. In this sense, “target genes” comprise those genes encoding enzymes, chaperonins, or structural proteins that affect the stress response. Variation of this sort could take the form of segregating polymorphisms in the promoter regions of target genes or of diversification of cis-regulatory variation associated with expansion or contraction of gene families via gene duplication followed by regulatory subfunctionalization (Force et al., 1999; Kliebenstein, 2010).
The Spring accession Sq-8 may be an interesting case study in this regard. Sq-8 was characterized by the highest number of stress-responsive genes in our study but exhibited one of the lowest values for statistically significant MapMan functional bins. Furthermore, it was the only accession that had significant upregulation of the biotic stress bin, the WRKY transcription factor bin, and the mitogen-activated protein kinase signaling bin. The downstream targets of the signaling proteins in these bins are largely unknown, but even a small number of such signaling genes uniquely expressed in response to stress in the Sq-8 accession might well explain the larger number genes that responded to the soil drying treatment in this accession. Three of the 10 most strongly induced genes in Sq-8 encode JAZ proteins (JAZ7, At2g34600; JAZ8, At1g30135; and JAZ10, At5g13220). JAZ proteins negatively regulate jasmonic acid signaling by binding the promoters of jasmonic acid–responsive genes (Chini et al., 2007), yet expression of JAZ genes is positively affected by jasmonic acid signaling via the MYC2 transcription factor (Chini et al., 2007; Pauwels et al., 2008) which is, itself, also ABA responsive (Abe et al., 2003). Therefore, differential expression of JAZ proteins in Sq-8 could potentially have affected the expression of hundreds of genes in response to the soil moisture deficit treatment. Physiologically Sq-8 was extreme in a number of ways, allowing a large decrease in leaf RWC with little change in leaf ψtot and almost no change in ψp (Figure 1) and exhibiting the greatest increase in leaf N% and SLA (Figures 2B and 2C).
Our data do not support the hypothesis that gene copy number differences cause spurious detection of transcript differences in this study because our probe filtering likely removed most segregating gene duplicates from the expression analyses. However, subfunctionalization of expression among gene copies fixed within Arabidopsis is a possible explanation for variation in treatment effects among accessions. The promoter enrichment analyses (Table 4) suggested that various forms of the ABRE motif played an important role in the observed soil moisture deficit responses. ABREs promote transcription through interaction with the ABRE binding factor (AREB/ABF) proteins, which constitute a small, functionally diverse gene family that are variously responsive to osmotic- and dehydration stress–mediated ABA signaling (Fujita et al., 2005; Yoshida et al., 2010). However, ABRE motifs cannot recruit AREBs without the close proximity of a second, distinct, coupling element (CE) (Narusaka et al., 2003). Novel drought-responsive promoter activity might arise if a point mutation creates a CE, thereby activating an otherwise dormant ABRE, or silences an ABRE by eliminating a CE. Therefore, de novo creation or activation of ABREs may constitute a large pool of drought-responsive target genes. Gene copy number variation may still be driving large transcriptional differences if, for example, there are deletions or duplications of master regulatory genes segregating in Arabidopsis. These genes themselves would likely have been removed from the analysis by our probe filtering, but their downstream effects could have a significant impact on the transcriptional differences that we observed.
In our analysis of genes that showed a significant accession by treatment effect, which suggest genotype-dependent response to environmental stimulus, we found that the DUF26 (also known as CRK) MapMan bin was highly enriched (see Supplemental Table 1 online). DUF26/CRK genes encode transmembrane receptors that, as a group, are transcriptionally responsive to diverse environmental stresses (Lehti-Shiu et al., 2009; Wrzaczek et al., 2010). Although the cellular functions of most DUF26 domain proteins remain uncertain, genes from this group have been shown previously to exhibit a high frequency of major-effect coding changes, such as premature stop codons, altered initiation codon position, and polymorphism in splice acceptor/donor sites (Clark et al., 2007). Many DUF26 genes also show the signature of directional (positive) selection, particularly in the extracellular DUF26 domain (Strain and Muse, 2005). The variation in treatment response that we observe between accessions suggests that these proteins may be involved in environment-specific adaptation.
Limitations
Our comparison of diverse natural accessions under natural stress conditions allowed us to identify many transcriptional responses and molecular pathways that are components of Arabidopsis drought stress response. We were limited, however, by two important factors. First, the ATH1 array does not include probes for nearly 4000 genes annotated in the most recent (TAIR10) release of the Arabidopsis genome nor can the ATH1 array identify small RNA species that almost certainly play a role in stress responses. We had to exclude an additional ~1000 genes from our analyses due to sequence polymorphism in probe sites. A second limitation of our approach is that we were only able to sample a single time point and a single tissue type during stress acclimation. Some of the genetic variation that we see may therefore represent variation in the level of soil drying that elicits a response in a particular accession or in the speed with which a response is deployed once soil drying has been detected by a particular accession (e.g., one accession may be barely responding to a 60% reduction of soil WC, while a second accession may have begun to acclimate at 50% reduction). Both of these limiting factors suggest that our analysis underestimates the magnitude of response to soil drying and that the precise dynamics of when and under what conditions these accessions deploy a response remain to be determined.
Evolution and Physiological Genomics
From an evolutionary perspective, the variation we observed may be significant for at least two reasons. First, because the accessions in this study come from a wide range of habitats throughout Europe, the constitutive and inducible transcriptional and physiological differences between accessions may be the signature of locally adapted genotypes. Constitutive differences (transcript traits with a significant accession term) likely represent slight variations in the molecular mechanisms used by the genotypes to grow normally in a benign environment or may reflect varying deviations between the genotypes’ natural environments and the environment used here (potting media and artificial light). Second, the variation in inducible responses (accession treatment interactions) represents the source material for the evolution of adaptive plastic responses. Fine-grained adaptation to multiple environments through plastic responses requires natural variation in the degree of response to the different environments (Via, 1987). Our results suggest that Arabidopsis varies in the degree of response for both physiology and transcript traits that could facilitate the evolution of adaptive drought responses
METHODS
Selection of Lines
We selected 17 natural accessions of Arabidopsis thaliana to maximize the ecogeographic and physiological diversity represented in our experiment (Table 1). Ten of these lines exhibit Spring-annual life history, as determined by their propensity to flower in the greenhouse in the absence of cold treatment, and seven exhibit a Winter-annual life history, which flower rapidly only after vernalization (see Supplemental Table 3 online). Climate diagrams depicting seasonal patterns of rainfall and temperature from the locations of origin of the seventeen accessions are given in Supplemental Figure 1 online.
Plant Growth and Soil Moisture Deficit Treatment
Plants were grown in 6 × 6 × 5-cm plastic pots filled with ~150 mL Profile porous ceramic rooting media (Profile Products). Because it was not possible to harvest enough replicates of both Spring and Winter accessions simultaneously, we performed separate experiments for each life history type, with germination staggered by 5 d. Prior to planting, the dry weight of each pot and soil (DWpotandsoil) was measured, and the media were saturated with a complete nutrient solution (Epstein and Bloom, 2005) for several days. Saturated pots were covered and allowed to drain by gravity for >24 h until a constant field capacity weight (FCpotandsoil) was obtained for each pot. Seeds were planted in each pot and dark stratified at 4°C for 6 d. The plants were then moved to a controlled environment chamber with a 12-h photoperiod of 330 μmol m−2 s−1 PPFD with daytime/nighttime temperatures of 23/20°C with 40/50% relative humidity. After germination, plants were thinned to one per pot. During growth, all plants were watered with distilled water every other day and fertilized with the nutrient solution weekly. Watering and fertilization were done by bottom watering for 1 h, followed by free drainage of the pots. Within each experiment (Winter and Spring life history types), there were eight to 12 replicate plants within each treatment (control and soil moisture deficit) randomly located in the controlled environment chamber.
The two life history types of plants have inherently different growth rates; thus, plants were 20 (Spring) or 30 (Winter) d old when the soil moisture deficit treatment began; at that time, the plants had rosettes just large enough for sampling for all physiological and gene expression parameters and were therefore at very similar developmental rosette stage (see below). During the treatment period, bottom watering ceased, each pot (both control and treatment) was weighed daily at predawn (WTpotandsoil), and distilled water was added by pipette at the base of the rosette to bring the percent field capacity water remaining (WatRem%) in each pot to the target level for that day until a final drying treatment level of ~40% was reached by all pots. WatRem% was calculated as 100*(WTpotandsoil × DWpotandsoil)/(FCpotandsoil − DWpotandsoil). Wet (control) pots were treated identically to soil moisture deficit treatment pots, except WatRem%, which was maintained near 96% of field capacity WC for the entire dry-down period, at 96% field capacity, Profile media is ~20% air space. For these experiments, the daily sequence of target soil moisture deficit levels was 80, 70, 60, 60, 50, 40, and 40%. This procedure assured that all accessions experienced daily the same level of soil moisture deficit despite differences in transpiration due to different leaf areas or stomatal opening. All accessions had 7 d of drying with the final level of soil moisture deficit held constant during the last 2 d of the dry-down. Also, during the last 3 d of dry-down, each pot was weighed and WatRem% adjusted to target level twice a day, at predawn and dusk, to ensure a nearly constant level of nonlethal, moderate water stress. Preliminary experiments found that when WatRem% was ≤34%, the plants wilted and did not recover upon rewatering. In addition to assuring equal soil moisture deficit treatments among all accessions, a major goal of this slow dry-down treatment was to allow the intact, growing plants to acclimate to a soil moisture deficit that developed slowly in relation to plant growth and development, mimicking typical patterns of soil moisture stress in natural conditions. At dusk, prior to the harvest day, plants were enclosed in covered trays to minimize evapotranspiration. Covered trays were transported to the lab the following morning, final pot and soil weights were taken, WatRem% at harvest calculated, and plants were harvested. Harvests occurred during the first several hours of the normal daily light period.
Harvests and Physiological Measurements
For each experiment, plants of all lines and both treatments in each block were harvested together. Rosettes were excised and fresh weight (FW) of rosette and any bolt stem were determined separately. Immediately, ~80 mg (~40% of the leaves on the rosette) of healthy, fully expanded leaves were detached from the rosette and placed in RNAlater (Ambion). For some plants, several additional leaves were removed and within 30 s of excision placed in a psychrometer chamber for water potential measurement (see below), while other plants had two to three leaves removed for WC and RWC measurements. For WC and RWC measurements, leaf FW was immediately determined on a microbalance, leaf bases were placed in distilled water in an open microfuge vial, and the vial and leaves were enclosed in a larger sealed vial to allow leaves to reach full hydration in the dark for ~18 h. Turgid weight (TW) of the hydrated leaves was then determined, and leaves were dried and reweighed (DW) using the same microbalance. Leaf WC was calculated as 100*(FW − DW)/DW) and leaf RWC as 100*(FW − DW)/(TW − DW) (Boyer, 1995). Remaining leaves of all plants were weighed fresh and after drying so total rosette and leaf weight, both fresh and dry, could be calculated by summing values from material used for each type of measurement. After rosette harvests, roots of a subset of plants were washed free of media, dried, and weighed for dry biomass. After drying, leaves were assayed for C and N concentration and C stable isotope composition at the University of California Davis Stable Isotope Facility. Carbon isotope composition (δ13C) is given relative to the PeeDee Belemnite standard, and composition is used rather than discrimination (Δ) because the isotopic composition of carbon dioxide in the ambient air, which is required to compute Δ, was extremely variable (McKay et al., 2003).
Leaf water potential (Ψtot) was measured using the excised leaves, individually calibrated (Brown and Bartos, 1982) thermocouple psychrometers (Merrill Specialty Equipment), and stainless steel chambers (Donovan et al., 2001, 2003). At harvest, several entire leaves were placed in chambers, and the chambers were sealed and suspended in a water bath to minimize temperature gradients during measurement. Mature leaves, similar to the leaves used for RNA extraction, were used to minimize any growth effects on leaf Ψtot (Boyer, 1995). Psychrometer outputs were logged hourly (CR7x data logger; Campbell Scientific), and leaf Ψtot was determined after equilibration (~24 h). After Ψtot was measured, the sealed chambers were immersed in liquid N to rupture cell membranes of the leaves and an estimate of bulk leaf osmotic potential (Ψsol) was determined after a second 24-h equilibration. For each plant, leaf turgor (Ψp) was calculated as Ψtot − Ψsol. Similar to leaf measurements, soil water potential (Ψsoil) was determined on a sample of soil, with roots, taken from the center of a subset of the pots and placed in stainless steel chambers with thermocouple psychrometers. All water potentials were calculated from psychrometer μV outputs using the Brown and Bartos (1982) model that accounts for zero offset (always between 0.2 and −0.2 μV) and temperature.
Genomic Hybridization to Arrays
Probes on the ATH1 array were designed from the Col-0 reference genome sequence. Comparative genomic hybridizations were completed following the protocols outlined by Borevitz (2005) and using the Invitrogen BioPrime labeling system as described by Juenger et al. (2010). Genomic hybridizations to the Affymetrix ATH1 array were completed for three samples of each accession for a total of 51 arrays.
Gene Expression Microarrays
We used a subset of replicate plants from the dry-down experiment in our studies of soil water deficit–regulated gene expression. We used a fully factorial experiment involving 17 accessions, two treatments (control and soil drying), with three biological replicates (17 × 2 × 3 = 102 arrays) from two life histories, Winter accessions and Spring accessions. Fully expanded rosette leaves were sampled on RNAlater (Ambion), and total RNA was extracted using Qiagen RNAeasy kits. Samples for mRNA profiling studies were processed by Asuragen according to the company’s standard operating procedures for the Affymetrix ATH1 array. The integrity of total RNA was qualified by Agilent Bioanalyzer 2100 capillary electrophoresis and prepared as described by Juenger et al. (2010). The arrays were scanned on an Affymetrix GeneChip Scanner 3000. A summary of the image signal data for every gene interrogated on the array was generated using the Affymetrix Statistical Algorithm MAS 5.0 (GCOS v1.3) algorithm.
Analyses of Treatment and Whole-Plant and Physiological Parameters
Two-way (accession and treatment) factorial ANOVA was used for analysis of each soil and plant response variable in the Winter and Spring experiments separately. When needed to meet assumptions of normality and homoscedasticity variables were ln transformed. For these univariate analyses for each experiment, the number of replicate plants used varied by parameter: 8 to 12 for WatRem%, soil gravimetric WC, and soil water potential; 6 to 8 for leaf N, C, and stable isotope analyses and for all leaf water potential measurements; 4 for leaf WC, RWC, and SLA; and 3 for root, total plant, and RWR. All results presented are least-square means ± 1 se. The values and standard errors were back-transformed for presentation. Statistical analyses of these data were conducted with JMP 9.0.0 (SAS Institute). Correlation analyses were conducted with SigmaPlot 11.0 (Systat Software).
Genomic Hybridization Analysis
Statistical analyses were performed using SAS procedures as called by JMP Genomics. Original .CEL files from genomic DNA hybridizations were imported and processed at the probe level using a RMA based background correction, log2 transformation, and quantile normalization of raw intensity values (Irizarry et al., 2003). We used a custom CDF file (ATH1_AT_TAIR.cdf) constructed from the TAIR version 10 of the Arabidopsis genome (available for download at http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/CDF_download.asp) (Dai et al., 2005). This custom CDF file was created using a series of searches to identify unique probes and filter probes with cross-hybridization to multiple genomic sites. We confirmed sample labeling by performing a hierarchical clustering of intensity data across all arrays. As expected, replicates of each accession consistently clustered.
One-way ANOVA models were subsequently fit for each probe with an ANOVA in JMP Genomics 5.0. An empirical Bayes approach was used to shrink the residual variance for each probe based on a prior distribution of the variance estimated from all probes using an inverted-gamma distribution. This approach resulted in increased power and sensitivity by improving the stability of the residual variance estimates. We controlled for multiple testing using a pFDR of 0.0001 using the Q value program in R (Storey and Tibshirani, 2003). This filtering resulted in the removal of 31,075 probes (of ~220,000 probes on the ATH1 array) from the Spring accession analysis and 17,256 probes from the Winter accession analysis. Because we restricted subsequent gene expression analyses to genes with at least three probes, this probe filtering resulted in the removal of 700 genes in the Spring data set and 264 genes from the Winter data set (from ~22,500 probe sets on the ATH1 array). Supplemental Data Set 1 online lists the genes excluded from expression analyses.
Gene Expression Analyses
We completed analyses of gene expression data using filtered RMA expression values (Irizarry et al., 2003). CEL files were imported into JMP Genomics, and gene expression measures were generated using the RMA function (background corrected, log2 transformed, quantile normalized, median-polish summary) with the custom CDF file described above. Expression measures were then processed by ANOVA. In this case, we fit a fixed effect general linear model, including a term for “accession,” “treatment,” and their interaction. In addition, we conducted a series of targeted ANOVAs testing for fixed “treatment” effect for each accession singly to further explore the common and unique responses of our sampled plant material to drying soil. In all analyses, we controlled for multiple testing using a pFDR of 0.05 (Storey and Tibshirani, 2003). Statistical analysis of Winter and Spring accessions was performed separately. Complete array results are shown in Supplemental Data Set 2 online.
Quantitative PCR Studies
In addition to our array studies, we used quantitative PCR assays (qPCR) of seven candidate genes (see Supplemental Table 2 online) picked from prior studies (Bray, 2004) to further evaluate gene expression responses. These genes include Response to Dessication20 (RD20) (At2g33380), which has been shown in previous studies to be drought responsive (Seki et al., 2002; Fujita et al., 2005); DDF1 (At1G12610), a member of the drought-responsive DREB transcription factor family; the putative ABA receptor Pyrabactin Resistance1 (At4g17870); and four genes that showed particularly strong responses in our analysis of the ATH1 arrays: a chlorophyll a/b binding protein (At1g19150), a gene involved in ethylene biosynthesis (At1g03400), and the gibberellic acid–responsive GAST1 Protein Homolog1 (GASA1) (At1g75750).
As described above, rosette leaf material of replicate plants was harvested on RNAlater and extracted as described above for each experimental plant. We used ProbeLibrary (Roche Applied Science) for our qPCR experiments with ABgene one-step qPCR reagents and the ABI 7900 HT real-time PCR machine. Samples were screened with duplicate technical replicates, and subsequent analyses were completed on the replicate averages. Relative mRNA abundance was determined on the basis of the threshold cycle (CT) value for each reaction. We used two reference genes (At2G43330 and At1G55850) as endogenous controls to normalize the quantity of input RNA in reactions; these genes were chosen based on screens of our microarray data for consistent and stable expression. The CT value for each target gene was subtracted from the geometric mean CT value for the control genes to obtain DCT values, which were used in subsequent statistical analyses. DCT values were analyzed using fixed-factor ANOVA in JMP (Littel et al., 1996) with block, accession, treatment, and accession × treatment interaction effects. We sequenced the probes and primers for each qPCR assay in all 17 accessions to evaluate the impact of sequence polymorphisms on expression measures and designed probes and primers free of polymorphisms.
The results of these analyses are largely congruent with the results of the array experiments (see Supplemental Table 2 online). All seven genes from the Spring analysis and six of seven genes from the Winter analysis showed a significant treatment effect in the same direction in the qPCR and ATH1 experiments. The two experiments were less congruent for the accession and accession by treatment (AxT) interaction effects. For the Spring analysis, two of seven genes showed consistent accession effects between the experiments and four of seven for the AxT interactions. In the Winter analysis, five of seven genes showed consistent accession effects, and six of seven showed consistent effects for the AxT interaction. We note that for three of the four genes for which we found a discrepancy between the two platforms in identifying AxT interactions, the qPCR assay identified an interaction that the ATH1 assay did not. This may be due to the greater sensitivity of qPCR to detect subtle differences in expression as well as the greater dynamic range offered by this platform. This finding may suggest that the lists of AxT genes we identified using the ATH1 array are conservative.
Annotation Analyses
\We used several annotation-based analyses to explore the biological underpinnings of the observed gene expression differences. We searched for overrepresented promoter motifs in various gene lists using the Web-based software Athena (http://www.bioinformatics2.wsu.edu/cgi-bin/Athena/cgi/home.pl) (O’Connor et al., 2005) at a statistical significance of P < 0.0001. In addition, we used the PageMan module of the MapMan software package (http://gabi.rzpd.de/projects/MapMan) to explore broad functional patterns in our transcript abundance data. We used differences in least-square mean RMA values as data input [log2(fold change values)]. Functional categories of the MapMan annotation (Thimm et al., 2004; Usadel et al., 2006) were tested for significance of expression change by applying a two-sided Wilcoxon rank tests with a Benjamin-Hochberg correction for multiple tests. In this test, the median log2 ratios for all genes in a particular MapMan annotation bin were compared with the median log2 ratios of all other MapMan bins. Because the TAIR10 mapping is not yet available, we used the ATH_AFFY_TAIR9 mapping. Additionally, we tested for statistical overrepresentation of bins with an accession treatment interaction by performing a Fisher’s exact test of each bin using MapMan categories. This analysis was implemented in JMP Genomics 5.0 and was corrected for multiple tests using a pFDR of 0.05 (Storey and Tibshirani, 2003). While the PageMan analysis included expression ratios for all genes in a given bin, regardless of whether a given gene showed a statistically significant expression change in our initial ANOVA, the Fisher’s test asked only whether a given bin has an overrepresentation of genes found to be significant in our initial expression ANOVA.
PC Analysis
We performed principal components analysis of centered means and standardized variances from the complete array data sets and nonredundant physiology data treatment response (dry control; total shoot dry weight, root dry weight, leaf RWC, leaf Ψtot, leaf Ψp, SLA, leaf C%, leaf N%, and leaf δ13C) using the prcomp function in R. In the Spring expression data, the first PC (expPC1) explains 23.6% of the variance, and 71.3% of the variance is explained by the first five PCs. The first PC of the Spring physiology data (physPC1) explains 43.0% of the variance and the first three components explain 79.8% of the variance. Using JMP Pro 9.0, we estimated Pearson’s correlations between these first five expression PCs and first three physiology PCs in the Spring data and then estimated correlations between expPC1 and expPC2 (which showed marginally nonsignificant associations with the physPCs) and individual physiology traits. In the Winter experiment, expPC1 explains 29.4% of the variance, while the first four PCs explain 77.4% of the variance. Winter physPC1explains 29.8% of the variance, and the first three PCs explain 74.6% of the variance. We then asked whether expPC1 (which showed marginally nonsignificant correlations with the physPCs) correlated with any of the individual physiology traits.
We assessed functional enrichment in the expression PCs by performing Fisher’s tests of MapMan categories, as described above for accession treatment interaction genes. The query sets for these analyses were the 5% tails of transcripts with the strongest loading (either positive or negative) on expression PC1 and PC2 from each experiment.
Accession Numbers
The genomic hybridization data from this study are accessible through Gene Expression Omnibus Series accession numbers GSE27549 and GSE27551. The cRNA hybridization data from this study are accessible through Gene Expression Omnibus Series accession numbers GSE27548 and GSE27550.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Climate Diagrams for Sites of Origin of 17 Arabidopsis Accessions Used in the Experiments.
Supplemental Figure 2. Interaction Plots for Effects of Treatments on Soil and Plant Parameters in Spring and Winter Accession Experiments.
Supplemental Figure 3. Complete Results for MapMan Analyses.
Supplemental Table 1. Enrichment of MapMan Functional Bins for Accession × Soil Drying Treatment Interaction Genes.
Supplemental Table 2. Quantitative PCR Results.
Supplemental Table 3. Results from an Unpublished Study Showing Flowering Times for Unvernalized Rosettes from Genotypes Used in This Study.
Supplemental Data Set 1. Genes Removed from the Analysis Due to Polymorphism as Detected by Genomic DNA Hybridization to ATH1 Arrays.
Supplemental Data Set 2. All Genes Showing Significant Effects (A, T, AxT) for Both Life Histories.
Acknowledgments
We thank Robin Hopkins, David Lowry, and Samuel Taylor for helpful comments on the manuscript and Elizabeth Bray for discussion of previously published data on transcriptome responses to drought treatments. Jesse Lasky assembled Supplemental Figure 1 online. This work was supported by National Science Foundation Arabidopsis 2010 funding to T.E.J. (DEB-0618347), J.H.R. (DEB-0618294), and J.K.M. (DEB-0618302). Support of the California Agricultural Experiment Station and the University of Texas Institute for Cellular and Molecular Biology is also acknowledged.
AUTHOR CONTRIBUTIONS
T.E.J., J.K.M., J.H.R., and S.S. designed the research. T.W. and J.H.R. performed the research. D.L.D., T.E.J., and J.H.R. analyzed the data. D.L.D., J.H.R., J.K.M., and T.E.J. wrote the article.
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirrezabal L., Bouchier-Combaud S., Radziejwoski A., Dauzat M., Cookson S.J., Granier C. (2006). Plasticity to soil water deficit in Arabidopsis thaliana: Dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ. 29: 2216–2227 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C., Aarts M.G.M., Bentsink L., Keurentjes J.J.B., Reymond M., Vreugdenhil D., Koornneef M. (2009). What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araus J.L., Slafer G.A., Reynolds M.P., Royo C. (2002). Plant breeding and drought in C3 cereals: What should we breed for? Ann. Bot. (Lond.) 89(Spec No): 925–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi R., Gantt J.S. (1997). A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol. Biol. 34: 629–641 [DOI] [PubMed] [Google Scholar]
- Bartels D., Sunkar R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24: 23–58 [Google Scholar]
- Berrington de Gonzalez A., Cox D.R. (2007). Interpretation of interaction: A review. Ann. Appl. Stat. 1: 371–385 [Google Scholar]
- Bohnert H.J., Nelson D.E., Jensen R.G. (1995). Adaptations to environmental stresses. Plant Cell 7: 1099–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J.O. (2006). Genotyping and mapping with high-density oligonucleotide arrays. In Arabidopsis Protocols, 2nd ed, Salinas J. Sanchez-Serrano J.J., (Totowa, NJ: Humana Press; ), pp. 137–145 [DOI] [PubMed] [Google Scholar]
- Bouchabke O., Chang F., Simon M., Voisin R., Pelletier G., Durand-Tardif M. (2008). Natural variation in Arabidopsis thaliana as a tool for highlighting differential drought reponses. PLoS One 3: e1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J.S. (1995). Measuring the Water Status of Plants and Soils. (San Diego, CA: Academic Press; ). [Google Scholar]
- Bray E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2: 48–54 [Google Scholar]
- Bray E.A. (2004). Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J. Exp. Bot. 55: 2331–2341 [DOI] [PubMed] [Google Scholar]
- Bressan R., Bohnert H., Zhu J.K. (2009). Abiotic stress tolerance: From gene discovery in model organisms to crop improvement. Mol. Plant 2: 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R.W., Bartos D.L. (1982). A calibration model for screen-caged Peltier thermocouple psychrometers. USDA Forest Service Research Paper INT-293
- Chaves M.M., Maroco J.P., Pereira J. (2003). Understanding plant responses to drought–from genes to the whole plant. Funct. Plant Biol. 30: 239–264 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Christman M.A., Richards J.H., McKay J.K., Stahl E.A., Juenger T.E., Donovan L.A. (2008). Genetic variation in Arabidopsis thaliana for night-time leaf conductance. Plant Cell Environ. 31: 1170–1178 [DOI] [PubMed] [Google Scholar]
- Clark R.M., et al. (2007). Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342 [DOI] [PubMed] [Google Scholar]
- Dai M., Wang P., Boyd A.D., Kostov G., Athey B., Jones E.G., Bunney W.E., Myers R.M., Speed T.P., Akil H., Watson S.J., Meng F. (2005). Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C., Pöschl Y., Raschke A., Ullrich K., Ettingshausen S., Hauptmann V., Grosse I., Quint M. (2010). Natural variation of transcriptional auxin response networks in Arabidopsis thaliana. Plant Cell 22: 2184–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais D.L., Juenger T.E. (2010). Pleiotropy, plasticity, and the evolution of plant abiotic stress tolerance. Ann. N. Y. Acad. Sci. 1206: 56–79 [DOI] [PubMed] [Google Scholar]
- Donovan L.A., Linton M.J., Richards J.H. (2001). Predawn plant water potential does not necessarily equilibrate with soil water potential under well-watered conditions. Oecologia 129: 328–335 [DOI] [PubMed] [Google Scholar]
- Donovan L.A., Richards J.H., Linton M.J. (2003). Magnitude and mechanisms of disequilibrium between predawn plant and soil water potentials. Ecology 84: 463–470 [Google Scholar]
- Epstein E., Bloom A.J. (2005). Mineral Nutrition of Plants: Principles and Perspectives, 2nd ed (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M.M., Seki M., Hiratsu K., Ohme-Takagi M., Shinozaki K., Yamaguchi-Shinozaki K. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X., et al. (2011). Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G., Pichersky E., Malik V.S., Timko M.P., Scolnik P.A., Cashmore A.R. (1988). An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 85: 7089–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah M.A., Wiese D., Freund S., Fiehn O., Heyer A.G., Hincha D.K. (2006). Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol. 142: 98–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann N.J., Juenger T.E., Sen S., Stowe K.A., Dawson T.E., Simms E.L. (2005). Quantitative trait loci affecting delta13C and response to differential water availibility in Arabidopsis thaliana. Evolution 59: 81–96 [PubMed] [Google Scholar]
- Hijmans R.J., Cameron S.E., Parra J.L., Jones P.G., Jarvis A. (2005). Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25: 1965–1978 [Google Scholar]
- Hsiao T.C. (1973). Plant responses to water stress. Annu. Rev. Plant Physiol. 24: 519–570 [Google Scholar]
- Huang D., Wu W., Abrams S.R., Cutler A.J. (2008). The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 59: 2991–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel I., Pantin F., Sulpice R., Piques M., Rolland G., Dauzat M., Christophe A., Pervent M., Bouteillé M., Stitt M., Gibon Y., Muller B. (2010). Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: An integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiol. 154: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger T., Pérez-Pérez J.M., Bernal S., Micol J.L. (2005a). Quantitative trait loci mapping of floral and leaf morphology traits in Arabidopsis thaliana: Evidence for modular genetic architecture. Evol. Dev. 7: 259–271 [DOI] [PubMed] [Google Scholar]
- Juenger T.E., McKay J., Hausmann N., Keurentjes J., Sen S., Stowe K., Dawson T., Simms E., Richards J. (2005b). Identification and characterization of QTL underlying whole-plant physiology in Arabidopsis thaliana: Delta C 13, stomatal conductance and transpiration efficiency. Plant Cell Environ. 28: 697–708 [Google Scholar]
- Juenger T.E., Sen S., Bray E., Stahl E.A., Wayne T., McKay J., Richards J.H. (2010). Exploring genetic and expression differences between physiologically extreme ecotypes: comparative genomic hybridization and gene expression studies of Kas-1 and Tsu-1 accessions of Arabidopsis thaliana. Plant Cell Environ. 33: 1268–1284 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R., Girke T., Bray E.A., Bailey-Serres J. (2004). Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim J.M., To T.K., Ishida J., Morosawa T., Kawashima M., Matsui A., Toyoda T., Kimura H., Shinozaki K., Seki M. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49: 1580–1588 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D.J. (2010). Systems biology uncovers the foundation of natural genetic diversity. Plant Physiol. 152: 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera E., Tasaka M., Shikanai T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Kramer P.J., Boyer J.S. (1995). Water Relations of Plants and Soils. (San Diego, CA: Academic Press; ). [Google Scholar]
- Kreps J.A., Wu Y., Chang H.S., Zhu T., Wang X., Harper J.F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Lambers H., Chapin F.S., III, Pons T.L. (2008). Plant Physiological Ecology. (New York: Springer; ). [Google Scholar]
- Lehti-Shiu M.D., Zou C., Hanada K., Shiu S.H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littel R.C., Milliken G.A., Stroup W.W., Wolfinger R.D. (1996). SAS System for Mixed Models. (Cary, NC: SAS Institute; ). [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow M.M. (1989). Strategies of response to water stress. In Structural and Functional Responses to Environmental Stresses, Kreeb K.H., Richter H., Hinckley T.M., (The Hague, The Netherlands: SPB Academic; ), pp. 269–281 [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay J.K., Richards J.H., Mitchell-Olds T. (2003). Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Mol. Ecol. 12: 1137–1151 [DOI] [PubMed] [Google Scholar]
- McKay J.K., Richards J.H., Nemali K.S., Sen S., Mitchell-Olds T., Boles S., Stahl E.A., Wayne T., Juenger T.E. (2008). Genetics of drought adaptation in Arabidopsis thaliana II. QTL analysis of a new mapping population, KAS-1 x TSU-1. Evolution 62: 3014–3026 [DOI] [PubMed] [Google Scholar]
- Meierhoff K., Felder S., Nakamura T., Bechtold N., Schuster G. (2003). HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15: 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., et al. (2008). Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monda K., Negi J., Iio A., Kusumi K., Kojima M., Hashimoto M., Sakakibara H., Iba K. (2011). Environmental regulation of stomatal response in the Arabidopsis Cvi-0 ecotype. Planta 234: 555–563 [DOI] [PubMed] [Google Scholar]
- Narusaka Y., Nakashima K., Shinwari Z.K., Sakuma Y., Furihata T., Abe H., Narusaka M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34: 137–148 [DOI] [PubMed] [Google Scholar]
- Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T.R., Dyreson C., Wyrick J.J. (2005). Athena: A resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Okuda K., Nakamura T., Sugita M., Shimizu T., Shikanai T. (2006). A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 281: 37661–37667 [DOI] [PubMed] [Google Scholar]
- Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inzé D., Goossens A. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro C., Chaves M.M. (2011). Photosynthesis and drought: Can we make metabolic connections from available data? J. Exp. Bot. 62: 869–882 [DOI] [PubMed] [Google Scholar]
- Rohlf F.J., Sokal R.R. (1981). Statistical Tables. (New York: W. H. Freeman & Company; ). [Google Scholar]
- Seki M., Narusaka M., Abe H., Kasuga M., Yamaguchi-Shinozaki K., Carninci P., Hayashizaki Y., Shinozaki K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Seki M., Umezawa T., Urano K., Shinozaki K. (2007). Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 10: 296–302 [DOI] [PubMed] [Google Scholar]
- Sokol A., Kwiatkowska A., Jerzmanowski A., Prymakowska-Bosak M. (2007). Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 227: 245–254 [DOI] [PubMed] [Google Scholar]
- Stebbins G.L. (1952). Aridity as a stimulus to plant evolution. Am. Nat. 86: 33–44 [Google Scholar]
- Stockinger E.J., Gilmour S.J., Thomashow M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain E., Muse S.V. (2005). Positively selected sites in the Arabidopsis receptor-like kinase gene family. J. Mol. Evol. 61: 325–332 [DOI] [PubMed] [Google Scholar]
- Taiz L., Zeiger E. (2010). Plant Physiology. (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Tessadori F., Schulkes R.K., van Driel R., Fransz P. (2007). Light-regulated large-scale reorganization of chromatin during the floral transition in Arabidopsis. Plant J. 50: 848–857 [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O.E., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tittel-Elmer M., Bucher E., Broger L., Mathieu O., Paszkowski J., Vaillant I. (2010). Stress-induced activation of heterochromatic transcription. PLoS Genet. 6: e1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.S., Urao T., Qin F., Maruyama K., Kakimoto T., Shinozaki K., Yamaguchi-Shinozaki K. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K., Maruyama K., Ogata Y., Morishita Y., Takeda M., Sakurai N., Suzuki H., Saito K., Shibata D., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. (2009). Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 57: 1065–1078 [DOI] [PubMed] [Google Scholar]
- Urao T., Yakubov B., Satoh R., Yamaguchi-Shinozaki K., Seki M., Hirayama T., Shinozaki K. (1999). A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Steinhauser D., Gibon Y., Bläsing O.E., Redestig H., Sreenivasulu N., Krall L., Hannah M.A., Poree F., Fernie A.R., Stitt M. (2006). PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen H., Kliebenstein D.J., West M.A., Kim K., van Poecke R., Katagiri F., Michelmore R.W., Doerge R.W., St Clair D.A. (2007). Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid. Plant Cell 19: 2099–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M., Tessadori F., McLoughlin F., Smith R., Millenaar F.F., van Driel R., Voesenek L.A., Peeters A.J., Fransz P. (2010). Photoreceptors CRYTOCHROME2 and phytochrome B control chromatin compaction in Arabidopsis. Plant Physiol. 154: 1686–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S. (1987). Genetic constraints on the evolution of phenotypic plasticity. In Genetic Constraints on Adaptive Evolution, Loeschcke V., (Berlin: Springer Verlag; ), pp. 147–156 [Google Scholar]
- Walter H. (1964). DieVegetation der Erde. (Jena, Germany: Gustav-Fischer Verlag; ). [Google Scholar]
- Walter H. (1968). DieVegetation der Erde. (Jena, Germany: Gutav-Fischer Verlag; ). [Google Scholar]
- Whittaker R.H. (1975). Communities and Ecosystems. (New York: Macmillan; ). [Google Scholar]
- Wrzaczek M., Brosché M., Salojärvi J., Kangasjärvi S., Idänheimo N., Mersmann S., Robatzek S., Karpiński S., Karpińska B., Kangasjärvi J. (2010). Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10: 88–94 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61: 672–685 [DOI] [PubMed] [Google Scholar]